Abstract
A hybrid analytic-systematic (HAS) coarse-grained (CG) lipid model is developed and employed in a large-scale simulation of a liposome. The methodology is termed hybrid analyticsystematic as one component of the interaction between CG sites is variationally determined from the multiscale coarse-graining (MS-CG) methodology, while the remaining component utilizes an analytic potential. The systematic component models the in-plane center of mass interaction of the lipids as determined from an atomistic-level MD simulation of a bilayer. The analytic component is based on the well known Gay-Berne ellipsoid of revolution liquid crystal model, and is designed to model the highly anisotropic interactions at a highly coarse-grained level. The HAS CG approach is the first step in an “aggressive” CG methodology designed to model multi-component biological membranes at very large length and timescales.
Keywords: Coarse-graining, membrane, multiscale, liposome, molecular dynamics
1. Introduction
Arguably, one of the greatest challenges facing the field of biomolecular simulation and modeling of lipid bilayers is making the connection to real biological membranes. Real membranes are highly inhomogenous and include multiple lipids, cholesterol, and numerous proteins. Furthermore, in contrast to the original fluid mosaic model,1 a new picture is emerging where the membrane is crowded, containing a large number of highly oligomerized proteins, and has a varying membrane thickness as well as lipid spatial organization.2,3 In some cases, upwards of 20% or higher of the surface area of a membrane is occupied by proteins.4 This scenario is quite far removed from model systems, for example, Giant Unilamellar Vesicles (GUV’s) consisting of a limited number of components.5 Many experiments have utilized model membrane systems (see, e.g., refs 6,7 for two examples) rather than examining actual in vivo processes as the inherent complexity of the real biological membrane does not allow for a fine control over various experimental conditions. Nonetheless, especially in terms of simulation and modeling, the challenge of moving towards examining real biological membranes and associated systems remains a critical priority.
From a simulation viewpoint, modeling real biological membranes presents a number of key challenges. First, as previously mentioned, real biological membranes contain multiple components that can include lipids, proteins, and non-lipid molecules such as cholesterol. Molecular dynamics (MD) simulations require that the interactions between every atom for each of the different components be rigorously specified within the MD force field. Here, the large number of components implies that a large number of interactions thus need to be specified, and even at the atomistic level this can be a challenge as a high level of accuracy in the simulation force fields maybe required. Second, and perhaps more importantly, real biological membranes contain multiple length and time-scales ranging from the atomistic (nm, ns), to the macroscopic (mm, ms). As a result, the behavior of a small, isolated component of a biological membrane can be quite different from that observed in the real membrane. Furthermore, MD simulation alone currently cannot even remotely access the longest length and time-scales that are characteristic of real membranes (e.g., a cell with dimensions on the order of µm and time-scales of ms). As such, from the onset, multiscale simulation methodologies are required where the different scales are somehow systematically bridged or coupled.8
An emerging and core component of an overall multiscale simulation methodology for biological membranes is coarse-grained (CG) MD simulation.8–47 This particular component bridges atomistic length and time-scales with more field-based mesoscopic scales.48–52 In fact, CG-MD has been used to examine emergent phenomena for relatively complex biomolecular systems at length and time-scales that are currently out of reach of traditional atomistic-level MD.8 For example CG-MD has been employed to model both proteins8,9,47,53,54 and lipid bilayers.8–47 In the case of lipids, CG lipid models generally contain around 10 to 15 CG sites,10–15 compared to 46 atoms in a united atom lipid model,55 or over 100 atoms for a fully atomistic representation. Since the number of CG sites is less than the original number of atoms and the interactions between them are generally simpler and shorter range, a significant computational speed-up is possible.
It should be noted that the particular CG scheme that is chosen can be flexible. That is, both the number and location of the CG sites required to describe a particular molecule can in principle vary depending on the problem at hand. In cases where very large membranes are to be examined (but not so large that field-based mesoscopic models are required48–52), the degree of coarse-graining may have to be quite aggressive. For example, in the case that a 200 nm diameter liposome containing upwards of half a million lipids is to be modeled, an entire lipid may have to be coarse-grained into one or two sites. It is also possible to develop so-called “solvent-free” CG membrane models,17–21,23 where the hydrophobic/hydrophilic lipid-solvent interactions are subsumed into the new CG lipid interactions. This latter approach, in particular, can result in significant computational speed-ups as a large part of the numerical effort is taken up accounting for the surrounding solvent and membrane solvent interactions. This important aspect of the problem will be revisited shortly.
Simulations of liposomes and vesicles have been performed with CG simulation;18,23,26–28,37,39,56 however, at this point in time the largest systems studied have been small vesicles with diameters in the range of 40 to 60 nm39 with typically less than 5000 lipids.18,23,26,28 Liposomes of these sizes are too small to model most biologically relevant systems where the liposome diameter is upwards of 200 nm.6 Thus, it is important to explore even more highly coarsegrained lipid models such that these large liposome length-scale can be achieved.
The aim of the present paper is not to introduce yet another CG lipid model. As has been previously discussed, there are a multitude of “higher resolution” CG lipids models that have already been developed. The goal here is to develop a methodology wherein highly coarse-grained, low resolution lipid models can utilized at very large length-scales while retaining at least a semi-quantitative agreement with the underlying molecular-scale system. The methodology presented here is general and can potentially be applied to a number of CG lipid models17,18,20,29 as its basis. However, the overall spirit of the present work is to start with a low resolution CG lipid model that has as few sites as possible as the difference, e.g., between a single site versus ten site CG model translates into a substantial increase in computational efficiency, thereby allowing the model to access much larger length scales. Moreover, as one begins to model larger scale structures such as a 200 nm diameter liposome, it is quite sensible that the resolution of the underlying CG model can become correspondingly lower.
An experimental example of a scenario where a highly coarse-grained membrane model could be utilized is protein mediated membrane remodeling,6,7,47,48,57 where, for example, whole liposomes are remodeled by proteins and protein modules (i.e., the BAR (Bin/amphiphysin/Rvs) domain) into tubules.6,48,58 Furthermore, experimentally, the process of tubulation often occurs over time-scales much longer than microseconds.59
It is important to note that CG-MD shares with MD its dependence on an effective force field to define the interactions between CG “sites”. It is not immediately obvious or trivial to determine these interactions, especially in multi-component systems such as real biological membranes. In practice, many current CG models rely on a more “top down” approach,10,11,13,20,21 where the interactions between CG sites are designed from the onset to reproduce some desired macroscopic behavior. Such techniques, for example, Inverse Monte Carlo, have even been aggressively employed to construct a two-dimensional model for a dipalmitoylphosphatidylcholine (DPPC)/cholesterol bilayer that mapped the entire lipid (and cholesterol molecules) onto a single site associated with the center of mass of the molecules.46 The two dimensional constraint significantly simplifies the interactions between the CG sites; since the CG lipids can not leave the plane of the membrane, only a pairwise additive, radially symmetric interaction was required. The CG interactions incorporated, in some effective way, the out-of-plane lipid interactions. However, by definition, the model misses critical long wavelength thermal undulations out of the plane of the membrane, and it would also be quite restrictive in the case of modeling liposomes and other closed surfaces that are more indicative of real biological membranes in cells.
Ideally, a CG model should be systematically derived from its underlying atomistic-level interactions. This is, in fact, one of the outstanding challenges in the field of CG simulation: To rigorously derive the force field for a CG model from the corresponding force field employed in a high-resolution atomistic level system. It is especially desirable to construct a CG model entirely from atomistic level information; in this case, all new emergent behavior that is observed is then predicted, rather than “tuned” or “engineered” based on empirical data. In this way, the behavior of the CG system can be traced back to the underlying atomistic level interactions that were used in developing the CG model.
One possible approach to systematically develop CG models from atomistic level information is the multiscale coarse-graining (MS-CG) methodology.15,16,60–68 This approach relies on a variational principle60,61 that determines the optimal CG force field for a given finite set of MD configurations (i.e., employing the Cartesian coordinates and forces of all the atoms for a large number of MD generated configurations), along with a pre-defined CG force-field basis set that contains Nd parameters, ϕ = {ϕ1,…,ϕNd}, that can be variationally determined from the available MD data.60–62
The process of variational fitting has been called “force matching”,15,60,61 and can be shown to guarantee the best fit of the available atomistic data to the CG force field basis set. The scheme only requires that the CG force is linear in its coefficients,61 and what results is a CG “fitted force field” whose degree of flexibility and/or complexity lies in the initial choice of the basis set.61 In the case of non-bonded interactions, the MS-CG methodology can readily generate pairwise additive radial force fields, where the radial dependence of the force field is modeled with, for example, spline coefficients15,16,64–66 or even a delta function basis.63
However, in the case of real biological membranes, a direct application of the MS-CG methodology can prove challenging due to the size and complexity of the initial MD simulation that is required to generate the MS-CG force field. Strictly speaking, the optimum force field obtained from the MS-CG methodology is reached only in the limit that the underlying MD simulation samples, essentially, all possible configurations. In that limit, all of the atoms that correspond to various CG sites can interact with all the others such that a set of converged MS-CG force fields for that system can be generated. However, atomistic MD simulations of large biological membranes can only sample a relatively small set of configurations. For example, consider an atomistic simulation of a multicomponent lipid bilayer, with cholesterol and multiple embedded proteins. Over the duration of the typical MD simulation (perhaps 100 ns), some interactions are hardly, if ever, sampled, for example a water-lipid tail interaction. Furthermore, within the duration of the MD simulation, long time-scale processes, such as the formation of domain structures, protein mediated lipid sequestering3 and other complex phenomena (e.g., membrane bound protein oligomerization2) may not be observed.
The option of running multiple, extended MD simulations in order to fully sample the entire system in many ways defeats the purpose of moving to a CG representation. Ideally, the atomistic MD simulations used to construct the MS-CG force field should be as small and efficient as possible. In fact, the full biological membrane system may be broken down into smaller sub-systems for the explicit purpose of sampling key interactions. The overall coarsegrained configurational space can be thought of as a vast domain that is highlighted by small regions that are well-sampled by the initial MD simulations used to generate the MS-CG force field. However, it also contains large domains where little or no atomistic-level sampling and information was obtained. The MS-CG methodology can therefore be used with confidence to obtain CG models that apply to those well-sampled domains, but some other approach is required in order to fill in the “gaps” between them.
The option explored in this paper is to develop a hybrid CG model that has two components: The first component is a systematically obtained MS-CG force field that is employed for CG configurations that correspond to well-sampled MD configurations, while the second component relies on an analytic model to describe the poorly sampled configurations. This approach will be denoted as a hybrid analytic-systematic (HAS) CG model. It is utilized in cases where the majority of the sampled CG configurations are related well-sampled MD configurations, so that only periodically is the analytic component of the CG model employed. For multicomponent membranes, the full CG configuration space can then be thought of as being populated by “islands” of CG configurations that correspond to underlying well-sampled atomistic MD configurations that are connected via the analytic component of the HAS CG model. This type of approach affords a operational solution to limited MD sampling in large biological membranes, yet retains a strong degree of systematically obtained CG interactions, which are critical in order for the CG model to be bridged to the underlying atomistic-scale.
The present implementation of the HAS CG approach is also an “aggressive” CG model that is capable of reaching significant length and time-scales; for example, a reasonably sized liposome of 200 nm diameter and half a million lipids. The HAS CG methodology thus aims to combine systematically obtained interactions obtained via the MS-CG methodology with very computationally efficient analytic models, where from the onset an aggressive CG scheme is employed such that large biological membrane systems can be modeled. Specifically, in this work a single site, solvent-free, lipid model will be explored; however, other options could be employed as the approach is general.
For this application of the HAS CG approach to membranes, the systematic component of the CG lipid bilayer will use the MS-CG approach to develop an in-plane, single site, CG membrane force field. There are a number of analytic models that can then be employed for the analytic component of the HAS CG model. One option could be the so-called “shape-based” CG model for lipids.47 Here, the lipid is modeled as a dimer with one bead representing the headgroup, while other bead represents the tail. Another possibility used here is the Gay-Berne (GB) liquid crystal model,69,70 which has the ability to model ellipsoids of revolution with varying aspect ratios, and reduces to a standard Lennard-Jones (LJ) form when two identical ellipsoids are parallel and side-by-side. A single GB ellipsoid requires one site at its center to designate its Cartesian location, along with a unit vector to designate its orientation. It has been used to model a variety of liquid crystal phases71 and even lipids at a very reduced level of resolution.35,36,38 For this work, the single-site GB analytic model will be employed as it ties in with the single-site MS-CG systematic component as previously introduced.
With this HAS CG approach, the GB interaction provides an anisotropic “core”, while the interactions found from the MS-CG methodology supply systematically determined force details arising from the actual average in-plane interactions as observed in the atomistic MD simulation. The MS-CG methodology only samples interactions over a limited set of MD configurations (e.g., an MD trajectory of a stable bilayer in the liquid crystal phase), where a large contribution to the total force acting on a lipid comes from the in-plane lipid interactions. As such, the GB “core” accounts for other interactions that were never fully sampled in the original MD trajectory. These include, for example, lipid-lipid interactions that might be observed in a completely isotropic lipid-water mixture.
This paper will describe a HAS CG lipid bilayer model that will be employed at a very large length-scale to simulate an entire liposome with a diameter on the scale of 200 nm. The next section will introduce how analytic force fields such as the GB interaction can be combined with the MS-CG methodology to yield a HAS CG model, and will discuss the MS-CG variational principle as used in the present context. Section 3 gives the results of both the atomistic and CG simulations; section 4 provides a brief summary.
2. Methods
2.1 Hybrid Analytic-Systematic (HAS) Lipid Bilayer Model
The goal is to construct a HAS CG, single-site, solvent free bilayer model by combining an analytic GB ellipsoidal liquid crystal model with a MS-CG force field. The MS-CG force field is employed in specific configurations where the force matching method can be applied with a good level confidence, while the GB force field is used otherwise. Specifically, the MS-CG force field will give the in-plane lipid center of mass (COM) interactions, while the GB component will model the inter-monolayer interactions, very close-range interactions, as well as any other out-of-plane interactions such that the bilayer stability is maintained in three dimensions. The parameterizations for the GB component will be empirically determined from all the available atomistic MD information on hand.
The resulting HAS CG model is a CG pair potential that depends not only on the Cartesian locations of the CG sites, Rα, where α is the label corresponding to that site, but also on the orientation of the CG site, given by a unit vector eα. The orientational dependence is critical in order to model the anisotropy of the lipid-lipid interaction with a single site model. The final CG pair potential is given by
| (1) |
where RC,1 and RC,2 are an inner and outer pre-set cutoff, RIJ = |RI − RJ|, and ΔU(RIJ) is a spherically symmetric correction term that is found from
| (2) |
Here is the MS-CG pair potential and is the GB interaction for a pair of perfectly aligned and in-plane GB ellipsoids. As such, the pair potential as given in eq (1) becomes the MS-CG pair interaction for two parallel and in-plane CG sites that are within the two inner and outer cut-offs. The hybrid nature of this approach is clear: The model gives exactly the MS-CG interaction when two CG particles are parallel and side-by-side. This particular lipidlipid configuration reasonably maps over to many of the configurations sampled in the atomistic MD bilayer simulation where the MS-CG in-plane force field was originally calculated. Of course, while the MD simulation remains in the bilayer phase throughout the atomistic simulation, the CG model can potentially explore new regions of phase space. When this happens, the anisotropic GB component of the interaction in eq (1) takes over. Since the full CG model contains both a GB ellipsoid component, as well as a MS-CG part, the CG particles will be referred to as “CG lipids”, versus, for example, GB ellipsoids.
Details of the MS-CG approach can be found elsewhere;15,16,60–66 here the focus will be on the HAS approach used to combine the GB liquid crystal model with the MS-CG force field. A detailed description of the HAS CG methodology is discussed in the following sub-sections, and consists of four steps:
Instantaneously calculate the centers of mass (COM) of the lipids in a fully solvated atomisticlipid bilayer MD simulation.
Force match this system using the COMs to give an in-plane, pairwise additive MS-CG force field between the COM locations of the lipids for each monolayer.
Use the average vertical separation of the COM sites, along with the atomistic area per lipid, to construct an analytic GB model of the bilayer.
Combine the two force fields into one CG lipid model where the MS-CG component is employed in configurations where the FM force field applies, and the GB component is used otherwise as given in eq (1).
2.2 Molecular Dynamics Simulation
An initial MD simulation of a fully solvated dimyristoylphosphatidylcholine (DMPC) bilayer was performed. The system consisted 64 lipids and was equilibrated for 6 ns under isothermal and isobaric conditions. Simulation details can be found elsewhere.72 Subsequent MS-CG calculations were performed under constant volume V, atom number n, and temperature T conditions (nVT) in order to map over to the MS-CG framework which is currently developed for the nVT ensemble.60,62 The Cartesian spatial coordinates of the n atoms are given by rn ≡ {r1,…,rn}. The center of mass (COM) of each lipid was calculated instantaneously via the mapping operator, , which is the set of N CG mapping operators where each one separately is given by60
| (3) |
where in this case cIi = PI,i mi/MI, is the mass of CG site I, and PI,i = 1 if atom i is part of CG site I, and is zero otherwise. The associated CG sites are denoted by RN ≡ {R1,…,RN} and are related to the mapping operator via the identity
| (4) |
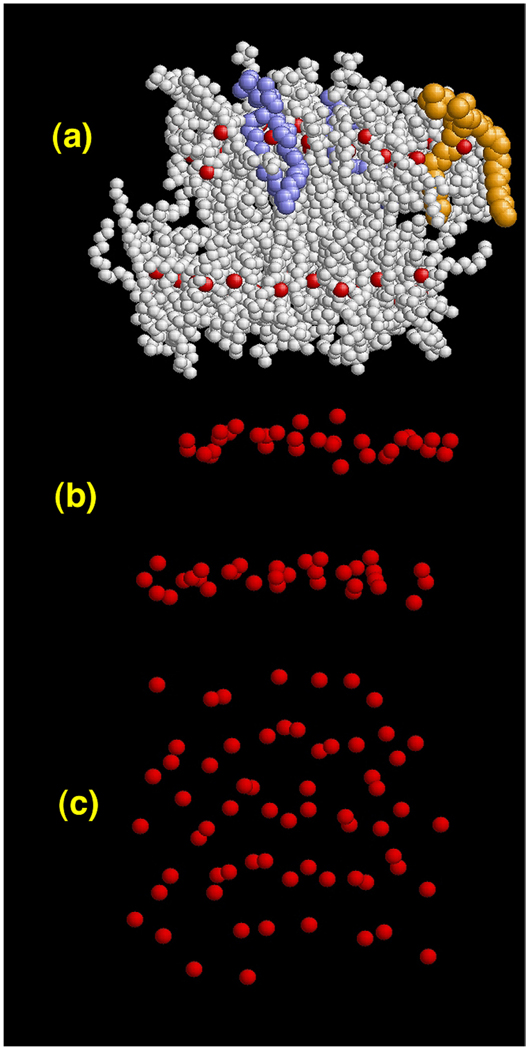
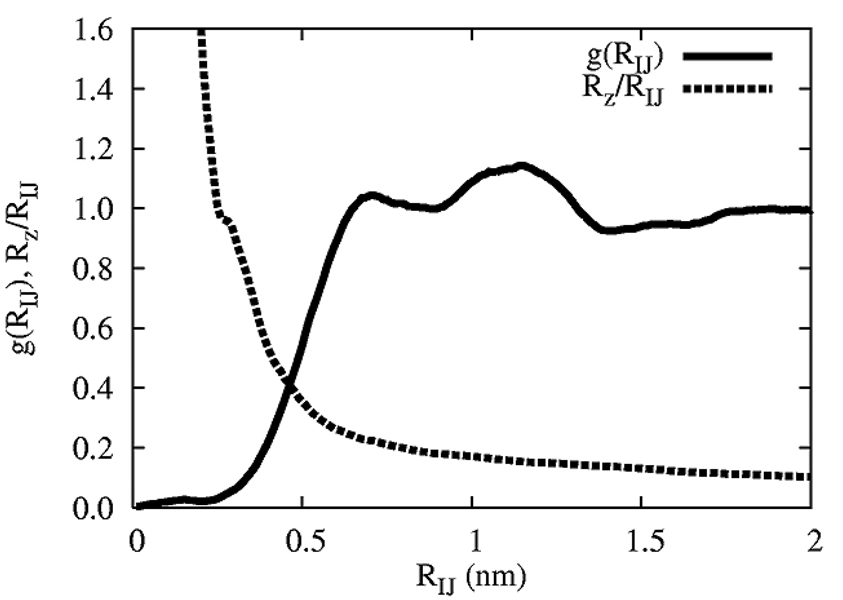
The distinction between the CG sites and the mapping operator is subtle, but must be appreciated. The mapping operator gives an instantaneous measured COM of the lipids from an MD simulation; in turn, the CG sites describe the Cartesian locations of the CG lipids in the corresponding CG simulation. However, in order to keep the notation relatively simple, RN ≡ {R1,…,RN} will sometimes be used to denote the COM location of the lipids in the MD simulation when appropriate. A snapshot of this instantaneous mapping is given in Figure 1, where it is clear from panels (a,b) that the COM resides near the middle of each lipid, and that two distinct monolayers result. However, the COM locations are not perfectly in the plane, and have a fair amount of variation in the z-direction. Panel (c) shows a top down view where, if anything, a complete lack of strong hexagonal symmetry is evident. In fact, some COM points almost lie on top of each other. This behavior can be traced back to the COM mapping in eq (3); the lipid chains (as highlighted by the blue and orange lipids in panel (a) of Figure 1) are quite disordered, which is typical of lipids in the liquid crystal phase. As such, the COM mapping can result in sites that may reside at locations that are quite close, or even on top of, other lipid COM’s. The resulting COM structure of the full MD system gives two monolayers, where each monolayer has, at best, weak correlations. This is more fully realized in Figure 2, where the in-plane COM radial distribution function (RDF) is shown. The solid line in Figure 2 shows the RDF, averaged over the COM points in both leaflets of the bilayer separately. The small bump near zero corresponds to lipid COM points that are almost “on top” of each other and hence have a small in-plane radial component. The two peaks around 1 nm exhibit weak correlations, consistent with the observations in Figure 1. The size of the MD cell did not allow for the RDF to be examined out to farther distances. However, larger systems would start to exhibit undulation modes that could affect the planar projection of the RDF. The dotted line in the same figure is the average ratio of Rz/RIJ where here RIJ = ((MRIx − MRJx)2 + (MRIy − MRJy)2)1/2, Rz = |MRIz − MRJz| and MRα = MRαxî + MRαyĵ + MRαzk̂. As long as Rz/RIJ, is less than 1, then the correlations in the RDF originate from lipids that are nearly planar; however, when Rz/RIJ is greater than 1 (i.e., around RIJ < 0.3 nm), the correlations arise from particles with a large Rz component. From Figure 2, it is therefore clear that the short-range correlations arise from lipid COM’s that contain large non-planer components, while the longer-ranged correlations arise from nearly planar structures. The two peaks around 1 nm can be loosely characterized as arising from in-plane COM correlations.
Figure 1.
Simulation snapshots of the original atomistic DMPC system. Panel (a) shows DMPC lipids with the CG sites shown as red spheres. The colored lipids highlight the highly two disordered tail conformations in the liquid crystal phase. Panel (b) is the instantaneous locations of the COM CG sites corresponding to (a). A distinct bilayer structure is observed. Panel (c) is a top down view of the CG sites; very little structural correlation is observed and some of the lipid COM CG sites are quite close together.
Figure 2.
The lipid COM radial distribution function (RDF) (solid line) along with the average of the ratio Rz/RIJ (dark dotted line) where here RIJ = |MRIx − MRIy|, Rz = |MRIz − MRJz| and is the α Cartesian component corresponding to the COM of lipid I.
2.3 In Plane Force Matching of the Lipid Bilayer
The COM mapping introduced previously can be used to force match the bilayer system. For this study, the delta function basis set was used,63 and the procedure is briefly described here. A more detailed account can be found elsewhere63 and in Appendix A. The effective MS-CG force for the Ith CG site can be expressed as a pair-wise sum over all J CG sites within a range delineated by the inner and outer pre-set cut-off distances RC,1,RC,2 as
| (5) |
where here the MS-CG pair force, , is linearly dependent on Nd coefficients, ϕ = {ϕ1,…ϕNd} The optimal value of the coefficients is found from the minimization of the χ2 residual given by 60–62
| (6) |
where the additional “t” subscript denotes a specific set of atomistic configurations from the MD trajectory, and
| (7) |
The instantaneous atomic force on atom i is given by fi and the average in eq (6) is taken over all CG sites and configurations.
It can be shown60,61 that the minimum value of the residual is found when , that is when the MS-CG force field is in fact the gradient of the exact potential of mean force, U(RN) , that governs the CG system.60,62 In a practical sense, the MS-CG force field will reflect any degree of limited sampling in the underlying atomistic MD simulation. As will be discussed later, the analytic component of the interaction is designed to enhance and fill in the overall sampling range. In this study, the force will only be calculated in the plane of the membrane. It may be possible to calculate the normal force, which would give the lipid tail-tail interaction. However, it is doubtful that either the head-head or head-tail cross interactions could be adequately sampled. Thus, the normal component will be modeled via the analytic component. The two inner and outer cut-offs, RC,1,RC,2 , control the radial range to be used in the force matching calculation. The inner cut-off, RC,1, in particular, excludes specific data (i.e., data accumulated where the corresponding Rz/RIJ is large or where the RDF is quite small) from the force-matching calculation. The outer cut-off , RC,2, is determined from the size of the MD simulation and is given by the range of the RDF (in this case 2 nm). The resulting MS-CG force field will then give the “best” force field subject to the range of inner and outer cut-offs employed. In previous work, the behavior of the CG potential at close range had to be interpolated.15 This approach could also, in principle, be used here; however, as will be discussed, the HAS CG approach circumvents this problem and adds additional flexibility to the CG model.
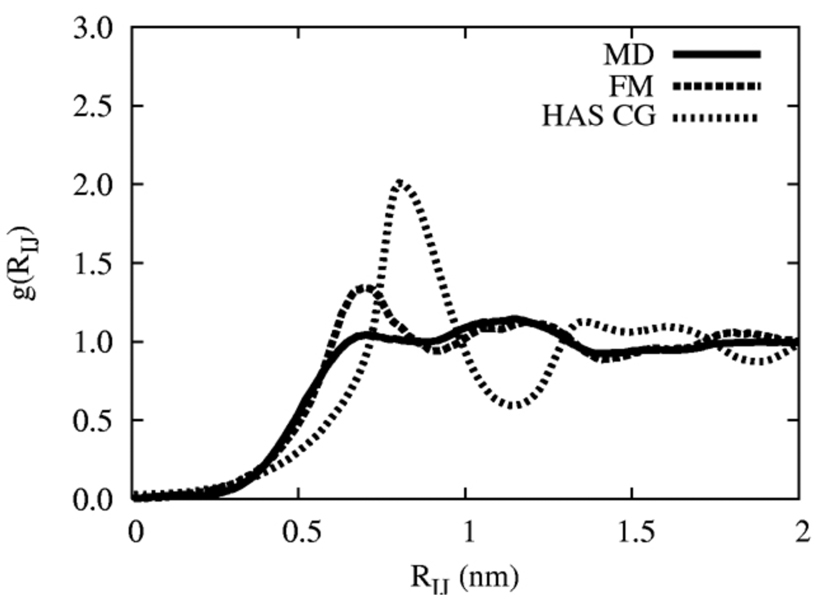
It was found that the MS-CG force field became increasingly unreliable (i.e., the condition number for the matrix inversion increased dramatically) when poorly sampled data obtained at close-range was used. Therefore, an initial MS-CG force-field using the delta function approach was found by only including data from 0.4 < RIJ < 2.0 nm. This discrete force field was then smoothed and directly employed in a 2D CG simulation, similar to previous work,46 in order to test the accuracy of this component of the HAS CG model relative to all-atom MD data. The resulting CG RDF is shown as the dark-dotted line in Figure 4. The slightly enhanced first peak is due to the sensitivity of the force field at close-range, and could be improved with a more elaborate treatment of the short-ranged interactions (i.e., versus simply omitting them in the force-matching calculation).
Figure 4.
The CG RDF. The solid line is as given in Figure 2, the dark-dotted line is the CG RDF using a constant area 2D system, while the light-dashed line is the RDF for the HAS CG system in 3D under zero surface tension.
However, the end goal here is not to develop a purely two dimensional force field, but to extend the model to three dimensions. As such, the focus will now shift to the analytic GB component of the HAS CG interaction.
2.4 A Modified GB Lipid Bilayer Model
In this section, some details of the GB component of the HAS CG bilayer model are discussed. It should be noted that the GB interaction was modified such that it possesses the symmetry of a lipid, i.e., the head is different from the tail. How this was done is given in Appendix B. The functional form of the GB pair potential is expressed as
| (8) |
where
| (9) |
In this expression, R̂IJ = RIJ/RIJ is the full three dimensional vector. As previously mentioned eI is an orientation unit vector that lies along the long axis of the Ith GB ellipsoid; likewise for J. The quantity σ0 is the width of the GB ellipsoid, while σ(R̂IJ,eI,eJ) gives the “distance of closest approach along R̂IJ for a pair of GB ellipsoids with orientations eI, eJ. See Appendix B for further details.
When two GB ellipsoids are constrained to be perfectly aligned and side-by-side, eq (8) reduces to
| (10) |
which is just the standard Lennard-Jones (LJ) interaction, and the additional superscript A refers to the fact that the GB ellipsoids are perfectly aligned and side-by-side. This is exactly the interaction that appears in eq (2). In this form, the interaction only contains an energy parameter, ε0 , as well as a fundamental unit of length, σ0 and depends just on the pair distance between I and J, RIJ . If the approximation is made that the lipids are perfectly aligned, then this form of the potential can used in combination with the MS-CG force as described in subsecs 2.2 and 2.3. It should be noted that explicitly including the angular dependence would drastically increase the complexity of the MS-CG mapping. Initially setting σ0 amounts to pre-defining the “size” of the CG particle, and it is empirically found from a knowledge of the atomistic area per lipid and the phase behavior of the GB ellipsoid system. However, this aspect falls under combining the GB and the MS-CG interaction and will be discussed next.
2.5 HAS CG Lipid Bilayer Model
The first step in constructing the full HAS CG lipid model is to introduce a GB “core”; this is shown graphically in Figure 3 as the ellipsoid of revolution. This component of the model accounts for the highly anisotropic shape of a lipid. Equation (8) is employed with the following parameterization scheme: The fundamental unit of length, σ0 , was chosen based on the tomistic area per lipid, AL, as found from the atomistic MD simulation ( AL ≈ 0.62 nm2 per lipid), combined with the known density of the fluid phase of a GB ellipsoidal system . A value of σ0 = 0.72 nm was therefore chosen. The length of the ellipsoid was chosen to be 3σ0 . The motivation for this 3:1 ratio draws on the extensive previous data on 3:1 GB ellipsoidal systems, and the fact that this aspect ratio is known to give stable liquid crystal as well as smectic phases.70 This aspect ratio also reasonably models the bilayer thickness. Other approaches could also be employed; however, this approach reasonably combines the behavior of the atomistic and CG systems. The value of ε0 was obtained based on an estimate of the MS-CG potential well-depth, and the requirement that the raw GB core should result in a stable bilayer. Values between 12 and 14 amu (nm/ps)2 gave reasonable results. The other parameters in the GB model were as commonly employed.70
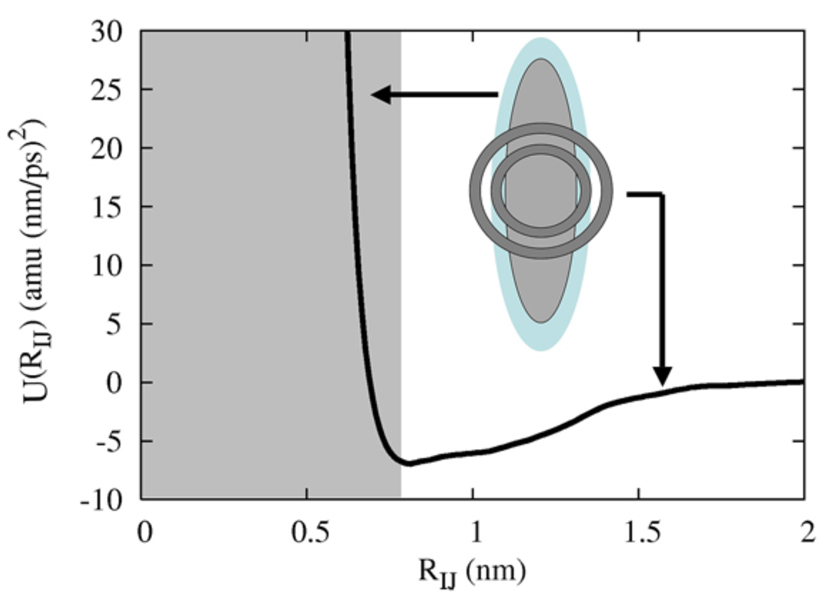
Figure 3.
The HAS CG potential. The inset shows a single CG particle where the anisotropic component (the ellipsoid) corresponds to the “core”, the short-ranged spherically symmetric component comes from the MS-CG method and gives the attractive component of the interaction. The shaded region on the left corresponds to the potential arising from the superimposed anisotropic core region.
At this point the nature of the HAS CG approach may seem rather ad-hoc. However, it should be re-stated that the full HAS CG lipid model is designed such that the MS-CG component takes over and in fact dominates for specific configurations as expressed in eq (1). As such, the GB component is mainly being utilized in configurations that were never fully sampled in the initial MS-CG force matching calculation.
The resulting full pair potential for two perfectly aligned and in-plane CG lipids is shown in Figure 3 and graphically depicts the pair interaction as given in eq (1). The “core” region is shaded, while the MS-CG component gives the critical attractive interaction. When two CG lipids are aligned and in-plane, the attractive component is exactly that as determined from the MS-CG calculation. When they are in other orientations, the full pair potential contains an anisotropic GB component as well as the spherically symmetric MS-CG part. However, since the MS-CG part is fairly short-ranged (and in fact is zero beyond RC,2), the end-to-end interaction of two CG lipids is entirely given by the GB component. As such, the sum of the two contributions can be illustrated by the inset in Figure 3; the GB component gives an analytic model for the anisotropic part of the system, while the MS-CG component gives a spherically symmetric part that comes fully into effect when two CG lipids are aligned and in-plane.
3. Results
The CG simulation results will be separated into two sets of simulations: A relatively small square “patch” membrane (N=5000 CG sites), and a much larger 190 nm diameter liposome (N=380,000 CG sites). The smaller system will be employed to measure a number of key membrane properties, while the liposome simulation will be used to demonstrate the feasibility of large-scale CG simulation employing the HAS CG approach. The HAS CG pair potential as graphically represented in Figure 3 and in eq (1) is employed throughout. An N=1250 CG site “raw GB system” that employs an interaction as in eq (1) but with ΔU(RIJ) = 0 will also be employed for comparison when appropriate.
3.1. N = 5000 CG Lipid System
A square patch of CG bilayer was constructed using a total of N = 5000 CG lipids, with each monolayer containing half of the CG lipids. This CG bilayer system is equivalent to a united atom lipid model with 230,000 sites, or a fully all-atom model with 588,000 lipid atoms and around 1.5 million total atoms with water solvent included. Multiple simulations were performed under zero surface tension for up to 200 ns. Key simulation parameters are given in Table 1. First, the in-plane RDF for the CG lipid model is shown as the light-dash line in Figure 4. Clearly, the inclusion of the GB core to the potential in three dimensions has altered the shortranged structure as compared to the original atomistic (solid) and 2D MS-CG (dark-dotted) results. However, the new correlations are not that strong, and importantly under zero surface tension, the correct area per lipid is recovered. It is to be expected that some degree of accuracy in the structural correlations will be lost in aggressive coarse-grained models such as this one.
Table 1.
Key CG simulation parameters
| Parameter | Symbol | Units | Value |
|---|---|---|---|
| GB width | σ0 | nm | 0.72 |
| GB length | h | nm | 2.16 |
| Temperature | T | K | 308 |
| timestep | δt | ps | 0.01 |
| GB mass | m | amu | 678 |
| GB energy | ε0 | amu (nm/ps)2 | 12–14 |
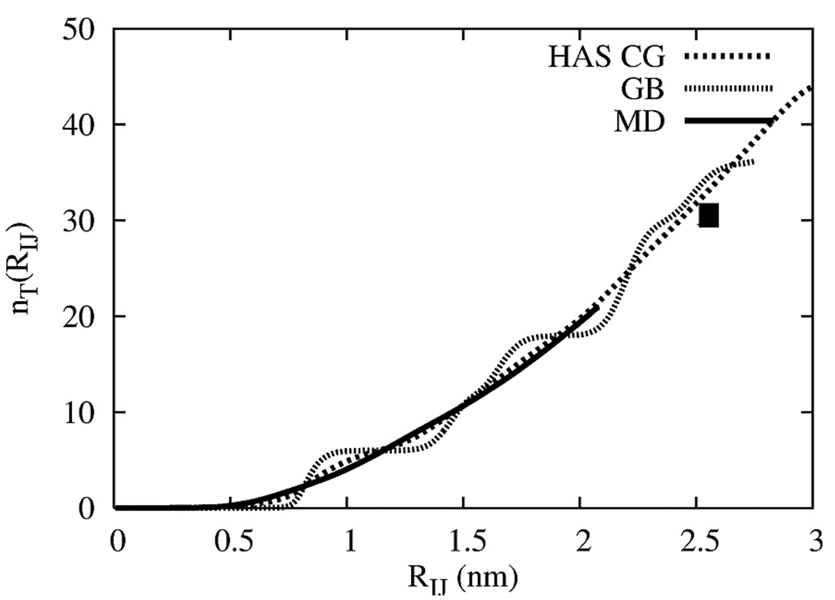
In Figure 5 is shown the accumulated number of CG lipids, nT(RIJ), around a central lipid in one of the monolayers. The solid line is the original atomistic result and is directly related to the area per lipid in the atomistic MD simulation. The dotted line is the CG lipid result using the HAS CG potential as given in eq (1). The accumulated CG lipid density around a central CG lipid is seen to track well to the atomistic result. Furthermore, it continues to accumulate density correctly. The solid square is the predicted number of CG lipids based on the atomistic area per lipid. Since the CG lipid simulation is under zero surface tension, it could easily elect to adopt a structure such that the CG area per lipid was quite different from the atomistic result. The raw GB result is also shown, and it exhibits pronounced oscillations due to the strong hexagonal correlations in that system. Thus, at short range the HAS interaction modulates the over-structured GB system such that it much better reproduces the atomistic MD result.
Figure 5.
The accumulated average number of lipids in one of the membrane leaflets for the N=5000 CG site HAS membrane (dotted), the “raw GB” (hatched), and the COM atomistic MD simulation (solid line). The solid square is the estimated number of lipids around a central one based on the atomistic area per lipid.
It should be noted that the CG area per lipid as calculated from the ratio of the CG simulation cell area to half the number of CG sites was found to be per lipid which is slightly less than the MD value of AL = 0.62 nm2 per lipid; this is due to undulation modes that begin to emerge in the larger system (cf. Figure 6). These undulation modes are clearly evident in Figure 6 (b) where the CG lipids are rendered as spherocylinders with an aspect ratio of h /σ0 = 3. The top view in Figure 6 (a) also shows a distinct lack of hexagonal correlations, similar to the atomistic structure in Figure 1. It should be noted that when only the GB interaction was used in eq (1) (i.e., with ΔU(RIJ) = 0 ) then the CG bilayer actually froze into a tightly packed hexagonal lattice. It is clear therefore what the MS-CG component is doing, above and beyond the analytic GB core of the HAS CG model; it is bringing back critical interactions that actually “melt” the otherwise solid membrane, and accordingly give it a structure much more reminiscent of the original atomistic MD membrane. Furthermore, returning to Figure 4, it is clear that in the CG lipid system that the close-range structure (i.e., configurations where lipids are close to being “on top” of each other) actually re-emerged in the CG system despite the inclusion of the GB core. When compared to the snapshots in Figure 6, the disordered nature of the system is consistent with the correlations in the RDF.
Figure 6.
Snapshots of the N=5000 CG site square patch HAS CG bilayer. Panel (a) shows a top down view; very little distinct structural correlation is evident. Panel (b) shows a side view where thermal undulations are clearly visible. Panel (c) shows two different self-assembled structures coming from an N=2048 CG site system. Bicellar as well as bilayer structures were observed depending on the volume of the system.
A key part of the HAS CG model relies on bridging the MS-CG force field to the GB interaction under the ideal configuration where the CG lipids are perfectly aligned and in the plane. The validity of this approximation can be tested by examining the orientational order of the CG lipid bilayer. If the CG lipids are highly aligned, then a large part of their interaction is coming from the systematically determined MS-CG component of the overall interaction. This can be measured via the P2 order parameter defined as , where θ gives the angle between the symmetry axis of the CG lipid and the director, d.73 Values of the P2 order parameter around 0 to 0.2 indicate basically an isotropic system, while values around 0.8 indicate a highly orientationally ordered phase. Here, the P2 order parameter was found to be P2 = 0.79 ± 0.004, indicating that the CG lipids were quite aligned. For reference, the same quantity calculated from the original MD simulation was found to be P2 = 0.76 ±0.010 (where the lipid orientation vector was found from the corresponding radius of gyration tensor). When combined with the fact that a stable bilayer structure is observed, it is reasonable to conclude that a large part of the CG lipid interaction is directly coming from the systematically force-matched MS-CG component.
Self-assembly has been observed in a number of CG models, resulting in bicelles, vesicles, and planar bilayers.19,20,26,27,37,44 Although the current study simulates liposomes at much larger lengthscales, it is interesting to examine the self-assembly properties of the single-site HAS CG model. A low density system (N/V= 0.2 nm−3) of N=2048 HAS CG particles was initially annealed at 400 K. This isotropic configuration (P2 ~ 0.02) was then used at various other densities up to (N/V= 0.27 nm−3). The resulting self-assembled structures are shown in Figure 6 (c). Bicelles as well as bilayers are seen to spontaneously form. If only the raw GB interaction was employed, then only lamellar bilayers spontaneously formed.
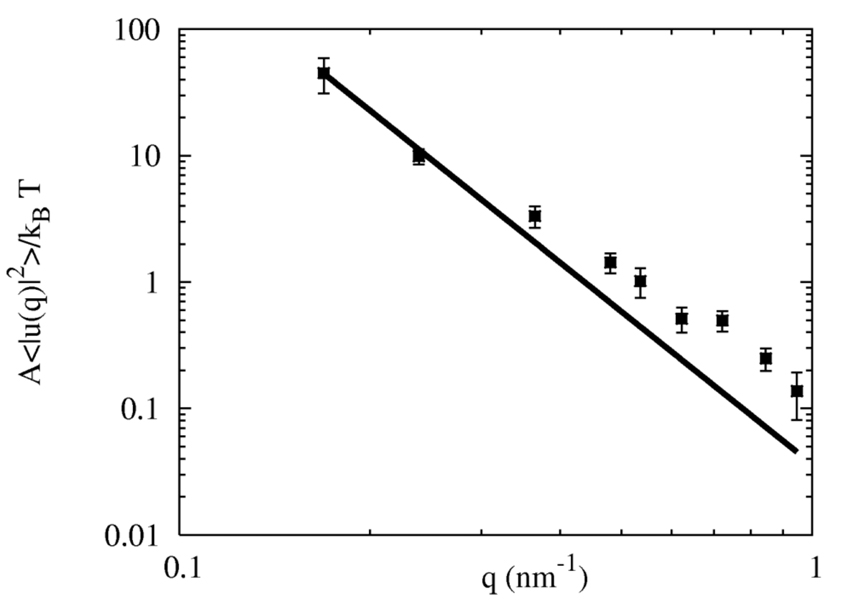
A critical membrane material property is the bending modulus, kc, which can in principle be found in the small wavevector, q, regime from membrane undulation fluctuations,20,29,74,75 though care must be taken in interpreting the resulting undulation spectrum.19,20,22,25 Recent studies have shown that wavevectors with (qσ0)2 ≤~ 0.1 need to be accessed in order for the 1/q4 scaling to be observed.19 Furthermore, it has been shown that membrane protrusions can also alter the undulation spectrum.22 Here, the boundary conditions of the CG lipid system are such that a square patch of membrane is bound in the xy plane, and thermal undulations, u(r, t) occur in the z-direction. Under these boundaries, and for (qσ0)2 ≤~ 0.1, the bending modulus can be measured using the undulation spectrum of 〈|u(q)|2〉 = 〈u(q,t)u(−q,t)〉, where q is a wavevector, u(q,t) is the Fourier mode of the undulation height u(r,t) and
| (11) |
Equipartition under zero surface tension gives
| (12) |
Figure 7 shows the quantity 〈|u(q)|2〉A/kBT versus q = |q|. For reference, the experimental measurement of the bending modulus for DMPC (3 ~ 6 × 10− 20J).76,77 The resulting low q CG lipid undulation magnitudes are quite reasonable. The bending modulus was estimated from fitting the undulation spectrum according to Eq. 12) using only those wavevectors that satisfy (qσ0)2 ≤ 0.1. The bending modulus for the HAS membrane was found to be kc = 4.6 ± 0.2 × 10−20 J , while the modulus for the raw GB membrane was significantly larger at around kc = 8 × 10−20 J . Deviations from the 1/q4 are expected to contribute at higher wavevectors, and a more complex analysis is required in this regime.22 It should be noted that the bending modulus from the atomistic MD system was not determined as it was too small to access low wavelength undulation modes, with a lowest q of about 1 nm−1. Thus, the bending modulus of the CG lipid membrane is entirely a predicted quantity and results from the HAS CG model proposed in eq (1). It should be noted that tests with different values of ε0 had little or no effect on the bending modulus. This results from the design of the HAS CG model and the fact that that most of the in-plane forces arise from the MS-CG component and thus appear to dominate the membrane’s material properties. However, the GB component is responsible for “holding” the structure of the CG lipid bilayer together in a non-trivial way. Interestingly, even though the atomistic MD system that was used for force matching was too small to bend, the resulting CG model, when extended to larger system sizes, resulted in a bending modulus that was consistent with the underlying MD DMPC model from which it was originally derived (assuming that the atomistic DMPC model would give the correct bending modulus for a large enough system). The undulation dynamics of this solvent free CG model would, of course, miss critical hydrodynamic dampening due to interactions of the membrane with the surrounding viscous solvent. This behavior is characteristic of any solvent-free model where the membrane essentially exists in a vacuum with effective interactions between the lipids mimicking the effects of the solvent. A much more involved model would be required to correctly incorporate membrane hydrodynamics.
Figure 7.
The undulation spectrum for an N=5000 CG site square patch of HAS (solid squares ) and a “raw GB” (open squares) CG membrane under zero surface tension. Error bars are from block averages. The bending modulus for the HAS membrane using the smallest q was found to be kc = 4.6 ± 0.2 × 10−20 J , while the modulus for the raw GB membrane was significantly larger at kc = 8.0 × 10−20 J. For comparison, the experimental bending modulus for DMPC is kc = 3 ~ 6 × 10−20 J.
The area expansion modulus, KA, defined via (∂γ/∂A) T = KA/A* where γ is the surface tension, A is the area, and A* is the average area found under a starting state of zero surface tension,78 was found to be ~ 138 mJ/m2, in good agreement with experimental values of 145 mJ/m2.77 It should be noted that the area compressibility modulus of the small atomistic system is estimated at ~ 158 mJ/m2. Apparently, the larger CG system under zero surface tension, with the appearance of soft thermal modes, slightly modifies the atomistic result. Given that KA can vary by as much as a factor of 6 for different membranes, it is encouraging that the present HAS model can incorporate this property at such a high degree of coarse-graining.
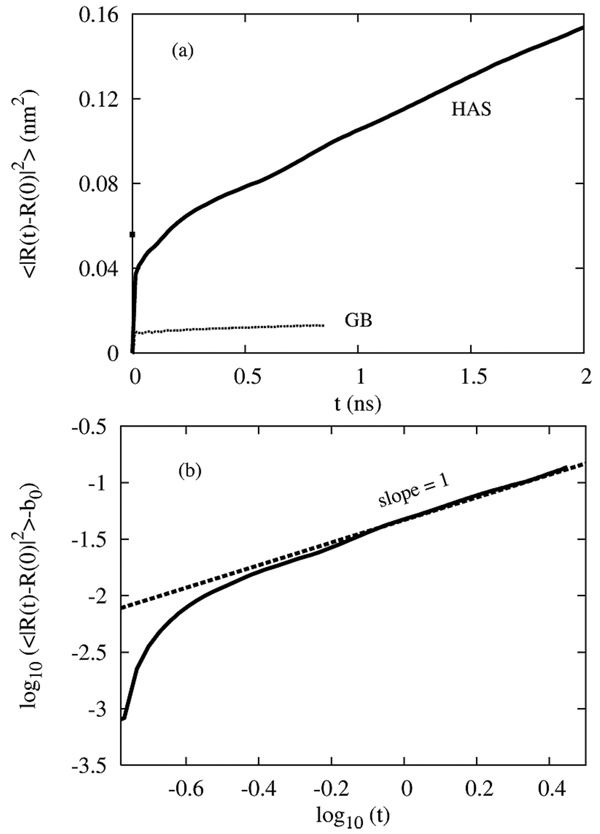
Figure 8a shows the two dimensional mean square displacement (MSD), 〈|R(t)−(0)|2〉, which is related to the two dimensional diffusion coefficient, D, via 〈|R(t)−(0)|2〉 = 4Dt. Results from both the HAS as well as the raw GB membrane are shown. It appears that the HAS CG bilayer is indeed fluid, with a diffusion coefficient of around 1.2 × 10−7 cm2/s, which is actually not far off the experimental value of around 1.10 × 10−7 cm2/s.77 The diffusion constant was found from a linear fit to the data above t = 0.8 ns. In contrast, the raw GB membrane is frozen, indicating once again that the effect of the HAS interaction is to melt the otherwise frozen GB bilayer. Panel b shows the quantity log10(〈|R(t)−(0)|2〉−b0) versus log10(t) which is expected to have a slope of 1 when 〈|R(t)−(0)|2〉 ~ 4Dt at long times. Indeed this behavior appears to recovered for t > 0.8 ns, indicating that the dynamics is diffusive. However, as has been noted previously, comparing CG and atomistic dynamical quantities only really has meaning when the correct frictional and drag forces are incorporated into the CG scheme;79 in this case, the fact that the system appears fluid (i.e., the MSD increased linearly) is sufficient. (Frictional forces would tend to decrease the diffusion constant and bring it into even better agreement with the experimental value.) We also note that we have made no attempt to “re-scale” time in these CG simulations.
Figure 8.
(a) The two dimensional mean square displacement (MSD), 〈|R(t) − R(0)|2〉 for the HAS (solid line) and “raw GB” (dotted) system. The corresponding diffusion coefficient is found from a linear fit, 〈|R(t) − R(0)|2〉 = 4Dt + b0, of the MSD for t > 0.8 ns for the HAS bilayer and is D = 1.2 × 10−7 cm2/s, compared to the experimental estimate of 1.10 × 10−7 cm2/s. The t = 0 intercept for this fit is b0 = 0.057 nm2, as shown by the small square. The corresponding raw GB bilayer is frozen. Panel (b) The quantity, log10(〈|R(t) − R(0)|2〉 − b0) versus log10(t) which is predicted to have a slope of unity at long times in the case that the linear fit proposed in (a) holds.
3.2 CG Simulation of a Liposome
With the in-plane fluid nature and the membrane bending modulus confirmed from the N=5000 simulation, a much larger HAS CG liposome simulation was performed. In this case, the liposome was placed in the center of a 220 × 220 × 220 nm3 simulation box and the initial liposome diameter was 190 nm. A total of 379,858 CG lipids were employed; the outer leaflet contained 195,312 CG lipids, while the inner leaflet contained 184,546 lipids. This HAS CG simulation is equivalent to a united atom lipid model with 17 million atoms, or about 1.1 × 109 atoms when a water solvent and an all-atom lipid model are employed. The difference between the two leaflets ensured that the CG lipid density in each monolayer was the same. The simulation was performed in a solvent-free mode over 512 processors using our in-house code TANTALUS.80 It was observed that about two nanoseconds of simulation time could be achieved over 10 wall clock hours even with this moderate level of parallelization. Given that fully solvated CG simulations at these length-scales are not even currently possible, this type of scaling is encouraging; scaling over 1000 processors is entirely possible with a more advanced parallelization scheme. The aim of these simulations was to assess the stability of the liposome, and to ensure that its membrane structure was similar to that observed with the smaller N=5000 simulation.
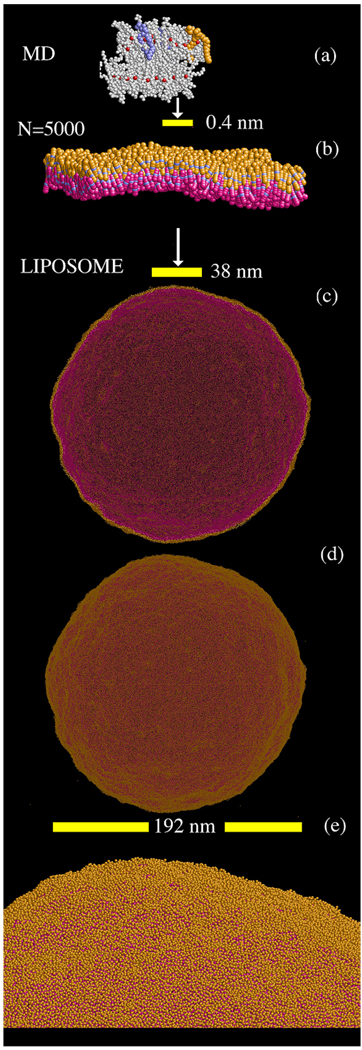
Figure 9, panels (c) and (d) show snapshots of the liposome after 60 ns of simulation. The first two panels, (a) and (b), give an estimate of the relative length-scales of the original atomistic system (as in Figure 1a) and the N=5000 CG lipid system (as in Figure 7), respectively, as compared to a close-up of the liposome surface. When the scale bars are compared relative to the entire liposome, it becomes clear how much of a length-scale jump has been achieved in the HAS CG simulation; the original MD system would be not much more than a small spot on the liposome surface. After 60 ns, the liposome structure remains intact, except for visible thermal undulations that have appeared all over the surface. The final average diameter of the liposome was 192 ± 2 nm, where the error arises from thermal undulations. The slightly larger diameter results from the CG lipids not being perfectly aligned at finite temperature; thus, the liposome swells slightly. The final area per CG lipid evaluated over the surface area of the liposome and averaged over the two leaflets was found to be 0.60 ± 0.02 nm2/CG lipid, indicating that the local structure in the liposome simulation was consistent with that in the smaller N=5000 HAS CG study.
Figure 9.
Relative lengthscales eventually reaching the HAS CG liposome with N = 379,858 total CG sites. Panel (a) shows the original MD system as in Figure 1, panel (b) is a snapshot of the N = 5000 HAS membrane. Panel (c) shows a slice of the HAS liposome after 60 ns of simulation, while (d) shows the entire surface. The lipids are colored as in Figure 6. The different scales are shown in the yellow scale bars. The liposome surface is clearly visible in the close-up in panel (e). The area per CG lipid evaluated over the surface area of the liposome was found to be 0.60 ± 0.02 nm2/CG lipid. The lipid flip flop rate was estimated at around 5 × 10−6 ns−1, based on counting the number of lipids that had moved from one leaflet to the other during the course of the simulation.
At this point at least the shorter-time stability of the HAS CG liposome over a reasonable simulation time-scale is confirmed. Longer simulations will be performed in the future when more complex scenarios (e.g., protein mediated membrane remodeling) are to be examined. The feasibility of the HAS CG approach is therefore established and the overall approach looks promising in systems where aggressive coarse-graining is required.
It should be noted that after 60 ns of CG simulation time around 60 lipids had swapped leaflets (out of the ~ 190,000 CG lipids per leaflet). It may be possible to pursue a more thorough lipid flip-flop rate analysis;19 however, given the ambiguity of what CG time actually means, the limited statistics, and the fact that some lipids actually leave the liposome, the fact that lipids do seems to swap leaflets over long time-scales is taken to be encouraging. A rough estimate for the lipid flip-flop rate is about 5 × 10−6 ns−1 , consistent with other CG models.19
A question arises as to whether 60 ns of CG simulation time is sufficient to equilibrate the liposome. As previously noted, CG time does not correspond to real time; the highly accelerated lipid flip flop rate supports this notion, and suggests that the system has effectively sampled longer timescales. Ideally, the duration of the simulation should be such that deformation modes corresponding to all accessible wavelengths of the liposome are fully sampled.81–83 Of course, the long wavelength undulation modes evolve much more slowly than the fast ones, and it is reasonable to assume that for a system of this size the long wavelength modes have not yet been fully sampled in the simulations presented here. At very large length scales, e.g., vesicles, the undulations due to thermal fluctuations become in fact sub-visible and act more as a reservoir for increases in the apparent surface area of the vesicle.84 However, within the context of specific biomolecular processes, e.g., membrane remodeling via BAR domains, simulations of the duration reported in this paper could be employed to examine to examine the early stages of protein induced membrane remodeling.
4. Conclusions
The aim of this paper was to develop an aggressively coarse-grained, yet systematically obtained CG lipid model. A CG approach was therefore presented where MS-CG with force matching was utilized to generate a CG force field that dominates in certain configurations that are well-sampled at the atomistic level. An analytic force field based on the Gay-Berne liquid crystal model is then used to model the anisotropic “core” of the CG lipid, and the final hybrid analytic-systematic (HAS) CG force field is obtained by combining the two components.
The resulting HAS CG lipid model is able to reproduce key bilayer quantities such as the bending modulus and lipid diffusion, all within a solvent-free CG simulation methodology that affords a substantial computational speed up as compared to the original atomistic MD model. When extended to much longer length-scales (i.e., a liposome) the HAS CG lipid model can be employed to simulate a liposome. Here, a 60 ns CG simulation of a liposome with a starting diameter of 190 nm was performed as a demonstration of the method. The bilayer structure of the liposome remained intact, including the emergence of thermal undulation modes.
The HAS CG methodology proposed here will become an integral part of a more fully enriched CG model in the future aimed at examining real biological membranes and processes such as membrane remodeling.
Acknowledgements
This research was supported by the National Institutes of Health (R01-GM063796). Computational resources were provided by the National Science Foundation through TeraGrid computing resources, specifically the Texas Advanced Computing Center.
Appendix A
The normal solution and the delta function basis
Begin with the residual as given in eq (6) where is given by
| (A1) |
and δC = 1 if Rd ≤ RIJ < Rd+1 and is zero otherwise. A set of linear equations is found from setting ∂χ2/∂ϕd' = 0 for each d' = 1…Nd . It should be noted that the d=1 bin at RC,1 could be set at any desired location. For example, RC,1 could be set at the distance corresponding to the first non-zero entry in the RDF (see Figure 2). It could also be set at other distances and a different solution will then found for that set of coefficients.
Appendix B
An anisotropic Gay-Berne lipid bilayer model
The Gay-Berne (GB) liquid crystal model has been discussed in detail elsewhere;69,70 here an anisotropic version is presented where the two “ends” of the GB ellipsoids interact differently. By breaking the symmetry of the GB model, molecules such as lipids, which have headgroups at one end, and hydrocarbon chains at the other, can be modeled.
The GB interaction energy for a pair of ellipsoids I and J was given by eq (8). The quantity σ0 is the width of the GB particle, while σ(RIJ,eI,eJ) gives the distance of closest approach along R̂IJfor a pair of GB particles with orientations eI, eI and is given by
| (B1) |
where χ = (κ2 − 1)/(κ2 + 1) and h = κσ0. Here h is the length of the GB particle along the symmetry axis. This component of the GB interaction models the ellipsoidal shape of the molecule. The energy component, ε(RIJ,eI,eJ), can be easily modified to break the symmetry of the GB model; one scheme is presented here. The energy component is given by
| (B2) |
where, in order to make the ends of two GB particles interact differently, the last term, ε2(RIJ,eI,eJ), can be slightly modified to
| (B3) |
and the last term is no longer squared (as it usually is in a regular GB interaction model). To see how this works, first consider two GB particles that are side-by-side and parallel: In this case ε2(R̂IJ,eI,eJ) = l. Now consider two GB particles end-to-end and with eI•eJ = (R̂IJ,eI,eJ = (1 − χ')/(1 + χ'). With RIJ = RI − RJ, a configuration with eI•eJ = −l and R̂•eI = 1 has ε2eI=1/(1+χ'), while with R̂•eI = −l, ε2(R̂IJ,eI,eJ = (1 + 2χ')/(1 + 2χ'). The commonly employed value of χ' is given by where εss/εee gives the ratio of the well depths for two side-by-side and end-to-end particles, and is typically given a value between 3 and 5.69,70 A value of 5 was chosen here to match over to previous studies.70 Thus, this broken-symmetry GB model gives the deepest end-to-end well depth when eI•eJ = −1 and R̂IJ•eI= −1. In terms of a lipid bilayer, this configuration corresponds to a configuration where the lipid tails are in contact, and the headgroups are at opposite ends.
References
- 1.Singer SJ, Nicolson GL. Science. 1972;175:720. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Engelman DM. Nature. 2005;438:578. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin S, Murray D. Nature. 2005;438:605. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 4.Dupuy AD, Engelman DM. Proc. Nat. Acad. Sci. 2008;105:2848. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veatch SL, Keller SL. Biophys. J. 2003;85:3074. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. Science. 2004;303:495. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 7.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, McMahon HT. EMBO J. 2006;25:2898. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayton GS, Noid WG, Voth GA. Curr. Opin. Struct. Bio. 2007;17:192. doi: 10.1016/j.sbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Voth GA, editor. Coarse-graining of condensed phase and biomolecular systems. Boca Raton: CRC Press/Taylor and Francis Group; 2009. [Google Scholar]
- 10.Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML. J. Phys. Chem. B. 2001;105:4464. [Google Scholar]
- 11.Marrink SJ, deVries AH, Mark AE. J. Phys. Chem. B. 2004;108:750. [Google Scholar]
- 12.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, deVries AH. J. Phys. Chem. B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 13.Stevens MJ. J. Chem. Phys. 2004;121:11942. doi: 10.1063/1.1814058. [DOI] [PubMed] [Google Scholar]
- 14.Faller R, Marrink SJ. Langmuir. 2004;20:7686. doi: 10.1021/la0492759. [DOI] [PubMed] [Google Scholar]
- 15.Izvekov S, Voth GA. J. Phys. Chem. B. 2005;109:2469. doi: 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- 16.Izvekov S, Voth GA. J. Chem. Theor. Comp. 2006;2:637. doi: 10.1021/ct050300c. [DOI] [PubMed] [Google Scholar]
- 17.Farago O. J. Chem. Phys. 2003;119:596. [Google Scholar]
- 18.Cooke IR, Kremer K, Deserno M. Phys. Rev. E. 2005;72:011506. doi: 10.1103/PhysRevE.72.011506. [DOI] [PubMed] [Google Scholar]
- 19.Cooke IR, Deserno M. J. Chem. Phys. 2005;123:224710. doi: 10.1063/1.2135785. [DOI] [PubMed] [Google Scholar]
- 20.Brannigan G, Brown FLH. J. Chem. Phys. 2004;120:1059. doi: 10.1063/1.1625913. [DOI] [PubMed] [Google Scholar]
- 21.Brannigan G, Lin LCL, Brown FLH. Eur. Biophys. J. 2006;35:104. doi: 10.1007/s00249-005-0013-y. [DOI] [PubMed] [Google Scholar]
- 22.Brannigan G, Brown FLH. Biophys. J. 2006;90:1501. doi: 10.1529/biophysj.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooke IR, Deserno M. Biophys. J. 2006;91:487. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Nature. 2007;447:461. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 25.Harmandaris VA, Deserno M. J. Chem. Phys. 2006;125:204905. doi: 10.1063/1.2372761. [DOI] [PubMed] [Google Scholar]
- 26.Lyubartsev AP. European Journal of Biophysics. 2005;35:53. doi: 10.1007/s00249-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 27.Markvoort AJ, Pieterse K, Steijaert MN, Spijker P, Hilbers PAJ. J. Phys. Chem. B. 2005;109:22649. doi: 10.1021/jp053038c. [DOI] [PubMed] [Google Scholar]
- 28.Markvoort AJ, van Santen RA, Hilbers PAJ. J. Phys. Chem. B. 2006;110:22780. doi: 10.1021/jp064888a. [DOI] [PubMed] [Google Scholar]
- 29.Goetz R, Gompper G, Lipowsky R. Phys. Rev. Lett. 1999;82:221. [Google Scholar]
- 30.Goetz R, Lipowsky R. J. Chem. Phys. 1998;108:7397. [Google Scholar]
- 31.Kumar PBS, Gompper G, Lipowsky R. Phys. Rev. Lett. 2001;86:3911. doi: 10.1103/PhysRevLett.86.3911. [DOI] [PubMed] [Google Scholar]
- 32.Shillcock JC, Lipowsky R. NIC Symposium Proceedings; 2001. p. 407. [Google Scholar]
- 33.Shillcock JC, Lipowsky R. J. Chem. Phys. 2002;117:5048. [Google Scholar]
- 34.Shillcock JC, Lipowsky R. Nat. Mater. 2005;4:225. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 35.Michel J, Orsi M, Essex JW. J. Phys. Chem. B. 2008;112:657. doi: 10.1021/jp076142y. [DOI] [PubMed] [Google Scholar]
- 36.Orsi M, Haubertin DY, Sanderson WE, Essex JW. J. Phys. Chem. B. 2008;112:802. doi: 10.1021/jp076139e. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto S, Maruyama Y, Hyodo S. J. Chem. Phys. 2002;116:5842. [Google Scholar]
- 38.Ayton G, Bardenhagen S, McMurtry P, Sulsky D, Voth GA. J. Chem. Phys. 2001;114:6913. [Google Scholar]
- 39.Risselada HJ, Mark AE, Marrink SJ. J. Phys. Chem. B. 2008;112:7438. doi: 10.1021/jp0758519. [DOI] [PubMed] [Google Scholar]
- 40.Marrink SJ, Risselada HJ, Mark AE. Chemistry and Physics of Lipids. 2005;135:223. doi: 10.1016/j.chemphyslip.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Risselada HJ, Marrink SJ. Proc. Nat. Acad. Sci. 2008;105:17367. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knecht V, Marrink SJ. Biophys. J. 2007;92:4254. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marrink SJ, Mark AE. J. Amer. Chem. Soc. 2003;125:11144. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 44.Marrink SJ, Mark AE. J. Amer. Chem. Soc. 2003;125:15233. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- 45.Murtola T, Falck E, Karttunen M, Vattulainen I. J. Chem. Phys. 2007;126:075101. doi: 10.1063/1.2646614. [DOI] [PubMed] [Google Scholar]
- 46.Murtola T, Falck E, Patra M, Karttunen M, Vattulainen I. J. Chem. Phys. 2004;121:9156. doi: 10.1063/1.1803537. [DOI] [PubMed] [Google Scholar]
- 47.Arkhipov A, Yin Y, Schulten K. Biophys. J. 2008;95:2806. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayton GS, Blood PD, Voth GA. Biophys. J. 2007;92:3595. doi: 10.1529/biophysj.106.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayton GS, Lyman E, Swenson RD, Voth GA. Biophys. J. 2008 (in press) [Google Scholar]
- 50.Brown FLH. Biophys. J. 2003;84:842. doi: 10.1016/S0006-3495(03)74903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin LC-L, Brown FLH. Biophys. J. 2004;86:764. doi: 10.1016/S0006-3495(04)74153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin LCL, Brown FLH. Phys. Rev. E. 2005;72:011910. doi: 10.1103/PhysRevE.72.011910. [DOI] [PubMed] [Google Scholar]
- 53.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. J. Chem. Theory Comput. 2008;4:819. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 54.Tozzini V. Curr. Opin. Struc. Bio. 2005;15:144. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Smondyrev AM, Berkowitz ML. J. Comp. Chem. 1999;20:531. [Google Scholar]
- 56.Hiroshi N, Gerhard G. J. Chem. Phys. 2006;125:164908. [Google Scholar]
- 57.Blood PD, Voth GA. Proc. Nat. Acad. Sci. 2006;103:15068. doi: 10.1073/pnas.0603917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takei K, Slepnev VI, Haucke V, De Camilli P. Nat Cell Biol. 1999;1:33. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 59.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. EMBO J. 2006;25:2889. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noid WG, Chu JW, Ayton GS, Krishna V, Izvekov S, Voth GA, Das A, Anderson HC. J. Chem. Phys. 2008;128:244114. doi: 10.1063/1.2938860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noid WG, Liu P, Wang Y, Chu J-W, Ayton GS, Izvekov S, Andersen HC, Voth GA. J. Chem. Phys. 2008;128:244115. doi: 10.1063/1.2938857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayton GS, Noid WG, Voth GA. MRS Bulletin. 2007;32:929. [Google Scholar]
- 63.Noid WG, Chu J-W, Ayton GS, Voth GA. J. Phys. Chem. B. 2007;111:4116. doi: 10.1021/jp068549t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Izvekov S, Voth GA. J. Chem. Phys. 2005;123:134105. doi: 10.1063/1.2038787. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Izvekov S, Yan T, Voth GA. J. Phys. Chem. B. 2006;110:3564. doi: 10.1021/jp0548220. [DOI] [PubMed] [Google Scholar]
- 66.Zhou J, Thorpe IF, Izvekov S, Voth GA. Biophys. J. 2007;92:4289. doi: 10.1529/biophysj.106.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Izvekov S, Voth GA. J. Phys. Chem. B. :2008. submitted. [Google Scholar]
- 68.Thorpe I, Zhou J, Voth GA. J. Phys. Chem. B. 2008;112:13079. doi: 10.1021/jp8015968. [DOI] [PubMed] [Google Scholar]
- 69.Gay JG, Berne BJ. J. Chem. Phys. 1981;74:3316. [Google Scholar]
- 70.Brown JT, Allen MP. Phys. Rev. E. 1998;57:6685. [Google Scholar]
- 71.De Miguel E, Rull LF, Chalam MK, Gubbins KE. Mol. Phys. 1991;74:405. [Google Scholar]
- 72.Ayton G, Smondyrev AM, Bardenhagen S, McMurtry P, Voth GA. Biophys. J. 2002;82:1226. doi: 10.1016/S0006-3495(02)75479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford: Clarendon; 1987. [Google Scholar]
- 74.Lindahl E, Edholm O. Biophys. J. 2000;79:426. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marrink SJ, Mark AE. J. Phys. Chem. B. 2001;105:6122. [Google Scholar]
- 76.Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Biophys. J. 2005;88:2626. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipowsky R, Sackmann E. Structure and Dynamics of Membranes. Vol. 1A. North-Holland: 1995. [Google Scholar]
- 78.Feller SE, Pastor RW. J. Chem. Phys. 1999;111:1281. [Google Scholar]
- 79.Izvekov S, Voth GA. J. Chem. Phys. 2006;125:151101. doi: 10.1063/1.2360580. [DOI] [PubMed] [Google Scholar]
- 80.Ayton GS, Voth GA. Int. J. Mult. Comp. Eng. 2004;2:291. [Google Scholar]
- 81.Drouffe J-M, Maggs AC, Leibler S. Science. 1991;254:1353. doi: 10.1126/science.1962193. [DOI] [PubMed] [Google Scholar]
- 82.Gompper G, Kroll DM. J. Phys.: Condens. Matter. 2000;12:A29. [Google Scholar]
- 83.Milner ST, Safran SA. Phys. Rev. A. 1987;36:4371. doi: 10.1103/physreva.36.4371. [DOI] [PubMed] [Google Scholar]
- 84.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Biophys. J. 2000;79:328. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]