Abstract
Background
The Tec-family kinase Itk plays an important role during T-cell activation and function, and controls also conventional versus innate-like T-cell development. We have characterized the transcriptome of Itk-deficient CD3+ T-cells, including CD4+ and CD8+ subsets, using Affymetrix microarrays.
Results
The largest difference between Itk-/- and Wt CD3+ T-cells was found in unstimulated cells, e.g. for killer cell lectin-like receptors. Compared to anti-CD3-stimulation, anti-CD3/CD28 significantly decreased the number of transcripts suggesting that the CD28 co-stimulatory pathway is mainly independent of Itk. The signatures of CD4+ and CD8+ T-cell subsets identified a greater differential expression than in total CD3+ cells. Cyclosporin A (CsA)-treatment had a stronger effect on transcriptional regulation than Itk-deficiency, suggesting that only a fraction of TCR-mediated calcineurin/NFAT-activation is dependent on Itk. Bioinformatic analysis of NFAT-sites of the group of transcripts similarly regulated by Itk-deficiency and CsA-treatment, followed by chromatin-immunoprecipitation, revealed NFATc1-binding to the Bub1, IL7R, Ctla2a, Ctla2b, and Schlafen1 genes. Finally, to identify transcripts that are regulated by Tec-family kinases in general, we compared the expression profile of Itk-deficient T-cells with that of Btk-deficient B-cells and a common set of transcripts was found.
Conclusion
Taken together, our study provides a general overview about the global transcriptional changes in the absence of Itk.
Background
The Tec family of non-receptor protein-tyrosine kinases consists of five members (Bmx, Btk, Itk, Rlk/Txk and Tec); T-cells express Itk, Rlk and Tec, and T-cell receptor (TCR) stimulation leads to the activation of Tec family kinases [1,2]. A large number of biochemical studies and the generation of mice that are single- or double-deficient for Itk, Tec or Rlk have identified important roles, in particular for Itk, during T-cell development and activation, and in Th2 effector differentiation. Itk-/- mice show impaired positive selection of CD4+ T-cells and it was suggested that Itk modulates signaling thresholds during T-cell development [3-5]. TCR signaling in naïve T-cells, and therefore activation and proliferation, is impaired in the absence of Itk, and Itk-/- T-cells show defective Th2 polarization [6]. Further, Itk regulates the actin cytoskeleton and is therefore necessary for proper synapse formation and for efficient T-cell activation [7,8]. More recent data indicate that Itk is involved in signaling pathways that regulate conventional versus innate-like T-cell development. The majority of CD8+ T-cells from Itk-/- as well as from Itk-/-Rlk-/- mice show a more "innate-like" T-cell phenotype, sharing characteristics with conventional memory T-cells, i.e. CD44hi, CD62L- and CD122hi [9-11]. These cells depend on IL-15, express TCRs specific for non-classical MHC class Ib molecules, and exhibit direct effector functions such as rapid IFNγ production upon PMA/ionomycin stimulation [9-12]. A significant fraction of innate-like CD44hiCD62L- T-cells has also been described for the CD4+ T-cell lineage in Itk-/- mice [13].
Biochemically, the defects in T-cell activation were linked to an impaired phospholipase C-γ (PLCγ) phosphorylation and activation [5]. PLCγ hydrolyzes phosphatidylinositol-4,5-biphosphate to produce inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 induces the release of intracellular calcium (Ca2+) thereby activating the serine/threonine phosphatase calcineurin. Itk activation results in high levels of IP3, which is required for Ca2+ entry via store-operated channels leading to increased Ca2+ in cells stimulated via the TCR [5]. Both Ca2+ and calmodulin will bind and activate calcineurin, which in turn dephosphorylates serines in the regulatory domain of cytosolic NFAT. This induces a conformational change in NFAT exposing nuclear localization signals allowing its transport into the nucleus [14]. In Itk-deficient T-cells the Ca2+-levels are reduced resulting in impaired NFAT translocation [6]. Mice deficient in NFAT family members share phenotypes with Tec kinase family-deficient mice, as described by Lucas et al. [15]. The NFAT family was first described as binding to and controlling the interleukin 2 (IL-2) promoter and other lymphokine promoters in T-cells [14]. The family consists of five members; NFATc1–4 and NFAT5 [16,17]. Efficient inhibitors for the activation of NFAT proteins have been developed. Two of these, Cyclosporin A (CsA) and FK506, indirectly inhibit NFAT by blocking the enzymatic activity of calcineurin.
In order to further decipher the role of Itk we have investigated changes in gene expression of CD3+ as well as CD4+ and CD8+ T-lymphocytes in normal and Itk-defective mice. The aim of the study was to (1) define the transcriptome in unstimulated cells, (2) elucidate the influence of anti-CD3 and anti-CD3/CD28-stimulation and (3) to dissect which part of the observed alterations in Itk-deficiency is dependent on the calcineurin/NFAT pathway.
Methods
Mice and generation of T-cells
CD3+ as well as CD4+ and CD8+ T-cells from pooled suspensions of spleen and lymph nodes of Wt and Itk-/- mice on C57BL/6 background were isolated by negative depletion; antibodies used are listed in Additional file 1. The cell suspensions were incubated with the antibodies in PBS supplemented with 2% FCS. Streptavidin beads (BD Pharmingen) were used for negative depletion according to manufacturer's instructions. The purity of the cells was assessed by flow cytometry and was routinely >90% CD3+, >96% CD4+ and >90% CD8+ T-cells. All animal experiments were approved by the Federal Ministry for Science and Research.
T-cell stimulations and Cyclosporin A treatment
Unstimulated as well as stimulated T-cells were studied. Stimulations were performed in 48-well plates, coated with anti-CD3 (1 μg/ml) with or without anti-CD28 (3 μg/ml) in the presence or absence of CsA (1 μg/ml) for 24 hrs. For each stimulus, at least duplicate samples were used in all but one experiment. For the CD4+ T-cells we collected triplicates from the Itk-/- mice and duplicates from the Wt group. For the CD8+ T-cells, we got duplicates from Itk-/-, while we obtained a single sample from Wt owing to the low cell yield for resting Wt CD8+ T-cells. After anti-CD3-stimulation we got a single sample from the CD8+ subset of both Wt and Itk-/-, while for the CD4+ subsets we collected duplicates. To control if the number of differentially expressed probe-sets was truthful in the CD8+ subset, and not due to the lack of replicates, we analyzed the CD4+ in the same way as the CD8+ T-cells. The results were consistent and we found the same number of differentially expressed transcripts when single CD4+ samples were studied separately.
RNA isolation and microarray processing
RNA isolations were done according to RNeasy Mini protocol (Qiagen, Valencia, CA, USA) and microarray processing as previously described [18]. The Affymetrix MOE430 2.0 chips were used. In total 37 arrays were analyzed. The microarray data are accessible through the Gene Expression Omnibus (GEO; GSE12466) [19,20].
Data and statistical analysis
The processing and primary data analysis was performed in DNA-Chip Analyzer (dChip) [21]. In short, the invariant set normalization method was used [22]. Thereafter, model-based expression values were calculated according to the perfect match (PM)-only model. The criterion for fold-change analysis was set to ≥ 2-fold between groups. Signal values were then used in further statistical analysis steps such as paired and unpaired Student t-test in Excel. Some comparisons were performed using the chi square (χ2) test. Immunoglobulin and histocompatibility transcripts were excluded from the tables, since changes in their expression may be secondary to events unrelated to Itk-deficiency. Also the Xist and Tsix transcripts, X-chromosome encoded and unique to females, as well as Y-specific mRNAs were removed owing to that we used mixed sexes of mice in the experiments. One probe set that corresponded to Itk (1456836_at) was found to also be complementary to an EST gene (recognized at Ensembl) [23] and was therefore only included in the Additional material. We manually annotated a group of genes on the basis of prior knowledge about their role in the immune system. The 900 probe-sets list of differentially expressed transcripts between unstimulated Itk-/- and Wt T-cells was used for this purpose. The classification resulted in 14 subgroups.
Validation of differentially expressed genes using quantitative RT-PCR
Total RNA (100 ng) was reversed-transcribed into cDNA with AMV reverse transcriptase using random hexamer primers (Roche Applied Science, IN, USA). TaqMan Gene Expression Assays from Applied Biosystems were used to confirm the microarray data and it was done as previously described [24]. The validated genes were Klrg1 (Mm00516879_m1), Klra3 (Mm01702813_m1) and Klra7 (Mm01183384_m1). 18S rRNA was used as endogenous control.
Chromatin-immunoprecipitation for detection of NFATc1-binding
Whole splenic and thymic cells were used for chromatin-immunoprecipitation (ChIP) assay. The cells were lysed by ammoniumchloride solution to remove erythrocytes, counted and divided into three groups each. One group was untreated, while the other two were treated with anti-CD3ε(1 μg/ml) with or without pre-treatment of CsA (1 μg/ml) for 1 hour. The stimulated cells were incubated for 24 hrs in 37°C with 5% CO2. The protocol for ChIP was described by Yu et al. [25] with the following modifications. After sonication and centrifugation, lysates were incubated with 1 μg of polyclonal anti-NFATc1 (K-18) antibody (sc-1149-R, Santa Cruz Biotechnology, Inc.) or rabbit normal Ig overnight at 4°C with rotation. Identification of targets was done by PCR using primers for the genes corresponding to IL2, IL7R, Schlafen1, Bub1, Ctla2a and Ctla2b (Additional file 2).
Results
Transcriptional changes in the absence of Itk in unstimulated CD3+ T-cells
In order to survey Itk-dependent transcriptional signatures we initially conducted microarray analysis on MACS-sorted CD3+ primary, unstimulated T-cells from Itk-/- and Wt mice. The number of probe-sets changing ≥ 2-fold in Itk-/- compared to Wt samples was 900 (2% of the total number of probe-sets on the MOE 430 2.0 chips), which is equivalent to 56% up- and 44% down-regulated probe-sets (33% with p < 0.05). From the 900-list we show the 60 most significantly up- and down-regulated transcripts in Itk-deficiency (Table 1). Most up-regulated were the killer cell lectin-like receptors Klra3 and Klra8, followed by granzyme M. Oligoadenylate synthetase-like 2 (Oasl2) was the most down-regulated transcript, next was actinin alpha 2 (Actn2). Furthermore, from the 900 probe-sets we have manually extracted 106 immune response-related genes and divided them into 14 different subgroups (Table 2). Of the 106 genes 10% were Klrs and 8.5% encoded transcription factors (Table 3). Additional file 3 shows the individual genes in each category.
Table 1.
The 60 most up- and down-regulated transcripts in Itk-deficiency (unstimulated cells)
| Probe set | Gene Symbol, Gene Title | Itk-/- vs Wt | t-test |
| 1453196_a_at | Oasl2, 2'-5' oligoadenylate synthetase-like 2 | -7.91 | 0.008 |
| 1448327_at | Actn2, actinin alpha 2 | -3.97 | 0.016 |
| 1437445_at | Trpm1, transient receptor potential cation channel, subfamily M, member 1 | -3.72 | 0.051 |
| 1445450_x_at | A530021J07Rik | -3.71 | 0.012 |
| 1421234_at | Hnf1a, HNF1 homeobox A | -3.45 | 0.044 |
| 1448485_at | Ggt1, gamma-glutamyltransferase 1 | -3.39 | 0.005 |
| 1445194_at | Cnksr2, connector enhancer of kinase suppressor of Ras 2 | -3.31 | 0.034 |
| 1418545_at | Wasf1, WASP family 1 | -3.01 | 0.016 |
| 1451548_at | Upp2, uridine phosphorylase 2 | -2.91 | 0.049 |
| 1434722_at | Ampd1, adenosine monophosphate deaminase 1 | -2.85 | 0.004 |
| 1436836_x_at | Cnn3, calponin 3, acidic | -2.79 | 0.005 |
| 1429274_at | 2310010M24Rik | -2.46 | 0.001 |
| 1455442_at | Slc6a19, solute carrier family 6 member 19 | -2.34 | 0.005 |
| 1432383_a_at | Armc9, armadillo repeat containing 9 | -2.29 | 0.020 |
| 1417928_at | Pdlim4, PDZ and LIM domain 4 | -2.28 | 0.055 |
| 1444801_at | 2900041M22Rik | -2.27 | 0.029 |
| 1443570_at | Cops3, COP9 (constitutive photomorphogenic) homolog, subunit 3 | -2.25 | 0.005 |
| 1421895_at | Eif2s3x, eukaryotic translation initiation factor 2, subunit 3 | -2.25 | 0.013 |
| 1418055_at | Neurod4, neurogenic differentiation 4 | -2.23 | 0.020 |
| 1453009_at | Gene name not assigned for this probe set | -2.22 | 0.014 |
| 1429350_at | Eid3, EP300 interacting inhibitor of differentiation 3 | -2.21 | 0.039 |
| 1420877_at | Sept6, septin 6 | -2.17 | 0.001 |
| 1438825_at | Calm3, Calmodulin 3 | -2.16 | 0.022 |
| 1434915_s_at | Lrrc19, leucine rich repeat containing 19 | -2.12 | 0.041 |
| 1436103_at | Rab3ip, RAB3A interacting protein | -2.12 | 0.006 |
| 1456751_x_at | A530021J07Rik | -2.12 | 0.000 |
| 1439254_at | Gene name not assigned for this probe set | -2.11 | 0.037 |
| 1418003_at | 1190002H23Rik | -2.11 | 0.037 |
| 1449634_a_at | Anks1b, ankyrin repeat and sterile alpha motif domain containing 1B | -2.09 | 0.047 |
| 1418990_at | Ms4a4d, membrane-spanning 4-domains, subfamily A, member 4D | -2.09 | 0.031 |
| 1421182_at | Clec1b, C-type lectin domain family 1, member b | 3.7 | 0.038 |
| 1424842_a_at | Arhgap24, Rho GTPase activating protein 24 | 3.74 | 0.014 |
| 1418340_at | Fcer1g, Fc receptor, IgE, high affinity I, gamma polypeptide | 3.76 | 0.016 |
| 1444214_at | Tubb1, tubulin, beta 1 | 3.79 | 0.044 |
| 1452666_a_at | Tmcc2, transmembrane and coiled-coil domains 2 | 3.88 | 0.033 |
| 1457001_at | Cenpk, centromere protein K | 3.9 | 0.004 |
| 1449340_at | Sostdc1, sclerostin domain containing 1 | 3.91 | 0.014 |
| 1434115_at | Cdh13, cadherin 13 | 3.95 | 0.054 |
| 1434955_at | March1, membrane-associated ring finger (C3HC4) 1 | 4.04 | 0.000 |
| 1439397_at | Fmn1, formin 1 | 4.06 | 0.026 |
| 1448749_at | Plek, pleckstrin | 4.13 | 0.010 |
| 1426171_x_at | Klra7, killer cell lectin-like receptor, subfamily A, member 7 | 4.14 | 0.003 |
| 1436778_at | Cybb, cytochrome b-245, beta polypeptide | 4.15 | 0.012 |
| 1448025_at | Sirpb1, signal-regulatory protein beta 1 | 4.2 | 0.046 |
| 1420789_at | Klra5, killer cell lectin-like receptor, subfamily A, member 5 | 4.22 | 0.018 |
| 1441887_x_at | EG622976 | 4.26 | 0.017 |
| 1438553_x_at | Gene name not assigned for this probe set | 4.28 | 0.011 |
| 1417765_a_at | Amy1, amylase 1, salivary | 4.29 | 0.016 |
| 1451263_a_at | Fabp4, fatty acid binding protein 4 | 4.32 | 0.020 |
| 1427866_x_at | Gene name not assigned for this probe set | 4.43 | 0.037 |
| 1454200_at | Zeb2, zinc finger E-box binding homeobox 2 | 4.57 | 0.022 |
| 1420492_s_at | Smr3a, submaxillary gland androgen regulated protein 3A | 4.79 | 0.006 |
| 1427503_at | AI324046 | 4.85 | 0.024 |
| 1437463_x_at | Tgfbi, transforming growth factor, beta induced | 5.04 | 0.005 |
| 1419348_at | Psp, parotid secretory protein | 5.11 | 0.019 |
| 1419874_x_at | Zbtb16, zinc finger and BTB domain containing 16 | 5.46 | 0.005 |
| 1442025_a_at | Gene name not assigned for this probe set | 5.48 | 0.002 |
| 1449501_a_at | Gzmm, granzyme M | 6.39 | 0.013 |
| 1425436_x_at | Klra3, killer cell lectin-like receptor, subfamily A, member 3 | 9.92 | 0.000 |
| 1425417_x_at | Klra8, killer cell lectin-like receptor, subfamily A, member 8 | 35.69 | 0.000 |
The down-regulated transcripts are shown with "-"
Table 2.
Groups of genes expressed in the immune response group
| Immune response groups | Number of genes involved | Immune response groups | Number of genes involved |
| Chemokine receptors | 5 | Interleukins | 4 |
| Chemokines | 8 | Intracellular signaling components | 7 |
| Colony stimulating factor receptors | 4 | Killer cell lectin-like receptors | 11 |
| Fc receptors | 5 | Miscellaneous | 24 |
| Granzymes | 4 | Other surface antigens with CD-designation | 17 |
| Interferon-related genes | 3 | Toll-like receptors | 2 |
| Interleukin receptors | 4 | Transcription factors | 9 |
Table 3.
The genes found in Killer cell lectin-like receptor and transcription factor groups from Table 2
| Killer cell lectin-like receptors | ||
| Probe set | Gene title | Itk-/- vs Wt |
| 1458642_at | killer cell lectin-like receptor family E member 1 (NKG2I) | 2.6 |
| 1451664_x_at | killer cell lectin-like receptor subfamily A, member 12 (Ly49L) | 2.13 |
| 1422065_at | killer cell lectin-like receptor subfamily B member 1B (Ly55B/Ly55D) | 3.16 |
| 1425005_at | killer cell lectin-like receptor subfamily C, member 1 (NKG2A/2B) | 2.13 |
| 1420790_x_at | killer cell lectin-like receptor, subfamily A, member 16 (Ly49P) | -2.62 |
| 1426127_x_at | killer cell lectin-like receptor, subfamily A, member 18 (Ly49R) | 2.99 |
| 1426140_x_at | killer cell lectin-like receptor, subfamily A, member 19 (Ly49S) | 2.73 |
| 1425436_x_at | killer cell lectin-like receptor, subfamily A, member 3 (Ly49C) | 9.92 |
| 1420789_at | killer cell lectin-like receptor, subfamily A, member 5 (Ly49E) | 4.22 |
| 1426171_x_at | killer cell lectin-like receptor, subfamily A, member 7 (Ly49G) | 4.14 |
| 1425417_x_at | killer cell lectin-like receptor, subfamily A, member 8 (Ly49H) | 35.69 |
| Transcription factors | ||
| Probe set | Gene title | Itk-/- vs Wt |
| 1416916_at | E74-like factor 3 | 2.97 |
| 1457441_at | early B-cell factor 1 | * |
| 1416301_a_at | early B-cell factor 1 | * |
| 1435172_at | eomesodermin homolog (Xenopus laevis) | 2.56 |
| 1426001_at | eomesodermin homolog (Xenopus laevis) | 3.07 |
| 1421303_at | IKAROS family zinc finger 1 | -2.2 |
| 1422537_a_at | inhibitor of DNA binding 2 | 2.03 |
| 1447640_s_at | pre B-cell leukemia transcription factor 3 | 2.05 |
| 1460407_at | Spi-B transcription factor (Spi-1/PU.1 related) | 2.38 |
| 1429427_s_at | transcription factor 7-like 2, T-cell specific, HMG-box | 2.41 |
| 1419874_x_at | zinc finger and BTB domain containing 16 | 5.46 |
* Early B-cell factor 1 showed variable expression changes for different probe sets
The down-regulated transcripts are shown with "-"
Transcriptional changes in the absence of Itk in stimulated CD3+ T-cells
Stimulating the Itk-/- and Wt T-cells with anti-CD3 resulted in 804 differentially expressed probe-sets in Itk-deficiency (74% up- and 26% down-regulated, 68% with p < 0.05), while after anti-CD3/CD28-stimulation the number was reduced to 409 (78% up- and 22% down-regulated; 58% with p < 0.05) as depicted in Figure 1a. Between CD3- and CD3/CD28-stimulations, the overlap was 252 probe-sets (see Table 4 for a list of the 60 most up- and down-regulated transcripts). We show there that Itk was the most down-regulated transcript in Itk-deficiency, followed by Crabp2, which encodes cellular retinoic acid binding protein 2. This is a 15 kD regulator of retinoic acid signaling recently reported to be differentially expressed in acute lymphoblastic leukaemia [26]. Other down-regulated transcripts were IL-2 and IL-3.
Figure 1.
The number of differentially expressed probe-sets in Itk-deficiency. a. Venn diagram showing overlapping probe-sets in CD3+ Itk-defective T-cells, unstimulated (upper), anti-CD3- (left) and anti-CD3/CD28-stimulated (right). All the comparisons were made against Wt and with the criterion ≥ 2-fold. b. Quantitative RT-PCR confirms up-regulated expression of Klra3, Klra7 and Klrg1 in Itk-deficiency. The bar charts show relative amount of Klr mRNA compared to unstimulated Wt CD3+ T-cells (Wt C).
Table 4.
The 60 most up- and down-regulated transcripts in Itk-deficiency after anti-CD3- (1) and anti-CD3/CD28-stimulation (2)
| Probe set | Gene Symbol, Gene Title | Itk-/- vs Wt (1) | t-test | Itk-/- vs Wt (2) | t-test |
| 1457120_at | Itk, IL2-inducible T-cell kinase | -6.96 | 0.007 | -6.63 | 0.009 |
| 1451191_at | Crabp2, cellular retinoic acid binding protein II | -5.31 | 0.013 | -2.79 | 0.054 |
| 1449990_at | Il2///LOC630222, interleukin 2 | -3.89 | 0.002 | -5.17 | 0.030 |
| 1436194_at | Prelid2, PRELI domain containing 2 | -3.54 | 0.036 | -2.42 | 0.077 |
| 1437935_at | 4930486G11Rik, RIKEN cDNA | -3.44 | 0.032 | -3.16 | 0.079 |
| 1439995_at | Nhedc2, Na+/H+ exchanger domain containing 2 | -2.92 | 0.021 | -2.71 | 0.028 |
| 1441971_at | Gene name not assigned for this probe set | -2.91 | 0.050 | -2.88 | 0.082 |
| 1426243_at | Cth, cystathionase | -2.9 | 0.000 | -2.8 | 0.072 |
| 1438380_at | Ddx47, DEAD box polypeptide 47 | -2.69 | 0.002 | 2.03 | 0.300 |
| 1450566_at | Il3, interleukin 3 | -2.68 | 0.031 | -2.81 | 0.005 |
| 1420843_at | Ptprf, protein tyrosine phosphatase, receptor type, | -2.49 | 0.027 | -2.07 | 0.222 |
| 1448788_at | Cd200, Cd200 antigen | -2.47 | 0.014 | -2.43 | 0.011 |
| 1427049_s_at | Smo, smoothened homolog (Drosophila) | -2.46 | 0.000 | -2.45 | 0.015 |
| 1422070_at | Adh4, alcohol dehydrogenase 4 (class II) | -2.33 | 0.015 | -2.26 | 0.157 |
| 1456226_x_at | Ddr1, discoidin domain receptor family, member 1 | -2.29 | 0.014 | -2.91 | 0.038 |
| 1419136_at | Akr1c18, aldo-keto reductase family 1, member C18 | -2.09 | 0.052 | 2.41 | 0.008 |
| 1433571_at | Serinc5, serine incorporator 5 | -2 | 0.011 | -2.05 | 0.125 |
| 1425832_a_at | Cxcr6, chemokine (C-X-C motif) receptor 6 | 5.34 | 0.000 | 3.27 | 0.085 |
| 1437463_x_at | Tgfbi, transforming growth factor, beta induced | 5.36 | 0.045 | 2.4 | 0.012 |
| 1421802_at | Ear1, eosinophil-associated, ribonuclease A family, member 1 | 5.38 | 0.017 | 2.8 | 0.000 |
| 1448620_at | Fcgr3, Fc receptor, IgG, low affinity III | 5.57 | 0.036 | 3.72 | 0.004 |
| 1438855_x_at | Tnfaip2, tumor necrosis factor, alpha-induced protein 2 | 5.6 | 0.0034 | 3.12 | 0.000 |
| 1450009_at | Ltf, lactotransferrin | 5.68 | 0.080 | 2.03 | 0.034 |
| 1416514_a_at | Fscn1, fascin homolog 1 | 5.7 | 0.000 | 3.69 | 0.096 |
| 1451948_at | Gm1409, gene model 1409 | 5.81 | 0.002 | 3.45 | 0.136 |
| 1451675_a_at | Alas2, aminolevulinic acid synthase 2, erythroid | 5.84 | 0.003 | 2.92 | 0.096 |
| 1420330_at | Clec4e, C-type lectin domain family 4, member e | 5.89 | 0.014 | 3.15 | 0.091 |
| 1420699_at | Clec7a, C-type lectin domain family 7, member a | 5.98 | 0.014 | 4.05 | 0.004 |
| 1427747_a_at | Lcn2, lipocalin 2 | 5.98 | 0.056 | 2.23 | 0.011 |
| 1427503_at | AI324046, expressed sequence AI324046 | 6.33 | 0.005 | 3.48 | 0.055 |
| 1419082_at | Serpinb2, serine (or cysteine) peptidase inhibitor, clade B, member 2 | 6.34 | 0.001 | 2.51 | 0.125 |
| 1419627_s_at | Clec4n, C-type lectin domain family 4, member n | 6.36 | 0.021 | 2.6 | 0.0129 |
| 1448213_at | Anxa1, annexin A1 | 6.43 | 0.069 | 3.07 | 0.029 |
| 1419874_x_at | Zbtb16, zinc finger and BTB domain containing 16 | 6.62 | 0.032 | 5.3 | 0.001 |
| 1417898_a_at | Gzma, granzyme A | 6.71 | 0.009 | 5.59 | 0.001 |
| 1419598_at | Ms4a6d, membrane-spanning 4-domains, subfamily A, member 6D | 6.73 | 0.005 | 3.27 | 0.083 |
| 1429889_at | Faim3, Fas apoptotic inhibitory molecule 3 | 6.73 | 0.047 | 3.01 | 0.041 |
| 1415904_at | Lpl, lipoprotein lipase | 6.76 | 0.054 | 4.1 | 0.026 |
| 1449254_at | Spp1, secreted phosphoprotein 1 | 6.8 | 0.028 | 3.79 | 0.020 |
| 1427910_at | Cst6, cystatin E/M | 7.1 | 0.023 | 4.17 | 7.43E-06 |
| 1438553_x_at | Gene name not assigned for this probe set | 7.11 | 0.011 | 4.63 | 0.055 |
| 1434150_a_at | Mettl7a///Ubie, methyltransferase like 7A | 7.14 | 0.000 | 4.04 | 0.185 |
| 1442025_a_at | AI467657, expressed sequence AI467657 | 7.48 | 0.041 | 6.52 | 0.000 |
| 1436778_at | Cybb, cytochrome b-245, beta polypeptide | 7.77 | 0.077 | 4.03 | 0.003 |
| 1439426_x_at | Lyz, lysozyme | 7.78 | 0.006 | 2.98 | 0.011 |
| 1449846_at | Ear2///Ear3, eosinophil-associated, ribonuclease A family, member 2 | 8.62 | 0.008 | 3.54 | 0.031 |
| 1434194_at | Mtap2, microtubule-associated protein 2 | 8.73 | 0.035 | 5.78 | 0.000 |
| 1427866_x_at | Beta globin | 9.29 | 0.001 | 5.44 | 0.074 |
| 1450912_at | Ms4a1, membrane-spanning 4-domains, subfamily A, member 1 | 9.4 | 0.087 | 4.31 | 0.036 |
| 1422873_at | Prg2, proteoglycan 2, bone marrow | 9.81 | 0.028 | 4.19 | 0.026 |
| 1419764_at | Chi3l3, chitinase 3-like 3 | 10.51 | 0.070 | 4.1 | 0.004 |
| 1422411_s_at | Ear1///Ear12///Ear2///Ear3, eosinophil-associated, ribonuclease A family, member 1 | 10.54 | 0.010 | 4.61 | 0.000 |
| 1418722_at | Ngp, neutrophilic granule protein | 11.39 | 0.031 | 3.74 | 0.006 |
| 1450989_at | LOC100047300///Tdgf1, teratocarcinoma-derived growth factor | 11.64 | 0.009 | 2.89 | 0.076 |
| 1419394_s_at | S100a8, S100 calcium binding protein A8 | 12.08 | 0.071 | 4.6 | 0.034 |
| 1425436_x_at | Klra3, killer cell lectin-like receptor, subfamily A, member 3 | 12.34 | 0.002 | 14.75 | 0.000 |
| 1415897_a_at | Mgst1, microsomal glutathione S-transferase 1 | 12.62 | 0.082 | 3.77 | 0.006 |
| 1448756_at | S100a9, S100 calcium binding protein A9 | 12.97 | 0.077 | 5.15 | 0.010 |
| 1426171_x_at | Klra7, killer cell lectin-like receptor, subfamily A, member 7 | 14.08 | 0.027 | 14.02 | 0.015 |
| 1425417_x_at | Klra8, killer cell lectin-like receptor, subfamily A, member 8 | 21.07 | 0.026 | 17.99 | 0.000 |
The down-regulated transcripts are shown with "-"
Stimulation affected the majority of the transcripts in the same direction as observed in unstimulated cells (p < 10-6) (only in 5/252 cases the CD3- or CD3/CD28-stimulations showed opposite fold-changes; Additional file 4). We show there that the most induced mRNA was Klra8 (Ly49H) (21-fold-change upon anti-CD3-stimulation). Other up-regulated Klrs were Klra3 (Ly49C), Klra5 (Ly49E), Klra7 (Ly49G), Klra19 (Ly49S), Klrc1 (NKG2A/2B), Klrd1 (CD94), Klre1 (NKG2I) and Klrg1 (2P1-Ag). By quantitative RT-PCR we confirmed the up-regulated expression of Klrg1, Klra3 and Klra7 in Itk-defective samples (Fig. 1b). Two transcription factors, inhibitor of DNA binding 2 (Id2) and eomesodermin, were also found up-regulated. Thus, in CD3+ cells, differential transcriptional signatures between Wt and Itk-deficient cells were more pronounced in unstimulated when compared to activated cells.
We continued to analyze the activation-dependent signatures in Wt and Itk-/- T-cells separately. The number of probe-sets changing ≥ 2-fold after anti-CD3-stimulation (compared to the unstimulated state) in the Wt samples was 4252, and the corresponding number after co-stimulation was 4385 (Figure 2). The overlap between the two stimulations was 3713 (87% and 85%, respectively; Additional file 5). However, the differences were significantly more pronounced in anti-CD3 versus anti-CD3/CD28 activated cells in the Itk-defective group, with only 50% of the transcripts in the co-stimulated group overlapping with the CD3-stimulated (p < 10-6) (Additional file 6). Thus, co-stimulation had much greater effect on Itk-deficient than on Wt cells. We further examined some of the immune response-related genes previously mentioned. Ten members of the Klr family were up-regulated in unstimulated Itk-defective compared to Wt samples (Table 3), while after stimulation the majority of Klrs were down-regulated in both Wt and Itk-defective T-cells. Down-regulation of Klrs were also reported for human cells from healthy individuals in a recent paper by Wang et al., where primary human T-cells were analyzed after anti-CD3/CD28-stimulation [27]. With respect to cytokine expression, IL-2 and IL-6 were found up-regulated upon anti-CD3-stimulation in both Wt and Itk-deficient samples when compared to the corresponding unstimulated cells. In contrast, IL-16 and IL-18 were down-regulated (Additional files 5 and 6). Two cytokines, whose expression was only altered in Itk-deficient cells upon anti-CD3-stimulation, were IL-10 (up-regulated) and IL-33 (down-regulated) (data not shown). IL-33 is a novel IL-1 family cytokine, IL1F11/IL-33, playing an important role in eosinophil-mediated inflammation [28]. Interestingly, Itk-deficient mice have previously been shown to have reduced lung inflammation, eosinophil infiltration and mucous production after induction of allergic asthma [29]. No other cytokines were differentially expressed. In the stimulated Wt samples we also observed altered expression of several transcription factors such as Zbtb16 (encoding the transcriptional regulator PLZF), Id2 and Spi-C, while in Itk-defective cells we found Zbtb16, Spi-C and Id3 to be differentially expressed upon stimulation.
Figure 2.
The amount of differentially expressed probe-sets in Wt and Itk-defective CD3+ T-cells following stimulation. The upper panel represents the Wt T-cells and the lower the Itk-defective T-cells. The left panel symbolizes the anti-CD3-stimulation and the right panel the anti-CD3/CD28-stimulation. All the stimulations were compared to the untreated condition. The arrows denote the number of overlapping probe-sets.
CD4+ and CD8+ T-cell signatures in Itk-deficiency
As T-cells can be divided into CD4+ and CD8+ subsets, and as these subsets have very distinct functions and gene regulations, we examined Itk-deficiency in MACS purified CD4+ and CD8+ Wt and Itk-deficient T-cells. The Itk-deficient CD4+ and CD8+ T-cells are known from previous studies to be of a memory-like phenotype, characterized by the markers CD44 and CD122 [9-11,13]. Since CD122 expression is enhanced by the transcription factors eomesodermin and T-box 21 (T-bet), we looked for their expression in our data. Eomesodermin was previously reported to be up-regulated in Itk-deficient T-cells [9] and we found the expression of eomesodermin much higher in the CD8+ T-cell population compared to CD4+ in unstimulated condition. The same was also seen with T-bet. Taken together, the observed expression pattern of eomesodermin and T-bet is in agreement with previously published studies and thus validates our microarray data. Moreover, our analysis also includes new knowledge related to these transcripts, namely how they respond to activation of T-cells as well as the effect of CsA (Additional file 7).
Both CD4+ and CD8+ T-cell subsets in Itk-deficient mice have been shown to differ in phenotype compared to the Wt mice. In the absence of Itk, a higher percentage of each subset expresses surface markers, typical for memory phenotype cells, such as CD44hi and CD122hi [9,10,13]. We sought to determine whether this was also reflected by their transcriptomes. The number of transcripts differentially expressed between unstimulated Itk-/- and Wt in the CD4+ population was 2050, while in the CD8+ population the number was higher (n = 6907). The 60 most up- and down-regulated transcripts from each subset are shown in Tables 5 and 6. Among those are genes already mentioned, e.g. eomesodermin, Klra3 and 8, T-bet and Granzyme M. In these groups we also found Zbtb16. Interestingly, Zbtb16 was 12-fold up-regulated in CD4+ cells and 45-fold down-regulated in CD8+ cells, also suggesting a highly efficient separation of the two subsets. Based on these findings PLZF was selected for further studies presented elsewhere [30]. The most pronounced changes were seen in the CD8+ population (Table 6), with 69-fold down-regulation of Clca1, which is a calcium-activated chloride-channel, of importance in airway epithelial cells. Two up-regulated transcripts were the PTB-domain containing MAP-kinase regulator Dok5 (29-fold) and α-tubulin (30-fold), whose expression in T-cells, to our knowledge, was not previously reported. After anti-CD3-stimulation we found the number of differentially expressed transcripts reduced in both subsets, approximately 30% and 47% fewer probe-sets in CD4+ and CD8+, respectively. The overlapping probe-sets between the unstimulated and the anti-CD3-stimulated conditions are shown in Figure 3. 82% of the transcripts in the CD4+ subset were also found in the CD8+ population in unstimulated cells. The percentage of overlapping transcripts decreased with stimulation.
Table 5.
The 60 most up- and down-regulated transcripts in Itk-defective CD4+ T-cells (unstimulated cells)
| Probe set | Gene Symbol, Gene Title | Itk-/- vs Wt | t-test |
| 1436386_x_at | OTTMUSG00000010671 | -7.51 | 0.012 |
| 1444708_at | Tmem29, transmembrane protein 29 | -5 | 0.014 |
| 1434418_at | Lass6, LAG1 homolog, ceramide synthase 6 | -4.93 | 0.001 |
| 1438354_x_at | Cnn3, Calponin 3, acidic | -4.81 | 0.030 |
| 1430988_at | 2810407C02Rik | -3.84 | 0.028 |
| 1430827_a_at | Ptk2, PTK2 protein tyrosine kinase 2 | -3.68 | 0.002 |
| 1458977_at | A530021J07Rik | -3.36 | 0.006 |
| 1439778_at | Cables1, Cdk5 and Abl enzyme substrate 1 | -3.26 | 0.018 |
| 1427675_at | V1ra2, vomeronasal 1 receptor, A2 | -3.22 | 0.041 |
| 1448338_at | Pgcp, plasma glutamate carboxypeptidase | -3.16 | 0.001 |
| 1458945_at | AU015148 | -3.16 | 0.038 |
| 1421507_at | Olfr78, olfactory receptor 78 | -3.07 | 0.021 |
| 1457120_at | Itk, IL2-inducible T-cell kinase | -2.96 | 0.002 |
| 1456178_at | Bambi-ps1, BMP and activin membrane-bound inhibitor, pseudogene (Xenopus laevis) | -2.94 | 0.000 |
| 1455907_x_at | Phox2b, paired-like homeobox 2b | -2.88 | 0.050 |
| 1452474_a_at | Art3, ADP-ribosyltransferase 3 | -2.86 | 0.026 |
| 1459508_at | C85600 | -2.79 | 0.007 |
| 1440761_at | 4833422C13Rik | -2.77 | 0.052 |
| 1446412_at | Gene name not assigned for this probe set | -2.71 | 0.012 |
| 1441221_at | Gene name not assigned for this probe set | -2.7 | 0.019 |
| 1427632_x_at | Cd55, CD55 antigen | -2.65 | 0.042 |
| 1439181_at | BC043301 | -2.63 | 0.032 |
| 1434473_at | Slc16a5, solute carrier family 16 (monocarboxylic acid transporters), member 5 | -2.59 | 0.001 |
| 1448002_x_at | 2610001J05Rik | -2.57 | 0.018 |
| 1455425_at | BB001228 | -2.57 | 0.016 |
| 1419620_at | Pttg1, pituitary tumor-transforming 1 | -2.53 | 0.000 |
| 1453009_at | Gene name not assigned for this probe set | -2.39 | 0.000 |
| 1455740_at | Hnrnpa1, heterogeneous nuclear ribonucleoprotein A1 | -2.31 | 0.048 |
| 1429413_at | Cpm, carboxypeptidase M | -2.28 | 0.003 |
| 1416441_at | Pgcp, plasma glutamate carboxypeptidase | 3.12 | 0.005 |
| 1425216_at | Ffar2, free fatty acid receptor 2 | 3.13 | 0.036 |
| 1448471_a_at | Ctla2a, cytotoxic T lymphocyte-associated protein 2 alpha | 3.13 | 0.006 |
| 1450334_at | Il21, interleukin 21 | 3.13 | 0.013 |
| 1423091_a_at | Gpm6b, glycoprotein m6b | 3.21 | 0.032 |
| 1435339_at | Kctd15, potassium channel tetramerisation domain containing 15 | 3.24 | 0.044 |
| 1428197_at | Tspan9, tetraspanin 9 | 3.37 | 0.001 |
| 1449036_at | Rnf128, ring finger protein 128 | 3.44 | 0.002 |
| 1449361_at | Tbx21, T-box 21 | 3.5 | 0.012 |
| 1447839_x_at | Adm, adrenomedullin | 3.62 | 0.031 |
| 1418318_at | Rnf128, ring finger protein 128 | 3.77 | 0.021 |
| 1419647_a_at | Ier3, immediate early response 3 | 3.78 | 0.005 |
| 1448961_at | Plscr2, phospholipid scramblase 2 | 4.06 | 0.012 |
| 1449280_at | Esm1, endothelial cell-specific molecule 1 | 4.15 | 0.013 |
| 1427445_a_at | Ttn, titin | 4.32 | 0.006 |
| 1425471_x_at | Gene name not assigned for this probe set | 4.33 | 0.050 |
| 1438553_x_at | Gene name not assigned for this probe set | 4.35 | 0.019 |
| 1423231_at | Nrgn, neurogranin | 4.66 | 0.002 |
| 1416846_a_at | Pdzrn3, PDZ domain containing RING finger 3 | 4.69 | 0.002 |
| 1430946_at | 2600014E21Rik | 4.82 | 0.031 |
| 1426001_at | Eomes, eomesodermin homolog (Xenopus laevis) | 5.43 | 0.024 |
| 1422280_at | Gzmk, granzyme K | 5.6 | 0.002 |
| 1427608_a_at | Tcrg-V1, T-cell receptor gamma, variable 1 | 5.62 | 0.044 |
| 1434194_at | Mtap2, microtubule-associated protein 2 | 5.71 | 0.040 |
| 1434115_at | Cdh13, cadherin 13 | 6.15 | 0.047 |
| 1455435_s_at | Chdh, choline dehydrogenase | 6.54 | 0.041 |
| 1449864_at | Il4, interleukin 4 | 6.77 | 0.031 |
| 1424011_at | Aqp9, aquaporin 9 | 7.23 | 0.011 |
| 1420678_a_at | Il17rb, interleukin 17 receptor B | 9.84 | 0.029 |
| 1442025_a_at | Gene name not assigned for this probe set | 11.69 | 0.008 |
| 1419874_x_at | Zbtb16, zinc finger and BTB domain containing 16 | 12.07 | 0.009 |
The down-regulated transcripts are shown with "-"
Table 6.
The 60 most up- and down-regulated transcripts in Itk-defective CD8+ T-cells (unstimulated cells)
| Probe set | Gene Symbol, Gene Title | Itk-/- vs Wt |
| 1417852_x_at | Clca1, chloride channel calcium activated 1 | -68.71 |
| 1419874_x_at | Zbtb16, zinc finger and BTB domain containing 16 | -45.13 |
| 1436759_x_at | Cnn3, calponin 3, acidic | -39.91 |
| 1454869_at | Wdr40b, WD repeat domain 40B | -34.02 |
| 1427054_s_at | Abi3bp, ABI gene family, member 3 (NESH) binding protein | -31.51 |
| 1437992_x_at | Gja1, gap junction protein, alpha 1 | -25.36 |
| 1416203_at | Aqp1, aquaporin 1 | -24.68 |
| 1437279_x_at | Sdc1, syndecan 1 | -24.33 |
| 1448182_a_at | Cd24a, CD24a antigen | -21.41 |
| 1456956_at | Ikzf2, IKAROS family zinc finger 2 | -19.8 |
| 1442025_a_at | Gene name not assigned for this probe set | -19.78 |
| 1439422_a_at | C1qdc2, C1q domain containing 2 | -19.69 |
| 1454086_a_at | Lmo2, LIM domain only 2 | -18.62 |
| 1416330_at | Cd81, CD81 antigen | -17.08 |
| 1451867_x_at | Arhgap6, Rho GTPase activating protein 6 | -17.08 |
| 1456060_at | Maf, avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog | -16.84 |
| 1419014_at | Rhag, Rhesus blood group-associated A glycoprotein | -16.79 |
| 1416193_at | Car1, carbonic anhydrase 1 | -16.49 |
| 1450744_at | Ell2, elongation factor RNA polymerase II 2 | -14.83 |
| 1456147_at | St8sia6, ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 | -14.78 |
| 1456475_s_at | Prkar2b, protein kinase, cAMP dependent regulatory, type II beta | -14.57 |
| 1460431_at | Gcnt1, glucosaminyl (N-acetyl) transferase 1, core 2 | -14.03 |
| 1437171_x_at | Gsn, gelsolin | -13.64 |
| 1417777_at | Ltb4dh, leukotriene B4 12-hydroxydehydrogenase | -13.54 |
| 1437935_at | 4930486G11Rik | -13.51 |
| 1434499_a_at | Ldhb, lactate dehydrogenase B | -13.44 |
| 1450333_a_at | Gata2, GATA binding protein 2 | -13.33 |
| 1423569_at | Gatm, glycine amidinotransferase (L-arginine:glycine amidinotransferase) | -12.94 |
| 1435884_at | Itsn1, intersectin 1 (SH3 domain protein 1A) | -12.72 |
| 1425145_at | Il1rl1, interleukin 1 receptor-like 1 | -12.6 |
| 1449313_at | Klk1b5, kallikrein 1-related peptidase b5 | 13.38 |
| 1454106_a_at | Cxxc1, CXXC finger 1 (PHD domain) | 13.41 |
| 1442639_at | Gene name not assigned for this probe set | 13.76 |
| 1443006_at | Gene name not assigned for this probe set | 13.93 |
| 1433449_at | Snx32, sorting nexin 32 | 14.78 |
| 1423603_at | Zfpm1, zinc finger protein, multitype 1 | 14.98 |
| 1441770_at | Ppat, phosphoribosyl pyrophosphate amidotransferase | 15.29 |
| 1420343_at | Gzmd, granzyme D | 15.42 |
| 1445596_at | Gene name not assigned for this probe set | 15.54 |
| 1452985_at | Uaca, uveal autoantigen with coiled-coil domains and ankyrin repeats | 16.55 |
| 1420233_at | Gene name not assigned for this probe set | 16.79 |
| 1423020_at | Gene name not assigned for this probe set | 17.39 |
| 1454481_at | Mif, macrophage migration inhibitory factor | 17.64 |
| 1427426_at | Kcnq5, potassium voltage-gated channel, subfamily Q, member 5 | 18.06 |
| 1424698_s_at | Gca, grancalcin | 18.46 |
| 1447574_s_at | Slc32a1, Solute carrier family 32 (GABA vesicular transporter), member 1 | 18.73 |
| 1431854_a_at | 4930452B06Rik | 18.94 |
| 1460267_at | Dmrt3, doublesex and mab-3 related transcription factor 3 | 19.76 |
| 1417197_at | Wwc2, WW, C2 and coiled-coil domain containing 2 | 20.76 |
| 1442788_at | Afap1, actin filament associated protein 1 | 20.91 |
| 1447292_at | Actr1b, ARP1 actin-related protein 1 homolog B (yeast) | 21.96 |
| 1431878_at | Grhl2, grainyhead-like 2 (Drosophila) | 22.18 |
| 1449501_a_at | Gzmm, granzyme M (lymphocyte met-ase 1) | 23.58 |
| 1436500_at | Rps24, Ribosomal protein S24 | 25.96 |
| 1425436_x_at | Klra3, killer cell lectin-like receptor, subfamily A, member 3 | 26.39 |
| 1456130_at | LOC553091 | 27.36 |
| 1454240_at | Nfe2l3, nuclear factor, erythroid derived 2, like 3 | 29.25 |
| 1422641_at | Dok5, docking protein 5 | 29.31 |
| 1417375_at | Tuba4a, tubulin, alpha 4A | 30.36 |
| 1425417_x_at | Klra8, killer cell lectin-like receptor, subfamily A, member 8 | 131.81 |
The down-regulated transcripts are shown with "-"
Figure 3.
Overlapping probe-sets in unstimulated and anti-CD3-stimulated Itk-deficient CD4+ (left) and CD8+ (right) T-cell populations. Each circle is a comparison between Itk-defective and Wt samples. All the comparisons were made with the criterion ≥ 2-fold.
The largest number of differentially expressed transcripts was observed in the unstimulated groups of Itk-deficient CD4+ and CD8+ T-cell subsets. By subtracting the overlapping 1495 probe-sets (Figure 3) from unstimulated Itk-deficient CD4+ and CD8+ cells, respectively, we characterized separate core groups of transcripts for each subset. The remaining number of probe-sets in the CD4+ population was 324 (Additional file 8), while it was more than 10-fold higher in the CD8+ group (Additional file 9). Interestingly, two members of the Klr family (Klrb1a (Ly55a) and Klrb1c (NK-1.1)) were present in the CD4+population, while four other members were found in the CD8+ subset (Klra4 (Ly49D), Klra19 (Ly49S), Klrc2 (NKG2C) and Klrk1 (NKG2D)). Five Klrs were in common between unstimulated CD4+ and CD8+ groups, they were Klra3, a7, a8, a22 and b1b (by comparing 2050 and 6907 in Fig. 3). The differentially expressed NK/innate cell-related transcripts were not limited to cell surface markers, since RNA for the cytotoxic protein Granzyme M was strongly enriched in the Itk-deficient population, again confirming that NK- and innate cells have overlapping transcriptomes [31].
Itk-deficiency mimics calcineurin inhibition
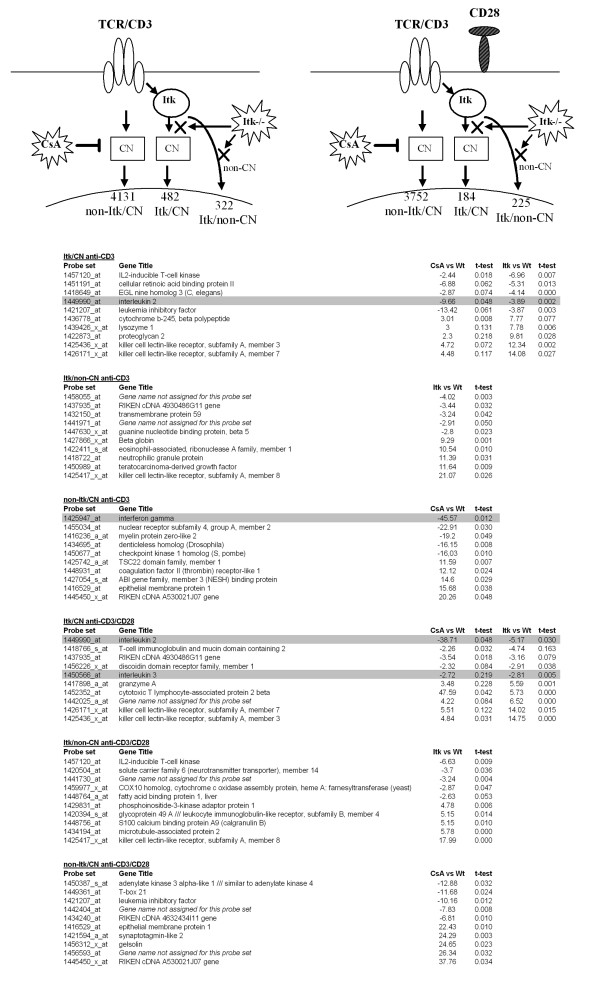
Tec-family kinases activate PLCγ and are therefore important regulators of Ca2+-mobilization and the calcineurin/NFAT pathway [5,32]. However, Tec-family kinases regulate also other signaling pathways. To investigate which of the Itk-related changes is the consequence of an impaired calcineurin/NFAT pathway, we compared the expression profiles of anti-CD3 ± CD28 stimulated Itk-/- CD3+ T-cells and of CsA-treated Wt T-cells. CsA specifically inhibits calcineurin and by that affects downstream signaling and the activation of the transcription factors of the NFAT-family. Based on the dependency of Itk and/or calcineurin, three groups of genes could be identified: Itk- and calcineurin-dependent (Itk/CN); Itk-dependent and calcineurin-independent (Itk/non-CN) and Itk-independent and calcineurin-dependent (non-Itk/CN).
Altogether, after anti-CD3-stimulation 4613 probe-sets were differentially expressed in CsA-treated cells compared to untreated, and after co-stimulation the number was reduced by 15% to 3936. The gene numbers observed in Itk-deficient compared to Wt cells were 804 and 409 after anti-CD3- and anti-CD3/CD28-stimulation, respectively (Figure 1a). About 60% of the probe-sets that were changed ≥ 2-fold in Itk-/- compared to Wt after anti-CD3-stimulation were also found in the CsA-treated samples (Figure 4, Itk/CN anti-CD3, showing the 10 most highly-regulated transcripts). In co-stimulated cells 45% of the probe-sets were the same (Figure 4, Itk/CN anti-CD3/CD28). When comparing the Itk-dependent probe-sets being calcineurin-dependent in both stimulations the overlap was 113 (Additional file 10). As expected, IL-2 was found in that group, confirming the biological relevance of our data, since IL-2 is known to be both Itk- and calcineurin-dependent [16,33]. Among other genes found in this group were Zbtb16 (up-regulated) and Crabp2 (down-regulated). Interestingly, the transcript for chemokine (C-motif) ligand 1 (Xcl1) was down-regulated in the CsA-treated cells while it showed increased expression in Itk-deficient samples. Furthermore, cytotoxic T lymphocyte-associated protein 2 alpha and beta (Ctla2a and Ctla2b) were up-regulated. Interestingly, their altered expression was more pronounced in the CsA-treated (>3 times higher) than in the Itk-deficient samples. Two granzyme-encoding genes, Gzma and Gzmk, were also found among those that were Itk- and calcineurin-dependent.
Figure 4.
Anti-CD3- and anti-CD3/CD28-stimulations leading to Itk- and calcineurin-dependent and -independent transcriptional signatures. Three different groups of genes exist in each stimulation condition. The groups are Itk- and calcineurin-dependent (Itk/CN), Itk-dependent and calcineurin-independent (Itk/non-CN) and Itk-independent and calcineurin-dependent (non-Itk/CN). 113 probe-sets are overlapping between Itk/CN groups in the two stimulations. In each group the 10 most highly regulated transcripts are presented. All the chosen genes passed the t-test criterion of p < 0.05. The down-regulated transcripts are shown with "-". The transcripts in the two Itk/CN groups passed the criterion in at least one of the two comparisons. Genes previously known to be calcineurin-regulated are grey-shaded [16]. Arrows denoting signal transduction from CD28 have been omitted for clarity.
The fractions of Itk/non-CN genes (322 vs 225- for anti-CD3- and anti-CD3/CD28-stimulated T-cells, respectively) shared 95 probe-sets, corresponding to 89 transcripts (Additional file 11). Among them were three up-regulated members of the Klrs; Klra5, Klra8 and Klre1. After co-stimulation, a much smaller number of probe-sets were Itk-dependent compared to anti-CD3-stimulation only (p < 10-6). The transcripts being calcineurin-dependent but Itk-independent (non-Itk/CN group) were 4131 and 3752 in anti-CD3- and co-stimulated cells, respectively. It is interesting to note that CsA-treatment, but not Itk-deficiency (the non-Itk/CN anti-CD3 group), results in severely reduced transcript levels for IFNγ. Previous studies show that an immediate IFNγ release is a hallmark of the innate CD8-population [9,10].
NFAT-binding genes that are Itk- and calcineurin- dependent
The comparison of Itk-deficient and CsA-treated Wt T-cells revealed 482 up- or down-regulated transcripts upon anti-CD3-stimulation. In order to identify putative NFAT-binding sites (GGAAA), we selected 24 genes for bioinformatic analyses. The genes were chosen as being highly regulated in the CsA or Itk-/- comparisons after anti-CD3-stimulation. 19/24 of these genes were also significantly regulated after co-stimulation with anti-CD3/CD28. 15/24 genes had putative NFAT-sites in the 500 bp region upstream of the transcriptional start site (Additional file 12). We identified 1 to 2 binding sites in 4 of those genes: IL7R, Bub1, Ctla2a and Ctla2b, as well as upstream of the translation initiation of Schlafen1 (Slfn1) gene (Figure 5a). To test whether NFAT binds to the promoter region of the genes in vivo, chromatin-immunoprecipitation experiments were performed. As a positive control, we used the IL-2 promoter region known to contain functional NFAT-sites bound by NFATc1 [34,35]. ChIP assays demonstrated NFATc1 binding to the IL-2 promoter region as expected and revealed anti-CD3 induced binding of NFATc1 to the selected regions of the five genes. They were also shown to be bona fide calcineurin-regulated genes owing to that the induced binding was reversed by CsA-treatment (Fig. 5b). A heat-map presenting the signal intensities of the above mentioned genes is shown in Figure 5c.
Figure 5.

NFAT-binding genes being Itk- and calcineurin-dependent. a. Promoter regions with NFAT-binding sites in IL2, IL7R, Bub1, Slfn1 (Schlafen1), Ctla2a and Ctla2b genes are shown with the binding site(s) represented by black boxes. This identification was done by bioinformatic analyses of the 500 bp region upstream of each gene's transcriptional start site. This approach identified 1–2 NFAT binding sites within the promoter regions of these genes as well as upstream of the translation initiation of Slfn1 gene. The numbers below each box represent the position of the binding site in correlation to the transcription start. The arrows indicate forward and reverse primers. b. NFATc1-binding in IL2, IL7R, Bub1, Slfn1, Ctla2a and Ctla2b genes. CD3+ T-cells were isolated from Wt thymus (thy) and spleen (spl) as described in experimental procedures. The PCR pictures were analyzed with Fluoro-S gel documentation equipment (BioRad Laboratories, CA) with a CCD camera, and further evaluated using the Quantity One software. Input; DNA before IP, NTC; no template control. c. Signal intensities of the six genes in Wt unstimulated, Wt anti-CD3-stimulated, Wt anti-CD3-stimulated + CsA-treated, Itk-defective unstimulated and Itk-defective anti-CD3- stimulated samples. The figure was made in dChip [21]. The color scale in the lower part of the picture corresponds to the mean expression of a gene. The red color represents expression level above mean expression of a gene across all samples, the white color is mean expression and the blue color represents expression lower than the mean.
Transcripts regulated by Tec-family kinases
Itk is crucial for T-cell development and activation. Similarly, Btk is essential for proper differentiation and activation of B-cells [36,37]. In a previous study, we investigated the genes modulated by Btk [24]. There was a pronounced overlap 18/38 (47%) between differentially expressed genes from the Itk-defective T-cells (900-list) and the previously published list of Btk-deficient genes from splenic B-cells as analyzed by the U74Av2 chip with approximately 12 000 genes [24]. The overlapping transcripts are shown in Table 7. Sixteen of the 18 genes were similarly regulated, which shows a highly significant co-variation (p = 0.01). Among these genes were those for transcription factors (Id2, Ikaros and Spi-C), cell membrane spanning (Csf1r, Mrc1 and Vcam1) and secreted proteins (Aif1, Igf1 and Tgfbi).
Table 7.
Overlapping transcripts between Btk-/- B-cells and Itk-/- T-cells
| U74Av2 | MOE 430 2.0 | Gene Symbol, Gene Title | Fold change Btk-/-/Itk-/- |
| 102330_at | 1418204_s_at | Aif1, allograft inflammatory factor 1 | 7.47/2.28 |
| 95546_g_at | 1419519_at | Igf1, insulin-like growth factor 1 | 6.5/2.46 |
| 103226_at | 1450430_at | Mrc1, mannose receptor, C type 1 | 5.42/6.06 |
| 95597_at | 1423414_at | Ptgs1, prostaglandin-endoperoxide synthase 1 | 4.68/2.43 ** |
| 95597_at | 1436448_a_at | Ptgs1, prostaglandin-endoperoxide synthase 1 | 4.68/2.19 ** |
| 96020_at | 1417063_at | C1qb, complement component 1, q subcomponent, beta polypeptide | 4.57/6.6 *** |
| 96020_at | 1434366_x_at | C1qb, complement component 1, q subcomponent, beta polypeptide | 4.57/5.87 *** |
| 96020_at | 1437726_x_at | C1qb, complement component 1, q subcomponent, beta polypeptide | 4.57/4.46 *** |
| 104354_at | 1419873_s_at | Csf1r, colony stimulating factor 1 receptor | 4.4/3.96 ** |
| 104354_at | 1423593_a_at | Csf1r, colony stimulating factor 1 receptor | 4.4/2.95 ** |
| 103736_at | 1448005_at | Sash1, SAM and SH3 domain containing 1 | 3.97/6.79 |
| 103454_at | 1418555_x_at | Spic, Spi-C transcription factor (Spi-1/PU.1 related) | 3.93/3.75 * |
| 103454_at | 1449134_s_at | Spic, Spi-C transcription factor (Spi-1/PU.1 related) | 3.93/8.5 * |
| 92877_at | 1415871_at | Tgfbi, transforming growth factor, beta induced | 3.9/3.46 *** |
| 92877_at | 1437463_x_at | Tgfbi, transforming growth factor, beta induced | 3.9/5.04 *** |
| 92877_at | 1448123_s_at | Tgfbi, transforming growth factor, beta induced | 3.9/4.52 *** |
| 92877_at | 1456250_x_at | Tgfbi, transforming growth factor, beta induced | 3.9/4.6 *** |
| 92223_at | 1449401_at | C1qc, complement component 1, q subcomponent, C chain | 3.73/5.47 |
| 92558_at | 1448162_at | Vcam1, vascular cell adhesion molecule 1 | 3.63/5.62 |
| 102065_at | 1418243_at | Fcna, ficolin A | 3.6/5.27 |
| 103070_at | 1416985_at | Sirpa, signal-regulatory protein alpha | 3.42/3.58 |
| 102860_at | 1424923_at | Serpina3g, serine (or cysteine) peptidase inhibitor, clade A, member 3G | 3.05/2.08 *** |
| 99476_at | 1453931_at | Col14a1, collagen, type XIV, alpha 1 | 2.95/(-)2.51 |
| 93013_at | 1422537_a_at | Id2, inhibitor of DNA binding 2 | 2.77/2.03 ** |
| 102293_at | 1421303_at | Ikzf1, IKAROS family zinc finger 1 | (-)2.7/(-)2.2 |
| 99413_at | 1419610_at | Ccr1, chemokine (C-C motif) receptor 1 | (-)4.13/2.62 * |
The down-regulated transcripts are shown with "(-)"
* Denotes significant changes in Itk-/- T-cells. * p ≤ 0.05 ** p ≤ 0.01 *** p ≤ 0.001
Discussion
For any analysis of individuals with defective genes there are important considerations related to the choice of accurate controls and the adequate interpretation of the data. This is nicely exemplified in Itk-deficiency. Thus, when mice with Itk-deficiency are immunized and generate impaired responses it is unclear to what extent the impairment is caused by the reduced numbers of mature T-lymphocytes as compared to the increased innate populations versus that the mature as well as the innate populations are deficient because they lack Itk. The net outcome is the sum of these alterations. The same is true in microarray experiments or when phenotypic markers are assayed by other means. In the likely event that the innate populations themselves are further altered owing to lack of Itk, the corresponding population may not even exist in the Wt. The same principle is true for any mutant gene, and it is important to be aware of this fact when interpreting data, including expression profiling, related to such defects. In this report we describe the phenotypic changes in Itk-deficiency and make comparisons to CsA-treatment. Owing to the very large number of genes with altered expression, we here provide an overview of the observed changes. We pinpoint some of the interesting findings obtained from this dataset. However, the original gene profiling data, available to any investigators at GEO, could be analyzed in different ways, depending on the biological question to be answered.
T-cells deficient for the Tec-family kinase Itk have severe impairment during T-cell activation. Furthermore, Itk has also been shown to be involved in signaling pathways that regulate the development decisions of conventional versus innate-like T-cell development [9-13], since CD8+ T-cells and a certain fraction of CD4+ T-cells have an innate-like T-cell phenotype. Collectively, these studies revealed that Itk has a crucial and important function in T-cells. In this study we performed an Affymetrix microarray expression analysis to investigate how Itk-deficiency affects the expression profile in T-cells. The effect of Itk-deficiency was investigated in CD3+ T-cells, as well as in the CD4+ and CD8+ T-cell subsets. These signatures for the first time reveal the transcriptome of Itk-deficiency.
The most pronounced changes were observed in resting Itk-deficient compared to Wt CD3+ T-cells. This is in agreement with the previous findings that more genes are expressed in untreated cells as compared to stimulated T-lymphocytes [38-40]. Thus, after anti-CD3/CD28-stimulation the number of differentially expressed transcripts was dramatically decreased in Itk-defective (down by approximately 50%) compared to Wt cells. This suggests that the CD28 co-stimulatory pathway is less dependent of Itk. It was previously shown that Itk was a negative regulator of CD28-signaling in CD4+ T-cells [41]. However, sorted naïve CD4+ T-cells from Itk-deficient mice had normal CD28 co-stimulatory responses when compared to Wt cells [42], showing that CD28-signaling is not dependent on Itk in these cells. Our result confirms that Itk is not essential for CD28-signaling and suggests that a great deal of the TCR signaling defects in Itk-/- T-cells is rescued by CD28 co-stimulation in vitro. However, expression of genes that is essential for T-cell proliferation like Il2 remain Itk-dependent after co-stimulation.
It was satisfying to observe the most pronounced transcriptional changes in CD8+ cells, since Itk-deficiency is known to predominantly affect this subpopulation [9-11]. The overlap between CD4+ and CD8+ subsets was highest in untreated cells indicating an innate-like pattern also of the CD4+ population. Moreover, a recent paper showed that CD4+ T-cells in Itk-deficient mice have a memory phenotype with expression of typical surface markers such as CD62Llow and CD44high [13]. When looking at the specific transcripts for each T-cell subset we found differences in expression of Klrs, two members in CD4+ and four in CD8+. Klrb1a (Ly55a) and Klrb1c (NK-1.1) were found in CD4+ T-cells. To our knowledge, only NK-1.1 was previously reported for Itk-deficient CD4+ T-cells [13]. Klrc2 (NKG2C) and Klrk1 (NKG2D) were previously reported in CD8+ T-cells [11]. In addition, we found Klra4 (Ly49D) and Klra19 (Ly49S), not previously described in this context. In unstimulated Itk-/- CD3+ T-cells eleven Klr members were found (shown in Table 3). Klra3, a7, a8 and b1b were shown to be common to the CD4+ and CD8+ subsets. Interestingly, we found Klra3 and Klra7 to also be calcineurin-dependent, while Klra8 was only Itk-dependent. Of note is also that a cytosolic protein known to characterize NK-cells, granzyme M, was present in the data. It has recently been shown to be expressed in NK-cells and cytotoxic T-cells with innate immune function [31]. Here, we show for the first time up-regulation of this transcript in CD8+ Itk-defective T-cells. As expected, more differentially expressed genes were revealed following separation into the CD4+ and CD8+ subsets. In a mixed population changes that affect both subsets in a similar way are preferentially detected.
Itk-deficiency partially mimicked CsA-treatment, since there was a large overlap of affected transcripts. However, we observed that CsA had a much greater effect on transcriptional regulation compared to loss of Itk, especially after co-stimulation. Approximately 4000 genes were affected by CsA following either anti-CD3- or anti-CD3/CD28-stimulation, while the corresponding numbers for those also affected by Itk was 482 and 184, respectively. 113 probe-sets were shared between Itk-defective and CsA-treated T-cells independent of stimulation. Among them we found Zbtb16, encoding the transcriptional regulator PLZF, and Xcl1, which is also called lymphotactin or ATAC, a chemokine mainly produced by activated CD8+ T-cells [43,44]. Also, Crabp2 was found in this comparison showing its calcineurin-dependency. Crabp2 is involved in regulating access of retinoic acid to its nuclear receptors, is developmentally regulated [45], and has been implicated in various forms of tumors. In addition, our analysis revealed that some of the Itk-induced changes are independent of the Ca2+/calcineurin pathway (322 and 225 transcripts after anti-CD3- and anti-CD3/CD28-stimulation, respectively). In this study we did not treat Itk-deficient cells with CsA. Such experiments could give further insights into the calcineurin-dependent regulation.
One interesting example of how different members of a gene family are differentially affected by Itk-deficiency and CsA-treatment is provided by the Granzyme family. Granzymes are serine proteases expressed in cytotoxic lymphocytes [46]. Interestingly, Granzymes A and K were both Itk- and calcineurin-dependent, while granzymes E and M were found to be Itk-dependent and calcineurin-independent after anti-CD3-stimulation. Both granzymes A and K induce caspase-independent cell death. Not much is known about granzyme E, while granzyme M is known to induce cell death in a caspase- and mitochondria-independent way [46]. Granzyme B was only affected in CsA-treated samples and has been shown to be involved in the induction of caspase-dependent apoptosis. Collectively, this demonstrates that expression of granzymes is differentially controlled.
The comparison of Itk-deficiency and CSA-treated CD3+ T-cells led also to the identification of novel NFAT target genes. The combination of a bioinformatics approach and chromatin-immunoprecipitation assays revealed that IL7R, Schlafen1, Bub1, Ctla2a and Ctla2b are novel Itk- and calcineurin-dependent genes with seemingly functional NFAT-binding sites. However, they can be regulated in different ways, e.g. Ctla2a and Ctla2b, IL7R and Slfn1 were negatively regulated, while IL-2 and Bub1 were positively regulated by CN-dependent pathways. The same regulation pattern was observed in unstimulated Itk-deficient samples, but after anti-CD3-stimulation IL7R and Slfn1 became positively regulated by Itk (Fig. 5c). Members of the Schlafen (Slfn) protein family have been implicated in the regulation of cell growth and T-cell development. Furthermore, similar to the Il2 gene [47], AP-1 and NF-κB are reported to regulate Slfn2 expression [48]. Bub1 (budding uninhibited by benomyl) is a serine/threonine kinase that has a function in the mitotic spindle checkpoint and is mutated in certain types of human cancer [49]. Ctla2a and Ctla2b are cysteine proteinase inhibitors expressed in activated T-cells and mast cells [50]. Both naïve and memory T-cells have high levels of IL7R, and IL7 is required for their homeostasis [51]. Furthermore, recently it was shown that Wt and Itk-deficient CD4+ T-cells express similar levels of IL7R (CD127) [13]. Certain genes in the Itk/CN group did not have bona fide NFAT-sites as determined by our bioinformatic approach. This could be due to that the current data base algorithms are not good enough to predict the putative sites or that the chromosomal stretches harboring NFAT-sites are located outside the 500 bp region that we choose to study. Future studies will aim to reveal a possible link between the altered expression of these genes and the phenotype of Itk-deficiency.
Finally, a comparative analysis of Itk-deficient T-cells and Btk-deficient B-cells revealed a significant overlap of transcripts, indicating that there is a common Tec family gene expression profile in lymphocytes. The fact that 16/18 genes had a similar fold-induction in T- and B-cells (p < 0.05) suggests common regulation of these genes by Itk and Btk. Conversely, the observation that two transcripts (Col14a1 and Ccr1) were differentially expressed may simply reflect that B- and T-cells represent different lineages, each characterized by unique features of their transcriptomes. Of the six most up-regulated genes in Btk-deficiency [24] all of them were >2-fold changed in cells lacking Itk, eight of which were also significantly altered in Itk-deficient T-cells (with p-values ranging from <0.05 to <3 × 10-5). Tgfbi, which was up-regulated in Btk-/- and confirmed as highly increased in Itk-/- (p < 3 × 10-5) samples, encodes an extracellular protein that mediates cell adhesion to collagen, laminin and fibronectin via its interaction with integrins [52]. Since these 16 genes are common to both Btk- and Itk-dependent transcriptomes it seems likely that the corresponding promoters could be activated through signaling components controlled by either pathway. Future identification of regulatory elements targeted by common factors could reveal the underlying mechanism.
Conclusion
This report is the first to define the transcriptional signature of cells from Itk-/- mice. The transcriptome of Itk-deficient cells revealed that there is a large overlap with regular CD4+ and CD8+ cells. Future studies analyzing different stages of innate, memory-like cells from Wt mice will aid in unraveling to what extent the innate population of Itk-deficiency also shows unique features, which differ from normal mice.
Authors' contributions
Contribution: KEMB. designed and performed the majority of the research, analyzed the data and wrote the paper; NB. did animal experiments and cell stimulations; JML. designed and helped analyzing the microarray data, and helped with writing; LY. performed the ChIP experiments; JR. helped with the animal experiments and cell stimulations; AB. helped with the animal experiments, cell stimulations and writing; WE. was involved in the planning and execution of the project, and helped with writing; CIES. conceived project, provided supervision throughout, interpreted data, and helped with writing.
Supplementary Material
Antibodies used in the isolation of CD3+ T-cells. Antibodies used in the isolation of CD3+ T-cells.
Primer sequences and PCR conditions used in the chromatin-immunoprecipitation assay. PCR conditions and primer sequences for the genes IL2, IL7R, Schlafen1, Bub1, Ctla2a and Ctla2b used in the chromatin-immunoprecipitation experiment. F, forward primer; R, reverse primer and T (a), annealing temperature.
106 immune response-related genes. The list of 900 probe-sets that were differentially expressed between unstimulated Itk-defective and Wt T-cells was used to manually annotate 106 immune response-related genes. The genes were further divided into 14 different subgroups. The down-regulated transcripts are shown with "-".
252 differentially expressed probe-sets in Itk-deficiency compared to Wt after anti-CD3- and anti-CD3/CD28-stimulations of CD3+ T-cells. All the comparisons were made against Wt and with the criterion ≥ 2-fold. Only 5/252 probe-sets showed opposite fold-changes in anti-CD3- and anti-CD3/CD28-stimulations for 24 h. These genes are marked with *. The down-regulated transcripts are shown with "-".
3713 probe-sets overlapping upon anti-CD3- and anti-CD3/CD28-stimulations in Wt CD3+ T-cells. 3713 differentially expressed probe-sets overlapped in Wt CD3+ T-cells after both anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were made against unstimulated Wt (Wt C) and with the criterion ≥ 2-fold. The paired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
2393 probe-sets overlapping upon anti-CD3- and anti-CD3/CD28-stimulations in Itk-defective CD3+ T-cells. 2393 differentially expressed probe-sets overlapped in Itk-/- CD3+ T-cells after both anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were made against unstimulated Itk-/- (Itk-/- C) and with the criterion ≥ 2-fold. The paired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
Eomesodermin and T-bet signal intensities. Bar charts showing the mean signal intensity (MSI) levels of Eomesodermin and T-bet in CD4+ and CD8+ T-cell populations (a and b) and in CD3+ T-cells (c and d) from Wt and Itk-deficient mice. Eomesodermin is represented by two probe-sets (grey and white bars). The two T-cell subsets are either unstimulated or anti-CD3-stimulated, while the CD3+ T-cells are either unstimulated, anti-CD3- or anti-CD3/CD28-stimulated.
324 differentially expressed probe-sets represent the core group of unstimulated Itk-defective CD4+ T-cells. The core group of probe-sets differentially expressed in unstimulated Itk-/- CD4+ T-cells (Itk-/- CD4+ C) compared to unstimulated Wt CD4+ T-cells (Wt CD4+ C). The comparisons were made with the criterion ≥ 2-fold. The unpaired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
4467 differentially expressed probe-sets represent the core group of unstimulated Itk-defective CD8+ T-cells. The core group of probe-sets differentially expressed in unstimulated Itk-/- CD8+ T-cells (Itk-/- CD8+ C) compared to unstimulated Wt CD8+ T-cells (Wt CD8+ C). The comparisons were made with the criterion ≥ 2-fold. The down-regulated transcripts are shown with "-".
113 probe-sets being both Itk- and calcineurin-dependent upon anti-CD3- and anti-CD3/CD28-stimulations in CD3+ T-cells. 113 probe-sets are Itk- and calcineurin-dependent upon anti-CD3- and anti-CD3/CD28-stimulations for 24 h. All the cells in the comparisons are CD3+ T-cells. The comparisons were done using the criterion ≥ 2-fold and the p-values were calculated using unpaired (for Itk-/- vs Wt) and paired (for Wt CsA vs Wt) Student t-tests. Cyclosporin A-treated samples are named CsA in the table. The down-regulated transcripts are shown with "-".
95 probe-sets being Itk-dependent and calcineurin-independent after anti-CD3- and anti-CD3/CD28-stimulations in CD3+ T-cells. 95 probe-sets, corresponding to 89 transcripts, are Itk-dependent but calcineurin-independent upon anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were done using the criterion ≥ 2-fold and the p-values were calculated using unpaired Student t-test. The down-regulated transcripts are shown with "-".
24 genes found in the bioinformatic searching for putative NFAT-sites. The 24 genes were selected as highly regulated from the list comparing Itk-/- anti-CD3- and Wt CsA anti-CD3-stimulated versus Wt anti-CD3-stimulated samples. 15/24 genes had NFAT-sites and six of them were verified to bind NFATc1. The genes that have NFAT-sites are grey-shaded and the genes verified to bind NFATc1 by chromatin-immunoprecipitation (ChIP) are in red. The genes marked with * are also found in the 113-list (Additional file 10) where genes are Itk- and calcineurin-dependent after both anti-CD3- and anti-CD3/CD28-stimulation. The down-regulated transcripts are shown with "-".
Acknowledgments
Acknowledgements
We would like to thank the Affymetrix core facility at Novum, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at the Karolinska Institutet and the research committee at the Karolinska hospital. This work was supported by the Swedish Science Council, the Swedish Cancer Fund, the European Council FP7 grant EURO-PADnet, the Wallenberg foundation, the Stockholm County Council (research grant ALF-projektmedel medicin), the Special Research Area SFB-F23 (project SFB-F2305) of the Austrian Science Fund (FWF), and by the START program (grant Y-163) of the FWF and the Austrian Ministry of Science and Research (BM:WF).
Contributor Information
K Emelie M Blomberg, Email: emelie.blomberg@ki.se.
Nicole Boucheron, Email: nicole.boucheron@meduniwien.ac.at.
Jessica M Lindvall, Email: jessli@ifi.uio.no.
Liang Yu, Email: liang.yu@ki.se.
Julia Raberger, Email: julia.raberger@meduniwien.ac.at.
Anna Berglöf, Email: anna.berglof@ki.se.
Wilfried Ellmeier, Email: Wilfried.Ellmeier@meduniwien.ac.at.
CI Edvard Smith, Email: edvard.smith@ki.se.
References
- Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- Smith CI, Islam TC, Mattsson PT, Mohamed AJ, Nore BF, Vihinen M. The Tec family of cytoplasmic tyrosine kinases: mammalian Btk, Bmx, Itk, Tec, Txk and homologs in other species. Bioessays. 2001;23:436–446. doi: 10.1002/bies.1062. [DOI] [PubMed] [Google Scholar]
- Liao XC, Littman DR. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Schaeffer EM, Broussard C, Debnath J, Anderson S, McVicar DW, Schwartzberg PL. Tec family kinases modulate thresholds for thymocyte development and selection. J Exp Med. 2000;192:987–1000. doi: 10.1084/jem.192.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowell DJ, Shinkai K, Liao XC, Beebe AM, Coffman RL, Littman DR, Locksley RM. Impaired NFATc translocation and failure of Th2 development in Itk-deficient CD4+ T cells. Immunity. 1999;11:399–409. doi: 10.1016/S1074-7613(00)80115-6. [DOI] [PubMed] [Google Scholar]
- Dombroski D, Houghtling RA, Labno CM, Precht P, Takesono A, Caplen NJ, Billadeau DD, Wange RL, Burkhardt JK, Schwartzberg PL. Kinase-independent functions for Itk in TCR-induced regulation of Vav and the actin cytoskeleton. J Immunol. 2005;174:1385–1392. doi: 10.4049/jimmunol.174.3.1385. [DOI] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Readinger JA, Viorritto IC, Mueller KL, Houghtling RA, Schwartzberg PL. Tec kinases, actin, and cell adhesion. Immunol Rev. 2007;218:45–64. doi: 10.1111/j.1600-065X.2007.00534.x. [DOI] [PubMed] [Google Scholar]
- Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Dubois S, Waldmann TA, Muller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci USA. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, August A. Naive and innate memory phenotype CD4+ T cells have different requirements for active Itk for their development. J Immunol. 2008;180:6544–6552. doi: 10.4049/jimmunol.180.10.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–79. doi: 10.1016/S0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Lucas JA, Miller AT, Atherly LO, Berg LJ. The role of Tec family kinases in T cell development and function. Immunol Rev. 2003;191:119–138. doi: 10.1034/j.1600-065X.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Lindvall JM, Blomberg KE, Berglof A, Smith CI. Distinct gene expression signature in Btk-defective T1 B-cells. Biochem Biophys Res Commun. 2006;346:461–469. doi: 10.1016/j.bbrc.2006.05.146. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/
- dChip Software http://biosun1.harvard.edu/complab/dchip/
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl http://www.ensembl.org/
- Lindvall JM, Blomberg KE, Berglof A, Yang Q, Smith CI, Islam TC. Gene expression profile of B cells from Xid mice and Btk knockout mice. Eur J Immunol. 2004;34:1981–1991. doi: 10.1002/eji.200324051. [DOI] [PubMed] [Google Scholar]
- Yu L, Mohamed AJ, Simonson OE, Vargas L, Blomberg KE, Bjorkstrand B, Arteaga HJ, Nore BF, Smith CI. Proteasome-dependent autoregulation of Bruton tyrosine kinase (Btk) promoter via NF-kappaB. Blood. 2008;111:4617–4626. doi: 10.1182/blood-2007-10-121137. [DOI] [PubMed] [Google Scholar]
- Parker H, An Q, Barber K, Case M, Davies T, Konn Z, Stewart A, Wright S, Griffiths M, Ross FM, et al. The complex genomic profile of ETV6-RUNX1 positive acute lymphoblastic leukemia highlights a recurrent deletion of TBL1XR1. Genes Chromosomes Cancer. 2008;47:1118–1125. doi: 10.1002/gcc.20613. [DOI] [PubMed] [Google Scholar]
- Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics. 2008;9:225. doi: 10.1186/1471-2164-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol. 2008;121:1484–1490. doi: 10.1016/j.jaci.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, August A. Attenuation of immunological symptoms of allergic asthma in mice lacking the tyrosine kinase ITK. J Immunol. 2003;170:5056–5063. doi: 10.4049/jimmunol.170.10.5056. [DOI] [PubMed] [Google Scholar]
- Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci USA. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morice WG, Jevremovic D, Hanson CA. The expression of the novel cytotoxic protein granzyme M by large granular lymphocytic leukaemias of both T-cell and NK-cell lineage: an unexpected finding with implications regarding the pathobiology of these disorders. Br J Haematol. 2007;137:237–239. doi: 10.1111/j.1365-2141.2007.06564.x. [DOI] [PubMed] [Google Scholar]
- Mohamed AJ, Yu L, Backesjo CM, Vargas L, Faryal R, Aints A, Christensson B, Berglof A, Vihinen M, Nore BF, et al. Bruton's tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228:58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Yang WC, Ghiotto M, Barbarat B, Olive D. The role of Tec protein-tyrosine kinase in T cell signaling. J Biol Chem. 1999;274:607–617. doi: 10.1074/jbc.274.2.607. [DOI] [PubMed] [Google Scholar]
- Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney JW, Sun YL, Glimcher LH, Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Sideras P, Smith CI, Vorechovsky I, Chapman V, Paul WE. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science. 1993;261:355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- Lindvall JM, Blomberg KE, Valiaho J, Vargas L, Heinonen JE, Berglof A, Mohamed AJ, Nore BF, Vihinen M, Smith CI. Bruton's tyrosine kinase: cell biology, sequence conservation, mutation spectrum, siRNA modifications, and expression profiling. Immunol Rev. 2005;203:200–215. doi: 10.1111/j.0105-2896.2005.00225.x. [DOI] [PubMed] [Google Scholar]
- Chtanova T, Newton R, Liu SM, Weininger L, Young TR, Silva DG, Bertoni F, Rinaldi A, Chappaz S, Sallusto F, et al. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- Teague TK, Hildeman D, Kedl RM, Mitchell T, Rees W, Schaefer BC, Bender J, Kappler J, Marrack P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc Natl Acad Sci USA. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D, Crabtree GR, Brown PO. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci USA. 2002;99:11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XC, Fournier S, Killeen N, Weiss A, Allison JP, Littman DR. Itk negatively regulates induction of T cell proliferation by CD28 costimulation. J Exp Med. 1997;186:221–228. doi: 10.1084/jem.186.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Berg LJ. Itk is not essential for CD28 signaling in naive T cells. J Immunol. 2005;174:4475–4479. doi: 10.4049/jimmunol.174.8.4475. [DOI] [PubMed] [Google Scholar]
- Dorner B, Muller S, Entschladen F, Schroder JM, Franke P, Kraft R, Friedl P, Clark-Lewis I, Kroczek RA. Purification, structural analysis, and function of natural ATAC, a cytokine secreted by CD8(+) T cells. J Biol Chem. 1997;272:8817–8823. doi: 10.1074/jbc.272.13.8817. [DOI] [PubMed] [Google Scholar]
- Ordway D, Higgins DM, Sanchez-Campillo J, Spencer JS, Henao-Tamayo M, Harton M, Orme IM, Gonzalez Juarrero M. XCL1 (lymphotactin) chemokine produced by activated CD8 T cells during the chronic stage of infection with Mycobacterium tuberculosis negatively affects production of IFN-gamma by CD4 T cells and participates in granuloma stability. J Leukoc Biol. 2007;82:1221–1229. doi: 10.1189/jlb.0607426. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Saxena V, Liu RZ, Thisse C, Thisse B, Denovan-Wright EM, Wright JM. Differential expression of the duplicated cellular retinoic acid-binding protein 2 genes (crabp2a and crabp2b) during zebrafish embryonic development. Gene Expr Patterns. 2005;5:371–379. doi: 10.1016/j.modgep.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Bots M, Medema JP. Granzymes at a glance. J Cell Sci. 2006;119:5011–5014. doi: 10.1242/jcs.03239. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Ward SB. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn WJ, Kim D, Lee KW, Kim MS, Kwon S, Lee Y, Kim DS, Kwon HJ. Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol. 2007;44:3273–3282. doi: 10.1016/j.molimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Jaffrey RG, Pritchard SC, Clark C, Murray GI, Cassidy J, Kerr KM, Nicolson MC, McLeod HL. Genomic instability at the BUB1 locus in colorectal cancer, but not in non-small cell lung cancer. Cancer Res. 2000;60:4349–4352. [PubMed] [Google Scholar]
- Denizot F, Brunet JF, Roustan P, Harper K, Suzan M, Luciani MF, Mattei MG, Golstein P. Novel structures CTLA-2 alpha and CTLA-2 beta expressed in mouse activated T cells and mast cells and homologous to cysteine proteinase proregions. Eur J Immunol. 1989;19:631–635. doi: 10.1002/eji.1830190409. [DOI] [PubMed] [Google Scholar]
- Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- Irigoyen M, Anso E, Salvo E, de las Herrerias JD, Martinez-Irujo JJ, Rouzaut A. TGFbeta-induced protein mediates lymphatic endothelial cell adhesion to the extracellular matrix under low oxygen conditions. Cell Mol Life Sci. 2008;65:2244–2255. doi: 10.1007/s00018-008-8071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibodies used in the isolation of CD3+ T-cells. Antibodies used in the isolation of CD3+ T-cells.
Primer sequences and PCR conditions used in the chromatin-immunoprecipitation assay. PCR conditions and primer sequences for the genes IL2, IL7R, Schlafen1, Bub1, Ctla2a and Ctla2b used in the chromatin-immunoprecipitation experiment. F, forward primer; R, reverse primer and T (a), annealing temperature.
106 immune response-related genes. The list of 900 probe-sets that were differentially expressed between unstimulated Itk-defective and Wt T-cells was used to manually annotate 106 immune response-related genes. The genes were further divided into 14 different subgroups. The down-regulated transcripts are shown with "-".
252 differentially expressed probe-sets in Itk-deficiency compared to Wt after anti-CD3- and anti-CD3/CD28-stimulations of CD3+ T-cells. All the comparisons were made against Wt and with the criterion ≥ 2-fold. Only 5/252 probe-sets showed opposite fold-changes in anti-CD3- and anti-CD3/CD28-stimulations for 24 h. These genes are marked with *. The down-regulated transcripts are shown with "-".
3713 probe-sets overlapping upon anti-CD3- and anti-CD3/CD28-stimulations in Wt CD3+ T-cells. 3713 differentially expressed probe-sets overlapped in Wt CD3+ T-cells after both anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were made against unstimulated Wt (Wt C) and with the criterion ≥ 2-fold. The paired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
2393 probe-sets overlapping upon anti-CD3- and anti-CD3/CD28-stimulations in Itk-defective CD3+ T-cells. 2393 differentially expressed probe-sets overlapped in Itk-/- CD3+ T-cells after both anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were made against unstimulated Itk-/- (Itk-/- C) and with the criterion ≥ 2-fold. The paired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
Eomesodermin and T-bet signal intensities. Bar charts showing the mean signal intensity (MSI) levels of Eomesodermin and T-bet in CD4+ and CD8+ T-cell populations (a and b) and in CD3+ T-cells (c and d) from Wt and Itk-deficient mice. Eomesodermin is represented by two probe-sets (grey and white bars). The two T-cell subsets are either unstimulated or anti-CD3-stimulated, while the CD3+ T-cells are either unstimulated, anti-CD3- or anti-CD3/CD28-stimulated.
324 differentially expressed probe-sets represent the core group of unstimulated Itk-defective CD4+ T-cells. The core group of probe-sets differentially expressed in unstimulated Itk-/- CD4+ T-cells (Itk-/- CD4+ C) compared to unstimulated Wt CD4+ T-cells (Wt CD4+ C). The comparisons were made with the criterion ≥ 2-fold. The unpaired Student t-test was used to calculate the p-values. The down-regulated transcripts are shown with "-".
4467 differentially expressed probe-sets represent the core group of unstimulated Itk-defective CD8+ T-cells. The core group of probe-sets differentially expressed in unstimulated Itk-/- CD8+ T-cells (Itk-/- CD8+ C) compared to unstimulated Wt CD8+ T-cells (Wt CD8+ C). The comparisons were made with the criterion ≥ 2-fold. The down-regulated transcripts are shown with "-".
113 probe-sets being both Itk- and calcineurin-dependent upon anti-CD3- and anti-CD3/CD28-stimulations in CD3+ T-cells. 113 probe-sets are Itk- and calcineurin-dependent upon anti-CD3- and anti-CD3/CD28-stimulations for 24 h. All the cells in the comparisons are CD3+ T-cells. The comparisons were done using the criterion ≥ 2-fold and the p-values were calculated using unpaired (for Itk-/- vs Wt) and paired (for Wt CsA vs Wt) Student t-tests. Cyclosporin A-treated samples are named CsA in the table. The down-regulated transcripts are shown with "-".
95 probe-sets being Itk-dependent and calcineurin-independent after anti-CD3- and anti-CD3/CD28-stimulations in CD3+ T-cells. 95 probe-sets, corresponding to 89 transcripts, are Itk-dependent but calcineurin-independent upon anti-CD3- and anti-CD3/CD28-stimulations for 24 h. The comparisons were done using the criterion ≥ 2-fold and the p-values were calculated using unpaired Student t-test. The down-regulated transcripts are shown with "-".
24 genes found in the bioinformatic searching for putative NFAT-sites. The 24 genes were selected as highly regulated from the list comparing Itk-/- anti-CD3- and Wt CsA anti-CD3-stimulated versus Wt anti-CD3-stimulated samples. 15/24 genes had NFAT-sites and six of them were verified to bind NFATc1. The genes that have NFAT-sites are grey-shaded and the genes verified to bind NFATc1 by chromatin-immunoprecipitation (ChIP) are in red. The genes marked with * are also found in the 113-list (Additional file 10) where genes are Itk- and calcineurin-dependent after both anti-CD3- and anti-CD3/CD28-stimulation. The down-regulated transcripts are shown with "-".