Abstract
We describe a multiplex nucleic acid assay that identifies and determines the abundance of four different pathogenic retroviruses (HIV-1, HIV-2, and human T-lymphotrophic virus types I and II). Retroviral DNA sequences are amplified in a single, sealed tube by simultaneous PCR assays, and the resulting amplicons are detected in real time by the hybridization of four differently colored, amplicon-specific molecular beacons. The color of the fluorescence generated in the course of amplification identifies which retroviruses are present, and the number of thermal cycles required for the intensity of each color to rise significantly above background provides an accurate measure of the number of copies of each retroviral sequence that were present originally in the sample. Fewer than 10 retroviral genomes can be detected. Moreover, 10 copies of a rare retrovirus can be detected in the presence of 100,000 copies of an abundant retrovirus. Ninety-six samples can be analyzed in 3 hr on a single plate, and the use of a closed-tube format eliminates crossover contamination. Utilizing previously well characterized clinical samples, we demonstrate that each of the pathogenic retroviruses can be identified correctly and no false positives occur. This assay enables the rapid and reliable screening of donated blood and transplantable tissues.
Keywords: real-time PCR, hairpin-forming probes, multicolor fluorescent labels, homogeneous assays, viral quantitation
Donated blood routinely is screened for the presence of pathogenic retroviruses. HIV-1 (1) and HIV-2 (2) cause acquired immunodeficiency syndrome (AIDS), and human T-lymphotropic virus types I and II (HTLV-I and HTLV-II) are associated with adult T cell lymphoma/leukemia (3), rare lymphocytic neoplasms (4), and progressive neurodegenerative disease (5, 6). Conventional serodiagnostic techniques used for detecting these retroviruses are not satisfactory because they fail to detect early infection when the immune response is developing (7). Furthermore, specific antibodies are not always present in HTLV-infected individuals (8). Assays that detect retroviral nucleic acid, instead of antibodies, do not suffer from these shortcomings, because integrated retroviral DNA is present in every infected cell. However, there can be as little as a single infected cell in a blood sample (9), so it is necessary to use a powerful amplification technique, such as PCR, to generate millions of copies of the target sequence, enabling reliable detection of the retroviral DNA.
Although extremely promising results have been obtained with PCR assays (10), they suffer from a number of debilitating drawbacks that have limited their clinical acceptance. The oligonucleotide primers that are designed to hybridize specifically to retroviral target sequences sometimes hybridize to human sequences, generating unintended amplicons that give false-positive signals; furthermore, the primers sometimes hybridize to each other, generating short “primer–dimer” amplicons (11). To improve primer specificity, nested sets of primers are added at different times during amplification (12). Alternatively, intended amplicons are distinguished from false amplicons by gel electrophoresis, which often is followed by Southern blot analysis. An undesirable consequence of opening the reaction tubes to carry out these manipulations is that amplicons are spread throughout the laboratory, contaminating untested samples and leading to false-positive results that cannot be distinguished from the results obtained from truly infected samples (13). Although “sterilization” schemes have been used to destroy contaminating amplicons in untested samples (14), and multicompartment assay chambers have been developed to carry out amplification, amplicon sorting, and amplicon detection in a hermetically sealed environment from which amplicons cannot escape (15), the high cost and complexity of these schemes have limited their utility. Practical clinical assays should be simple, fast, inexpensive, sensitive, utilize a high-throughput format that enables the testing of many samples simultaneously, and, ideally, allow the detection of a series of different pathogenic agents in the same assay tube. Heretofore, nucleic acid detection assays have been far too complex to achieve all of these goals in a single assay. In this paper, we report the development of an extremely sensitive PCR assay that ignores false amplicons, detects intended amplicons in real time, is carried out in a sealed reaction tube, and is able to identify simultaneously four different pathogenic retroviruses. All of these advantageous properties are enabled by the use of remarkable fluorogenic hybridization probes called “molecular beacons.”
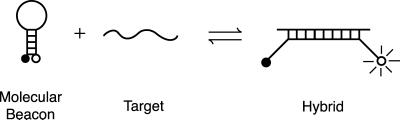
Molecular beacons are single-stranded oligonucleotide detector probes that form a stem-and-loop structure (Fig. 1). The loop contains a probe sequence that is complementary to the target amplicon, and the stem is formed by the annealing of complementary arm sequences that are located on either side of the probe sequence. A fluorophore is linked covalently to the end of one arm and a quencher is linked covalently to the end of the other arm. Molecular beacons do not fluoresce when free in solution. However, when they hybridize to a target strand they undergo a conformational change that enables them to fluoresce brightly. In the absence of target, the stem keeps the fluorophore in close proximity to the quencher, causing the fluorescence of the fluorophore to be quenched by energy transfer. The quencher is a nonfluorescent chromophore that dissipates the energy that it receives from the fluorophore as heat. When the probe encounters a target molecule, it forms a probe–target hybrid that is longer and more stable than the stem hybrid. The rigidity and length of the probe–target hybrid precludes the simultaneous existence of the stem hybrid. Consequently, the molecular beacon undergoes a spontaneous conformational reorganization that forces the stem hybrid to dissociate and the fluorophore and the quencher to move away from each other, restoring fluorescence (16).
Figure 1.
Principal of operation of molecular beacons. Free molecular beacons are nonfluorescent because the hairpin stem keeps the fluorophore close to the quencher. When the probe sequence in the hairpin loop hybridizes to its target, forming a rigid double helix, a conformational change occurs that removes the quencher from the vicinity of the fluorophore, thereby restoring fluorescence.
Because nonhybridized molecular beacons are dark, it is not necessary to isolate the probe–target hybrids to determine the number of amplicons synthesized during an assay. Molecular beacons are added to the assay mixture before carrying out amplification, and fluorescence is measured in real time. The assay tube remains sealed, and carryover contamination does not occur. Furthermore, the use of molecular beacons provides an additional level of specificity. Because it is very unlikely that false amplicons or primer dimers possess target sequences for the molecular beacons, the generation of a fluorescent signal is exclusively due to the synthesis of the intended amplicons. Moreover, molecular beacons can be synthesized that possess differently colored fluorophores (17), enabling assays to be designed that simultaneously detect different pathogenic retroviruses in the same reaction. In these multiplex assays, a number of different primer sets are present, each set enabling the amplification of a unique gene sequence from a different retrovirus, and a number of different molecular beacons are present, each specific for one of the retroviral amplicons and each labeled with a differently colored fluorophore. The color of the resulting fluorescence, if any, identifies the retrovirus that is present in the sample, and the number of amplification cycles required to generate a detectable fluorescent signal indicates the number of retroviral targets that were present originally.
The multiplex PCR assay that we report here uses differently colored molecular beacons to simultaneously detect HIV-1, HIV-2, HTLV-I, and HTLV-II, in a rapid, high-throughput format, carried out in a 96-well spectrofluorometric thermal cycler. Using each of the four retroviral DNAs as templates, we demonstrate the extraordinary specificity and sensitivity of the assay. We also show that, in a sample that contains two different retroviruses, a rare retrovirus can be distinguished from an abundant retrovirus without interfering with the ability to quantitate each. Finally, we used a diverse series of well characterized human blood samples to demonstrate that the assay correctly identifies each of the retroviruses in clinical samples and does so without producing false-positive results.
MATERIALS AND METHODS
Primers.
Conserved sequences of the gag gene of HIV-1, the env gene of HIV-2, the tax gene of HTLV-I, and the pol gene of HTLV-II were amplified. Compatible primer sets were selected to ensure efficient amplification and detection of most subtypes of each retrovirus (18). The sequences of the primers were: gagF, 5′-ATAATCCACCTATCCCAGTAGGAGAAAT-3′, and gagR, 5′-TTTGGTCCTTGTCTTATGTCCAGAATG-3′ (19); envF, 5′-CTCCAGGCAAGAGTCACTGCTAT-3′, and envR, 5′-CCCATGGTACAGTAGTGTGGCAG-3′ (20); taxF, 5′-CAATCACTCATACAACCCCCAA-3′, and taxR, 5′-TCTGGAAAAGACAGGGTTGGG-3′ (21); and polF, 5′-CAGGGCAAGACCATCTACCT-3′, and polR, 5′-TCAGGGGAACAAGGGGAGC-3′ (22). We chose the sequence of polF to enable the detection of both the A and B subtypes of HTLV-II.
Molecular Beacons.
Four molecular beacons were used to detect the PCR amplicons. Each possessed 6-nt arm sequences and either a 25- or a 33-nt probe sequence. The arm sequences were designed to form a stable stem hybrid at the annealing temperature of the PCR, ensuring that nonhybridized molecular beacons would be dark. The probe sequences were designed to be complementary to a conserved region within their target amplicon. The length of each probe sequence was chosen to ensure that fluorescent probe–target hybrids would form at the annealing temperature, even if the target sequence is from a retroviral subtype that contains mutations in the target sequence. The 25-nt probe sequences could tolerate one mismatched base pair, and the 33-nt probe sequence could tolerate two mismatched base pairs. Each molecular beacon was labeled with a differently colored fluorophore: fluorescein (FAM) was used to label the molecular beacon that was specific for the HIV-1 amplicon, tetrachloro-6-carboxyfluorescein (TET) was used for the HIV-2-specific molecular beacon, tetramethylrhodamine (TMR) was used for the HTLV-I-specific molecular beacon, and 5-carboxyrhodamine 6G (RHD) was used for the HTLV-II-specific molecular beacon. The emission maxima of the four fluorophores were well spaced across the visible spectrum. All of the molecular beacons possessed the nonfluorescent quencher DABCYL (or a DABCYL analog). The sequences of the molecular beacons were: HIV-1/FAM, 5′-GCGAGCCTGGGATTAAATAAAATAGTAAGAATGTATAGCGCTCGC-3′; HIV-2/TET, 5′-GCGAGCAAAGGACCAGGCGCAACTAAATTCAGCTCGC-3′; HTLV-I/TMR, 5′-GCGAGCTCCTCCAGGCCATGCGCAAATACTCGCTCGC-3′; and HTLV-II/RHD, 5′-CGCTCGCTCCCCGACCCAATTTCCACCTTCACGAGCG-3′, where underlines identify the probe sequences.
Molecular beacons HIV-1/FAM and HTLV-I/TMR possessed 4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL) at their 3′ ends and were prepared from synthetic oligonucleotides that contained a primary amino group at their 3′ end and a thiol group at their 5′ end, using a protocol that is available at http://www.molecular-beacons.org (17). This protocol was modified for the synthesis of molecular beacon HTLV-II/RHD. The succinimidyl ester of 5-carboxyrhodamine 6G was coupled to the 3′ amino group, and 4-dimethylaminophenylazophenyl-4′-maleimide (DABMI) was coupled to the 5′ thiol group. All of the fluorophores and quenchers were obtained from Molecular Probes. Molecular beacon HIV-2/TET was synthesized completely on an Applied Biosystems 394 DNA synthesizer (Perkin–Elmer), using a controlled-pore glass column to introduce a 4-dimethylaminoazobenzene-4′-sulfonyl moiety (DABSYL; Glen Research) at the 3′ end of the oligonucleotide and a tetrachloro-6-carboxyfluorescein phosphoramidite (Glen Research) to add the fluorophore to the 5′ end of the oligonucleotide. Each molecular beacon was purified by HPLC through a C-18 reverse-phase column.
PCR Assays.
Each 50-μl reaction contained the relevant template DNA, 1.00 μM of each HIV-1 primer, 0.25 μM of each HIV-2 primer, 0.50 μM of each HTLV-I primer, 0.25 μM of each HTLV-II primer, 0.14 μM HIV-1/FAM, 0.50 μM HIV-2/TET, 0.41 μM HTLV-I/TMR, 0.23 μM HTLV-II/RHD, 250 μM dATP, 250 μM dCTP, 250 μM dGTP, 500 μM dUTP, 2.5 units of AmpliTaq Gold DNA polymerase (Perkin–Elmer), 4 mM MgCl2, 50 mM KCl, and 10 mM Tris⋅HCl, pH 8.3. After activating the DNA polymerase by incubation for 10 min at 95°C, 35–45 cycles of amplification (94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec) were carried out in a 96-well spectrofluorometric thermal cycler (Applied Biosystems Prism 7700). Fluorescence was monitored during every thermal cycle at the 55°C annealing step. Before carrying out the reactions, the emission spectrum obtained from each molecular beacon hybridized to an excess of perfectly complementary target in the PCR buffer at 55°C was stored in the memory of the computer that controls the spectrofluorometric thermal cycler. These stored reference spectra were utilized by the computer to decompose the complex fluorescence emission spectra generated during the reactions into the spectral contributions of each of the four differently colored molecular beacons that were present in each reaction. To compare the fluorescent signals resulting from the synthesis of each target amplicon, the fluorescence intensity (F) of each molecular beacon was normalized by using the formula (F − Fmin)/(Fmax − Fmin), so that 0 represents fluorescence before its target was synthesized, and 1 represents the maximum level of fluorescence after its target was synthesized.
Template DNAs.
Plasmids BH10 (HIV-1) (23) and pJSP4–27/H6 (HIV-2) (24) and DNA extracted from cell lines MT-2 (HTLV-I) (25) and Mo (HTLV-II) (26) were used as templates for exploring the characteristics of the PCR assay. Plasmid K30 (HTLV-I) (27) was used as template to compare the sensitivity of our homogeneous detection method with the sensitivity of a conventional Southern blot detection method.
Clinical Samples.
Fifty-three well characterized blood samples containing either HIV-1, HTLV-I, HTLV-II, or no retroviral DNA and four well characterized cell lines infected with HIV-2 [HIV-2/ST (24), CBL-23 (28), HUT 78/HIV-2/D194 (29), and CEM-LAV2 (2)], were tested for the presence of each of the four retroviruses. These samples had been tested previously for the presence of retroviruses by using conventional serodiagnostic assays (30) and PCR amplification assays in which the target amplicons were detected with a solid-phase, colorimetric hybridization procedure (31). DNA was extracted from each blood sample by the quick lysis method (32), except for samples infected with HTLV-II, which were extracted organically (33), and 2–4 μg of the extracted DNA (originating from approximately 500,000 cells) was used as template in each PCR assay.
RESULTS
Design of the Assay.
To detect HIV-1, HIV-2, HTLV-I, and HTLV-II equally well in the same assay, it was necessary to select primer sets and molecular beacons that are compatible with each other. We chose primers that generate relatively short amplicons (100–130 bp), which produce brighter fluorescent signals, because molecular beacons are better able to compete with complementary strands for binding to target strands when the amplicons are shorter. The targets of the four primer sets were located in the gag gene of HIV-1, the env gene of HIV-2, the tax gene of HTLV-I, and the pol gene of HTLV-II. The primers were chosen so that they would not interact with each other and their target sequences would be unique to each retrovirus, highly conserved, present in most clinical subtypes, and not found in the human genome (18). In preliminary experiments, the concentration of each primer set was adjusted so that the rate of amplification of each of the four types of amplicons was approximately equal during the exponential phase of the reaction.
The probe sequence of each molecular beacon was chosen so that it would hybridize to a sequence within its target amplicon that is conserved among most clinical subtypes. The length of the probe sequence in each molecular beacon was chosen so that the stability of the resulting probe–target hybrid at the PCR annealing temperature would be about the same for all four molecular beacons, irrespective of the sequence of each probe. Although the length of the probe sequence can be shortened to ensure that it will bind only to a perfectly complementary target sequence (17), we used longer probe sequences so that probe–target hybrids were likely to form even if the retroviral target sequence was from a subtype that contained one or two nucleotide substitutions. Molecular beacon HIV-1/FAM was designed to detect HIV-1 subtypes A, B, C, D, F, and G; molecular beacon HIV-2/TET was designed to detect HIV-2 subtypes A, D, and SD; molecular beacon HTLV-I/TMR was designed to detect all HTLV-I subtypes; and molecular beacon HTLV-II/RHD was designed to detect HTLV-II subtypes A and B. The arm sequences of each molecular beacon were chosen so that they would hybridize to each other at the PCR annealing temperature but not to the probe sequence, and the entire sequence of each molecular beacon was analyzed by a computer program (34) to ensure that it would form a hairpin structure containing an unstructured probe sequence.
The concentrations of the molecular beacons were chosen to be about the same as the concentrations of the primers, so that they would hybridize to the amplicons at about the same rate as the primers; these concentrations were always greater than the maximum expected amplicon concentrations. However, each of the four fluorophores that was used to label the molecular beacons is stimulated to a different extent by the blue argon–ion laser of the spectrofluorometric thermal cycler. Thus, the signals generated by some molecular beacons were weaker than others. However, we found that the fraction of amplicons bound to molecular beacons could be increased by increasing the molecular beacon concentration. Therefore, we carried out preliminary experiments to adjust the concentration of each molecular beacon so that all four probes would provide a comparable fluorescent signal.
Each assay contained four primer sets, four molecular beacons, the DNA to be tested, nucleotides, and a thermostable DNA polymerase. The assay tubes were sealed permanently and then subjected to repeated cycles of DNA denaturation at 94°C, primer and molecular beacon annealing at 55°C, and primer extension at 72°C. The visible fluorescence emission spectrum (500–650 nM) was recorded from each assay tube during the annealing stage of each thermal cycle and stored in the memory of the computer controlling the assay instrument. The computer program then analyzed each stored spectrum and decomposed it into the fluorescent contribution that results from the binding of each type of molecular beacon to its target amplicon. The intensity of the decomposed signal at the end of the annealing stage was linearly proportional to the amount of target amplicon that was present.
Specificity of the Assay.
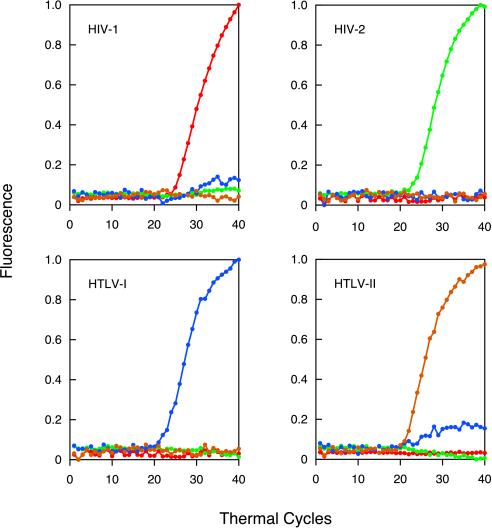
To determine how well individual retroviruses can be distinguished from one another in this multiplex format, we carried out four assays in parallel, each initiated with 100,000 molecules of one of the four retroviral DNAs. The results are shown in Fig. 2. The intensity of the fluorescence of each of the four molecular beacons (normalized on a scale from 0 to 1 to aid in their comparison) is plotted as a function of the number of thermal cycles completed. The only significant fluorescence that appeared in the course of the amplification reactions carried out in each assay tube was fluorescence from the molecular beacon that was complementary to the sequence of the expected amplicon. The color of the fluorescence identified the retroviral DNA that was originally added to the assay mixture. No significant fluorescence developed in a control assay that did not contain any template DNA. These results demonstrate that each molecular beacon binds only to its intended target amplicon, and the emission spectra of the four molecular beacons can be distinguished from each other.
Figure 2.
Real-time detection of four different retroviral DNAs in a multiplex format. Four assays were carried out in sealed tubes, each initiated with 100,000 molecules of a different retroviral DNA. Each reaction contained four sets of PCR primers specific for unique HIV-1, HIV-2, HTLV-I, and HTLV-II nucleotide sequences and four molecular beacons, each specific for one of the four amplicons and labeled with a differently colored fluorophore. Fluorescence from the fluorescein-labeled molecular beacon (HIV-1-specific) is plotted in red, fluorescence from the tetrachlorofluorescein-labeled molecular beacon (HIV-2-specific) is plotted in green, fluorescence from the tetramethylrhodamine-labeled molecular beacon (HTLV-I-specific) is plotted in blue, and fluorescence from the rhodamine-labeled molecular beacon (HTLV-II-specific) is plotted in brown. The slight HTLV-I signal seen in the assay initiated with HTLV-II DNA is an artifact that resulted from a portion of the rhodamine fluorescence being interpreted by the spectrofluorometric thermal cycler as tetramethylrhodamine fluorescence.
Simultaneous Detection of Two Different Retroviral Targets.
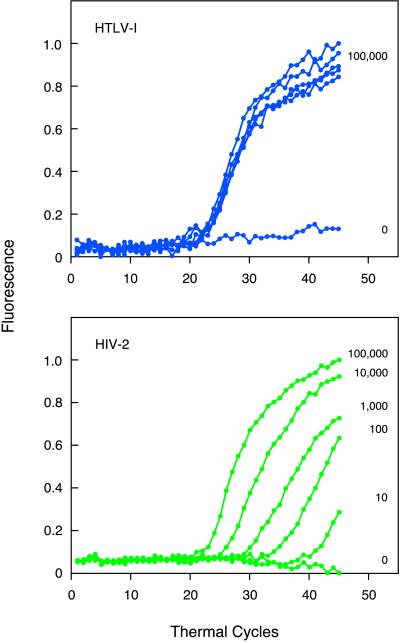
We evaluated the ability of the assay to detect two different retroviral DNAs in the same sample. We tested all six pairwise combinations of the four retroviral DNAs. The results showed that, irrespective of which two retroviral DNAs were present, the fluorescent signal that developed was due to fluorescence from the two molecular beacons that were complementary to the expected amplicons. In these experiments, both retroviral DNAs were present in the same concentration. We also assessed the ability of the assay to detect a rare retroviral DNA in the presence of an abundant retroviral DNA. We carried out five assays, in parallel, in which each tube initially contained 105 molecules of HTLV-I DNA and either 105, 104, 103, 102, or 101 molecules of HIV-2 DNA. The results (Fig. 3) show that both a fluorescent signal from tetramethylrhodamine (indicative of the presence of HTLV-I amplicons) and a fluorescent signal from tetrachlorofluorescein (indicative of the presence of HIV-2 amplicons) developed in every tube. The tetramethylrhodamine signal developed at the same time in all of the tubes, whereas the tetrachlorofluorescein signal developed at different times, depending on how many molecules of HIV-2 DNA were present originally. The fewer the number of molecules of HIV-2 DNA that were present initially, the longer it took for the fluorescent signal to appear. If a multiplex assay such as this one is to be useful for the simultaneous quantitation of different targets, the signal generated from one target must be independent of signals generated from other targets. The results of this experiment show that the number of thermal cycles required for a significant tetramethylrhodamine signal to develop from the 100,000 HTLV-I target molecules was unaffected by the number of HIV-2 target molecules that were present, and the number of thermal cycles required for a significant tetrachlorofluorescein signal to develop was indicative of the number of HIV-2 target molecules, irrespective of the presence of a relatively large number of HTLV-I target molecules. Moreover, the sensitivity of the assay was not affected significantly by the presence of the abundant HTLV-I DNA, because 10 molecules of HIV-2 DNA generated a clearly detectable signal.
Figure 3.
Detection of a rare retroviral target in the presence of an abundant retroviral target. Five multiplex assays were initiated with 105 molecules of HTLV-I DNA and either 105, 104, 103, 102, or 101 molecules of HIV-2 DNA, and a sixth multiplex assay, which served as a control, did not contain any template DNA. Each assay contained four sets of PCR primers and four differently colored molecular beacons. (Upper) Fluorescence from the tetramethylrhodamine-labeled molecular beacons, which is due to the synthesis of HTLV-I amplicons. (Lower) Fluorescence from the tetrachlorofluorescein-labeled molecular beacons, which is due to the synthesis of HIV-2 amplicons. The number of molecules of each retroviral DNA that were present originally in each assay tube is indicated to the right of each curve.
Quantitation and Sensitivity.
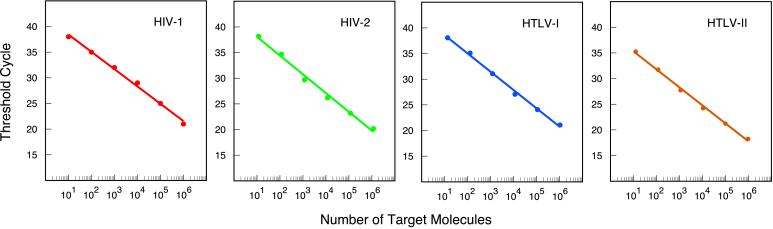
In assays that use PCR to exponentially amplify a target sequence, there is an inverse linear relationship between the number of thermal cycles it takes for enough amplicons to be synthesized for a fluorescent signal to appear and the logarithm of the number of target molecules that were present initially (35). To see whether our multiplex assay displays the same quantitative relationship, we carried out 24 reactions in parallel, in which each set of six assay tubes was initiated with 101, 102, 103, 104, 105, or 106 molecules of one of the four retroviral DNA templates. The results are shown in Fig. 4. The number of thermal cycles required to generate a significant fluorescent signal above the fluorescent background (threshold cycle) was plotted as a function of the logarithm of the number of retroviral DNA molecules placed in each assay tube. No matter which of the four retroviral DNAs was used, the results were linear and the slopes were similar (indicating that all four amplicons are amplified at a similar rate). Furthermore, these results demonstrate that, despite the presence of four different primer sets and four different molecular beacons, the assay provides quantitative results over an extremely wide range of target concentrations. Moreover, the assay reliably detects as little as 10 target molecules, irrespective of the identity of the retroviral target.
Figure 4.
Inverse linear relationship between the number of thermal cycles it takes for enough amplicons to be synthesized for a significant fluorescent signal to appear (threshold cycle) and the logarithm of the number of retroviral target molecules originally present in a sample. The threshold cycle for each fluorophore was reached when the intensity of the fluorescent signal was 10 times as great as the standard deviation of the background fluorescence. The results demonstrate that quantitative determinations can be made over an extremely wide range of target concentrations, and they show that the assay is sufficiently sensitive to detect as little as 10 molecules of retroviral DNA.
We compared the sensitivity of our multiplex assay with the sensitivity of an extremely sensitive, conventional PCR assay (33) in which the retroviral amplicons are analyzed by gel electrophoresis, transferred to a membrane, and identified by hybridization with a radioactively labeled probe. Serial dilutions containing 70,000, 7,000, 700, 70, 7, and 0 molecules of HTLV-I DNA were used as template for both assays. Neither assay gave a signal when no template molecules were present; yet, both assays were sufficiently sensitive to detect seven molecules of HTLV-I DNA. However, the homogeneous multiplex assay was much easier to perform and required only 3 hr to complete, whereas the conventional assay required 3 days. Moreover, the homogeneous assay could detect four different retroviruses and did not pose a risk of contaminating the laboratory with escaped amplicons.
Detection of Retroviruses in Clinical Samples.
To demonstrate that our multiplex assay works equally well with clinical samples, we tested 53 previously well characterized human blood samples. Eleven blood samples were from HIV-1-positive pregnant women, blood donors, and AIDS patients under treatment. These samples had a characteristically low target-copy number. Fifteen samples were from patients infected with HTLV-I and 17 samples were from patients infected with HTLV-II. The samples from both HTLV groups had a higher target-copy number than the samples from patients infected with HIV-1, because their blood is characterized by monoclonal expansion of HTLV-infected lymphocytes (36, 37). We also included 10 samples from healthy blood donors to serve as negative controls. Although we did not have any blood samples from HIV-2-infected patients, we prepared four additional samples from four different cultured human cell lines infected with HIV-2 at a characteristically high retroviral titer. The results are summarized in Table 1. The assay correctly identified all 11 HIV-1-infected blood samples, all 15 HTLV-I-infected blood samples, and all 17 HTLV-II-infected blood samples. The assay also correctly gave a negative result for all 10 uninfected blood samples. Although the assay correctly identified only two of the HIV-2 infected cell lines and incorrectly identified the other two as being uninfected, the HIV-2 target sequences in the positive cell lines were perfectly complementary to the probe sequence in the molecular beacon, whereas the target sequences in the “negative” cell lines each contained two nucleotide substitutions that caused the probe–target hybrids to be unstable. The inclusion of a longer probe sequence in the HIV-2-specific molecular beacon would have increased the stability of these hybrids, enabling the detection of HIV-2 in all four cell lines. The threshold cycles observed for the HTLV-I, HTLV-II, and HIV-2 samples were between 25 and 32, whereas the threshold cycles observed for the HIV-1 samples were 35 and higher, reflecting the characteristic lower abundance of retroviral targets in HIV-1-infected blood. Taken together, the results demonstrate that multiplex PCR assays in which amplicons are detected with molecular beacons are simple, sensitive, and fast, and they enable the reliable screening of donated blood and transplantable tissues for the presence of pathogenic retroviruses.
Table 1.
Detection of pathogenic retroviruses in clinical samples using multiplex PCR assays, each containing four primer sets and four differently colored molecular beacons
| Retrovirus in sample | No. of samples | Assay results
|
||||
|---|---|---|---|---|---|---|
| HIV-1 (+) | HIV-2 (+) | HTLV-I (+) | HTLV-II (+) | (−) | ||
| HIV-1 | 11 | 11 | — | — | — | — |
| HIV-2 | 4 | — | 2 | — | — | 2 |
| HTLV-I | 15 | — | — | 15 | — | — |
| HTLV-II | 17 | — | — | — | 17 | — |
| Control | 10 | — | — | — | — | 10 |
DISCUSSION
We have described a multiplex assay in which molecular beacons are used to detect amplicons in real time under homogeneous conditions. In this assay, the hairpin stem of the molecular beacons keeps the quencher and the fluorophore so close to each other that quenching occurs as a result of physical contact between the two label moieties (17). Consequently, a universal, nonfluorescent quencher, such as DABCYL, was incorporated into all of the molecular beacons, and only a single fluorophore needed to be present in each probe. There is sufficient room in the visible spectrum to distinguish four differently colored molecular beacons. Another way to detect amplicons in real time would have been to use the 5′ endonucleolytic activity of DNA polymerase to cleave a fluorophore from a doubly labeled oligonucleotide probe (38). Unlike molecular beacons, these “TaqMan” probes do not form hairpin stems, and the fluorophore and quencher interact with each other across a distance by fluorescence resonance energy transfer. Consequently, TaqMan probes must be labeled with two different fluorophores, and the signal that is measured is a change in the relative intensity of the fluorescence of each fluorophore. In multiplex assays, a different pair of fluorophores must be used to label each TaqMan probe. Consequently, fewer TaqMan probes can be distinguished from each other in an assay. Moreover, the fluorescence of nonhybridized molecular beacons is quenched much more efficiently than is the fluorescence of uncleaved TaqMan probes, significantly enhancing the ability to distinguish the fluorescence of each hybridized molecular beacon.
The number of different probes that can be accommodated in a homogeneous assay depends on the design of the assay instrument as well as the design of the probes. In the assay described here, we used an instrument that possessed a monochromatic light source for the excitation of the fluorophores, which limited the fluorophores to those that could be efficiently excited by its blue laser, preventing us from using molecular beacons labeled with fluorophores that fluoresce in the far-red wavelengths. If we had used an instrument with a variable-wavelength light source and a high-resolution detector, then a wider range of fluorophores could have been used for labeling the molecular beacons and more probes could have been accommodated in the same assay. However, assay instruments with these characteristics would be very expensive, limiting their wide acceptance. As an alternative, we have synthesized “wavelength-shifting” molecular beacons (unpublished data), which absorb the blue light from the laser but fluoresce strongly in the far-red wavelengths, thus increasing the number of probes that can be included in an assay. The use of these modified molecular beacons will enable us to detect simultaneously hepatitis B and hepatitis C, as well as HIV-1, HIV-2, HTLV-I, and HTLV-II.
Multiplex assays that utilize molecular beacons can be designed to detect infectious agents (39) and to discriminate genetic alleles (40–42). Moreover, the use of molecular beacons is not limited to assays that employ PCR, which require temperature cycling. Molecular beacons can be used in assays that employ isothermal nucleic acid amplification schemes (43, 44), such as strand-displacement amplification (45), nucleic acid sequence-based amplification (46), and rolling-circle amplification (47, 48). These assays do not require a thermal cycler, so they can be carried out in laboratories with limited resources. No matter which amplification scheme is employed, the addition of molecular beacons to enable multiple target detection will improve the reliability, speed, and ease of use of diagnostic clinical assays.
Acknowledgments
We thank Dong-Hun Lee, Amy Piatek, and Maria Rios for their assistance and advice. This work was supported by National Institutes of Health Grant HL-43521, Public Health Service Contract HB-67131, and the Barbara Kopp Cancer Research Fund.
ABBREVIATIONS
- HTLV-I and HTLV-II
types I and II human T-lymphotrophic virus
- DABCYL
4-(4′-dimethylaminophenylazo)benzoic acid
- FAM
fluorescein
- TET
tetrachloro-6-carboxyfluorescein
- RHD
5-carboxyrhodamine 6G
- TMR
tetramethylrhodamine
References
- 1.Popovic M, Sarngadharan M G, Read E, Gallo R C. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 2.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey M A, Santos-Ferreira M O, Laurent A G, Dauguet C, Katlama C, Rouzioux C, et al. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 3.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalyanaraman V S, Sarngadharan M G, Robert-Guroff M, Miyoshi I, Golde D, Gallo R C. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 5.Bhagavati S, Ehrlich G, Kula R W, Kwok S, Sninsky J, Udani V, Poiesz B J. N Engl J Med. 1988;318:1141–1147. doi: 10.1056/NEJM198805053181801. [DOI] [PubMed] [Google Scholar]
- 6.Hjelle B, Appenzeller O, Mills R, Alexander S, Torrez-Martinez N, Jahnke R, Ross G D. Lancet. 1992;339:645–646. doi: 10.1016/0140-6736(92)90797-7. [DOI] [PubMed] [Google Scholar]
- 7.Pantaleo G, Graziosi C, Fauci A S. N Engl J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 8.Zucker-Franklin D, Pancake B A, Marmor M, Legler P M. Proc Natl Acad Sci USA. 1997;94:6403–6407. doi: 10.1073/pnas.94.12.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho D D, Moudgil T, Alam M. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 10.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlich H E, Gelfand D, Sninsky J J. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 12.Mullis K B, Faloona F A. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 13.Kwok S, Higuchi R. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 14.Longo M C, Berninger M S, Hartley J L. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 15.Findlay J B, Atwood S M, Bergmeyer L, Chemelli J, Christy K, Cummins T, Donish W, Ekeze T, Falvo J, Patterson D, et al. Clin Chem. 1993;39:1927–1933. [PubMed] [Google Scholar]
- 16.Tyagi S, Kramer F R. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi S, Bratu D P, Kramer F R. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 18.Heredia A, Soriano V, Weiss S H, Bravo R, Vallejo A, Denny T N, Epstein J S, Hewlett I K. Clin Diag Virol. 1996;7:85–92. doi: 10.1016/s0928-0197(96)00255-3. [DOI] [PubMed] [Google Scholar]
- 19.Abbott M A, Poiesz B J, Byrne B C, Kwok S, Sninsky J J, Ehrlich G D. J Infect Dis. 1988;158:1158–1169. doi: 10.1093/infdis/158.6.1158. [DOI] [PubMed] [Google Scholar]
- 20.Udaykumar V, Heredia A, Soriano V, Bravo R, Epstein J S, Hewlett I K. J Virol Methods. 1994;49:37–46. doi: 10.1016/0166-0934(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 21.DeFreitas E, Hilliard B, Cheney P R, Bell D S, Kiggundu E, Sankey D, Wroblewska Z, Palladino M, Woodward J P, Koprowski H. Proc Natl Acad Sci USA. 1991;88:2922–2926. doi: 10.1073/pnas.88.7.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok S, Kellogg D, Ehrlich G, Poiesz B, Bhagavati S, Sninsky J J. J Infect Dis. 1988;158:1193–1197. doi: 10.1093/infdis/158.6.1193. [DOI] [PubMed] [Google Scholar]
- 23.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 24.Kong L I, Lee S W, Kappes J C, Parkin J S, Decker D, Hoxie J A, Hahn B H, Shaw G M. Science. 1988;240:1525–1529. doi: 10.1126/science.3375832. [DOI] [PubMed] [Google Scholar]
- 25.Popovic M, Sarin P S, Robert-Gurroff M, Kalyanaraman V S, Mann D, Minowada J, Gallo R C. Science. 1983;219:856–859. doi: 10.1126/science.6600519. [DOI] [PubMed] [Google Scholar]
- 26.Chen I S, McLaughlin J, Gasson J C, Clark S C, Golde D W. Nature (London) 1983;305:502–505. doi: 10.1038/305502a0. [DOI] [PubMed] [Google Scholar]
- 27.Zhao T M, Robinson M A, Bowers F S, Kindt T J. J Virol. 1995;69:2024–2030. doi: 10.1128/jvi.69.4.2024-2030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz T F, Whitby D, Hoad J G, Corrah T, Whittle H, Weiss R A. J Virol. 1990;64:5177–5182. doi: 10.1128/jvi.64.10.5177-5182.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhnel H, von Briesen H, Dietrich U, Adamski M, Mix D, Biesert L, Kreutz R, Immelmann A, Henco K, Meichsner C, et al. Proc Natl Acad Sci USA. 1989;86:2383–2387. doi: 10.1073/pnas.86.7.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dube D K, Dube S, Erensoy S, Jones B, Bryz-Gornia V, Spicer T, Love J, Saksena N, Lechat M F, Shrager D I, et al. Virology. 1994;202:379–389. doi: 10.1006/viro.1994.1354. [DOI] [PubMed] [Google Scholar]
- 31.Dyster L M, Abbott L, Bryz-Gornia V, Poiesz B J, Papsidero L D. J Clin Microbiol. 1994;32:547–550. doi: 10.1128/jcm.32.2.547-550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. AIDS Res Hum Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- 33.Dube D K, Sherman M P, Saksena N K, Bryz-Gornia V, Mendelson J, Love J, Arnold C B, Spicer T, Dube S, Glaser J B, et al. J Virol. 1993;67:1175–1184. doi: 10.1128/jvi.67.3.1175-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 35.Higuchi R, Fockler C, Dollinger G, Watson R. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 36.Ehrlich G D, Davey F R, Kirshner J J, Sninsky J J, Kwok S, Slamon D J, Kalish R, Poiesz B J. Am J Hematol. 1989;30:128–139. doi: 10.1002/ajh.2830300304. [DOI] [PubMed] [Google Scholar]
- 37.Love J L, Marchioli C C, Dube S, Bryz-Gornia V, Loughran T P, Jr, Glaser J B, Esteban E, Feldman L, Ferrer J F, Poiesz B J. J AIDS Hum Retrovirol. 1998;18:178–185. doi: 10.1097/00042560-199806010-00010. [DOI] [PubMed] [Google Scholar]
- 38.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 39.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Kramer F R, Alland D. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 40.Kostrikis L G, Tyagi S, Mhlangha M M, Ho D D, Kramer F R. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 41.Giesendorf B A J, Vet J A M, Tyagi S, Mensink E J M G, Trijbels F J M, Blom H J. Clin Chem. 1998;44:482–486. [PubMed] [Google Scholar]
- 42.Marras S A E, Kramer F R, Tyagi S. Genet Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 43.Guatelli J C, Whitfield K M, Kwoh D Y, Barringer K J, Richman D D, Gingeras T R. Proc Natl Acad Sci USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi S, Landegren U, Tazi M, Lizardi P, Kramer F R. Proc Natl Acad Sci USA. 1996;93:5395–5400. doi: 10.1073/pnas.93.11.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker G T, Fraiser M S, Schramm J L, Little M C, Nadeau J G, Malinowski D P. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leone G, van Schijndel H, van Gemen B, Kramer F R, Schoen C D. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D Y, Brandwein M, Hsuih T C, Li H. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 48.Lizardi P M, Huang X, Zhu Z, Bray-Ward P, Thomas D C, Ward D C. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]