Abstract
Background
N-arachidonoyl glycine (NAGly) is an endogenous signaling lipid with a wide variety of biological activity whose biosynthesis is poorly understood. Two primary biosynthetic pathways have been proposed. One suggests that NAGly is formed via an enzymatically regulated conjugation of arachidonic acid (AA) and glycine. The other suggests that NAGly is an oxidative metabolite of the endogenous cannabinoid, anandamide (AEA), through an alcohol dehydrogenase. Here using both in vitro and in vivo assays measuring metabolites with LC/MS/MS we test the hypothesis that both pathways are present in mammalian cells.
Results
The metabolic products of deuterium-labeled AEA, D4AEA (deuterium on ethanolamine), indicated that NAGly is formed by the oxidation of the ethanolamine creating a D2NAGly product in both RAW 264.7 and C6 glioma cells. Significantly, D4AEA produced a D0NAGly product only in C6 glioma cells suggesting that the hydrolysis of AEA yielded AA that was used preferentially in a conjugation reaction. Addition of the fatty acid amide (FAAH) inhibitor URB 597 blocked the production of D0NAGly in these cells. Incubation with D8AA in C6 glioma cells likewise produced D8NAGly; however, with significantly less efficacy leading to the hypothesis that FAAH-initiated AEA-released AA conjugation with glycine predominates in these cells. Furthermore, the levels of AEA in the brain were significantly increased, whereas those of NAGly were significantly decreased after systemic injection of URB 597 in rats and in FAAH KO mice further supporting a role for FAAH in endogenous NAGly biosynthesis. Incubations of NAGly and recombinant FAAH demonstrated that NAGly is a significantly less efficacious substrate for FAAH with only ~50% hydrolysis at 30 minutes compared to 100% hydrolysis of AEA. Co-incubations of AEA and glycine with recombinant FAAH did not, however, produce NAGly.
Conclusion
These data support the hypothesis that the signaling lipid NAGly is a metabolic product of AEA by both oxidative metabolism of the AEA ethanolamine moiety and through the conjugation of glycine to AA that is released during AEA hydrolysis by FAAH.
Background
N-arachidonoyl glycine (NAGly) was synthesized as part of a structure activity relationship study of the endocannabinoid anandamide (N-arachidonoyl ethanolamine; AEA; Fig. 1A) differing from AEA by the oxidation state of the carbon beta to the amido nitrogen (Fig. 1B); a modification that drastically reduces its activity at both cannabinoid receptors [1]. Nevertheless, NAGly produces antinociceptive and anti-inflammatory effects in mice and rats [2-5]. These findings gained physiological relevance when Huang et al. [3] demonstrated that NAGly is formed in numerous mammalian tissues including the brain. Subsequent studies by Kohno and colleagues [6] found that low concentrations (EC50 ~20 nM) of NAGly activate GPR18, an orphan G protein-coupled receptor. Consistent with the anti-inflammatory effects of NAGly, GPR18 is highly expressed in peripheral blood leukocytes and several hematopoietic cell lines. In pancreatic beta cells, NAGly caused intracellular calcium mobilization and insulin release [7]. NAGly inhibited the glycine transporter, GLYT2a through direct, non-competitive interactions [8] and more recently was reported as a partial agonist of Gq/11-coupled GPR92 receptors [9]. These data support the hypothesis that NAGly is an endogenous signaling molecule with multiple biological activities.
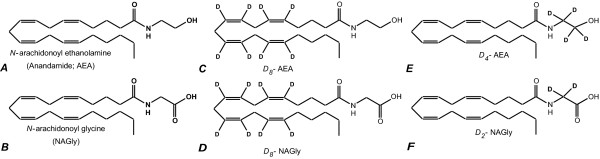
Figure 1.
Structures of AEA and NAGly. A) the endocannabinoid, N-arachidonoyl ethanolamine (anandamide; AEA) and B) the related signaling lipid, N-arachidonoyl glycine (NAGly); C) deuterium-labeled AEA with eight deuteriums on the arachidonic acid moiety; D) deuterium-labeled NAGly with eight deuteriums on the arachidonic acid moiety; E) deuterium-labeled AEA with four deuteriums on the ethanolamine moiety; F) deuterium-labeled NAGly with 2 deuteriums on the glycine moiety.
The biosynthesis and regulation of NAGly are only partially understood. Unlike 2-arachidonoyl glycerol and AEA, the biosynthesis of NAGly cannot logically be derived from phospholipid biochemistry. Two primary pathways for the biosynthesis of NAGly, have been proposed: 1) conjugation of arachidonic acid and glycine [2,3,10] and 2) oxygenation of AEA via the sequential enzymatic reaction of alcohol dehydrogenase (ADH) and aldehyde dehydrogenase [2,11].
Huang et al. [3] proposed that NAGly is synthesized by the condensation of arachidonic acid (AA) with glycine based upon the formation of deuterated NAGly following incubations of brain membranes with deuterated AA and deuterated glycine. McCue and colleagues [10] demonstrated that NAGly is formed via cytochrome C acting on arachidonoyl CoA and glycine in support of this conjugation pathway. Fatty acid amide hydrolase (FAAH), the primary hydrolyzing enzyme of AEA and other N-acyl amides [12], could potentially be involved in this reaction by acting in the biosynthetic pathway. In addition to being a hydrolytic enzyme, FAAH was suggested to play a role in the conjugation pathway of the biosynthesis of AEA from AA and ethanolamine [13] and was recently reported to participate in the synthesis of N-arachidonoyl 4-aminophenol (AM404) by conjugation of AA to exogenously administered p-acetamidophenol [14]. NAGly inhibited the hydrolysis of AEA by FAAH [3,15] indicating that it likely interacts with FAAH, presumably as a competitive substrate, though this interaction has not been fully examined.
An alternative pathway was proposed by Burstein et al. [2] who speculated that NAGly is produced by the oxidation of the ethanolamine in AEA, most likely through an ADH. Recent evidence using in vitro studies with human ADH confirmed this hypothesis by demonstrating that AEA is the precursor to NAGly through an aldehyde intermediate, N-arachidonoyl glycinal, which was synthesized and measured throughout the reaction [11]. These experiments show that after the reaction proceeds through to the carboxylic acid it cannot be then formed into the aldehyde supporting the hypothesis that AEA is a precursor for NAGly via ADH and further showing that the reaction cannot proceed in the opposite direction.
The potent actions of NAGly in a number of biological systems magnify the need to better understand its biosynthesis. Here using both in vitro and in vivo biochemical assays and measuring metabolites with LC/MS/MS we show that AEA serves as a precursor to NAGly by both oxidative metabolism of the ethanolamine moiety and through an additional conjugation pathway involving FAAH activity.
Results
Oxidative Metabolism of deuterium-labeled AEA in RAW 264.7 cells produces deuterium-labeled NAGly
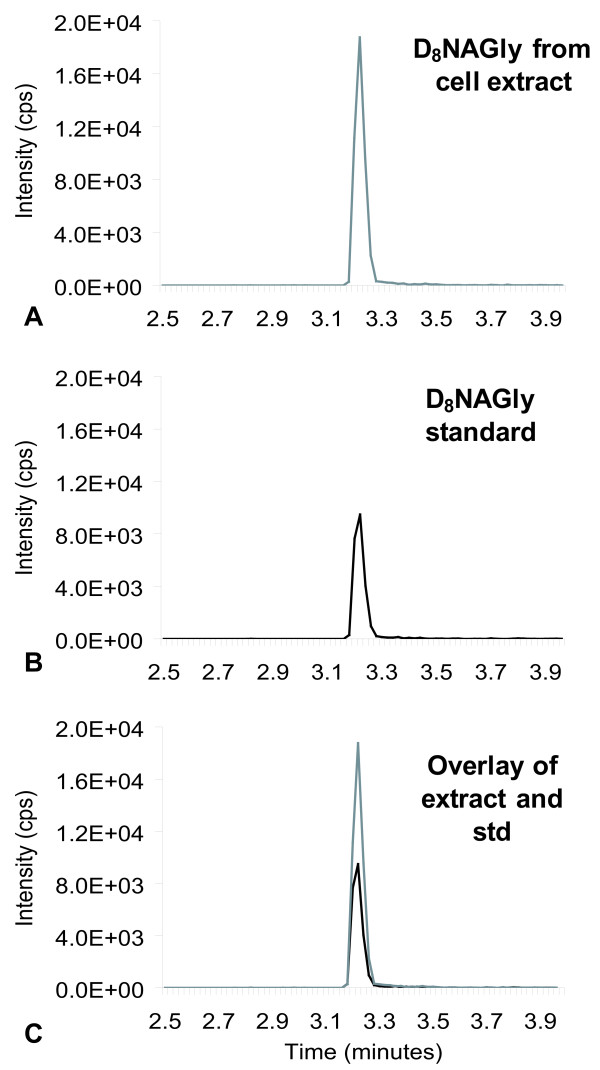
Incubation of deuterium-labeled AEA, (D8AEA, deuterium labeled on the arachidonic acid chain; Fig 1C) with RAW 264.7 murine macrophage-like cells resulted in the production of deuterium-labeled NAGly (D8NAGly, deuterium labeled on the arachidonic acid chain; Fig 1D). This was demonstrated by the isolation and measurement of a product that has the exact retention-time and parent mass/fragment pairing as synthetic D8NAGly (368.3/74.2; Fig 2). Incubation of RAW 264.7 with D8NAGly did not result in the production of a compound with the chromatographic or mass spectrometric properties of D8AEA, nor did incubation of D8AA produce either D8AEA or D8NAGly (data not shown). Because D8AA incubation alone did not produce D8NAGly, this suggested that conjugation of D8AA to glycine was not the biosynthetic pathway in this cell type. Furthermore, pre-incubation of RAW 264.7 cells with the FAAH inhibitor URB 597 for one hour followed by incubation with D8AEA did not block the production of D8NAGly. Therefore, we hypothesized that the conversion was on the ethanolamine moiety in AEA to form the glycine as was previously suggested [2,11].
Figure 2.
Chromatograms of the tandem mass spectrometric (MS) method for deuterium-labeled NAGly (D8NAGly) in which the parent mass in negative mode [368.3]- is paired fragment mass of glycine [74.0]-. A) Chromatogram of an MS scan for the 368.3/74 pair from RAW 267.4 cell extracts that were incubated with deuterium-labeled anandamide. B) Chromatogram of a scan for the 368.3/74 pair with the synthesized D8NAGly standard (std). C) Overlay of the two independent scans.
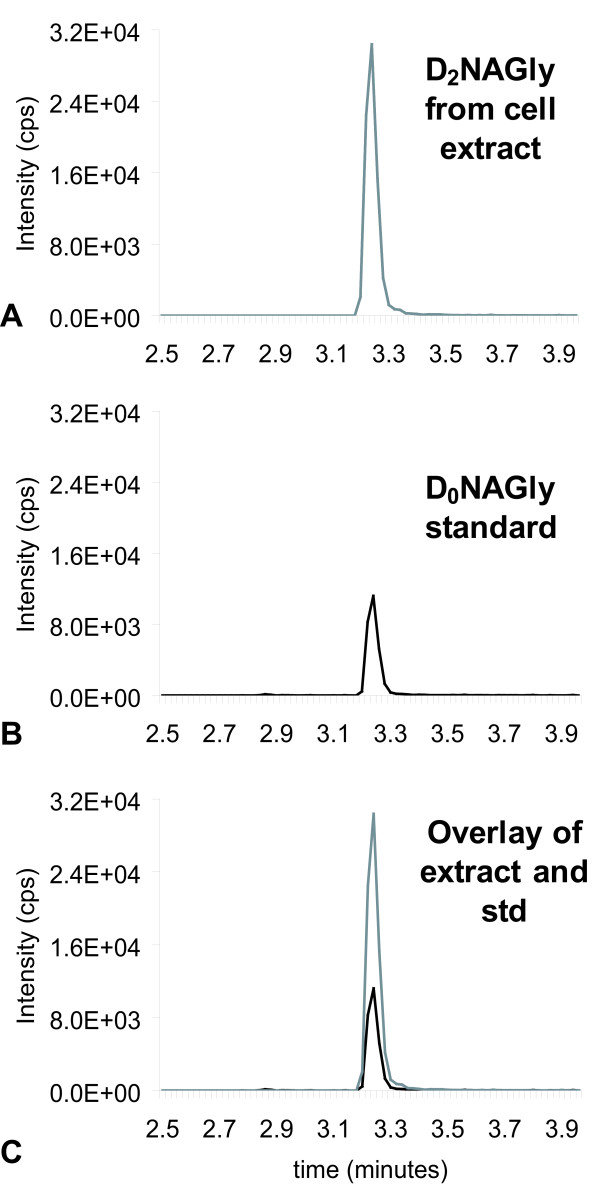
To test this hypothesis, the same series of incubations were performed using D4AEA (Fig. 1E), with the reasoning that the actions of an ADH on the D4ethanolamine moiety would yield a glycine with two deuterium atoms and would, therefore, produce D2NAGly (Fig. 1F). Incubation of D4AEA with RAW 264.7 cells yielded a product that matched the characteristics of the proposed D2NAGly (Figs. 3, 4). Chromatographic matches using HPLC/MS/MS showed that a molecule with the parent mass of the predicted D2NAGly (363.2 in negative ion mode) and a fragment that was 2 atomic mass units (amu) greater than the glycine fragment (76.1 and 74.1 respectively; Fig. 3A) had the identical retention time as the non-deuterated (D0NAGly) standard (Fig. 3B, C).
Figure 3.
Chromatograms of the tandem mass spectrometric (MS) method for deuterium-labeled N-arachidonoyl glycine (D2NAGly) in which the parent mass in negative mode [362.3]- is paired fragment mass of deuterium-labeled glycine [76.0]- or the non-deuterium-labeled N-arachidonoyl glycine (D0NAGly) that has a parent mass in negative ion mode of [360.3]- and a paired fragment mass of glycine [74.0]-. A) Chromatogram of an MS scan for the 362.3/76 pair from RAW 267.4 cell extracts that were incubated with deuterium-labeled anandamide (D4AEA). B) Chromatogram of the MS scan for the 360.3/74 pair with the synthesized D0NAGly standard (std). C) Overlay of the two independent scans.
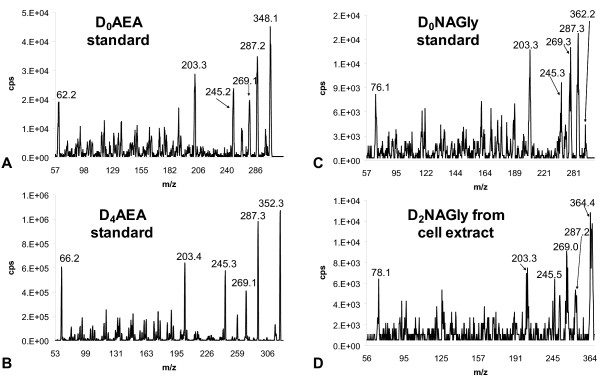
Figure 4.
Mass spectrometric product ion scans of synthesized standards and RAW 264.7 lipid extracts. The numbers above the peaks are the calculated centroid mass. A) Positive ion mode product ion scan of the mass [348.3]+ of the synthesized standard of non-deuterium labeled N-arachidonoyl ethanolamine (D0AEA). B) Positive ion mode product ion scans of the mass [352.3]+ of the synthesized standard of deuterium-labeled AEA (D4AEA). C) Positive ion mode product ion scan of the mass [362.3]+ of the synthesized standard of non-deuterium labeled NAGly (D0NAGly). D) Positive ion mode product ion scan of the mass [364.3]+ of the RAW 264.7 cell extract that was incubated with D4AEA.
Product ion scans in positive ion mode (used to facilitate the generation of more fragments) established that the fragmentation pattern of non-deuterated AEA (D0AEA; Fig. 4A) and the deuterium-labeled AEA synthesized here, D4AEA (Fig. 4B), are identical with the exception of the parent mass increasing by 4 amu from [348.1]+ to [352.3]+ and ethanolamine fragment from [62.2]+ and [66.2]+ respectively. Likewise, positive ion scans of NAGly standard (Fig. 4C) are identical to the molecule produced after D4AEA incubation with the predicted addition of 2 amu on the parent mass [362.2]+ and [364.4]+ respectively and glycine fragments [76.1]+ and [78.1]+ respectively (Fig 4D). Major product ions (287, 269, 245, and 203 m/z), which are associated with the fragmentation of AA, were likewise the same in each of the four scans (Fig 4A–D). These findings provide further evidence that NAGly is produced via an ADH from AEA in RAW 264.7 cells by activity on the ethanolamine moiety rather than cleavage to AA which is subsequently conjugated with glycine.
The Role of FAAH in the Biosynthesis of Deuterium-labeled-NAGly from Deuterium-labeled AEA in C6 glioma cells
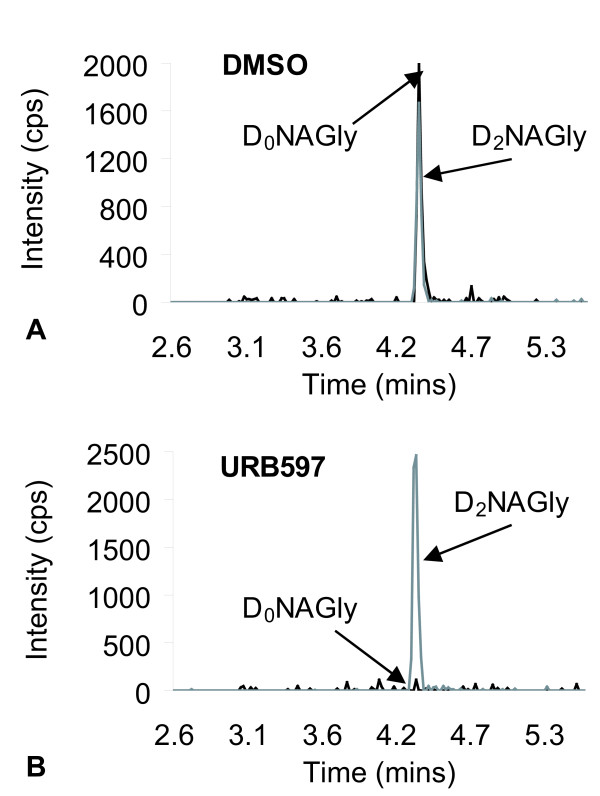
FAAH plays a role in the biosynthesis of N-arachidonyl-p-aminophenol following treatment of rats with acetaminophen [14]. Therefore, we hypothesized that FAAH may catalyze the biosynthesis of NAGly in C6 glioma cells, a murine cell line that exhibits robust FAAH activity [16,17]. D8AEA and D4AEA were incubated with C6 glioma cells using the treatment protocols described above for RAW 264.7 cells. Compounds matching the retention times and mass spectrometric properties of both D8NAGly and D2NAGly were present in the respective cell extracts. Unlike the RAW 264.7 cells, C6 glioma cells also produced excess D0NAGly after incubation with D4AEA (Fig 5A). The production of D0NAGly was prevented by pre-incubation with the FAAH inhibitor, URB 597 (Fig. 5B), however, like with the RAW 264.7 cells, D2NAGly was still produced (Fig 5B).
Figure 5.
Chromatograms of lipid extracts of C6 Glioma of the tandem mass spectrometric (MS) methods for both deuterium-labeled NAGly (D2NAGly) in which the parent mass in negative mode [362.3]- is paired fragment mass of deuterium-labeled glycine [76.0]- and the non-deuterium-labeled N-arachidonoyl glycine (D0NAGly) that has a parent mass in negative ion mode of [360.3]- and a paired fragment mass of glycine [74.0]-. A) Chromatogram of MS scans for D2NAGly and D0NAGly from the lipid extract of C6 Glioma cells that were incubated with DMSO followed by D4AEA. B) Chromatogram of MS scans for D2NAGly and D0NAGly from the lipid extract of C6 Glioma cells that were incubated with 1 μM URB 597 followed by D4AEA.
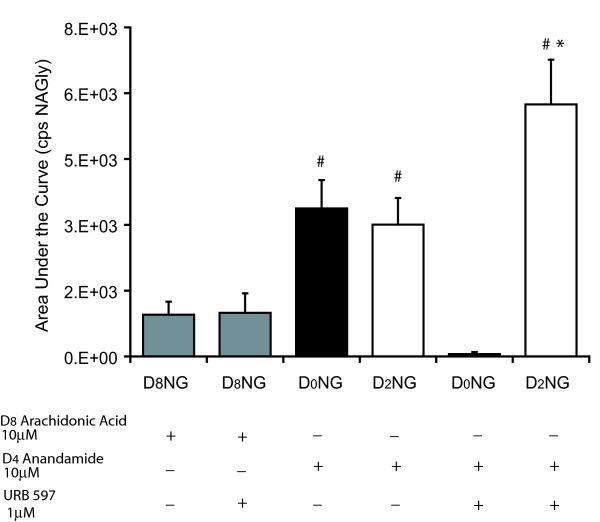
In contrast to RAW 264.7 cells, D8AA incubated with C6 glioma cells yielded D8NAGly, however, this product was not blocked by the addition of URB 597 (Fig. 6). Furthermore, a comparison of D8AA versus D4AEA indicated that mole-for-mole AEA is a significantly better substrate for the biosynthesis of NAGly in C6 glioma cells than AA (Fig 6). Significantly, blocking FAAH-dependent production of NAGly with URB 597 produced an increase in the amount of D2NAGly in these cells (Fig 6). Indeed, the shunting of substrate to the ADH pathway was very efficient, evidenced by the observation that the total production of NAGly was the same with and without URB 597 (Fig. 6).
Figure 6.
Comparison of NAGly production in C6 glioma cells after incubation with deuterium-labeled arachidonic acid (D8AA) or ethanolamine moiety deuterium-labeled AEA (D4AEA). The products measured were arachidonoyl chain deuterium-labeled NAGly (D8NG); non-deuterium-labeled NAGly (D0NG); and glycine moiety deuterium-labeled NAGly (D2NG). + denotes an addition of the compound to the cell media for 1 hour before lipid extraction. -denotes compounds that were not present during the incubation. # p ≤ 0.05 compared to levels of D8NG in D8AA treatment group; * p ≤ 0.05 compared to levels of D2NG in D4AEA+ URB 597 treatment group; n = 6–8 per group.
Brain levels of AEA and NAGly after URB 597 injections in rats and in FAAH knockout (KO) and wild-type (WT) mice
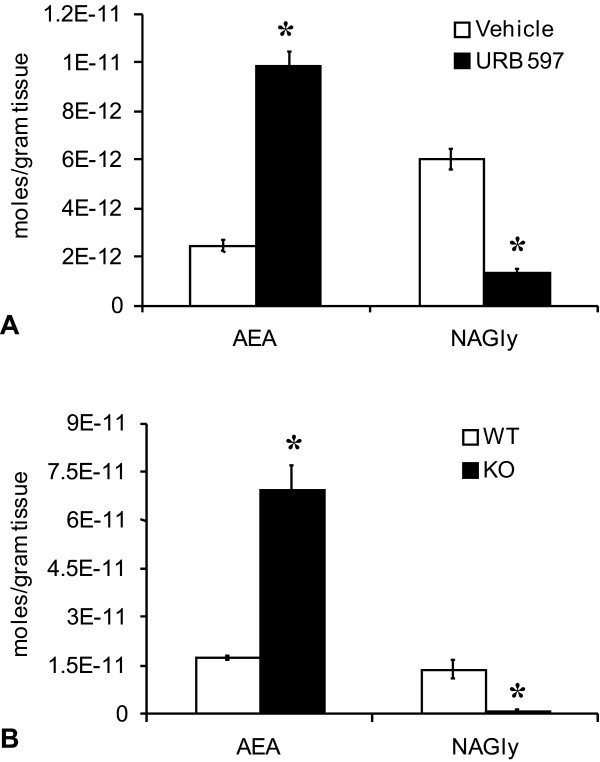
After examining the biosynthesis of NAGly in a FAAH-rich in-vitro cellular model (C6 glioma), we sought to determine whether FAAH-dependent biosynthesis of NAGly occurs also in vivo. We examined the levels of AEA and NAGly in brains of rats treated with URB597 or vehicle as well as in FAAH KO and WT mice. The levels of AEA in rat whole brain significantly increased 2 hours after systemic injection of URB 597 (0.3 mg/kg) compared to vehicle controls (Fig 7A). In contrast, levels of NAGly significantly decreased (Fig 7A). The same pattern was shown in the levels of AEA and NAGly in FAAH KO and WT mice: AEA levels were significantly higher in FAAH KO mice, whereas, NAGly levels were significantly lower (Fig. 7B).
Figure 7.
Production of N-arachidonoyl ethanolamine (AEA) and N-arachidonoyl glycine (NAGly) in whole brain. A) Levels of AEA and NAGly in rat whole brain two hours after vehicle (white bar) or 3 μg/kg URB 597 (black bar). * p ≤ 0.05 B) Levels of AEA and NAGly in mouse whole brain of WT (white bar) or FAAH KO (black bar). * p ≤ 0.05.
NAGly hydrolysis by recombinant FAAH
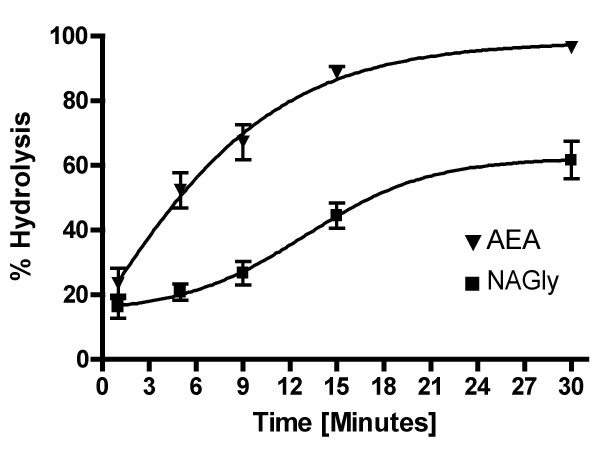
The evidence that NAGly levels were significantly decreased with FAAH inhibition and in FAAH KO mice led us to test the hypothesis that: 1) FAAH is acting as a biosynthetic enzyme with AEA and glycine as precursors and 2) FAAH has a low efficacy for NAGly. If FAAH is highly efficacious for producing robust levels of NAGly hydrolysis, then it is unlikely to play a role in its biosynthesis. Conversely, if AEA is converted to NAGly during hydrolysis in the presence of glycine, then FAAH would be a candidate enzyme for NAGly biosynthesis. Our results show that when recombinant FAAH was incubated with AEA and glycine, AEA was measured via HPLC/MS/MS and was shown to be rapidly hydrolyzed as expected (Fig 8). At 5 minutes ~60% of all AEA was hydrolyzed and by 15 minutes over 95%. At 30 minutes there was almost no detectable AEA present in the solution. Conversely, at 5 minutes less than 20% of NAGly was hydrolyzed and at 30 minutes there was still over 45% of NAGly still left in the solution. There was no detectable NAGly produced in incubations of AEA, glycine and recombinant FAAH indicating that FAAH is not a direct biosynthetic enzyme in the recombinant form.
Figure 8.
Comparison of hydrolysis rates of N-arachidonoyl ethanolamine (AEA) and N-arachidonoyl glycine (NAGly) via recombinant FAAH. n = 4–6 per time point. Time points (minutes): 1, 3, 5, 9, 15, 30.
Discussion
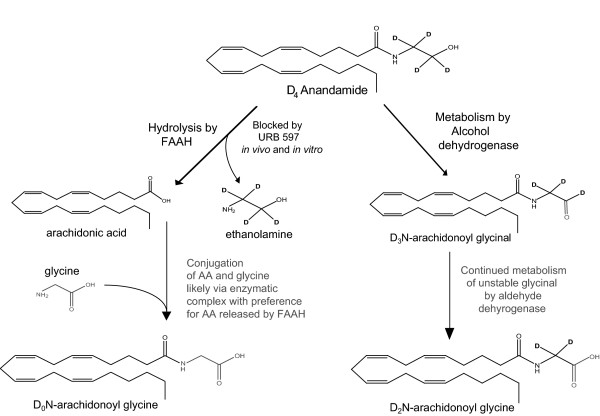
The data presented here supports the hypothesis that the endogenous cannabinoid AEA acts as a precursor in the biosynthesis pathways of the signaling lipid NAGly. One pathway is a FAAH-dependent conjugation of glycine to AEA-released AA and the second is by oxidation of the ethanolamine moiety in AEA, likely by an ADH (Fig. 9).
Figure 9.
Two pathways for N-arachidonoyl glycine (NAGly) biosynthesis from N-arachidonoyl ethanolamine (AEA).
The conjugation of AA with glycine to form NAGly demonstrated by Huang and colleagues [3] was confirmed here in the incubation with C6 glioma cells. The previous assay was performed with brain membranes and is more comparable to the brain-derived C6 glioma cells than the macrophage RAW 264.7 cell line. As shown here, AEA had a 4-fold greater efficacy as a substrate than AA in C6 glioma cells and the production of NAGly by conjugation of AA and glycine was not blocked by URB 597. Therefore, the present data suggest that AEA is a substrate for NAGly biosynthesis through the URB 597-sensitive pathway in C6 glioma cells and brain. Through this pathway, AEA must undergo hydrolysis by FAAH to be a substrate for NAGly biosynthesis via conjugation.
Previously, we demonstrated that incubation of D8AEA in a neuronal cell line (F-11) resulted in accumulation of D8AEA in lipid rafts while its metabolite D8AA was found mostly in non-lipid raft fractions [18]. It is possible that the trafficking of AEA from specialized membrane compartments such as lipid rafts to compartments rich in FAAH [19] may position the AA precursor in proximity to additional enzymes that are involved in the conjugation reaction to glycine. In essence, this would make the arachidonic acid in AEA more bio-available to form NAGly. Recently, McHue and colleagues [10] proposed that cytochrome c catalyzes the synthesis of NAGly from arachidonoyl CoA and glycine in the presence of hydrogen peroxide suggesting yet another conjugation pathway for the production of NAGly. These enzymes and FAAH are present in mitochondrial membranes, which may be the site of NAGly biosynthesis.
The conversion of AEA to NAGly through an ADH pathway in both RAW 264.7 macrophage and C6 glioma cell lines suggests a more ubiquitous biosynthesis reaction. Given that there are multiple members of the protein family of ADHs that act with different affinity to different substrates [20] it would also not be surprising that the level of NAGly production would be different with different cell types through this pathway.
We observed that incubation of D8NAGly with RAW 264.7 and C6 glioma cells did not lead to the production D8AEA. This finding is at variance with the report by Burstein et al. [21] in which incubation of NAGly with RAW 264.7 cells yielded increased levels of AEA. In the present study we used 5 cm2 flasks for each assay which was an amount of cells that fell below the limit to detect endogenous AEA. In addition, the concentration of AEA in the incubations was 10-times less than in the earlier study. In light of the data presented here Burstein et al.'s result [21] may represent a case where NAGly at higher concentrations may be activating a pathway for AEA production or sufficiently blocking its enzymatic hydrolysis to cause a build-up of native AEA and not acting as a substrate for AEA synthesis.
That the levels of NAGly were dramatically decreased in brain after URB 597 and in the FAAH KO mice suggests that the compensatory NAGly production shown in the C6 glioma cells may be an acute phenomenon specific for these cells. If NAGly production were driven by ADH in the brain then the excess AEA generated by URB 597 should have produced an increase in NAGly. The lack of increase in this experiment and the FAAH KO mice suggests that the brain-derived NAGly is primarily through the FAAH-dependent conjugation biosynthesis pathway.
Finally, the evidence that recombinant FAAH has a significantly lower efficacy for NAGly hydrolysis than AEA suggests that it would not be readily hydrolyzed in an enzyme complex including FAAH permitting time for production and trafficking of the signaling molecule to its site of action, which is likely to be at plasma membrane receptors.
Conclusion
Growing evidence supports a mechanism for non-CB1, non-CB2 activity of AEA [22-24]. The hypotheses generated from those studies were that AEA is acting on a separate receptor or receptors or through metabolites of AEA [24]. Here, we provide evidence that AEA is metabolized into the signaling lipid NAGly that activates GPR18 [6] and GPR92 [9] suggesting the hypothesis that non-CB receptor effects of AEA are potentially through this bioactive metabolite.
Methods
Subjects
Twelve male (300–450 g) Sprague-Dawley (Harlan, Indianapolis, IN) rats were used. We also used brain tissue from six FAAH WT and six FAAH KO mice, which were littermates from the thirteenth generation offspring from intercrosses of 129SvJ-C57BL/6 FAAH (±) mice [25]. All protocols were approved by the Indiana University Institutional Animal Care and Use Committee.
Cell Culture
The RAW 267.4 and C6 glioma cell lines were purchased from ATCC (Manassas, VA). Both cell lines were cultured in DMEM (Mediatech, VA) with 10% fetal bovine serum (Hyclone, Logan, UT) and 1% penicillin-streptomycin (Gibco-Invitrogen, Carlsbad, CA).
Drugs and reagents
HPLC-grade methanol and acetonitrile used for mass spectrometric studies were purchased from VWR international (Plainview, NY). HPLC grade water, mass spectrometry/HPLC grade acetic acid, formic acid, and ammonium acetate were purchased from Sigma-Aldrich (St. Louis, MO). N- [2H8]arachidonoyl glycine (D8NAGly), N- [2H8]arachidonoyl ethanolamine, D8AEA, and [2H8] arachidonic acid (D8AA) were purchased from Cayman Chemical (Ann Arbor, MI). URB 597 was purchased from BIOMOL International (Plymouth Meeting, PA). Arachidonic acid was purchased from Nu-Chek Prep (Elysian, MN). Ethanol-1,1,2,2-d4-amine was purchased from CDN Isotopes Inc. (Pointe-Claire, Quebec).
Synthesis of deuterium-labeled N-arachidonoyl ethanolamine (D4-AEA)
D4AEA was synthesized as previously described with the exception that the ethanolamine was deuterium labeled instead of the arachidonic acid [26].
Characterization of metabolites of D8AEA and D4AEA in cell culture systems
RAW 264.7 and C6 glioma cell lines were used at 70% confluence. Cells were washed twice with dPBS then either 1 μM of either D8AEA, D4AEA, D8AA, D8NAGly or vehicle, DMSO (10 μl) were added to serum-free media and incubated for 1 hour. In experiments in which the FAAH inhibitor URB 597 was used, the cells were first incubated with 1 μM URB 597 or DMSO vehicle (10 μl) in serum-free media for one hour prior to D8AEA or D4AEA that was added directly to this media. Then, equal volumes of methanol were added to the flasks; cells were scraped, aspirated, and centrifuged at 2000 × g for 15 min at 24°C. Supernatants were collected and HPLC grade water was added to make a 30% organic solution. Lipids were partially purified on C18 solid phase extraction columns as previously described [27]. In brief, each 500 mg column was conditioned with 5 ml methanol and 2.5 ml water followed by loading of the water/supernatant solution. Columns were then washed with 2 ml water and 1.5 ml 55% methanol. Compounds were eluted with 1.5 ml methanol. Eluants were vortexed at maximum speed prior to mass spectrometric analysis.
Rapid separation of analytes was obtained using 10 μl injections (Agilent 1100 series autosampler, Wilmington, DE) onto a Zorbax eclipse XDB 2.1 × 50 mm reversed phase column. Gradient elution (200 μl/min) was formed under pressure on a pair of Shimadzu (Columbia, Maryland) 10AdVP pumps. Mass spectrometric analysis was performed with an Applied Biosystems/MDS Sciex (Foster City, CA) API 3000 triple quadrupole mass spectrometer equipped with an electrospray ionization source. Levels of each compound were analyzed by multiple reactions monitoring (MRM) on the LC/MS/MS system. Mass spectrometric conditions were optimized for each compound using direct flow injection of synthetic standards of each compound. Product ion scans were performed with identical chromatographic conditions as the MRM scans with identical ionization and collision energy.
Eluants were tested for levels of D8AEA, D4AEA, D8NAGly, D2NAGly, (Fig. 1) and D8AA using MRM tandem mass spectrometric methods with parent and fragment ions as follows: in positive ion mode, AEA 348.3 → 62.3, D8AEA 356.3 → 62.3 and D4AEA 352.3 → 66.3; in negative ion mode, NAGly 360.2 → 76.2, D8NAGly 368.2 → 76.2, D2NAGly 362.2 → 78.2, and D8AA 311.5 → 267.3. Product ion scans were performed for D0NAGly and D2NAGly using the API 3000 scanning in positive ion mode for the products of 362.2 and 364.2 respectively.
Quantification of tissue levels of AEA and NAGly
Each of the analytes was extracted and quantified using methods reported [27]. In brief, whole brains were dissected and flash-frozen in liquid nitrogen prior to lipid extraction, at which time twenty volumes of ice-cold methanol and 100 pmol D8NAGly (internal standard) were added to the methanol-tissue sample. The samples were maintained on ice and homogenized via polytron for 2 min and centrifuged for 20 min at 40,000 × g at 24°C. Supernatants were transferred to polypropylene 50 ml centrifuge tubes (VWR, Plainview, NY) and HPLC grade water was added to each sample to create a 70:30 (water:supernatant) mixture. Partial purification on C18 solid phase extraction and mass spectrometric analyses were identical to that described above.
Effects of inhibition of FAAH on brain levels of AEA and NAGly
Animals were injected with either URB 597 (0.3 mg/kg, in 1% DMSO, i.p.) or vehicle. After two hours animals were decapitated and brains were dissected and flash-frozen in liquid nitrogen, extracted, purified and analyzed as described above.
Analysis of brain extracts from FAAH KO and WT mice
FAAH KO and WT mice were sacrificed when they were 6 weeks old. The brains were dissected and stored at -80°C until used. Lipid extraction, partial purification, and quantitation were performed using the methods described above.
Recombinant FAAH cell-free assay
To determine the rates of AEA and NAGly hydrolysis a solution of ethanol and compound (400 μM, 10 μl) was added to a solution of recombinant FAAH protein (10 μl, 1.3 μg/μl in 20 mM HEPES (pH 7.8), 150 mM NaCl, 10% glycerol, 1% Triton X-100) in buffer (Tris/EDTA, 380 μl, pH 9) at room temperature. A 40 μl aliquot of the reaction mixture was taken at appropriate time points and quenched with 1 ml of MeOH. To control for loss of AEA and NAGly to the sides of the tube and into micelles in the aqueous buffer, equal numbers of controls were run at the same time without FAAH. 1 μl of the quenched solution from each (FAAH incubations and controls) was analyzed by LC/MS/MS mass spectrometry as discussed above. Hydrolysis rates were determined by the average values of the analyte measured from the FAAH incubations subtracted from the average values of the controls (incubations with buffer and no FAAH) at each time point.
Data Analysis
Mass spectrometric quantitation
The quantitation of analytes was achieved using Analyst software (Applied Biosystems-MDS Sciex; Framingham MA), which quantifies the amount of analyte in the sample based upon a power fit of a linear regression of known concentrations of synthetic standards. Those data were then analyzed as vehicle verses drug in the case of the URB 597 and as FAAH KO verses WT. Statistical differences were determined using ANOVA with post-hoc Fisher's LSD using a 95% confidence interval for the mean (SPSS software, Chicago, IL). Data are presented as mean ± standard error of the mean where p ≤ 0.05 was considered statistically significant
List of abbreviations
(D8NAGly): N- [2H8]arachidonoyl glycine; (D8AEA): N- [2H8]arachidonoyl ethanolamine; (D8AA): [2H8] arachidonic acid; (D2NAGly): N- [2H2]arachidonoyl glycine; (D4AEA): N- [2H4]arachidonoyl ethanolamine; (AEA): arachidonoyl ethanolamide; (NAGly): N-arachidonoyl glycine; (AA): arachidonic acid; (FAAH): fatty acid amide hydrolase.
Authors' contributions
HB conceived of the project, developed culture and MS method, analyzed data and wrote the manuscript, NR performed C6 glioma culture experiments and edited the manuscript, S S-J H performed in vivo URB 597 and FAAH KO/WT MS experiments, VB and JS performed recombinant FAAH MS experiments, KM genotyped the FAAH KO and WT mice, BC created and maintained the FAAH KO mice in his laboratory, DO synthesized deuterium-labeled anandamide, JMW took part in planning and project development. This work was supported by DA01822, J Michael Walker.
Acknowledgements
We dedicate this manuscript to J Michael Walker (1950–2008) whose love of science and life was an inspiration to us all.
Contributor Information
Heather B Bradshaw, Email: hbbradsh@indiana.edu.
Neta Rimmerman, Email: neta.rimmerm@gmail.com.
Sherry Shu-Jung Hu, Email: shujunghu@gmail.com.
Valery M Benton, Email: howardvm@indiana.edu.
Jordyn M Stuart, Email: jmstuart@indiana.edu.
Kim Masuda, Email: masuda@scripps.edu.
Benjamin F Cravatt, Email: cravatt@scripps.edu.
David K O'Dell, Email: alchemancer@gmail.com.
J Michael Walker, Email: mtheodor@indiana.edu.
References
- Sheskin T, Hanus L, Slager J, Vogel Z, Mechoulam R. Structural requirements for binding of anandamide-type compounds to the brain cannabinoid receptor. J Med Chem. 1997;40:659–667. doi: 10.1021/jm960752x. [DOI] [PubMed] [Google Scholar]
- Burstein SH, Rossetti RG, Yagen B, Zurier RB. Oxidative metabolism of anandamide. Prostaglandins Other Lipid Mediat. 2000;61:29–41. doi: 10.1016/S0090-6980(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Huang SM, Bisogno T, Petros TJ, Chang SY, Zavitsanos PA, Zipkin RE, Sivakumar R, Coop A, Maeda DY, De Petrocellis L, et al. Identification of a new class of molecules, the arachidonyl amino acids, and characterization of one member that inhibits pain. J Biol Chem. 2001;276:42639–42644. doi: 10.1074/jbc.M107351200. [DOI] [PubMed] [Google Scholar]
- Succar R, Mitchell VA, Vaughan CW. Actions of N-arachidonyl-glycine in a rat inflammatory pain model. Mol Pain. 2007;3:24. doi: 10.1186/1744-8069-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong LA, Mitchell VA, Vaughan CW. Actions of N-arachidonyl-glycine in a rat neuropathic pain model. Neuropharmacology. 2008;54:189–193. doi: 10.1016/j.neuropharm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Kohno M, Hasegawa H, Inoue A, Muraoka M, Miyazaki T, Oka K, Yasukawa M. Identification of N-arachidonylglycine as the endogenous ligand for orphan G-protein-coupled receptor GPR18. Biochem Biophys Res Commun. 2006;347:827–832. doi: 10.1016/j.bbrc.2006.06.175. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Iguchi H, Nakata M, Ioka RX, Tanaka T, Iwasaki S, Magoori K, Takayasu S, Yamamoto TT, Kodama T, et al. Identification of N-arachidonylglycine, U18666A, and 4-androstene-3,17-dione as novel insulin Secretagogues. Biochem Biophys Res Commun. 2005;333:778–786. doi: 10.1016/j.bbrc.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Wiles AL, Pearlman RJ, Rosvall M, Aubrey KR, Vandenberg RJ. N-Arachidonyl-glycine inhibits the glycine transporter, GLYT2a. J Neurochem. 2006;99:781–786. doi: 10.1111/j.1471-4159.2006.04107.x. [DOI] [PubMed] [Google Scholar]
- Oh da Y, Yoon JM, Moon MJ, Hwang JI, Choe H, Lee JY, Kim JI, Kim S, Rhim H, O'Dell DK, et al. Identification of farnesyl pyrophosphate and N-arachidonylglycine as endogenous ligands for GPR92. J Biol Chem. 2008;283:21054–21064. doi: 10.1074/jbc.M708908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue JM, Driscoll WJ, Mueller GP. Cytochrome c catalyzes the in vitro synthesis of arachidonoyl glycine. Biochem Biophys Res Commun. 2008;365:322–327. doi: 10.1016/j.bbrc.2007.10.175. [DOI] [PubMed] [Google Scholar]
- Aneetha H, O'Dell DK, Tan B, Walker JM, Hurley TD. Alcohol dehydrogenase-catalyzed in vitro oxidation of anandamide to N-arachidonoyl glycine, a lipid mediator: synthesis of N-acyl glycinals. Bioorg Med Chem Lett. 2009;19:237–241. doi: 10.1016/j.bmcl.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- Arreaza G, Devane WA, Omeir RL, Sajnani G, Kunz J, Cravatt BF, Deutsch DG. The cloned rat hydrolytic enzyme responsible for the breakdown of anandamide also catalyzes its formation via the condensation of arachidonic acid and ethanolamine. Neuroscience letters. 1997;234:59–62. doi: 10.1016/S0304-3940(97)00673-3. [DOI] [PubMed] [Google Scholar]
- Hogestatt ED, Jonsson BA, Ermund A, Andersson DA, Bjork H, Alexander JP, Cravatt BF, Basbaum AI, Zygmunt PM. Conversion of acetaminophen to the bioactive N-acylphenolamine AM404 via fatty acid amide hydrolase-dependent arachidonic acid conjugation in the nervous system. J Biol Chem. 2005;280:31405–31412. doi: 10.1074/jbc.M501489200. [DOI] [PubMed] [Google Scholar]
- Grazia Cascio M, Minassi A, Ligresti A, Appendino G, Burstein S, Di Marzo V. A structure-activity relationship study on N-arachidonoyl-amino acids as possible endogenous inhibitors of fatty acid amide hydrolase. Biochem Biophys Res Commun. 2004;314:192–196. doi: 10.1016/j.bbrc.2003.12.075. [DOI] [PubMed] [Google Scholar]
- Deutsch DG, Glaser ST, Howell JM, Kunz JS, Puffenbarger RA, Hillard CJ, Abumrad N. The cellular uptake of anandamide is coupled to its breakdown by fatty-acid amide hydrolase. J Biol Chem. 2001;276:6967–6973. doi: 10.1074/jbc.M003161200. [DOI] [PubMed] [Google Scholar]
- Holt S, Paylor B, Boldrup L, Alajakku K, Vandevoorde S, Sundstrom A, Cocco MT, Onnis V, Fowler CJ. Inhibition of fatty acid amide hydrolase, a key endocannabinoid metabolizing enzyme, by analogues of ibuprofen and indomethacin. European journal of pharmacology. 2007;565:26–36. doi: 10.1016/j.ejphar.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Rimmerman N, Hughes HV, Bradshaw HB, Pazos MX, Mackie K, Prieto AL, Walker JM. Compartmentalization of endocannabinoids into lipid rafts in a dorsal root ganglion cell line. British journal of pharmacology. 2008;153:380–389. doi: 10.1038/sj.bjp.0707561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland MJ, Porter AC, Rakhshan FR, Rawat DS, Gibbs RA, Barker EL. A role for caveolae/lipid rafts in the uptake and recycling of the endogenous cannabinoid anandamide. J Biol Chem. 2004;279:41991–41997. doi: 10.1074/jbc.M407250200. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Burstein SH, Huang SM, Petros TJ, Rossetti RG, Walker JM, Zurier RB. Regulation of anandamide tissue levels by N-arachidonylglycine. Biochem Pharmacol. 2002;64:1147–1150. doi: 10.1016/S0006-2952(02)01301-1. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Breivogel CS, Tao Q, Bridgen DT, Razdan RK, Zimmer AM, Zimmer A, Martin BR. Levels, metabolism, and pharmacological activity of anandamide in CB(1) cannabinoid receptor knockout mice: evidence for non-CB(1), non-CB(2) receptor-mediated actions of anandamide in mouse brain. J Neurochem. 2000;75:2434–2444. doi: 10.1046/j.1471-4159.2000.0752434.x. [DOI] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Evans RM, Wease KN, MacDonald CJ, Khairy HA, Ross RA, Scott RH. Modulation of sensory neuron potassium conductances by anandamide indicates roles for metabolites. British journal of pharmacology. 2008;154:480–492. doi: 10.1038/bjp.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida A, Piomelli D. Isotope dilution GC/MS determination of anandamide and other fatty acylethanolamides in rat blood plasma. FEBS letters. 1998;422:373–376. doi: 10.1016/S0014-5793(98)00046-5. [DOI] [PubMed] [Google Scholar]
- Bradshaw HB, Rimmerman N, Krey JF, Walker JM. Sex and hormonal cycle differences in rat brain levels of pain-related cannabimimetic lipid mediators. Am J Physiol Regul Integr Comp Physiol. 2006;291:R349–358. doi: 10.1152/ajpregu.00933.2005. [DOI] [PubMed] [Google Scholar]