Abstract
Detection of internal body time (BT) via a few-time-point assay has been a longstanding challenge in medicine, because BT information can be exploited to maximize potency and minimize toxicity during drug administration and thus will enable highly optimized medication. To address this challenge, we previously developed the concept, “molecular-timetable method,” which was originally inspired by Linné's flower clock. In Linné's flower clock, one can estimate the time of the day by watching the opening and closing pattern of various flowers. Similarly, in the molecular-timetable method, one can measure the BT of the day by profiling the up and down patterns of substances in the molecular timetable. To make this method clinically feasible, we now performed blood metabolome analysis and here report the successful quantification of hundreds of clock-controlled metabolites in mouse plasma. Based on circadian blood metabolomics, we can detect individual BT under various conditions, demonstrating its robustness against genetic background, sex, age, and feeding differences. The power of this method is also demonstrated by the sensitive and accurate detection of circadian rhythm disorder in jet-lagged mice. These results suggest the potential for metabolomics-based detection of BT (“metabolite-timetable method”), which will lead to the realization of chronotherapy and personalized medicine.
Keywords: chronotherapy, circadian, metabolome, jet-lag, LC-MS
In the 18th century, the Swedish botanist Karl von Linné designed a “flower clock” comprising a series of plant species arranged according to the respective time of the day their flowers open or close. Watching this flower clock, one can estimate the time of the day by noting the pattern of flower opening and closing. Since Linné's early times, it has been a well known fact that plants have an internal clock and thereby can open or close their flowers at a precise time of the day. Similarly, animals possess an internal molecular mechanism, a “circadian clock,” which underlies endogenous, self-sustained oscillations with a period of ≈24 h manifest in diverse physiological and metabolic processes (1). In mammals, several clock genes, including Clock, Bmal1, Per1, Per2, Cry1, Cry2, RevErbA, Rora, Csnk1e, Csnk1d, and Fbxl3, regulate, at least in part, gene expression in central and/or peripheral clock tissues (2–4). Reflecting the temporal changes in gene expression in central and peripheral clock tissues (5–8), the potency and/or toxicity of administered drugs depend on the individual's present body time (BT) (9–13). It has been suggested that administrating a drug at a specific BT improves the outcome of pharmacotherapy by maximizing its potency and minimizing its toxicity (14). In contrast, administrating a drug at an inappropriate BT can result in severe side effects (15). Despite the importance of such BT-dependent therapy (also known as “chronotherapy”) (9–13), its application to clinical practice has been obstructed by a lack of clinically feasible methods for measuring BT.
To overcome this problem, we previously developed the concept of a “molecular-timetable method (16),” which was originally inspired by Linné's flower clock. In Linné's flower clock, one can estimate the time of the day by watching the opening and closing pattern of various flowers. Similarly, in molecular-timetable method, one can measure the BT of the day by profiling the up and down pattern of substances in the molecular timetable. This concept was proven using the expression profile of clock-controlled genes in a target organ (16). However, estimates of BT from the expression profile of oscillating substances within a target organ (in this case, the liver) are hard to apply directly to clinical situations. To make the molecular-timetable method more clinically relevant, we decided to determine BT from blood samples, which are more available in clinical practice.
In the blood of mammals, several small chemical substances such as metabolites and hormones have been reported to exhibit circadian oscillations. For example, the concentration of the steroid hormone, corticosterone, is rhythmically controlled by circadian clock with a peak in the evening (17), and an amine-derived hormone, melatonin, show circadian rhythm with a peak in the early morning in mice (18). In humans, several peptide hormone levels show daily variations; growth hormone increases during sleep (19), leptin increases during the evening (20), and prolactin increases at night (21). Concentrations of amino acids, including tryptophan, tyrosine, phenylalanine (22), methionine (23), cysteine, glutathione (24), and homocysteine (25), also exhibit daily variations in human blood plasma. Despite these findings, comprehensive profiling of circadian dynamics of chemical substances in mammalian blood has not yet been reported, and until now a comprehensive molecular timetable of such chemical substances has not been constructed.
Metabolomics technology aims to comprehensively identify and/or quantify the dynamic chemical substances present in biological samples. It is gaining interest in the fields of drug discovery, disease diagnostics, and treatment (26–28). The present metabolomics technology was developed rapidly by coupling advanced separation technology with highly sensitive and selective mass spectrometry–gas chromatography mass spectrometry (GC/MS) (29–31), liquid chromatography mass spectrometry (LC-MS) (32–34), and capillary electrophoresis mass spectrometry (CE-MS) (35, 36). To construct the molecular timetable from clinically available samples, we have performed blood metabolome analysis in this study. Using the LC-MS technique, we quantified hundreds of clock-controlled metabolites in mouse plasma and successfully constructed a molecular timetable of blood metabolites. This metabolite timetable allowed us to measure individual BT under various conditions and was robust enough to be used in mice with different genetic backgrounds, sex, age, and feeding conditions. It was also sensitive and accurate enough to detect circadian rhythm disorders in jet-lagged mice. Our preliminary results suggest that other metabolomics techniques such as CE-MS can also be applied to the molecular-timetable method, demonstrated by the quantification of hundreds of clock-controlled metabolites, the identification of substantial portion of these metabolites, and successful measurement of BT from independent blood samples. Thus, metabolomics-based measurement of BT will contribute to the potential areas of chronotherapy and personalized medication regimens.
Results
Construction of the Metabolite Timetable from Blood Plasma by LC-MS.
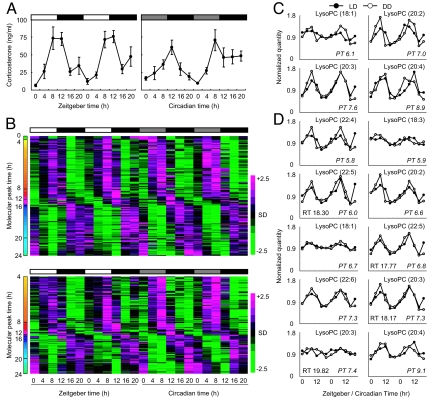
Samples of blood plasma were taken from young male CBA/N mice every 4 hours over 2 days during light–dark (LD) or constant darkness (DD) conditions. Plasma corticosterone was used as a quality control because it exhibits a clear circadian oscillation when quantified by radio immunoassay (Fig. 1A). Small chemical substances in the plasma were quantified by LC-MS analysis to construct the metabolite timetable. LC-MS analysis detected 695 negative ion and 938 positive ion peaks in the plasma. Of these, 176 negative and 142 positive ion peaks exhibited significant circadian oscillations in LD and DD conditions [Fig. 1B; false discovery rate (FDR) <0.01; see also Materials and Methods]; these peaks accounted for the ≈19.5% of the peaks detected in mouse plasma. These substances served as “time-indicating metabolites,” because they oscillate considerably even under constant environmental conditions (DD). For instance, at zeitgeber time 0 (ZT0; the beginning of day) or circadian time 0 (CT0; the beginning of a subjective day), concentrations of dawn-indicating metabolites, which peak at approximately ZT0 or CT0 (Fig. 1B, green color bars in the molecular peak time), are high, whereas those of dusk-indicating metabolites, which peak at approximately ZT12 or CT12 (Fig. 1B, red color bars in the molecular peak time), are low. Conversely, at ZT12 or CT12, concentrations of dawn-indicating metabolites are low, whereas those of dusk-indicating metabolites are high; this suggests that time-indicating metabolites can represent BT (BT), the endogenous state of circadian clock. In fact, the oscillations of these time-indicating metabolites are directly or indirectly controlled by circadian clock, because the disruption of functional molecular clock in Cry1−/−, Cry2−/− mice (37) results in the alteration of circadian oscillations of these metabolites (Fig. S1). We used these LC-MS data to construct the molecular timetable of time-indicating metabolites (a “metabolite timetable”) in mouse plasma (Table S1 online). We also note that, among these time-indicating metabolites (i.e., oscillatory peaks detected by LC-MS), 14 oscillatory peaks were identified as various types of lysophosphatidylcholines with different unsaturated fatty acids (Fig. 1 C and D).
Fig. 1.
Circadian patterns of metabolites in mouse plasma. (A) Circadian changes in corticosterone levels in the plasma of CBA/N mice under LD (Left) and DD, Center) conditions. All values are mean ± SEM. The white bars above the graph indicate day, gray bars indicate subjective day, and black bars indicate night/subjective night. ZT0 is the time of light on, and CT0 is the time the light used to be turned on. (B) Circadian oscillatory metabolites in the plasma of CBA/N mice [negative ions (up); positive ions (down)]. On the heat maps, magenta tiles indicate a high quantity of substances and green tiles indicate a low quantity in plasma. Metabolites are sorted according to their molecular peak time (molecular peak times are indicated as colors). (C and D) Identified oscillatory peaks measured by negative ion mode (C) and positive ion mode (D). Mean value was set to 1.0.
Measurement of BT from Independent Samples.
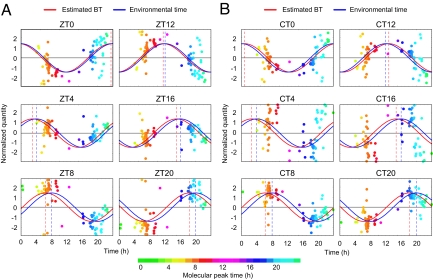
To verify whether the metabolite timetable was a good indicator of BT, we attempted to estimate the BT from the metabolite profiles of independently sampled mice. We collected fresh blood plasma from individual young male CBA/N mice every 4 h over 24 h under both LD or DD conditions because of the possibility that sampling time and/or light conditions would affect the accuracy of BT estimation. LC-MS analysis was performed to profile the time-indicating metabolites in the plasma samples (Fig. 2 A and B). After measured profile of the time-indicating metabolites was normalized by using the metabolite timetable, we filtered out outliers, fitted the normalized profile to cosine curve, and calculated the significance of its fitness (see also Materials and Methods). This metabolite-timetable method could successfully detect significant circadian rhythmicity in all metabolite profiles of these samples (P < 0.01, Fig. 2 A and B). The estimated BT closely matched with the environmental time when sampled (ZT under LD condition or CT under LD condition) with estimation errors of 1.0 ± 0.49 h for LD and 1.3 ± 0.45 h for DD (mean ± SD, Table S2). Estimation error was here defined as time difference between estimated BT and sampling time (environmental time). These results suggest that BT can be accurately determined from the metabolite profiles of independently sampled mice.
Fig. 2.
BT estimation. BT measurements of mice kept under LD (A) and DD (B) conditions. Colors of the dots indicate the molecular peak times of each substance (Table S1). Peak time of the red cosine curves indicates estimated BT and peak time of the blue indicates the time the sample was taken (“environmental time”). The greater the degree of overlap of the red and blue curves, the greater the accuracy of the measurement. The dashed vertical lines show the BT (red) or environmental time (ZT/CT, blue). See Table S2 for statistics.
Differences in Genetic Backgrounds.
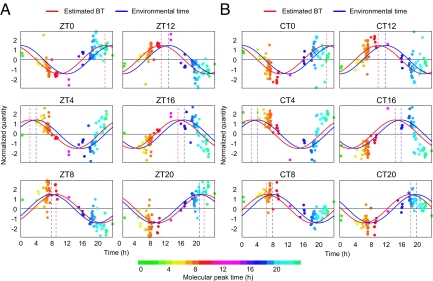
In clinical situations, methods for BT detection must apply to populations with heterogeneous genetic backgrounds. To demonstrate the suitability of the metabolite-timetable method for individuals with different genetic backgrounds, we applied the method to other inbred mouse strain with genetic backgrounds that differed from the original CBA/N strain. We chose C57BL/6, because C57BL/6 and CBA/N are genetically remote from each other and classified into 2 completely different clusters among 55 mice strains according to SNP-based study (38). We collected the blood plasma samples from individual young male C57BL/6 mice every 4 h over 24 h under LD and DD conditions and quantified the time-indicating metabolites in the plasma by LC-MS (Fig. 3 A and B). The metabolite-timetable method detected significant circadian rhythmicity (P < 0.01) in all metabolite profiles both under LD (Fig. 3A) and DD conditions (Fig. 3B) even if we used the metabolite timetable constructed from CBA/N mice. The estimated BT closely matched with the environmental time with the estimation errors of 1.6 ± 0.36 h for LD and 1.7 ± 0.24 h for DD (mean ± SD, Table S2). These results suggest that BT can be accurately determined from the metabolite profiles of mice with heterogeneous genetic backgrounds.
Fig. 3.
Genetic background. BT measurement using C57BL/6 mice plasma collected under LD (A) and DD (B) conditions. Colors of the dots indicate the molecular peak times of each substance (Table S1). Peak time of the red cosine curves indicate estimated BT and peak time of the blue indicate the environmental time. The dashed vertical lines show the BT (red) or environmental time (ZT/CT, blue). See Table S2 for statistics.
Differences in Age and Sex.
We constructed the metabolite timetable from young male mice only, so it was possible that age and sex factors might affect the accuracy of the metabolite-timetable method. To determine the influence of age and sex, we also applied the metabolite-timetable method to aged male and young female mice of the same strain. Blood plasma from individual aged male or young female CBA/N mice was sampled at 2 time points, ZT0 (the beginning of the day, i.e., time of light on) and ZT12 (the end of the day, i.e., time of the light off) under LD condition. These time points were considered as 2 “noisiest” time points, because light conditions were dramatically changed at these points. Time-indicating metabolites in the plasma were quantified by LC-MS (Fig. 4A) and significant circadian rhythmicity (P < 0.01) was detected in all metabolite profiles of both the aged male mice and the young female CBA/N mice (Fig. 4A). The estimated BT from individual mice sampled at ZT0 and ZT12 were BT23.0 and BT11.0 in aged male mice and BT1.2 and BT13.2 in young female mice (Table S2). These results demonstrate that BT can be accurately determined from the metabolite profiles of mice of different age and sex.
Fig. 4.
Age, sex differences and feeding condition. (A) BT measurement of young male (Top), aged male (Middle), and young female mice plasma (Bottom) harvested at ZT0 (Left) and ZT12 (Right). (B) BT measurement of young male mice kept under food-deprivation conditions. Colors of the dots indicate the molecular peak times of each substance (Table S1). Peak time of the red cosine curves indicates estimated BT, and peak time of the blue indicates the environmental time. The dashed vertical lines show the BT (red) or environmental time (ZT, blue). Results for young male mice (A) are replotted from Fig. 2A for comparison. See also Table S2 for statistics.
Differences in Feeding Conditions.
The circadian rhythmicity of food intake is well known (39); therefore, feeding conditions may severely affect the accuracy of the metabolite-timetable method. To validate the use of the metabolite timetable in individuals with different feeding conditions, we applied the metabolite-timetable method to CBA/N mice deprived of food (food deprivation). This feeding condition differed greatly from the original feeding condition where CBA/N mice were allowed ad lib feeding. We collected the blood plasma from individual young, male, food-deprived CBA/N mice every 4 hours over 24 h under LD condition. LC-MS analysis was performed to quantify the plasma metabolites (Fig. 4B). The metabolite-timetable method detected significant circadian rhythmicity (P < 0.03) in all metabolite profiles. The estimated BT matched with the environmental time with the estimation errors of 2.2 ± 0.50 h (mean ± SD, Table S2). These results suggest that BT can be determined from the metabolite profiles of mice even under severe feeding conditions.
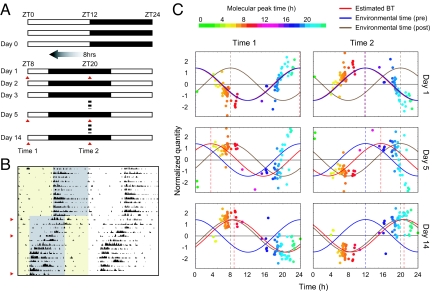
Detection of Jet Lag.
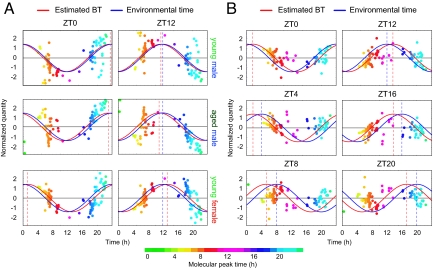
The final stage of the study was to evaluate the use of the metabolite-timetable in the diagnosis of circadian rhythm disorders. Jet lag is a common disorder of circadian rhythm, in which there is a difference between the internal BT and environmental time. To mimic jet lag, we kept the mice for 2 weeks under normal LD conditions and then rapidly advanced the lighting schedule by 8 h (40). Plasma samples were analyzed at 2 time points (ZT0 and ZT12 of the original LD cycle, termed as “Time 1” and “Time 2”) on 3 separate days: on day 1 (before entrainment to the new cycle), day 5 (during entrainment), and day 14 (after entrainment) (Fig. 5 A and B). On day 1, the estimated BTs were 23.8 h (Time 1) and 11.8 h (Time 2), suggesting that the internal BTs still follow the original LD cycle. By day 14, estimated BTs were 8.8 h (Time 1) and 20.8 h (Time 2), suggesting that the internal BTs had shifted by ≈8 h from the original LD cycle and had therefore become entrained completely to the advanced cycle. Notably, on day 5, estimated BTs were 3.5 h (Time 1) and 15.5 h (Time 2), a shift of 3.5 h from the original LD cycle, indicating incomplete entrainment to the advanced cycle, i.e., jet lag (Fig. 5C and Table S2). These results suggest that the metabolite-timetable method can accurately detect circadian rhythm disorders. Another set of BT estimation data for jet-lagged mice supports this conclusion (Fig. S2 and Table S2).
Fig. 5.
Detection of jet lag. (A) Schematic view of lighting conditions. White bars indicate light on, and black bars indicate light off. On day 1, the light was turned off 8 h earlier. Samples were collected at 2 time points on days 1, 5, and 14 after the LD shift (red triangles). (B) The actogram of a single mouse, showing that it was experiencing “jet lag” induced by the LD shift. Yellow shading indicates periods of light on, and gray shading indicates periods of light off. The red triangles indicate days 1 (Top), 5 (Middle), and 14 (Bottom). (C) BT measurement from mouse plasma collected before (day 1, Top), during (day 5, Middle), and after entrainment to the new LD cycle (day 14, Lower). Colors of the dots indicate the molecular peak time of each substance (Table S1). The red cosine curve is the estimation, the blue curve is the environmental time (pre LD condition shift), and the brown cosine curve is the environmental time (post shift). See also Table S2 for statistics.
General Applicability of the Metabolite-Timetable Method: CE-MS-Based Method.
As described above, the metabolite-timetable method based on LC-MS analysis can accurately measure the individual's BT and sensitively diagnose circadian rhythm disorders such as jet lag. The method depends entirely on the oscillations of numerous chemical substances and time-indicating metabolites; therefore, it can be applied to other metabolomics technologies such as CE-MS analysis. With CE-MS analysis, it is possible to separate charged compounds. So this is a complementary technology to LC-MS analysis (35). To demonstrate the general applicability of the metabolite-timetable method to other metabolomics technology, blood plasma was sampled and pooled from young male CBA/N mice every 4 hours under LD or DD conditions over 2 days. Small positively charged chemical substances in these samples were quantified by CE-MS, which detected 953 peaks. Of these peaks, 153 exhibited significant circadian oscillations under LD and DD conditions (Fig. S3A). We used these CE-MS data to construct the metabolite timetable in mouse plasma (Table S3). Notably, 28 peaks (18.3% of the total) were identified as known metabolites (Fig. S3B).
To confirm whether the CE-MS-based metabolite timetable was a good indicator of an individual's BT, we estimated BT from the metabolite profiles of independently sampled mice. Fresh plasma from young male CBA/N mice was collected every 4 hours over 24 h both under LD and DD conditions. CE-MS analysis was performed to profile the time-indicating metabolites (Fig. S3 C and D). The CE-MS-based metabolite-timetable method detected significant circadian rhythmicity in all metabolite profiles of these samples (P < 0.01, Fig. S3 C and D). The estimated BT was close to the environmental time with estimation errors of 0.6 ± 0.29 h for LD and 0.6 ± 0.54 h for DD (mean ± SD, Table S4). These results suggest the metabolite-timetable method is generally applicable to other metabolomics technologies such as CE-MS. See Fig. S3 E–H and SI Text for the result at a more stringent criterion (FDR < 0.01).
Discussion
We identified 14 and 28 oscillatory peaks in mouse blood as known metabolites in LC-MS and CE-MS analysis, respectively. For example, a different type of lysophsophatidylcolines exhibit significant circadian oscillations in LC-MS analysis (Fig. 1 C and D). The genes for key enzymes synthesizing lysophosphatidylcoline are Lcat (lecitine:cholesterol acyltransferase), Lipc (hepatic lipase), and Lipg (endothelial lipase). Among these genes, Lcat and Lipc are mainly expressed in liver. We found that Lipc mRNA are rhythmically expressed in the mouse liver with the peak time around peak time (PT) 5 (8), which slightly proceeds with the peak time of identified oscillatory lysophosphatidylcolines (PT5.8–9.1). In CE-MS analysis, many amino acids exhibit significant circadian rhythmicity. For example, glutamine (Gln), threonine (Thr), proline (Pro), valine (Val), phenylalanine (Phe), methionine (Met), isoleucine (Ile), leucine (Leu), and tryptophan (Trp) peak at around midnight (≈PT18), whereas glycine (Gly) peaks in the evening (PT12.1) (Fig. S4). In the urea cycle, metabolites such as ornithine (PT18.6), citrulline (PT19.9), and 4-guanidino-butyrate (PT20.1) exhibit significant circadian rhythmicity (FDR <0.1). Arginine (Arg), which plays an important role in the urea cycle, also exhibits suggestive circadian rhythmicity (FDR = 0.215; PT0.6) in our CE-MS data. It is also noteworthy that the final product—Urea—is reported to vary over 24 h in the blood of certain species such as rabbits (41) and rats (42). Interestingly, Reddy et al. (43) showed that 3 key enzymes involved in the urea cycle, carbamoyl-phosphate synthetase 1 (CPS1), argininosuccinate synthetase 1 (ASS1), and arginase 1 (ARG1), show circadian rhythms in the liver, the center of the urea cycle and urea formation (43). In the creatine pathway and neighboring glycine and threonine metabolism, metabolites such as guanidoacetate (PT6.2), Creatine (PT14.7), creatinine (PT18.7), sarcosine (PT18.0), and dimethylglycine (PT16.5) exhibit significant circadian rhythmicity (FDR <0.1). Arginine (PT0.6) first converts to guanidoacetate. Guanidoacetatete (CT6.2) then converts to creatine. Creatine (PT14.7) finally converts to creatinine (PT18.7) or sarcosine (PT18.0), which is also converted from dimethylglycine (PT16.5). The differences in the peak times of these metabolites may reflect successive processing throughout the day in the creatine pathway and neighboring glycine and threonine metabolism (Fig. S4).
Our results suggest that metabolite-timetable method can detect circadian rhythm disorders in vivo. In a normal situation, patients live under the zeitgeber (e.g., light). Notably, our method successfully diagnoses the jet-lag state under LD conditions (Fig. 5 and Fig. S2), and this strongly suggests that endogenous abnormal clock state can be diagnosed by our method, even if there is external time information such as light. Circadian rhythm disorders are caused by environmental factors (such as jet lag) and/or inherited factors (as in familial advanced sleep-phase syndrome). Brown et al. (44, 45) reported detecting circadian rhythm disorders by characterizing the feature of molecular circadian clocks in the isolated cells. They collected skin samples from human subjects, cultured the cells, and transfected clock-controlled reporter into the cells. The features of the molecular circadian clock in the isolated cells correlated with the chronotypes (i.e., the feature of organismal circadian clock) of the subjects, suggesting that the method should also allow detection of inherited circadian rhythm disorders. Our method can detect both inherited and acquired circadian rhythm disorders but cannot distinguish between them, whereas Brown et al.'s (44, 45) method can detect inherited but not acquired disorders. These 2 methods are therefore complementary for detecting circadian rhythm disorders.
Although our results suggest the metabolite-timetable method can successfully estimate BT, keeping MS and hiring a specialized operator in each hospital seems difficult. Establishing a special center for “detecting BT” performing MS analysis is 1 possibility to solve this. Another possibility is detecting time-indicating metabolites in a specific way (e.g., making an ELISA kit for detecting BT using a specific antibody for target time-indicating metabolites). To achieve the latter possibility, the assignment of oscillatory peaks to known metabolites is important, and we already identified 14 (LC-MS) and 28 (CE-MS) oscillatory peaks as known metabolites (Figs. 1 and S3). We also examined the effect of peak numbers on BT estimation. Fig. S5 shows the accuracy of the BT estimation using the different number of oscillatory metabolites. If we set the statistical error rate P < 0.05 and estimation error between environmental and estimated time <2 h, the minimum number of time-indicating metabolites was ≈20. In addition, the effect of feeding is an important issue, especially in humans, because humans eat different amounts of food at entirely different times. Further analysis on food intake conditions would be a great help for applying this method in clinical situations.
In this study, we showed that a metabolite-timetable method based on LC–MS analysis is able to estimate individuals' BTs with a high degree of accuracy throughout the time of the day, under different lighting conditions (LD and DD), and in individuals with different genetic backgrounds (CBA/N and C57BL/6 mice) (Figs. 2 and 3). We also found that the LC–MS-derived metabolite timetable is robust despite differences in age, sex, and feeding (Fig. 4); in addition, it is a sensitive and accurate detector of disordered circadian rhythm in jet-lagged mice. Our preliminary results suggest that the metabolite-timetable method can be also applied to other metabolomics techniques such as CE–MS; it allowed quantification of hundreds of clock-controlled metabolites, of which many could be identified, enabling successful measurement of BT from independent blood samples. The next step is to construct a metabolite timetable for human blood plasma, which will help measurement of BTs for humans and diagnosis of circadian rhythm disorders and facilitate the development of chronotherapy and tailored medication regimens.
Materials and Methods
BT Measurement.
Metabolomics-based measurement of BT is performed as described for expression-based measurement of BT (16), except that 2 samples are used for an estimation of BT. In the metabolite-timetable method, we used 2 samples with 12-h sampling time interval (e.g., ZT0 and ZT12 are used for 1 measurement of BT) to calibrate measurement-to-measurement experimental fluctuations of detection sensitivity, which usually differs among metabolites. We define the area in a certain sample as Asi and the mean areas of 2 samples (of 12-h time interval) as Msi for metabolite i. We also define the mean, standard deviation, and peak time of metabolite i in the timetable as Mti, Sti, and pti, respectively. For estimation of BT, we did not use outlying metabolites that do not satisfy the condition (|(Mti − Msi)/Sti| < 2). By changing b to 0, 0.1, …, 23.9, we searched for b with a maximum Pearson's correlation between { cos(2π(Pti −b)/24)} and {(Asi − Msi)/Sti}, and we predicted b as BT of the target sample. To estimate the P value of the prediction, we applied a permutation test to the maximum correlations.
Ethics.
All experiments were performed with the permission of Kobe Animal Experiment Supervisory Panel (permission IDs are AH15-10 and AH18-01).
Supporting Information.
More Materials and Methods information is available in SI Text.
Supplementary Material
Acknowledgments.
We thank Sachie Satoh and Yuki Ueno for technical support. We also thank Shigenobu Shibata, Masamitsu Iino, Kenichiro Uno, Rikuhiro Yamada, Hideki Ukai, Guojun Sheng, Yasushi Isojima, and Sato Honma for helpful discussions and Erik Kuld for proofreading of the manuscript. The Laboratory for Animal Resources and Genetic Engineering in the Center for Developmental Biology is noted for assistance with mouse work. This work is partly supported by New Energy Developing Organization (NEDO) (H.R.U.), a grant on Priority Aeas “Ensuring Chemical Hazard” and “Biomarker Discovery for Drug Development” from the Health and Labor Sciences Research Grants (T.S.), and a grant “Functional Genomics on Fundamental Brain Functions” from Utsunomiya University (M.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900617106/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland, MS: Sinauer; 2004. [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Ueda HR. Systems biology of mammalian circadian clocks. Cold Spring Harb Symp Quant Biol. 2007;72:365–380. doi: 10.1101/sqb.2007.72.047. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cdna microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 8.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 9.Halberg F. Chronobiology. Annu Rev Physiol. 1969;31:675–725. doi: 10.1146/annurev.ph.31.030169.003331. [DOI] [PubMed] [Google Scholar]
- 10.Labrecque G, Belanger PM. Biological rhythms in the absorption, distribution, metabolism and excretion of drugs. Pharmacol Ther. 1991;52:95–107. doi: 10.1016/0163-7258(91)90088-4. [DOI] [PubMed] [Google Scholar]
- 11.Lemmer B, Scheidel B, Behne S. Chronopharmacokinetics and chronopharmacodynamics of cardiovascular active drugs. Propranolol, organic nitrates, nifedipine. Ann NY Acad Sci. 1991;618:166–181. doi: 10.1111/j.1749-6632.1991.tb27245.x. [DOI] [PubMed] [Google Scholar]
- 12.Reinberg A, Halberg F. Circadian chronopharmacology. Annu Rev Pharmacol. 1971;11:455–492. doi: 10.1146/annurev.pa.11.040171.002323. [DOI] [PubMed] [Google Scholar]
- 13.Reinberg A, Smolensky M, Levi F. Aspects of clinical chronopharmacology. Cephalalgia 3 Suppl. 1983;1:69–78. doi: 10.1177/03331024830030S108. [DOI] [PubMed] [Google Scholar]
- 14.Levi F, Zidani R, Misset JL. Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. International organization for cancer chronotherapy. Lancet. 1997;350:681–686. doi: 10.1016/s0140-6736(97)03358-8. [DOI] [PubMed] [Google Scholar]
- 15.Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H. Changing the dosing schedule minimizes the disruptive effects of interferon on clock function. Nat Med. 2001;7:356–360. doi: 10.1038/85507. [DOI] [PubMed] [Google Scholar]
- 16.Ueda HR, et al. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci USA. 2004;101:11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennaway DJ, Owens JA, Voultsios A, Varcoe TJ. Functional central rhythmicity and light entrainment, but not liver and muscle rhythmicity, are clock independent. Am J Physiol. 2006;291:R1172–R1180. doi: 10.1152/ajpregu.00223.2006. [DOI] [PubMed] [Google Scholar]
- 18.Kennaway DJ, Voultsios A, Varcoe TJ, Moyer RW. Melatonin in mice: Rhythms, response to light, adrenergic stimulation, and metabolism. Am J Physiol. 2002;282:R358–R365. doi: 10.1152/ajpregu.00360.2001. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Kipnis DM, Daughaday WH. Growth hormone secretion during sleep. J Clin Invest. 1968;47:2079–2090. doi: 10.1172/JCI105893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoeller DA, Cella LK, Sinha MK, Caro JF. Entrainment of the diurnal rhythm of plasma leptin to meal timing. J Clin Invest. 1997;100:1882–1887. doi: 10.1172/JCI119717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok P, et al. Increased circadian prolactin release is blunted after body weight loss in obese premenopausal women. Am J Physiol. 2006;290:E218–E224. doi: 10.1152/ajpendo.00156.2005. [DOI] [PubMed] [Google Scholar]
- 22.Breum L, Rasmussen MH, Hilsted J, Fernstrom JD. Twenty-four-hour plasma tryptophan concentrations and ratios are below normal in obese subjects and are not normalized by substantial weight reduction. Am J Clin Nutr. 2003;77:1112–1118. doi: 10.1093/ajcn/77.5.1112. [DOI] [PubMed] [Google Scholar]
- 23.Forslund AH, et al. Inverse relationship between protein intake and plasma free amino acids in healthy men at physical exercise. Am J Physiol Endocrinol Metab. 2000;278:E857–E867. doi: 10.1152/ajpendo.2000.278.5.E857. [DOI] [PubMed] [Google Scholar]
- 24.Blanco RA, et al. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr. 2007;86:1016–1023. doi: 10.1093/ajcn/86.4.1016. [DOI] [PubMed] [Google Scholar]
- 25.Bonsch D, et al. Daily variations of homocysteine concentration may influence methylation of DNA in normal healthy individuals. Chronobiol Int. 2007;24:315–326. doi: 10.1080/07420520701290565. [DOI] [PubMed] [Google Scholar]
- 26.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: From diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 27.Kell DB, et al. Metabolic footprinting and systems biology: The medium is the message. Nat Rev Microbiol. 2005;3:557–565. doi: 10.1038/nrmicro1177. [DOI] [PubMed] [Google Scholar]
- 28.Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom Rev. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- 29.Fiehn O, et al. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 30.Fiehn O, Kopka J, Trethewey RN, Willmitzer L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal Chem. 2000;72:3573–3580. doi: 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- 31.Schauer N, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- 32.Plumb R, et al. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (lc-tofms): Detection of strain, diurnal and gender differences. Analyst. 2003;128:819–823. doi: 10.1039/b304296k. [DOI] [PubMed] [Google Scholar]
- 33.Wilson ID, et al. High resolution “Ultra performance” Liquid chromatography coupled to oa-tof mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 34.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal Chem. 2003;75:6737–6740. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 35.Soga T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 36.Soga T, et al. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 37.van der Horst GT, et al. Mammalian cry1 and cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 38.Tsang S, et al. A comprehensive snp-based genetic analysis of inbred mouse strains. Mamm Genome. 2005;16:476–480. doi: 10.1007/s00335-005-0001-7. [DOI] [PubMed] [Google Scholar]
- 39.Froy O, Miskin R. The interrelations among feeding, circadian rhythms and ageing. Prog Neurobiol. 2007;82:142–150. doi: 10.1016/j.pneurobio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Moriya T, et al. Potentiating action of mkc-242, a selective 5-ht1a receptor agonist, on the photic entrainment of the circadian activity rhythm in hamsters. Br J Pharmacol. 1998;125:1281–1287. doi: 10.1038/sj.bjp.0702176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccione G, Caola G, Refinetti R. Daily rhythms of liver-function indicators in rabbits. J Physiol Sci. 2007;57:101–105. doi: 10.2170/physiolsci.RP014606. [DOI] [PubMed] [Google Scholar]
- 42.Kato H, Mizutani-Funahashi M, Shiosaka S, Nakagawa H. Circadian rhythms of urea formation and argininosuccinate synthetase activity in rat liver. J Nutr. 1978;108:1071–1077. doi: 10.1093/jn/108.7.1071. [DOI] [PubMed] [Google Scholar]
- 43.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 44.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown SA, et al. Molecular insights into human daily behavior. Proc Natl Acad Sci USA. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.