Abstract
Rotaviruses, major causes of childhood gastroenteritis, are nonenveloped, icosahedral particles with double-strand RNA genomes. By the use of electron cryomicroscopy and single-particle reconstruction, we have visualized a rotavirus particle comprising the inner capsid coated with the trimeric outer-layer protein, VP7, at a resolution (4 Å) comparable with that of X-ray crystallography. We have traced the VP7 polypeptide chain, including parts not seen in its X-ray crystal structure. The 3 well-ordered, 30-residue, N-terminal “arms” of each VP7 trimer grip the underlying trimer of VP6, an inner-capsid protein. Structural differences between free and particle-bound VP7 and between free and VP7-coated inner capsids may regulate mRNA transcription and release. The Ca2+-stabilized VP7 intratrimer contact region, which presents important neutralizing epitopes, is unaltered upon capsid binding.
Keywords: electron microscopy, rotavirus VP7, virus assembly, viral entry, vaccines
The rotaviruses are a group of human and animal pathogens with multilayered, nonenveloped, icosahedral virions containing the 11 segments of their dsRNA genomes (1). These viruses have an unusual assembly pathway and a still more puzzling cell-penetration mechanism, both of which require disruption of a cellular membrane. During viral infection, an inner capsid particle [the so-called “double-layered particle” (DLP)] assembles in the cytosol and buds into the endoplasmic reticulum (ER), where it sheds its transiently acquired membrane and gains a surface glycoprotein layer (1–4). The resulting “triple-layered particle” (TLP) escapes from the infected cell by an uncharacterized transport route thought to bypass the Golgi apparatus (5).
The surface of a mature virion (Fig. 1A) includes 2 protein species—the VP4 spike protein, made in the cytosol, and the VP7 glycoprotein, made and acquired in the ER (1). A virally encoded “ER receptor”, NSP4, mediates the budding step (6, 7); it is lost from the assembling particle when VP7 displaces the transient envelope. Both VP4 and VP7 display epitopes for neutralizing antibodies (1). VP4 is the cell-surface receptor-binding protein (8) that must undergo tryptic cleavage to acquire efficient infectivity for susceptible cells. The cleavage generates an N-terminal, receptor-binding fragment called VP8* and a C-terminal fragment called VP5* (9). Cell penetration, which likely occurs in an endosomal compartment, delivers the DLP into the cytosol of the new host cell. Conformational changes in VP4 appear to mediate this process (10). The DLP contains the viral polymerase, VP1; the viral capping enzyme, VP3; and the genomic RNA, all enclosed by VP2 and VP6 (the 2 “layers” of the DLP). Without disassembly or further uncoating, the DLP synthesizes, caps, and extrudes multiple copies of the 11 mRNA species, some of which ultimately incorporate into new DLPs and become templates for synthesis of the genomic (-)-strand (reviewed in ref. 11).
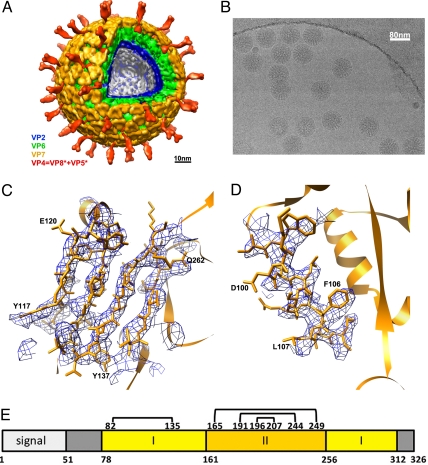
Fig. 1.
Rotavirus structure determined by cryo-EM. (A) Structure of the complete virion filtered at 25-Å resolution. The segmentation of the structure is based on reconstructions of the complete virion and the VP7-coated DLP and on published work of others (21–23). (B) Cryo-EM image of VP7-coated DLPs (7RP). (C and D) Regions of the VP7 density (filtered to 4.2-Å resolution) corresponding to strands β2, β1, β3, and β11 and helix αB. (E) Domain organization of VP7 (17). Domain I (light yellow) is a Rossmann fold; domain II (dark yellow), a β-jelly roll. The N- and C-terminal extensions (gray) become ordered when VP7 associates with the DLP. Numbers below the bar are those of the first residue in each segment. Disulfide bonds are indicated as residue numbers joined by lines across the top of the bar. The signal peptide is 50 residues long. Numbering corresponds to rhesus rotavirus, serotype G3.
The molecular structures of rotavirus particles and their various components inform us about both the mechanisms at each stage of the infectious cycle and the properties of ER and endosomes that the virus exploits. We have described the structure of the DLP determined by X-ray crystallography and single-particle cryo-EM (12), both at ≈3.8-Å resolution—a level of detail suitable for tracing polypeptide chains. The crystal structure of an isolated VP6 trimer has been determined by others (13). The newly developed power of cryo-EM and computational methods to achieve near-atomic resolution, as illustrated by our work on the DLP and by the work of others on bacteriophage and plant viruses (14, 15), has prompted us to extend our studies to particles that are even more difficult than the DLP to crystallize—the DLP recoated with recombinant VP7 (a particle we call “7RP”, for “VP7 recoated particle”) and the virion itself. We describe the former structure here and compare the structure of VP7 in the outer coat with the recently determined crystal structure of VP7 complexed with the Fab fragment of a neutralizing antibody (16). The cryo-EM structure of the 7RP has allowed us to trace the VP7 N- and C-terminal “arms,” disordered in the crystal structure, to describe conformational changes in VP7 that accompany association with VP6, and to analyze functionally significant differences between free DLPs, which are transcriptionally active, and VP7-associated DLPs, which are not. The structure shows that the VP7 N-terminal arms make the principal contacts with the DLP as well as important contacts between VP7 trimers. The results illustrate that cryo-EM can achieve a resolution comparable to that of X-ray crystallography, allowing de novo tracing of a polypeptide chain and assignment of many side-chain contacts. These advances greatly expand opportunities for understanding large-scale macromolecular assemblies.
Results
By use of the procedure recently applied to determine the structure of the DLP (12), we calculated an initial density map for the 7RP at a resolution of ≈6 Å (Fig. S1). Local averaging, using the redundancy afforded by the T = 13 surface lattice, enhanced the sharpness of the map and improved the estimated resolution to ≈5 Å (Fig. S1). The axial location of individual particles can vary between the top and bottom of the embedding ice layer (the ice thickness for our preparations was measured as 120–150 nm). The uncertainty in practice was as much as 100 nm (comparable with the particle diameter). Because an accurate evaluation of the contrast transfer function (CTF), which depends on axial position, is critical for correcting high-resolution contributions of the signal, we investigated a procedure to improve defocus parameters for individual particles, starting with the estimate given by the CTFTILT computer program used in our earlier work (12, 17). Because of the large mass of the 7RP (≈60 MDa), each particle produced sufficient contrast to allow refinement of defocus (see Materials and Methods and SI Text). After defocus refinement including data up to 9-Å resolution, the resolution of the resulting map improved to ≈4 Å (Fig. S1). Samples of the map are shown in Fig. 1 C and D and in Fig. S2.
We interpreted the map with the aid of the atomic structure of the DLP (12) and of a recently determined crystal structure of VP7 in complex with the Fab from a neutralizing antibody known as “4F8” (16). VP7 is a Rossmann-fold domain with N- and C-terminal extensions (arms) and with a β-jelly-roll domain inserted into a loop of the Rossmann fold (Fig. 1E). A Ca2+-stabilized VP7 trimer caps each VP6 trimer in the DLP (Fig. 2A). An interdomain conformational shift, with respect to its crystal structure, accompanies VP7 binding to VP6 (Fig. 2 C and D). The subunit flattens onto the VP6 surface; the β-jelly-roll domain moves by ≈8 Å with respect to the Rossmann-fold domain; the central helix (residues 149–161) gains an additional N-terminal turn and shifts by ≈3 Å axially toward VP6. The subunit–subunit interface within the VP7 trimer remains unchanged. The latter contact is the position of the epitopes for a number of neutralizing antibodies, including 4F8, as well as the location of calcium binding sites. The differences between VP7 conformations in the 7RP and in the crystals would therefore not affect our conclusions about how antibodies like 4F8 neutralize viral infectivity (16). The 4F8 epitope is fully exposed on the 7RP (and hence, by inference, on the virion), but the Fab arm of an antibody bound to one such epitope would block access by a second Fab (from the same or from a different IgG) to the nearest equivalent site on an adjacent trimer. We recently demonstrated that the Fab fragment of 4F8 neutralizes rhesus rotavirus (16) and concluded that crosslinking of VP7 is not the mechanism of neutralization. We can further conclude from the modeling here that a substoichiometric number of anti-VP7 Fab fragments is sufficient to block infection.
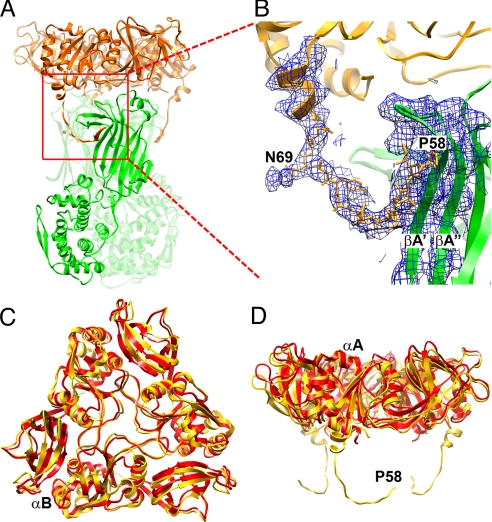
Fig. 2.
VP7 outer protein layer. (A) Structure of a VP6–VP7 heterohexamer, derived from crystal structures of VP6 (green) (14) and VP7 (gold) (16), docked into the cryo-EM density, and from the model built into cryo-EM density for the N-terminal arm. Two of the three VP7 N termini forming tight interactions with VP6 can be seen on the left and right side of the VP6 trimer. The red square indicates the area shown in more detail in B. (B) VP7 N terminus (residues 58–78) and corresponding cryo-EM density, showing its interaction with VP6. Pro-58 and Asn-69 are labeled; the latter bears a glycan for which density is present. Strands A′ and A″ of VP6 are also labeled. The density corresponding to residues 61–68 is weaker than in other parts of the map because this part of the structure lacks a clearly defined secondary structure and makes no contact with other parts of the structure. The map was filtered at 4.5-Å resolution to display a continuous density trace in this region. (C and D) Conformational differences between bound VP7 (gold) and the VP7 crystal structure (red) (16). The VP7 trimer is viewed from the outside of the virus along its symmetry axis (C) and from the “side”, normal to its symmetry axis (D). The way in which the domain hinge displacement “flattens” the subunit when the trimer binds VP6 is evident in the latter view. The alignment is based on a superposition of the Rossmann-fold domains.
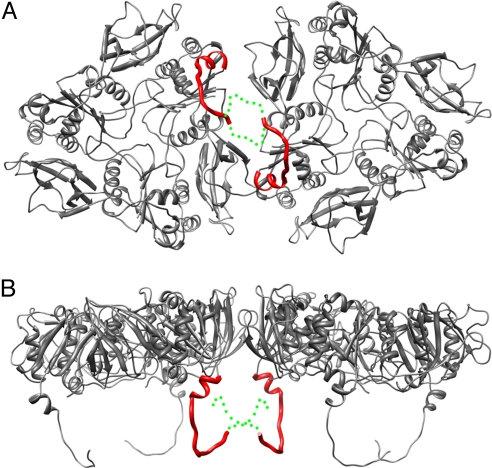
A more striking change in VP7, with respect to the crystal structure, is the conformation of its N-terminal arm, which is disordered in the crystal structure and presumably on free VP7. The arm includes residues 51–77; residues 1–50 are a long signal sequence removed from the mature protein. We have followed the polypeptide chain back almost to its N terminus and built a full model (with side chains) from residue 58 to 77. The arm extends away from the compact core of the VP7 subunit and inward along the surface of a VP6 subunit so that, in a claw-like configuration, the 3 arms of a VP7 trimer grip the VP6 trimer beneath it (Fig. 2 A and B). Residues 58–62 form a strand that augments the A′A″BIDG β-sheet of the VP6 jelly roll (13), and residues 72–78 form a 2-turn helix. There is clear density for the glycan on Asn-69 (>1 sugar unit is clearly visible, see Fig. 2B and Fig. S3). This carbohydrate moiety is recessed between VP6 trimers and not exposed on the virion surface. The contact point of each arm with VP6 is close to the local 2-fold that relates 1 VP6 trimer to its neighbor in the T = 13 lattice (Fig. 3). Near the N terminus of the polypeptide chain, arms from VP7 trimers related by the local 2-fold approach each other, and the N termini probably interact (Fig. 3). Intertrimer contact of the N-terminal arms would create some degree of continuity, and potentially cooperativity, within the VP7 shell. The C-terminal extension of VP7 (residues 313–325, which, like the N-terminal arm, are disordered in the VP7:4F8 crystal structure) may also interact across the local dyad with its counterpart from another trimer, but most of its contacts appear to be within its trimer of origin.
Fig. 3.
Interface between 2 VP7 trimers. The interface is shown from the outside of the virus (A) and from the “side”, normal to its symmetry axes (B). The N-terminal arms between residues 58 and 78 are shown as red curves; the approximate positions of the arms between residues 51 (the N terminus) and 57 are shown as dotted green curves, illustrating that the N termini may contribute to intertrimer contacts.
The grip by the VP7 N-terminal arm accounts for nearly all of the contacts between VP7 and VP6 (Table S1). The hydrated interface between the outward-facing surface of VP6 and the inward-facing surface of VP7 is devoid of protein interaction, except for 1 pair of van der Waals contacts (between Pro-279 and Thr-281 in VP7 and Pro-313 in VP6) and 1 side-chain hydrogen bond (Gln-305 to Asn-310).
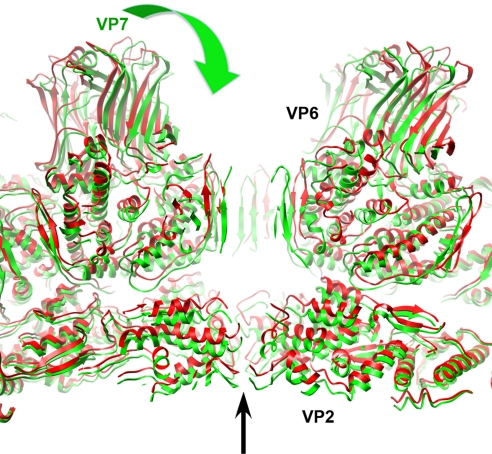
Most of the DLP remains unperturbed upon addition of VP7, except for a distinct shift in orientation of the VP6 trimers surrounding 5-fold positions and a smaller shift in their immediate neighbors (Fig. 4). VP7 binding tilts these trimers (as rigid bodies) toward the 5-fold axis, decreasing the diameter of the 5-fold channel from ≈25 Å (DLP) to 20 Å (7RP). Without the tilts, VP7 could not cap the VP6 trimers in question without colliding with neighboring VP7s. A similar shift in the VP6 trimers adjacent to the 5-fold channels has been observed at lower resolution by comparing VP2–VP6 with VP2–VP6–VP7 recombinant particles (18) and by examining DLPs bound by transcription-inhibiting Fabs (19, 20). Probably as a consequence of the VP6 shifts, the tips of VP2, which converge around the 5-fold axis, move inward by ≈5 Å when VP7 binds, closing the narrow pore through which nascent mRNA emerges. The converse change—the opening of the 5-fold channels when VP7 uncoats— may be related to activation of RNA synthesis in the DLP. In this way, a VP7 calcium “sensor” could transmit a signal to the polymerase and capping enzyme complex to coordinate the onset of transcription with entry into the host-cell cytoplasm.
Fig. 4.
Conformational differences between DLP and 7RP. The structures of VP2 and VP6 docked into the 7RP cryo-EM density are shown in green whereas the DLP structure (12) is shown in red. A narrowing of the central channel on the icosahedral 5-fold axis and an inward movement of the VP2 and VP6 layers in the 7RP, compared with the DLP, are the principal differences. The VP7 layer of the 7RP is not shown (for clarity); the green arrow indicates the “clamping down” of the VP6 and VP2 layers that accompanies VP7 binding.
In the virion, VP4 (when cleaved by trypsin to VP8* and VP5*, a normal step in viral activation) projects from the 6 coordinated positions in the T = 13 lattice that are adjacent to the 5-fold positions (Fig. 1A). The most prominent part of the spike has strong 2-fold symmetry, but there is good evidence that the complete spike contains 3 VP4 subunits (10, 21). The 2-fold symmetric part appears as if cantilevered out over the edge of an adjacent VP7, and the “foot” of the structure, anchored in the VP6 layer and fixed in place by VP7, has 3-fold symmetry in lower resolution reconstructions of virions (21, 22). The spikes adopt this curiously asymmetric conformation only after tryptic cleavage: Uncleaved VP4 is either more flexible or splayed out against the VP7 shell (23). The arrangement of VP6 and VP7 in the T = 13 icosahedral lattice is such that the different positions and orientations of these neighbors around the local 6-fold axis can determine the asymmetric orientation of the reorganized spike after cleavage. In particular, the diagonal “cantilever” will clearly pass across the tip of one of the surrounding VP7 trimers, with potential contacts near one of the calcium sites.
Discussion
Several factors have contributed to the resolution of our 7RP reconstruction, in addition to the alignment and stability of the microscope and to the power and data storage capacity of the computing cluster. The sample is homogeneous and stable. The icosahedral symmetry affords robust initial determination of orientation parameters for each particle (or class of particles), independent of cross-correlation with others, and the icosahedral symmetry and the T = 13 local symmetry effectively increase the size of the dataset by 780-fold. Recording images on photographic film preserves the high-resolution signal better than using currently available electronic devices. Several features of FREALIGN (24), notably the use of a weighted correlation coefficient (25), appear to avoid overfitting and noise amplification during refinement of the orientations and positions of individual images. Finally, a newly introduced, restrained refinement of image defocus and particle positions (see Materials and Methods, SI Text, and Fig. S4) has improved the quality of the final reconstruction.
The resolution we have obtained has been essential for identifying the most striking and informative features of the 7RP structure—the conformation and interactions of the VP7 N-terminal arms. The arms are both adaptors, allowing preformed VP7 trimers to grip the underlying array of VP6 trimers, and network formers, creating VP7 lattice contacts and generating cooperativity. The latter property probably accounts for the relatively sharp calcium dependence of uncoating (26) and for the predominance of continuous patches of VP7 when rotavirus DLPs are recoated with substoichiometric quantities of VP7 (27). The lattice contacts between N termini of adjacent VP7 trimers and the interactions between VP7 and VP6 together contribute to the high affinity of VP7 trimers for DLPs, despite the relatively weak trimerization of free VP7 even in calcium-containing buffers (27, 28). Once bound to VP6, VP7 will uncoat (under nondenaturing conditions) only when withdrawal of Ca2+ induces trimer dissociation.
Rotavirus appears to enter cells by a dynamin-dependent route (29). There are no systematic measurements of calcium concentrations in various endosomal compartments, but conditions that probably elevate intraendosomal calcium concentrations (increasing the extracellular calcium reserve with CaEGTA or inhibiting the endosomal H+-ATPase with bafilomycin A) block rotavirus infectivity and α-sarcin coentry (30). This evidence suggests that a drop in calcium concentration in an endosomal compartment is probably a characteristic of the infectious route and an important trigger for the molecular events that cause membrane penetration. Because VP7 locks VP4 into the assembled virion, VP7 uncoating probably precedes the VP5* conformational changes thought to mediate the penetration step (10).
Enveloped intermediate particles accumulate in the ER during rotavirus infection if VP7 expression is inhibited by siRNA (31). Calcium depletion from the ER also causes the intermediates to accumulate, suggesting that VP7 must trimerize to displace the transient membrane (32, 33). ER retention of VP7, a prerequisite for these steps, requires both the VP7 signal peptide and the first ≈31 residues of the mature protein (34, 35). The latter segment is precisely the N-terminal arm, shown here, to grip VP6. Cleavage of the signal peptide occurs rapidly during synthesis, and some noncovalent interaction between the signal region and the N-terminal arm has been proposed as a retention mechanism (36). These elements are also sufficient to retain a heterologous, chimeric protein in the ER, even in the absence of NSP4 (35). Thus, the retention mechanism, and perhaps the membrane displacement mechanism, is likely to involve one or more resident ER proteins, such as components of the translocation channel or proteins associated with it.
Materials and Methods
Sample Preparation.
Rhesus rotavirus VP7 was prepared as described previously (28). Bovine United Kingdom DLPs were also prepared as described (12, 37). The 7RPs were generated as described in ref. 28, with minor adjustments. Briefly, we added VP7 trimers to DLPs in a 1.4:1 molar ratio (VP7:VP6) at 0.56 mg/mL in 50 mM sodium acetate (pH 5.2), 1 mM CaCl2, and incubated at room temperature for 1 h. Particles were isolated by centrifugation at 288,000 × g for 2 h (SW41 rotor, Beckman) and resuspended at 5 mg/mL in 20 mM Tris·HCl (pH 8.0), 50 mM NaCl, and 2 mM CaCl2. Authentic TLPs were purified as described in ref. 27.
Electron Microscopy and Image Processing.
7RPs were embedded in thin vitreous ice on holey-carbon grids by plunge-freezing to ≈90K. Single-particle image data (Fig. 1B) were collected at 59,000× nominal magnification, 300keV beam energy on an FEI Tecnai F30 electron microscope. One-hundred-forty-eight micrographs were recorded on Kodak ISO163 film, at a dose of ≈20 e−/Å2 and under defocus ranging between 1.2 μm and 3.5 μm. The micrographs were digitized on a Zeiss SCAI scanner at 7 μm step-size, a scanning resolution of 1.233 Å/pixel after magnification calibration. From these micrographs, 3,786 particles were selected by using the program SIGNATURE (38). The defocus parameter of each particle was evaluated by the computer program CTFTILT (17), which determines the specimen tilt axis, tilt angle, and the image astigmatism, then assigns the CTF parameter for each particle according to its location in the micrograph. These parameters were subsequently used in the computer program FREALIGN (24) for precise particle alignment and density map reconstruction. A previously reconstructed EM density map of the DLP (12) was used as the reference for initial particle alignment through a systematic search at 1 ° angular interval. The alignment parameters were subsequently subjected to iterative refinement by using the density map reconstructed from the previous cycle as the reference. For the refinement, the density corresponding to the RNA genome was masked in the reference structure to enhance the signal of the ordered part of the structure. Although the 7RP and DLP particles differ in diameter by ≈90 Å (12% of the DLP particle size), the well-ordered icosahedral symmetry substantially reduced misalignment and enabled rapid convergence in the refinement. To improve the model resolution, particle images with defocus >3.0 μm and alignment phase residual in the bottom 10th percentile were not included in the reconstruction. The resolution of the icosahedral reconstruction was ≈6 Å as measured by the Fourier shell correlation (FSC) (5.5 Å at FSC = 0.143, 6.8 Å at FSC = 0.5; see Fig. S1).
In addition to the icosahedral symmetry, the rotavirus capsid contains T = 13 quasi-equivalence in the VP6 and VP7 layers. This 13-fold structural redundancy can be harnessed in local averaging to further improve the density resolution. Following the procedure implemented in the previous work on the EM reconstruction of DLP (12), we conducted a local search to identify the optimal transformation matrices for VP6 and VP7, respectively, in the 7RP EM map. The 2 sets of transformations were found to be equivalent within numerical error. Thus, the VP6 and VP7 trimers share the same 3-fold axis and form a rigid capsomer that constitutes the middle and outer layers of the 7RP. Local averaging extended the resolution of the map to ≈4 Å (4.2 Å at FSC = 0.143, 4.5 Å at FSC = 0.5; see Fig. S1). After sharpening with a negative B-factor of 300 Å2 (see Fig. S5) and figure-of-merit weighting (39), the map revealed most of the backbone tracing of VP7 and its interaction with VP6. The FSC resolution estimate used soft masks with a cosine edge (12). The maps were low-pass filtered at 4.2-Å (Fig. 1 C and D) or 4.5-Å resolution (Fig. 2B).
We calculated a low-resolution structure of the complete virion from 4,187 particle images recorded on 110 micrographs and processed in the same way as the 7RP data. At ≈25-Å resolution, the structure of the VP7 layer appears to be the same in the 7RP and the full virion.
Model Building.
The computer program Chimera (40) was used to fit the VP7 model derived from the crystal structure of VP7 (16) into the 7RP reconstruction. We divided the protein at the domain boundary (residues 71–160 and 256–312 in one fragment; residues 161–255 in the other) and fit each half as a rigid body into the averaged density. Segments that did not match (e.g., at the domain interface) were adjusted and refined by using the program O (41); residues in the N-terminal arm and C-terminal tail were likewise built and refined by using O. The glycan at Asn-69 provided a strong constraint on the sequence register of the N-terminal segment. For certain copies of VP7, density at the N terminus (residue 51) was consistent with the pyroglutamate known to be the product of processing by signal peptidase (44), and well-defined density extended to the second-to-last residue at the C terminus. After fitting 1 VP7 subunit, we generated the remaining 12 within the icosahedral asymmetric unit by the appropriate transformations, followed by rigid-body refinement in Chimera and visual inspection in O. The coordinates for VP6 (13) were fit into corresponding density by inspection and rigid-body refinement using Chimera, with the DLP model (12) as a starting point.
Supplementary Material
Acknowledgments.
We thank Marina Babyonyshev for assistance with protein and DLP preparation, Alexandra Deaconescu for advice on atomic model building, and Chen Xu for maintaining the Brandeis electron microscope facility. This work was supported by National Institutes of Health Grants GM-62580 (to N.G. and S.C.H.), AI-053174 (to P.R.D.), and CA-13202 (to S.C.H.); by an Ellison Medical Foundation New Scholars in Global Infectious Diseases Award (to P.R.D.); and by grants from the Health Research Council of New Zealand (to A.R.B.). N.G. and S.C.H. are investigators at the Howard Hughes Medical Institute. P.R.D. and E.C.S. are employees and shareholders of Novartis Vaccines and Diagnostics, Inc.
Footnotes
Conflict of interest statement: E.C.S. and P.R.D. are employees and shareholders of Novartis Vaccines and Diagnostics, Inc.
Data deposition: Density maps of 7RP and the nonicosahedrally averaged VP6–VP7 complex structure have been deposited in the Macromolecular Structure Database at the European Bioinformatics Institute (accession codes EMD-1517 and EMD-1609, respectively). The atomic structure of the 7RP capsid has been deposited in the Protein Data Bank, www.pdb.org, as 2 separate entries, VP2–VP6 (PDB ID code 3GZU) and VP7 (PDB ID code 3GZT).
See Commentary on page 10398.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904024106/DCSupplemental.
References
- 1.Estes MK, Kapikian AZ. Rotaviruses. In: Howley DMKPM, editor. Fields Virology. 5th ed. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. 1918–1974. [Google Scholar]
- 2.Adams WR, Kraft LM. Epizootic diarrhea of infant mice: Indentification of the etiologic agent. Science. 1963;141:359–360. doi: 10.1126/science.141.3578.359. [DOI] [PubMed] [Google Scholar]
- 3.Holmes IH, Ruck BJ, Bishop RF, Davidson GP. Infantile enteritis viruses: Morphogenesis and morphology. J Virol. 1975;16:937–943. doi: 10.1128/jvi.16.4.937-943.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altenburg BC, Graham DY, Estes MK. Ultrastructural study of rotavirus replication in cultured cells. J Gen Virol. 1980;46:75–85. doi: 10.1099/0022-1317-46-1-75. [DOI] [PubMed] [Google Scholar]
- 5.Jourdan N, et al. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol. 1997;71:8268–8278. doi: 10.1128/jvi.71.11.8268-8278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Au KS, Chan WK, Burns JW, Estes MK. Receptor activity of rotavirus nonstructural glycoprotein NS28. J Virol. 1989;63:4553–4562. doi: 10.1128/jvi.63.11.4553-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer JC, Bergmann CC, Bellamy AR. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology. 1989;171:98–107. doi: 10.1016/0042-6822(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 8.Fiore L, Greenberg HB, Mackow ER. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 9.Estes MK, Graham DY, Mason BB. Proteolytic enhancement of rotavirus infectivity: Molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton JT, Vasquez-Del Carpio R, Tortorici MA, Taraporewala ZF. Rotavirus genome replication and capsid assembly. Adv Virus Res. 2007;69:167–201. doi: 10.1016/S0065-3527(06)69004-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieu M, et al. Atomic structure of the major capsid protein of rotavirus: Implications for the architecture of the virion. EMBO J. 2001;20:1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, et al. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki ST, et al. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science. 2009 doi: 10.1126/science.1170481. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 18.Libersou S, et al. Geometric mismatches within the concentric layers of rotavirus particles: A potential regulatory switch of viral particle transcription activity. J Virol. 2008;82:2844–2852. doi: 10.1128/JVI.02268-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng N, et al. Inhibition of rotavirus replication by a non-neutralizing, rotavirus VP6-specific IgA mAb. J Clin Invest. 2002;109:1203–1213. doi: 10.1172/JCI14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawton JA, Estes MK, Prasad BV. Comparative structural analysis of transcriptionally competent and incompetent rotavirus-antibody complexes. Proc Natl Acad Sci USA. 1999;96:5428–5433. doi: 10.1073/pnas.96.10.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeager M, Berriman JA, Baker TS, Bellamy AR. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford SE, et al. Trypsin cleavage stabilizes the rotavirus VP4 spike. J Virol. 2001;75:6052–6061. doi: 10.1128/JVI.75.13.6052-6061.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Gajardo R, Vende P, Poncet D, Cohen J. Two proline residues are essential in the calcium-binding activity of rotavirus VP7 outer capsid protein. J Virol. 1997;71:2211–2216. doi: 10.1128/jvi.71.3.2211-2216.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trask SD, Dormitzer PR. Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J Virol. 2006;80:11293–11304. doi: 10.1128/JVI.01346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dormitzer PR, Greenberg HB, Harrison SC. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology. 2000;277:420–428. doi: 10.1006/viro.2000.0625. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-San Martin C, Lopez T, Arias CF, Lopez S. Characterization of rotavirus cell entry. J Virol. 2004;78:2310–2318. doi: 10.1128/JVI.78.5.2310-2318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chemello ME, Aristimuno OC, Michelangeli F, Ruiz MC. Requirement for vacuolar H+ -ATPase activity and Ca2+ gradient during entry of rotavirus into MA104 cells. J Virol. 2002;76:13083–13087. doi: 10.1128/JVI.76.24.13083-13087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez T, et al. Silencing the morphogenesis of rotavirus. J Virol. 2005;79:184–192. doi: 10.1128/JVI.79.1.184-192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michelangeli F, Liprandi F, Chemello ME, Ciarlet M, Ruiz MC. Selective depletion of stored calcium by thapsigargin blocks rotavirus maturation but not the cytopathic effect. J Virol. 1995;69:3838–3847. doi: 10.1128/jvi.69.6.3838-3847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poruchynsky MS, Maass DR, Atkinson PH. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J Cell Biol. 1991;114:651–656. doi: 10.1083/jcb.114.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stirzaker SC, Both GW. The signal peptide of the rotavirus glycoprotein VP7 is essential for its retention in the ER as an integral membrane protein. Cell. 1989;56:741–747. doi: 10.1016/0092-8674(89)90677-6. [DOI] [PubMed] [Google Scholar]
- 35.Maass DR, Atkinson PH. Retention by the endoplasmic reticulum of rotavirus VP7 is controlled by three adjacent amino-terminal residues. J Virol. 1994;68:366–378. doi: 10.1128/jvi.68.1.366-378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stirzaker SC, Poncet D, Both GW. Sequences in rotavirus glycoprotein VP7 that mediate delayed translocation and retention of the protein in the endoplasmic reticulum. J Cell Biol. 1990;111:1343–1350. doi: 10.1083/jcb.111.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Street JE, Croxson MC, Chadderton WF, Bellamy AR. Sequence diversity of human rotavirus strains investigated by northern blot hybridization analysis. J Virol. 1982;43:369–378. doi: 10.1128/jvi.43.2.369-378.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 42.Stirzaker SC, Whitfeld PL, Christie DL, Bellamy AR, Both GW. Processing of rotavirus glycoprotein VP7: Implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987;105:2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.