Abstract
Sulfide:quinone oxidoreductase (SQR) is a flavoprotein with homologues in all domains of life except plants. It plays a physiological role both in sulfide detoxification and in energy transduction. We isolated the protein from native membranes of the hyperthermophilic bacterium Aquifex aeolicus, and we determined its X-ray structure in the “as-purified,” substrate-bound, and inhibitor-bound forms at resolutions of 2.3, 2.0, and 2.9 Å, respectively. The structure is composed of 2 Rossmann domains and 1 attachment domain, with an overall monomeric architecture typical of disulfide oxidoreductase flavoproteins. A. aeolicus SQR is a surprisingly trimeric, periplasmic integral monotopic membrane protein that inserts about 12 Å into the lipidic bilayer through an amphipathic helix–turn–helix tripodal motif. The quinone is located in a channel that extends from the si side of the FAD to the membrane. The quinone ring is sandwiched between the conserved amino acids Phe-385 and Ile-346, and it is possibly protonated upon reduction via Glu-318 and/or neighboring water molecules. Sulfide polymerization occurs on the re side of FAD, where the invariant Cys-156 and Cys-347 appear to be covalently bound to polysulfur fragments. The structure suggests that FAD is covalently linked to the polypeptide in an unusual way, via a disulfide bridge between the 8-methyl group and Cys-124. The applicability of this disulfide bridge for transferring electrons from sulfide to FAD, 2 mechanisms for sulfide polymerization and channeling of the substrate, S2−, and of the product, Sn, in and out of the active site are discussed.
Keywords: flavoprotein, monotopic membrane protein, sulfide metabolism, X-ray crystallography, extremophilic organism
Sulfide attracts increasing attention because this compound is—although toxic—involved in a variety of important physiological processes. It is found in marine or soil environments populated by microorganisms, and it is endogenously produced by eukaryotic cells, including those of several tissues in the human body, as a product of the catabolism of cysteines (1) and possibly of iron–sulfur clusters (2). In bacteria, sulfide can act as an electron donor (2, 3). In yeast, complexes of phytochelatines and heavy metal sulfide are formed to be disposed by vacuole ATP-dependent transporters (2). In animals, it has been proposed that sulfide act as a “gasotransmitter” (4). Finally, the medical and pharmacological relevance of the sulfide metabolism is related to the fact that its incorrect regulation may contribute to severe pathologies, such as Alzheimer's disease and Down syndrome (1).
A key enzyme in maintaining sulfide homeostasis is the sulfide:quinone oxidoreductase (SQR), a ubiquitous protein present in all domains of life except plants (2). The SQRs have a monomeric molecular mass of around 50 kDa and have been described as dimeric membrane-associated proteins harboring 1 noncovalently bound FAD cofactor per monomer. The enzyme oxidizes sulfide ions (S2−, HS−) to zero-valent sulfur, thought to be released from the protein as a soluble but water-labile polysulfide chain of up to 10 sulfur atoms (5). The electrons are transferred via the FAD to the quinone pool in the membrane.
The SQRs are members of the disulfide oxidoreductase flavoprotein (DiSR) superfamily, like other well-characterized pyridine nucleotide:disulfide flavoproteins (6). The flavocytochrome c:sulfide dehydrogenase (FCC) from Allochromatium vinosum [Protein Data Bank (PDB) ID code 1fcd; ref. 7] is the most closely related enzyme of known structure to the SQR from Aquifex aeolicus, the sequence identity between the 2 enzymes being 24%. In general, sequence identity to the other members of the superfamily is low, and even within the SQR subfamily the sequences are not well-conserved. Based on alignments, however, the SQRs have been classified into 3 groups (6). Type I SQRs are found in many bacterial species, including A. aeolicus, and are involved in the cellular respiration pathway or in anaerobic photosynthesis (3, 5, 8). They have high catalytic rates and micromolar substrate affinities. Type II SQRs occur in other bacterial species (i.e., Chloroflexus, Bacilli, and a cyanobacterium, Synechocystis PCC 6803) and in many eukaryotes (where the enzyme is denoted by the acronym SQRDL, or “sulfide:quinone reductase-like”). In vitro, type II SQRs require cyanide as a cosubstrate to produce thiocyanate or more physiological reaction partners, such as thioredoxin and sulfite to produce disulfides (or polysulfides) (9, 10). Yeast SQR is involved in heavy metal tolerance (i.e., in Saccharomyces pombe; ref. 11). Interestingly, an SQRDL gene is also present in humans, where it could help regulate the cellular sulfide concentration, thus influencing the processes of sulfide signaling (2). Type III SQRs, which include enzymes from archaea and green sulfur bacteria, can less clearly be defined from evolutionary trees (2) and will not be discussed in this work.
Experimental information for many properties of SQR has been missing. This includes the topology of the enzyme, the way it interacts with the lipid bilayer, its cellular localization, its oligomeric state, and the mode of FAD binding. In addition, there has been a lack of experimental evidence in support of the previously postulated mechanism of action (5). To address these questions, we present the 3D structure of the SQR in the “as-purified,” substrate-bound, and inhibitor-bound states at 2.3-, 2.0-, and 2.9-Å resolution, respectively.
Results
Overall Structure.
The SQR was isolated from A. aeolicus, a Gram-negative hyperthermophilic bacterium (12), and it was purified to homogeneity in an active form. We refer to such a protein sample as the “as-purified” protein because of the specific structural features described and discussed below (i.e., the presence of unexplained electron density in the quinone-binding pocket and the fact that the structure might represent the conformation of an advanced stage of the catalytic cycle). The structure of the SQR was determined by a combination of multiple isomorphous replacement and anomalous scattering (MIRAS method; Table S1) using crystals soaked with osmium and gold complex ion salts. The initial model was refined to a final Rfree of 23.5% (Rwork is 19.2%) in the resolution range of 20.0–2.30 Å by using translation libration screw-rotation (TLS) refinement and noncrystallographic symmetry (NCS) averaging of the 6 monomers composing the asymmetric unit. Final rounds of refinements were done without NCS.
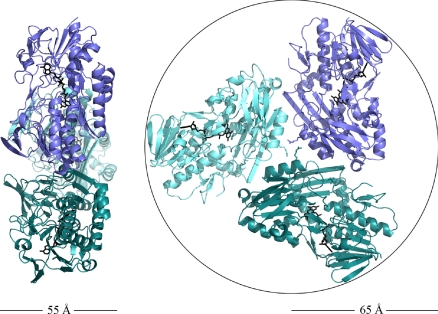
A. aeolicus SQR is, surprisingly, a homotrimer of pronounced ellipticity (Fig. 1) and not a homodimer, as was found for other proteins of the same superfamily (13). From sequence comparison studies, however, we could not conclude whether all SQRs share this oligomerization state.
Fig. 1.
Dimensions of the SQR trimer (cartoon representation with each monomer colored in a different shade of blue). The trimer has a thickness of ≈55 Å (Left), whereas when seen from its soluble face (Right), it can be inscribed in a circle with a radius of about 65 Å. All figures showing the structure were generated with PyMOL (www.pymol.org).
The structures of 6 monomers in the asymmetric unit are nearly identical. Two Rossmann fold domains bind the FAD and are typical of the DiSR proteins. The C-terminal domain is instead distinctive in each subfamily of DiSR. In A. aeolicus SQR, it consists of (i) 2 amphipathic helices (Lys-376–Asn-395 and Pro-400–Lys-412) mediating the membrane attachment and (ii) an 18-aa C-terminal loop that forms oligomerization contacts. Two disulfide bridges (Fig. S1) stabilizing the C-terminal domain help explain the heat stability of the enzyme (14) and corroborate the idea that the protein is localized in a periplasmic oxidizing environment (15).
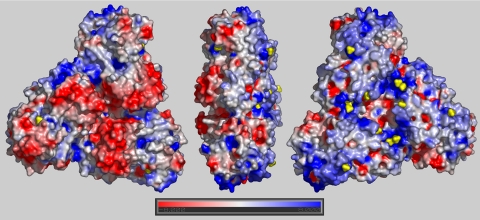
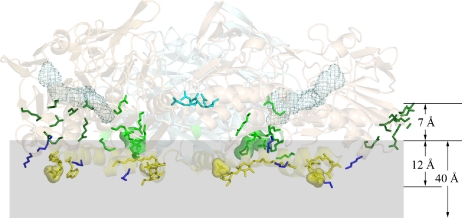
Protein surface analysis reveals a pronounced polarity of the SQR trimer on the basis of electrostatic potential calculations performed by using GRASP (16) (Fig. 2). One side of the trimer, formed by the 2 Rossmann fold domains, shows an overall negative surface potential, whereas the other side is characterized by an excess of positive charges and a pronounced hydrophobic patch in each monomer. The latter side, considered to be the membrane interaction domain, can be further subdivided into 4 structural motifs involved in potential interactions with the phospholipid bilayer (Fig. 3). First, the N-terminal surface residues (Ala-2, Lys-3, Arg-22, and Lys-29) and the spatially adjacent Arg-333 and Asn-334 of each subunit are distributed on a layer perpendicular to the central trimeric threefold axis. Within the same layer but on the inner central region of the trimer, the subunits form a positively charged ring with a diameter of about 25 Å that circles around the threefold axis. Its positive potential is due to 3 lysines (Lys-172, Lys-173, Lys-412) and 3 arginines (Arg-177, Arg-213, Arg-369) and a few other main-chain N atoms. Two putative sulfate groups form ion pairs with protein atoms, compensating for the excess of acidic residues that would weaken the interactions between the monomers. Both are present as strong electron density peaks in all 6 monomers of the asymmetric unit and appear to originate from 1 sulfate ion and 1 MES molecule (Fig. S2A). Third, 4 lysine residues (Lys-54, Lys-68, Lys-387, and Lys-405), oriented toward the bulk solvent, are conserved in type I SQRs. Finally, the base of the trimer body is the part of the protein most deeply inserted into the membrane and is ≈12 Å below the layer of the N-terminal domain and the sulfate groups. It consists of a helix–turn–helix motif (residues 376–412) and a dodecyl β-d-maltoside (DDM) molecule lying between the helices. The detergent mimics both the physiological lipids as well as the substrate quinone that presumably enters SQR at this place, as reported below. The maltose polar head group of DDM presents its oxygen atoms at hydrogen bond distances (3.0–3.5 Å) from residues Lys-373 and Trp-377 and from 1 putative sulfate ion, whereas its acyl chain is in hydrophobic contact (3.5–4.2 Å) with residues Phe-357, Phe-381, Leu-407, Phe-410, and Leu-411. The 2 helices are amphipathic, as is graphically illustrated in the scheme of Fig. S3. Their positive net charge (+4) and their relative hydrophobicity (1.21 kcal/mol) are comparable with the values of other amphipathic helical domains of monotopic membrane proteins (17). In addition, at the positive N-terminal end of helix 376–395, the indole group of conserved Trp-377 “snorkels up” (18) in a characteristic manner (Fig. S2B), and at the C terminus, Trp-391, Arg-394, and Asn-395 bind a putative sulfate ion (Fig. S2C).
Fig. 2.
Electrostatic surface potential of the trimeric unit, calculated by the software GRASP. (Left) The view from the solvent side shows the overall negative (red) surface of the Rossmann fold domains. Rotation of the trimer by 180° (Right) shows the domain that mediates the interaction with the membrane. It has an overall intense positive charge (blue), with helices 376–395 and 400–412 more neutral (white). Sulfate ions and the Mes molecules are shown in yellow. Solvent molecules were not included in the calculation of the electrostatic potential.
Fig. 3.
Membrane-binding motifs of the SQR trimer. The membrane is indicated in gray. The overall trimer is in a cartoon semitransparent representation. The side chains of Arg-204 of all monomers are represented in cyan sticks to highlight the central trimerization contacts. Other residues and molecules are shown only for the 2 subunits in the foreground for better clarity of visualization. The FAD is in cyan mesh. The side chains of the residues and the molecules belonging to the different membrane-interacting motifs are shown as sticks. For sulfate groups a semitransparent surface is also shown. Different colors are attributed to the different structural motifs. In particular the N-terminal domain is dark green, the inner domain binding 1 sulfate ion and 1 MES molecule is light green, the 4 conserved lysines are dark blue, and the base of the trimer body is yellow. The distances were calculated from the plane of the sulfur atoms of the MES molecules, respectively, to the Cα atom of Arg-204 and the C3B atom of the maltose head of DDM (chain A). An approximate value (40 Å) indicates the membrane thickness according to White and Wimley (23).
FAD and Substrate-Binding Sites.
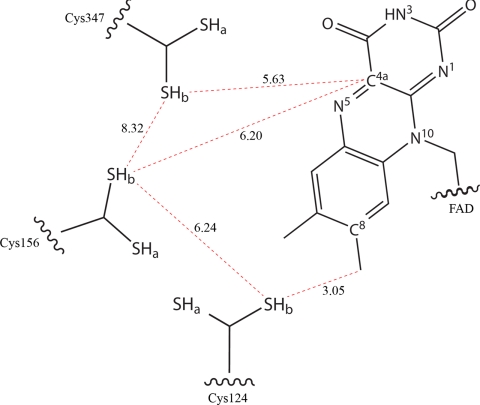
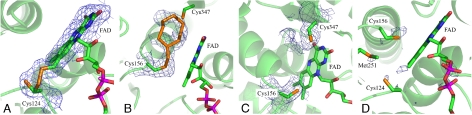
SQR binds FAD in the same position as in other DiSR proteins. The isoalloxazine ring is planar, in contrast to the more bent conformation found in the related FCC structure. Most remarkably, Cys-124 is connected to C8M (the methyl group in position C8 of the FAD isoalloxazine ring), but their distance is too long for a direct covalent linkage (Fig. 4). The connection is provided by a relatively heavy atom, as identified in an anomalous difference map calculated on the basis of a 2.30-Å resolution dataset collected at a wavelength of 1.907 Å (Fig. 5D). We therefore interpreted it as an S atom possibly contributed by the substrate, S2− (see Discussion). Thus, SQR binds FAD through a labile persulfide and not through a stable thioether bond (Fig. 5A).
Fig. 4.
Schematic representation of the distances (red dotted lines) between relevant Cys and FAD atoms in the catalytic site. Distances are in angstroms. The side chains of Cys-124, Cys-156, and Cys-347 are in a double conformation (-SHa/-SHb).
Fig. 5.
Electron densities in the sulfide oxidation site. The protein monomer is in a cartoon semitransparent representation in green. The FAD and the side chains of the relevant residues are in sticks, color-coded according to the atom type (C, green; N, blue; O, red; S, orange; and P, magenta). (A–C) Simulated annealing 2Fo − Fc electron density omit maps drawn at 1.0 σ contour level in blue mesh. (A) The connection between the sulfhydryl group of Cys-124 (chain A) in one conformation and the C8M group of FAD through a putative S atom. (B) The electron density extending from the sulfhydryl group of Cys-156 (chain A) and interpreted as a covalently bound cyclooctasulfur ring. The second S atom of the chain would be trivalent, as explained in the text. (C) The sulfide oxidation site of chain D. An elongated electron density peak is prolonging the sulfhydryl group of Cys-347, whereas an only weaker density is connected to Cys-156. (D) Anomalous difference map around the catalytic site calculated from a dataset collected at 6.5 keV and shown at 3.0 σ contour level in blue mesh. Anomalous peaks are visible for S atoms of cysteines and methionines and for the phosphate groups of the FAD. One additional peak is present between the sulfhydryl group of Cys-124 and the C8M group of FAD, indicating that a relatively heavy atom with residual anomalous scattering at 6.5 keV, possibly S, is present in the structure.
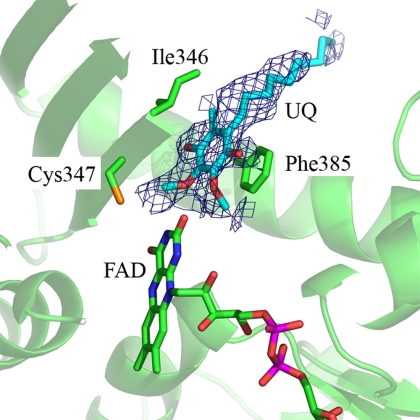
The quinone-binding site is on the si face (19) of the cofactor FAD, and it is accessible from the membrane attachment domain described. Crystals soaked with decylubiquinone are isomorphous to the “as-purified” protein crystals. The quinone-bound structure at 2.0-Å resolution does not show significant domain movements with respect to the structure “as-purified” (overall rmsd = 0.455 Å). The quinone is localized in the 2Fo − Fc-simulated annealing omit map at 0.8 σ (Fig. 6). In the same region, the structure of the protein as-purified shows a density peak of different shape, possibly corresponding to a copurified hydrophobic molecule. In the quinone-bound structure, the binding site is not fully occupied, which correlates with the low affinity (in the micromolar range) measured by kinetic experiments (5). However, it is possible to locate the aromatic ring and the hydrophobic side chain unequivocally. The aromatic ring is bound between the benzene ring of Phe-385 and the side chain of Ile-346 (both are conserved residues among SQRs but not in FCCs). There is a network of solvent molecules and charged amino acids, including Glu-318 and Lys-382 (conserved among SQRs) and Gln-319, Lys-392, and Glu-403 around the benzoquinone ring. The aliphatic side chain extends toward the membrane attachment domain, and it interacts with the hydrophobic side chains of amino acids Met-315, Ile-348, Phe-381, Tyr-388, Phe-402, Val-406, and Leu-407. The position of the quinone-binding site is confirmed also by the structure of the SQR in an inhibitor-bound state, solved at 2.9-Å resolution and also not significantly different from the structure of the protein “as-purified”. Aurachin C, a quinone analogue reported to be a potent competitive inhibitor of A. aeolicus SQR (3), was soaked into the crystals, and corresponding additional electron density was found in the same binding pocket.
Fig. 6.
The quinone-binding site (color code as in Fig. 5). The simulated annealing 2Fo − Fc electron density omit map is shown in blue at 0.8 σ contour level, and a model for the quinone molecule is in light blue sticks.
On the other hand, the re face of FAD and the protein segment Ser-155–Pro-159 delimit a cavity that extends ≈15 Å parallel and 4 Å perpendicular to the isoalloxazine plane. The cavity is accessible from the bulk solvent through a channel characterized by a positive electrostatic surface potential. Four solvent molecules occupy the channel at consecutive sites from the periplasm to Ser-155 (conserved only in type I SQR) and might mimic the trace of the substrate, S2−. At the end of the channel sits Val-294, also conserved only in type I SQR and essential for activity (5) (Fig. S4A). Interestingly, the loop Val-294–Lys-312, delimiting the channel on one side, and the segment Pro-152–Pro-159 opposite the isoalloxazine ring are linked by hydrogen bonds and exhibit a different conformation than in the FCC structure (7) (Fig. S5).
Three cysteines essential for activity (5) face the cavity, as schematically depicted in Fig. 4. First, Cys-124 binds the FAD dimethylbenzyl ring, as described above. Second, Cys-347 is part of a rigid β-sheet on the pyrimidine-2,4-dione side. Finally, Cys-156, localized in the loop Pro-152–Phe-157, is among the 3 cysteines the residue with the highest potential degree of mobility.
The sulfide oxidation pocket is, unexpectedly, not empty in the structure. The density of the SH group of Cys-156 in one conformation extends to form a long chain on one side (Fig. 5B). The chain is oriented toward Cys-347, it is surrounded by conserved hydrophobic amino acids (Phe-194, Val-196, Ile-199, Ala-349, and Phe-358), and it is near the couple Tyr-161–Glu-162 that was proposed to be implicated in proton exchange during the reaction (2, 3). In some monomers (i.e., monomers A and F in the structure of the as-purified protein), the density closes to form a cycle. In some other monomers (i.e., monomer D) the density connected to Cys-156 is shorter, but a chain is also observable connected to Cys-347 (Fig. 5C). We currently have no evidence for the crystallographic and possibly functional reasons for such differences between the densities of the active site of the single monomers. Because polysulfur is known to be the product of the reaction (5) and because sulfur has the property of forming chains, we have interpreted 2Fo − Fc density regions higher than 1 σ with chains of S atoms adjusting the occupancy (30%) to have Fo − Fc difference maps with positive and negative signals lower than 3 σ and B factors close to the average values of the whole protein chain (Boverall = 38 Å2). A few additional electron density peaks still remain unexplained. The geometry of the S chains modeled into the electron density is compatible with that of sulfur species known from inorganic chemistry [see, for instance, the description of the structure of the S8 ring in Rettig and Trotter (20), of the allotropic forms of elemental sulfur in Meyer (21), and of trivalent S atoms in Wong et al. (22)]. The presence of a polysulfur chain is additionally supported by preliminary total X-ray fluorescence (TXRF) experiments, which resulted in an S content of SQR higher than expected from the protein sequence (S. Metz, C. Rittmeyer, B. Kolbesen, personal communication).
Discussion
Membrane Insertion.
Membrane insertion of SQR is functionally necessary to allow the access of the hydrophobic quinone to its binding site, and perhaps also the release of the water-labile polysulfur product to the membrane via an apolar environment (see below).
SQR presumably inserts into the lipid bilayer to a maximal depth of about 12 Å (Fig. 3), which implicates its complete penetration through the lipidic polar head group region (17, 23) and contacts between the exposed hydrophobic side chains of the amphipathic helices and the hydrocarbon core of the membrane. We therefore classify the A. aeolicus SQR as an integral monotopic membrane protein (18). An important determinant for assessing the depth of membrane insertion is the position of several bound sulfate groups in the SQR structure, because sulfate chemically mimics the phosphate head groups of cellular lipids. The observed binding sites of some sulfates between the subunits (i.e., 1 sulfate ion bridges Lys-172 of one monomer and Arg-369 of a neighboring monomer; Fig. S2A) suggest that membrane insertion stabilizes oligomerization and, vice versa, the trimer creates an appropriate surface for binding lipid phosphate groups. The coupling between membrane insertion, trimerization, and enzymatic activity might ensure that SQR exclusively reduces hydrophobic quinones and not other soluble cytoplasmic electron acceptors [i.e., NAD(P)H, as it is the general case for the pyridine-nucleotide:disulfide flavoproteins]. SQR and other monotopic membrane proteins [i.e., the prostaglandin H2 synthase-1 (PDB ID code 1prh; ref. 24) and the squalene-hopene cyclase (PDB ID code 2SQC; ref. 25)] share common features of membrane association, such as characteristic tryptophans, exposed hydrophobic residues, amphipathic segments, and positively charged surface areas. However, the fold of the inserted segments differs substantially. Even more divergent are the membrane insertion motifs of the structurally and functionally related flavoproteins ETF-ubiquinone oxidoreductase (PDB ID code 2gmh; ref. 26) and glycerol-3-phosphate dehydrogenase (PDB ID code 2qcu; ref. 27) (Table S2), suggesting that membrane attachment was developed in a later stage of protein evolution after enzyme differentiation to a specific catalytic activity.
Enzymatic Reaction.
The SQR reaction couples sulfide oxidation with quinone reduction to provide reducing equivalents for redox processes in the membrane. Electron flow between the 2 half-reactions is accomplished via FAD by exploiting its capability to accept an electron pair from a transiently bound sulfide adduct and to donate these 2 electrons one by one to the neighbored quinone via the space.
Both half-reactions are spatially separated. The quinone is located at the si side of FAD and reaches its binding site by “desorption” (28) through a hydrophobic channel connected with the membrane. The quinone-bound SQR structure determines the shortest distance between quinone and FAD of about 3.5 Å, which allows a fast electron transfer. Invariant Glu-218 and water molecules in the binding site are proposed as proton donors to the quinol ring.
Sulfide oxidation takes place on the re side of FAD, and sulfide attains the catalytic site through a hydrophilic channel with the entrance in the periplasm (see Results and Fig. S4A).
SQR, as does FCC, catalyzes a reaction of sulfur polymerization that requires more complicated structural and mechanistic features than for the established reactions of the other DiSR proteins. The chemistry of sulfur in the binding pocket of a protein and—in the physiological state—at the high temperatures at which A. aeolicus grows is complex, and its description certainly requires a deeper investigation. However, the presented structural data of SQR from A. aeolicus provide an insight into this enigmatic multistep process composed of an initiation, an elongation, and a termination step.
A typical reaction scenario established for DiSR flavoproteins involves a redox-active cysteine/cystine couple that is reduced/oxidized by FAD via a Cys-S-C4A-FAD charge-transfer intermediate. This mechanism requires a distance between the sulfur atom of one of the cysteines and the C4A atom in the range of 3–4 Å (13). Surprisingly, in the presented structure of SQR, the key players Cys-156, Cys-347, and the C4A atom of FAD are 5.6–8 Å apart, a distance which is too big for a direct reaction (Fig. 4). Assuming substantial conformational changes, a hypothetical reaction scenario is outlined in Fig. S6A. It would include a Cys-156-S-S-C4A-FAD adduct and the Cys-156-S-S-S-Cys-347 trisulfide bridge proposed by Griesbeck et al. (8) as a central intermediate. The third crucial cysteine, Cys-124, which covalently binds to FAD via a disulfide bridge, would certainly influence the FAD position and its electronic properties (i.e., its redox potential) but would not directly participate in polymerization.
The A. aeolicus SQR structures, however, also suggest an alternative mechanism to convey electron pairs from sulfide to FAD which, of course, also implies a different elongation cycle. The connection between the Cys-124 Sγ and FAD C8M atoms via a labile persulfide bridge inspired us to a fairly speculative reaction scheme (Fig. S6B) that recalls the description (29) of FAD incorporation into flavoproteins that covalently bind their cofactor. In our proposed reaction scheme, one sulfur atom of the persulfide is the Sγ atom of Cys-124, and the other one originates from the substrate, S2−, after reduction of a Cys-124–Cys-156 disulfide resting state. The species Cys-124-S-S− attacks the FAD quinone-methide tautomer to form the “C8M adduct” observed in the structure. After electron transfer, a transient Cys-124-S-S+ state is generated, and the trisulfide bridge (8) is formed by a nucleophilic attack from Cys-156-S− to Cys-124-S-S+. Cys-347 is involved in taking over the growing sulfur chain and pulling it into a hydrophobic pocket, thereby recovering Cys-156-S− for the next reaction cycle. This alternative mechanism attributes a central role to Cys-124, and it is impaired by the fact that Cys-124 is only conserved in type I but not in type II SQR sequences, where it is replaced by a tyrosine. However, the active site and the mechanism of sulfide oxidation appear to be substantially different in type I and type II SQRs. This conclusion is based first on the effect of site-specific exchanges (V294D and C124Y) on the enzyme kinetics and on the mode of FAD binding (see Results and ref. 5), and second on the necessity of type II SQRs to have reaction partners, such as cyanide or thioredoxin and sulfite, to oxidize sulfide efficiently (10).
In both postulated mechanisms, the reaction is spontaneously initiated by sulfide, an oxidized FAD, quinone, and a disulfide bond. The following polymerization cycle is difficult to postulate because in the structure, the distances between Cys-124, Cys-156, and Cys-347 are too long for direct sulfur–sulfur coupling (Fig. 4). It appears attractive to conclude that the distances between the cysteines have been increased stepwise during the elongation process but are significantly reduced in an empty active site. This conclusion is supported by the observed polysulfur chains, visible in all monomers, suggesting that the reaction is trapped in an advanced stage of the cycle. Although the analysis of their B factors does not suggest flexibility, a concerted rotation of loops 294–312 and 152–157 toward a conformation closer to that observed for the equivalent FCC segments (Fig. S5) could be the basis of such movements. However, the Sγ–Sγ distance between Cys-156 and Cys-347 would be reduced to only ≈4.5 Å.
Independently of the mechanism, the termination of the elongation is presumably triggered by the limited space of the active site, which can only provide a catalytic productive environment for up to a 9-S atom chain, which is finally cleaved to an S8 ring, as found in the electron density. The release of the stable S8 ring might proceed via the sulfide channel. However, an alternative hydrophobic channel directly formed through the inner membrane-facing side of the trimer (only closed by the contacting side chains of surface residues Leu-209 and Met-374) appears to be attractive (Fig. S4B). By using this exit, the apolar and water-unstable S8 chain would move directly toward the membrane interior, and it could finally be incorporated into sulfur globules. In A. aeolicus, these are accumulated in the cytoplasm (30), so that despite being a monotopic and not a transmembrane protein, SQR would participate in cellular uptake of sulfur in an efficient manner (Fig. S7).
In conclusion, we could isolate an active SQR from the membranes of A. aeolicus in a state that seems to form a covalent Cys-124-S-S-C8M-FAD adduct and to harbor an elongating, covalently bound polysulfur chain in the active site. The quinone-binding site contains an unknown molecule in the as-purified enzyme that can be replaced by a quinone in the crystalline state. The reaction of SQR as a sulfur polymerase implies a complicated, multicycle mechanism of action. Our data allow drawing of an approximate picture of this fascinating reaction, and they stimulate further genetic, biochemical, and structural investigations.
Materials and Methods
Purification, Crystallization, and Heavy Atom Soaking.
A. aeolicus cells were obtained from the Archaeenzentrum (Regensburg, Germany). Membranes were prepared and solubilized as described previously (31). The supernatant was fractionated by anionic affinity chromatography onto a Mono Q HR 10/10 column (Amersham Biosciences) in 20 mM Tris·HCl buffer, pH 7.4; 0.05% sodium azide; 0.05% DDM; and a linear gradient of NaCl. The fractions containing the SQR were concentrated and separated further onto a TSK 4000 (Tosoh) in the same buffer with 150 mM NaCl. After desalting, the detergent was first exchanged to Zwittergent 3-10 to increase protein purity, and then shifted back to 0.05% DDM, which is more suitable for 3D crystallization. A TSK 4000 size-exclusion column was finally used as a polishing and desalting step. The homogeneous protein solution was concentrated and mixed with an equimolar amount of commercial FAD dissolved in water to a final protein concentration of 8–10 mg/mL. The protein was crystallized by the hanging drop vapor diffusion method at 18 °C in the condition JB6_D3 (Jena Bioscience classic screen 6), which was then refined to a solution containing 0.1 M buffer (sodium MES, pH 6.5 for the “as-purified” protein crystal, and sodium acetate, pH 5.6 for the crystals soaked with quinone analogues); 2.0 M ammonium sulfate; and 4% vol/vol PEG 400. The crystals started growing after 3–6 weeks and reached their full size within 1–2 days. The crystals were flash-cooled under a cryojet after washing in a cryoprotecting solution with 0.1 M buffer and 40% vol/vol PEG 400. The phases were obtained by the MIRAS method using Os and Au derivatives. After crystal growth, a 10-fold concentrated aqueous solution of K2OsCl6 or AuCl3 (Hampton Research heavy atom screens) was added to the crystallization drop to a final heavy atom concentration of 5 mM. After 24 h, the crystals were washed in the cryobuffer and frozen. A decylubiquinone stock in 100% ethanol was stored at −20 °C. A minimal amount was mixed homogeneously with 100% PEG 400 and diluted with water and sodium acetate to a final concentration of 0.1 M sodium acetate, pH 5.6; 40% vol/vol PEG 400; and 1 mM decylubiquinone. The crystals were soaked for 24 h in this solution and directly frozen. Similarly, crystals were soaked with 1 mM aurachin C (methanol stock; the compound was a kind gift from the Helmholtz Zentrum für Infektionsforschung (Braunschweig, Germany). All datasets were collected at the Swiss Light Source, beamlines X10SA and X06SA (Villigen, Switzerland).
Structure Determination.
SQR formed needle crystals of a size of typically 0.02 × 0.05 × 0.2 mm3 in the space group P212121, with unit cell dimensions of 112 × 154 × 178 Å3, a solvent content of 53%, and 6 molecules in the asymmetric unit. All datasets were reduced and scaled by using the XDS suite (32). Data collection and processing statistics are reported in Table S1. Heavy atom positions were determined with SHELX (33) and refined with SHARP (34) to obtain initial phases. Solvent flattening was performed with the software DM (35), and NCS averaging was performed in DM by using the NSC matrix derived from LSQ in the program O (36) and refined by IMP (37). An initial model was obtained with the automated program ARP/wARP (38), and subsequent model building was done manually in Coot (39). The initial model had an Rwork of 47% and an Rfree of 50%. Initial refinement was performed with CNS (40) to an Rfree of 30%. Further refinement was done with Refmac5 and by using TLS and imposing NCS restraints on the protein chains (41, 42). Final rounds of refinement were done without NCS restraints. The datasets of the decylubiquinone- and aurachin C-soaked crystals were directly refined with Refmac5 without the need for a molecular replacement run. Solvent, cofactor, and substrate molecules were added by using Coot and Refmac5. Omit maps were calculated by using CNS, excluding the atoms of interest, and avoiding model bias by simulated annealing refinement.
Supplementary Material
Acknowledgments.
We thank Tanja Hedderich for excellent technical assistance, Dr. Elena Olkhova for help in the calculation of the electrostatic surface potential, Dr. Eberhard Warkentin for discussion and help in refinement, Helga Volk and Paolo Lastrico for help in preparing the figures, the scientists at the macromolecular crystallography beamlines X10SA and X06SA at the Swiss Light Source for assistance during data collection, and the Archaeenzentrum (Regensburg, Germany), where A. aeolicus cells were grown. Aurachin C was a kind gift from the Helmholtz Zentrum für Infektionsforschung (Braunschweig, Germany). This work was supported by the Deutsche Forschungsgemeinschaft (SFB 472), the Fonds der Chemischen Industrie, the Max Planck Gesellschaft, the Cluster of Excellence “Macromolecular Complexes” (Frankfurt am Main, Germany), and the International Max Planck Research School on Structure and Function of Biological Membranes (Frankfurt am Main, Germany).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3H27 (“as-purified” protein), 3H28 (protein in complex with decylubiquinone), and 3H29 (protein in complex with aurachin C)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0904165106/DCSupplemental.
References
- 1.Kimura H. Hydrogen sulfide as a neuromodulator. Mol Neurobiol. 2002;26:13–19. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- 2.Shahak Y, Hauska G. Sulfide oxidation from cyanobacteria to humans: Sulfide-quinone oxidoreductase (SQR) In: Hell R, Dahl C, Knaff DB, Leustek T, editors. Advances in Photosynthesis and Respiration. Vol 27. Heidelberg: Springer; 2008. pp. 319–335. [Google Scholar]
- 3.Nübel T, Klughammer C, Huber R, Hauska G, Schütz M. Sulfide:quinone oxidoreductase in membranes of the hyperthermophilic bacterium Aquifex aeolicus (VF5) Arch Microbiol. 2000;173:233–244. doi: 10.1007/s002030000135. [DOI] [PubMed] [Google Scholar]
- 4.Wang R. Two's company, three's a crowd: Can H2S be the third endogenous gaseous transmitter? FASEB J. 2002;16:1792–1798. doi: 10.1096/fj.02-0211hyp. [DOI] [PubMed] [Google Scholar]
- 5.Griesbeck C, et al. Mechanism of sulfide-quinone reductase investigated using site-directed mutagenesis and sulfur analysis. Biochemistry. 2002;41:11552–11565. doi: 10.1021/bi026032b. [DOI] [PubMed] [Google Scholar]
- 6.Theissen U, Hoffmeister M, Grieshaber M, Martin W. Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol Biol Evol. 2003;20:1564–1574. doi: 10.1093/molbev/msg174. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZW, et al. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 8.Griesbeck C, Hauska G, Schütz M. Biological sulfide oxidation: Sulfide-quinone reductase (SQR), the primary reaction. In: Pandalai SG, editor. Recent Research Developments in Microbiology. Vol 4. Trivadrum, India: Research Signpost; 2000. pp. 179–203. [Google Scholar]
- 9.Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 10.Theissen U, Martin W. Sulfide:quinone oxidoreductase (SQR) from the lugworm Arenicola marina shows cyanide- and thioredoxin-dependent activity. FEBS J. 2008;275:1131–1139. doi: 10.1111/j.1742-4658.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 11.Vande Weghe JG, Ow DW. A fission yeast gene for mitochondrial sulfide oxidation. J Biol Chem. 1999;274:13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]
- 12.Deckert G, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: Advances in chemistry and function. Prog Nucleic Acid Res Mol Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 14.Schödl T. Germany: Universität Regensburg, Regensburg; 2003. Sulfid-Chinon Reductase (SQR) aus Aquifex aeolicus: Gensynthese, Expression, Reinigung und biochemische Characterisierung. Sulfide-quinone reductase (SQR) from Aquifex aeolicus: Gene synthesis, expression, purification and biochemical characterization (Translated from German) PhD thesis. [Google Scholar]
- 15.Schütz M, Maldener I, Griesbeck C, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: Requirement for growth, periplasmic localization, and extension of gene sequence analysis. J Bacteriol. 1999;181:6516–6523. doi: 10.1128/jb.181.20.6516-6523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JE, Cornell RB. Amphitropic proteins: Regulation by reversible membrane interactions. Mol Membr Biol. 1999;16:217–235. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- 18.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct Biol. 2007;7:44. doi: 10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNaught AD, Wilkinson A. IUPAC Compendium of Chemical Terminology (Golden Book) 2nd Ed. Cambridge, UK: Blackwell Synergy; 1997. [Google Scholar]
- 20.Rettig SJ, Trotter J. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 21.Meyer B. Solid allotropes of sulfur. Chem Rev. 1964;64:429–451. [Google Scholar]
- 22.Wong MW, Steudel Y, Steudel R. Novel species for the sulfur zoo: Isomers of S-8. Chem Phys Lett. 2002;364:387–392. [Google Scholar]
- 23.White SH, Wimley WC. Peptides in lipid bilayers: Structural and thermodynamic basis for partitioning and folding. Curr Opin Struct Biol. 1994;4:79–86. [Google Scholar]
- 24.Picot D, Loll PJ, Garavito RM. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 25.Wendt KU, Lenhart A, Schulz GE. The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J Mol Biol. 1999;286:175–187. doi: 10.1006/jmbi.1998.2470. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Frerman FE, Kim JJ. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci USA. 2006;103:16212–16217. doi: 10.1073/pnas.0604567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci USA. 2008;105:3280–3285. doi: 10.1073/pnas.0712331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forneris F, Mattevi A. Enzymes without borders: Mobilizing substrates, delivering products. Science. 2008;321:213–216. doi: 10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
- 29.Edmondson DE, Newton-Vinson P. The covalent FAD of monoamine oxidase: Structural and functional role and mechanism of the flavinylation reaction. Antioxid Redox Signal. 2001;3:789–806. doi: 10.1089/15230860152664984. [DOI] [PubMed] [Google Scholar]
- 30.Guiral M, et al. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem. 2005;280:42004–42015. doi: 10.1074/jbc.M508034200. [DOI] [PubMed] [Google Scholar]
- 31.Peng G, et al. Isolation, characterization and electron microscopic single particle analysis of the NADH:ubiquinone oxidoreductase (complex I) from the hyperthermophilic eubacterium Aquifex aeolicus. Biochemistry. 2003;42:3032–3039. doi: 10.1021/bi026876v. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 33.Sheldrick GM, Hauptman HA, Weeks CM, Miller R, Uson I. In: Ab initio phasing. International Tables for Crystallography. Rossmann MG, Arnold E, editors. Vol F. The Netherlands: Kluwer Academic, Dordrecht; 2001. pp. 333–351. [Google Scholar]
- 34.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multi-wavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 35.Cowtan K. DM: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 36.Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt GJ, Jones TA. Halloween masks and bones. In: Bailey S, Hubbard R, Waller R, editors. From First Map to Final Model. Warrington, UK: SERC Daresbury Laboratory; 1994. pp. 59–66. [Google Scholar]
- 38.Lamzin VS, Perrakis A, Wilson KS. The ARP/wARP suite for automated construction and refinement of protein models. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. Vol F. The Netherlands: Kluwer Academic, Dordrecht; 2001. pp. 720–722. [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.