Abstract
Both plants and animals require the activity of proteins containing nucleotide binding (NB) domain and leucine-rich repeat (LRR) domains for proper immune system function. NB-LRR proteins in plants (NLR proteins in animals) also require conserved regulation via the proteins SGT1 and cytosolic HSP90. RAR1, a protein specifically required for plant innate immunity, interacts with SGT1 and HSP90 to maintain proper NB-LRR protein steady-state levels. Here, we present the identification and characterization of specific mutations in Arabidopsis HSP90.2 that suppress all known phenotypes of rar1. These mutations are unique with respect to the many mutant alleles of HSP90 identified in all systems in that they can bypass the requirement for a cochaperone and result in the recovery of client protein accumulation and function. Additionally, these mutations separate HSP90 ATP hydrolysis from HSP90 function in client protein folding and/or accumulation. By recapitulating the activity of RAR1, these novel hsp90 alleles allow us to propose that RAR1 regulates the physical open–close cycling of a known “lid structure” that is used as a dynamic regulatory HSP90 mechanism. Thus, in rar1, lid cycling is locked into a conformation favoring NB-LRR client degradation, likely via SGT1 and the proteasome.
Keywords: innate immunity, Pseudomonas syringae, SGT1, STAND ATPase protein
Plants have evolved a highly complex immune system centered on pathogen recognition via the evolutionarily-conserved NB-LRR proteins. Pathogen-triggered activation of NB-LRR proteins leads to several responses, including cell wall strengthening, transcriptional reprogramming, and a form of programmed cell death termed the hypersensitive response (HR). Because their function often results in cell death, proper maintenance of NB-LRR protein levels and activation state are vital to the health of the plant (1).
NB-LRR proteins can be divided into 2 structural subgroups based on the presence of either a likely coiled-coil (CC) or Toll interleukin-1 receptor (TIR) domain at their N termini. Either of these N-terminal domains is followed in both subgroups by a middle nucleotide binding (NB) site and a C-terminal leucine-rich repeat (LRR). This general structure is not only conserved across all plants but extends to NOD/Caterpiller/NLR proteins that mediate various processes in mammalian innate immunity (2).
Just as the domain composition of these intracellular receptors is conserved from plants to animals, so is the regulation of their steady-state accumulation. Cytosolic HSP90 and the cochaperone SGT1 have been previously demonstrated to not only be important for regulation of NB-LRR proteins in plants, but also in regulation of NLR function in animals (3). A third protein called RAR1 appears to play a role in innate immunity specifically in plants (4).
All 3 of these proteins can independently interact with one another; the CS domain of SGT1b, or the CHORDI domain of RAR1, can interact with the N-terminal ATPase domain of HSP90; the CHORDII domain of RAR1 also interacts with the CS domain of SGT1 (5). The interaction of SGT1 with HSP90 has been shown to be required for SGT1 function (6). Mutation of SGT1 can suppress rar1 for some NB-LRR functions, but not all (7). However, the relationship between RAR1 and HSP90 is less understood.
We present and characterize specific missense alleles of HSP90.2 in the reference plant, Arabidopsis, that suppress rar1. These hsp90.2 alleles are uniquely interesting in that they can bypass the requirement for a cochaperone and result in recovery of client protein accumulation and function.
We used genetic and biochemical analyses to demonstrate that these hsp90.2 mutant proteins act on NB-LRR proteins affected by rar1, suppressing all identified rar1 phenotypes. We further show that these mutations are functionally distinct from previously-identified hsp90.2 mutations (8), including a null allele. These specific missense changes in hsp90.2 enable a separation of HSP90 ATP hydrolysis activity and HSP90 function in client protein accumulation. By recapitulating the activity of RAR1 in its absence, the phenotypes of these hsp90.2 mutants strongly suggest that RAR1 physically enhances the transition state of HSP90 as it moves from a “lid open” ADP-bound conformation to a “lid closed” ATP-bound conformation.
Results
Identification of Alleles of RAR1 and HSP90.
To identify new genes required for RPM1 function in Arabidopsis, we performed 2 genetic screens. Both took advantage of sensitized genetic backgrounds. The first was a modification of a previous screen (9), using a β-estradiol-inducible copy of the avrRpm1 bacterial type III effector gene whose product is recognized in Arabidopsis by the RPM1 NB-LRR protein (Fig. S1A). Given the very high recovery ratio of rpm1 alleles compared with second-site loci isolated previously (9), we modified the screen by crossing into this background a well-characterized transgenic, myc-epitope tagged copy of RPM1 expressed from the native promoter (Fig. S1A and ref. 10). Approximately 1 million M2 plants were screened from 200 ethyl methanesulfonate (EMS)-mutagenized seed lots. Putative surviving mutants were then assayed for loss of disease resistance in response to pathogen-delivered AvrRpm1 to eliminate mutations in the estradiol-inducible system (see Materials and Methods).
Various candidate genes previously implicated in RPM1 function were then sequenced in the remaining putative mutants. They included the endogenous copy of RPM1, the transgenic copy of RPM1-myc, RAR1, and all 4 genes encoding Arabidopsis cytosolic HSP90 (8–11). Four mutations in RAR1 and 1 allele of HSP90.2 were found, hsp90.2–6 (Fig. S2). The rar1 alleles are consistent with previous mutations: premature stops, splicing defects, and disruption of zinc-coordinating residues (12, 13). The allele of hsp90.2 displayed intermediate susceptibility and full penetrance, as previously found for hsp90.2–1 and hsp90.2–3 (8). Mutations were not found in these loci in the remaining mutants.
The second screen was a rar1 suppressor screen, aimed at identifying loci that would restore the loss of NB-LRR protein accumulation, and the consequent loss of NB-LRR function, that are the principal rar1 phenotypes (13). Approximately 200,000 EMS-mutagenized M2 individuals from 50 M1 seed lots of rar1–21 were spray-inoculated with Pto DC3000(avrPphB) (see Materials and Methods). This strain is recognized in Arabidopsis by the RPS5 NB-LRR protein (14). We used RPS5 as the read-out in this screen because rar1 exhibits a strong and uniform disease susceptibility phenotype to Pto DC3000(avrPphB). We reasoned that a suppressor would be obviously disease resistant against this susceptible background.
We identified 5 independent second-site mutants defining 3 loci in this screen. Although hsp90.2 has previously been shown to have no effect on RPS5 (8), 2 of the mutants are missense mutations in the hsp90.2 gene based on map-based cloning and subsequent sequencing of both mutant alleles (Fig. S2B). The other 2 loci will be discussed elsewhere. To avoid confusion, we will henceforth refer to hsp90.2 alleles that lose RPM1 function by the original notation, lra (loss of recognition of avrRpm1) (13), and alleles that suppress rar1 as rsp (rar1 suppressor). Like all of the lra alleles, hsp90.2–7rsp is completely recessive. However, based on disease symptoms after bacterial inoculation, hsp90.2–8rsp behaves as a weak semidominant allele (see Materials and Methods).
hsp90.2rsp Alleles Suppress all Known rar1 Phenotypes.
Inexplicably, hsp90.2lra alleles were previously shown to specifically impact RPM1 function, and not the function of other tested NB-LRR proteins (8). Conversely, rar1 affects the steady-state accumulation of all tested NB-LRR proteins, and the function of many, by lowering their accumulation below a functional threshold (7, 15). Hence, we did not expect to identify hsp90.2 alleles in our RPS5-based rar1 suppressor screen. We determined whether the rsp alleles suppressed rar1 phenotypes of other NB-LRR-dependent disease resistance specificities. The hsp90.2rsp alleles variably suppressed rar1 with respect to RPS5 function (Fig. 1A), RPM1 function (Fig. 1B), and RPS2 function (Fig. 1C).
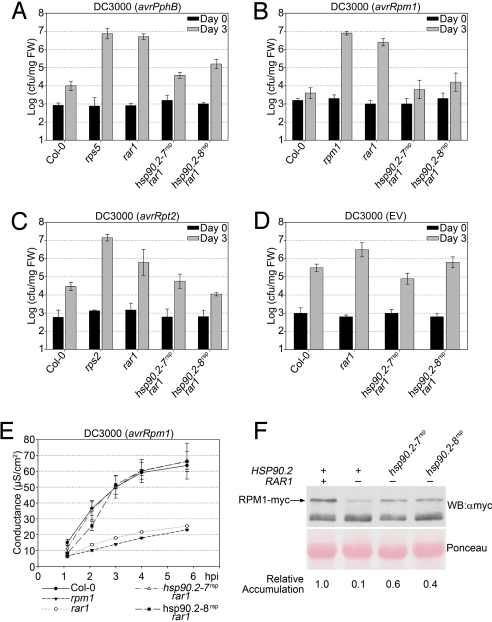
Fig. 1.
rsp alleles suppress all known rar1 phenotypes. (A–D) Bacterial growth assays comparing rar1 mutants to hsp90.2rsp rar1 double mutants. Note the logarithmic scale. hsp90.2rsp mutants suppress rar1 phenotypes for disease resistance mediated by RPS5 (A), RPM1 (B), and RPS2 (C). (D) hsp90.2rsp mutants suppress the rar1 phenotype of decreased basal resistance to Pto DC3000 (7). (E) hsp90.2rsp alleles suppress the rar1 phenotype of loss of RPM1-mediated HR. An increase in conductivity is indicative of the release of ions from cells undergoing HR. (F) hsp90.2rsp alleles suppress the rar1 phenotype of lowered steady-state accumulation of RPM1-myc protein.
rar1 mutants express decreased basal disease resistance to the virulent pathogen Pto DC3000 (7). The only molecular phenotype ever ascribed to rar1 is diminution of steady-state NB-LRR protein accumulation as noted above. Thus, this phenotype suggests an as-yet-undocumented role for RAR1 on NB-LRR proteins that might either function in basal defense and/or weak recognition of the type III effectors delivered by Pto DC3000 (16). Notably, both hsp90.2rsp alleles also suppress this phenotype (Fig. 1D).
We used the release of ions into solution by inoculated plant leaf discs to measure the ability of hsp90.2rsp to suppress the loss of HR associated with rar1 (17). Although neither rpm1 nor rar1 are able to generate an HR upon delivery of AvrRpm1, rar1 hsp90.2rsp double mutants display the same level of HR as wild-type Col-0 plants (Fig. 1E). The suppression of this particular rar1 phenotype is in marked contrast to results obtained with sgt1b as a rar1 suppressor (7).
We next assayed whether the hsp90.2rsp alleles were able to suppress the most direct rar1 mutant phenotype, a decrease in NB-LRR protein accumulation (13). We introgressed a transgenic RPM1-myc-epitope-tagged derivative driven from its native promoter (10) into each hsp90.2rsp rar1 mutant. In these double mutants, the hsp90.2rsp alleles suppressed the very low RPM1 accumulation observed in rar1 (Fig. 1F). Hence, the hsp90.2rsp alleles suppress the key biochemical phenotype of rar1, at least with respect to RPM1 and probably more generally, given the pathology data presented in Fig. 1.
Because RPM1, RPS2, and RPS5 are CC-NB-LRR proteins, we addressed whether a RAR1-dependent, TIR-NB-LRR protein is also suppressed by the hsp90.2rsp alleles. RPP4 conditions disease resistance to the oomycete pathogen, Hyaloperonospora arabidopsidis (Hpa) (18). In this case, the HR is likely to be required for disease resistance, whereas it is likely to be dispensable for resistance to bacterial pathogens. We noted that both hsp90.2rsp rar1 lines expressed higher RPP4 function than rar1 (Fig. S3A) and exhibited higher levels of HR (Fig. S3B). Hence, the hsp90.2rsp alleles also suppress rar1 for a TIR-NB-LRR and in a context where HR is likely to be the key mechanism of disease resistance.
The possibility existed that the recovery of disease resistance observed in hsp90.2rsp rar1 double mutants is not specific, but rather a result of general metabolic perturbation resulting in disease resistance. Such perturbations are typically accompanied by an increase in the levels of defense marker proteins such as PR-1 (19). However, we did not observe an obvious increase in PR-1 levels in hsp90.2rsp lines.
We previously demonstrated that a presumed truncated protein product made by the rar1–21 allele used as the parent in this screen, which would express only CHORD-I, is, surprisingly, able to coimmunoprecipitate HSP90. This coimmunoprecipitation was not observed with the rar1–20 null allele or the W47stop allele, rar1–28 (ref. 8 and Y. Belkhadir, personal communication). We constructed rar1–28 hsp90.2rsp combinations for both rsp alleles. We assayed for RPM1 function to rule out the possibility of a rar1 allele-specific effect. We clearly observed suppression of the rar1 phenotype in rar1–28 hsp90.2rsp double mutants; hence the effect of the rsp alleles on RPM1 function is not rar1–21 allele specific.
hsp90.2rsp Alleles Have No Phenotype in RAR1.
We isolated the hsp90.2rsp single mutants by backcrossing and assayed them for RPM1 function. As seen in Fig. 2A, the hsp90.2rsp single mutants express wild-type RPM1 function. Hence, the hsp90.2rsp single mutants are phenotypically distinct from the hsp90.2lra alleles (8), which all express partial loss of RPM1-mediated disease resistance. We also monitored HR in the hsp90.2rsp single mutants. hsp90.2rsp alleles again expressed wild-type phenotypes (Fig. 2B).
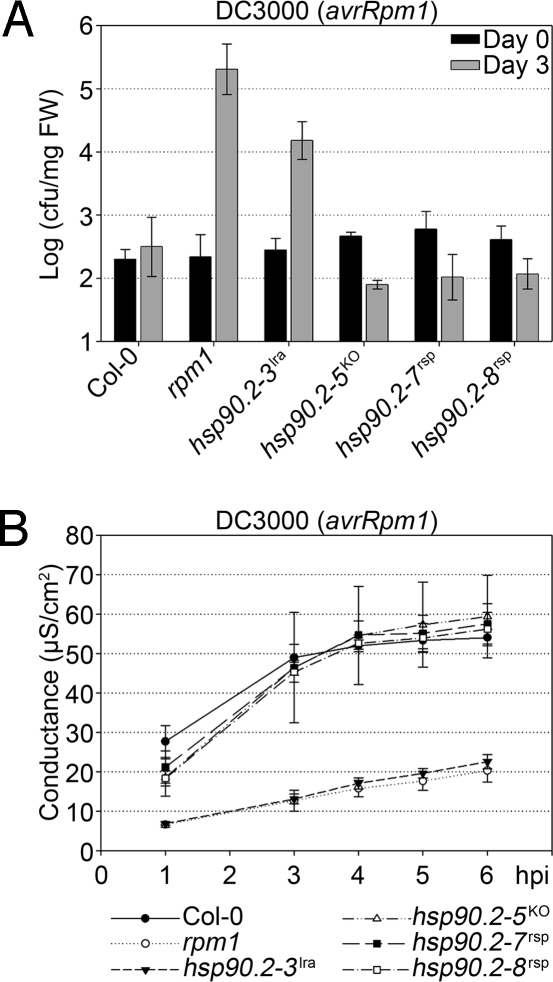
Fig. 2.
hsp90.2rsp mutants are phenotypically distinct from an hsp90.2lra single mutant and an hsp90.2 T-DNA insertion null mutant. hsp90.2rsp, hsp90.2lra, and hsp90.2KO single mutant plants are compared with each other and Col-0 and rpm1 plants. (A) Bacterial growth assay for recognition of Pto DC3000(avrRpm1) by RPM1. (B) Conductivity assay measuring the HR triggered by RPM1 activation after recognition of AvrRpm1.
The HSP90.2rsp proteins might counterbalance the decrease in NB-LRR protein accumulation observed in rar1 by conditioning “hyperaccumulation” above wild-type levels. We thus introgressed RPM1-myc into each hsp90.2rsp mutant and assayed for RPM1-myc protein accumulation. We detected wild-type RPM1-myc protein levels in the single mutant lines. We conclude that there is no increased NB-LRR protein activity indicative of “hyperchaperoning” by the hsp90.2rsp alleles. Thus, these alleles are true suppressors of the loss of RAR1 molecular activity and are not merely overcoming the rar1 phenotype by increased overall expression of client protein.
hsp90.2lra and hsp90.2KO Alleles Do Not Suppress rar1.
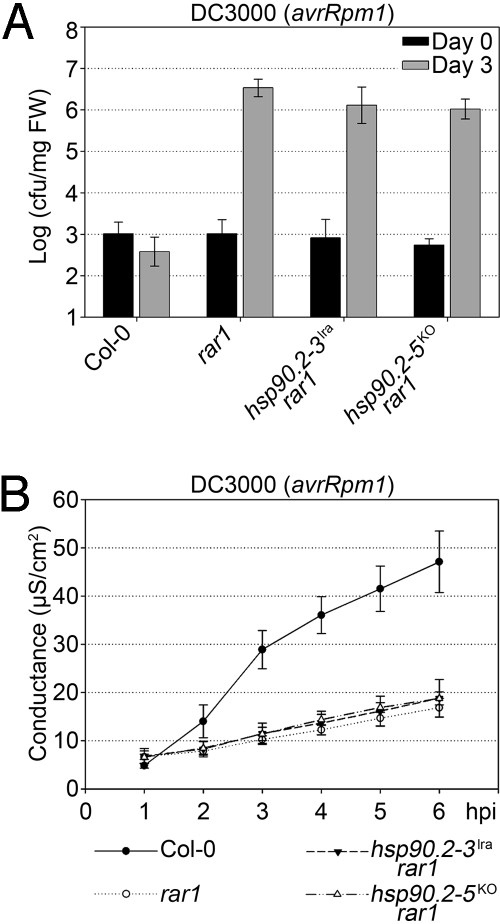
We next tested whether either hsp90.2KO or a reference hsp90.2–3lra allele (encoding D80N, a mutation analogous to the well-studied yeast D79N; ref. 20) could suppress rar1 for RPM1 function. We constructed the appropriate double mutants and noted that both were as susceptible to Pto DC3000(avrRpm1) as rar1 (Fig. 3A). Conductivity measurements of RPM1-mediated HR in these double mutants (Fig. 3B) gave similar results, supporting the conclusion that neither hsp90.2KO nor an hsp90.2–3lra allele can suppress rar1. Thus, the hsp90.2rsp alleles are also phenotypically distinct from both a null allele (Fig. 3) and the classic ATPase dead hsp90.2–3lra (Figs. 2 and 3).
Fig. 3.
Neither an hsp90.2lra allele nor the hsp90.2KO null allele suppress rar1. (A) Bacterial growth assay measuring disease resistance to Pto DC3000 (avrRpm1) mediated by RPM1. Wild-type Col-0 and rar1 mutant plants are compared with hsp90.2–3lra rar1 and hsp90.2KO rar1 double mutants. (B) Conductivity assay measuring the HR to Pto DC3000 (avrRpm1).
hsp90.2rsp Alleles Are Not Null Alleles.
Although both hsp90.2rsp alleles encode missense changes, there remained a possibility that they are functionally null. If so, then 1 of the 3 remaining cytosolic HSP90 proteins might compensate for the loss of HSP90.2 in these alleles, as previously noted for hsp90.2KO (8). Arabidopsis has 4 genes encoding cytosolic HSP90, 3 of which, including HSP90.2, reside in a cluster. The fourth, HSP90.1, lies ≈1.3 Mbp away on the same chromosome. We wanted to establish whether stepwise elimination of HSP90 function would reveal null phenotypes, which we could then compare with hsp90.2–3lra and hsp90.2–7rsp alleles. We were unable to recover hsp90.2–5KO hsp90.1KO double mutants. However, we did identify plants homozygous for hsp90.1KO and heterozygous for hsp90.2–5KO. These were stunted and expressed high accumulation of anthocyanins, loss of apical dominance, and very low fecundity. Selfed progeny segregated lethals. Hence, HSP90.1 is synthetically lethal with HSP90.2, suggesting that the overall level of cytosolic HSP90 has a minimum threshold for viability. Importantly, we were able to obtain hsp90.2–3lra hsp90.1KO, hsp90.2–7rsp hsp90.1KO, and hsp90.2–8rsp hsp90.1KO double mutants. These were viable and as healthy as either single mutant. Hence, hsp90.2–3lra, hsp90.2–7rsp, and hsp90.2–8rsp maintained the HSP90 activity required to support proper growth and development in the absence of HSP90.1.
We thus conclude that none of the tested rsp or lra alleles are null for HSP90 activity. Our collected genetic data strongly suggest that the hsp90.2rsp alleles are active, and that they recapitulate the molecular activity of RAR1 on client NB-LRR accumulation.
Analysis of Interactions Between hsp90.2 Mutant Proteins with RAR1 and SGT1.
Given the correlation between the region of HSP90 mutated in both of our genetic screens and the region of HSP90 that physically interacts with RAR1 and SGT1 (see Introduction), we were interested in finding out whether our HSP90 mutants were affected in their ability to interact with RAR1 and SGT1 in the yeast 2-hybrid system. As shown in Fig. 4, wild-type HSP90.2 can interact with both RAR1 and SGT1b, but not SGT1a, which is consistent with previously-published coimmunoprecipitation experiments (8). Hence, this system is likely to accurately reflect in vivo interactions in Arabidopsis.
Fig. 4.
Interactions between hsp90.2 mutant proteins and RAR1 or SGT1b does not correlate with phenotype. (A–C) β-Galactosidase assay quantification of the results of yeast 2-hybrid interaction measurements between HSP90.2 and mutant variants with RAR1 (A), SGT1a (B), or SGT1b (C). (D) HSP90.2 and mutant variants accumulate to equivalent levels in yeast as measured by Western blot. RAR1 interacts normally with SGT1a in this assay.
We found that all 4 of the hsp90.2lra proteins lost interaction with SGT1b in yeast 2-hybrid (Fig. 4C). Three lost interaction with RAR1 (Fig. 4A). The exception was S100F (hsp90.2–2lra), previously noted (8) to be partially penetrant, which maintained a strong interaction with RAR1. The 2 rsp mutant proteins exhibited opposing RAR1 and SGT1b interactions. The A11T (hsp90.2–7rsp) protein maintained strong interactions with both RAR1 and SGT1b. However, the R337C (hsp90.2–8rsp) protein lost the ability to interact with both RAR1 and SGT1b. None interacted with SGT1a (Fig. 4B), indicating that this protein is likely to be irrelevant to HSP90.2 function. Western blot analysis showed that all mutant proteins were expressed equally well in yeast (Fig. 4D). The loss of interaction between R337C (hsp90.2–8rsp) and both RAR1 and SGT1b suggests that these interactions are not necessary for restoration of NB-LRR function in this allele. This result is consistent with restoration of several different RAR1-dependent NB-LRR functions in rar1 sgtb double mutants (7).
We were unable to observe an interaction between HSP90.2 and any tested fragment of RPM1 by yeast 2-hybrid analysis. However, we did see a strong interaction between GST-HSP90 fusion purified from Escherichia coli and an HA epitope-tagged version of RPM1 produced via in vitro transcription and translation in wheat germ lysates (21). Using this system, we did not observe any difference in the ability of lra or rsp mutant HSP90.2 proteins to interact with RPM1. Hence, it is unlikely that an overall change in NB-LRR protein interaction with HSP90 causes the various hsp90.2 mutant phenotypes.
ATPase Activity Is Not Predictive of HSP90 Activity in NB-LRR Function.
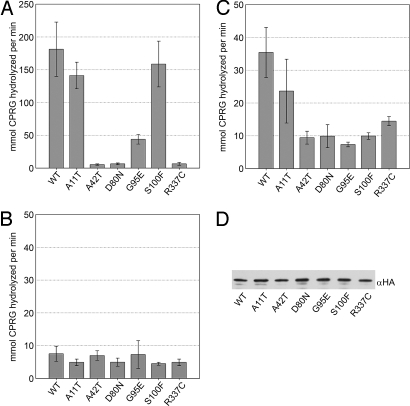
All hsp90.2 missense alleles obtained from our 2 screens were either located in the ATPase domain itself or in the case of R337Crsp in a part of the middle domain physically adjacent to the ATPase domain in the HSP90 crystal structure (Fig. S4). Thus, differences in ATPase activity associated with the N-terminal HSP90 domain (22) might also explain the different properties of the mutant HSP90.2 proteins. We purified recombinant wild-type HSP90.2 and all of the lra and rsp variants (see Materials and Methods). Circular dichroism analyses of the purified proteins showed that all variants had an equivalent proportion of α-helix and β-sheet indicative of proper folding. The ability of these proteins to hydrolyze ATP was measured in an ATP-regenerating system (see Materials and Methods).
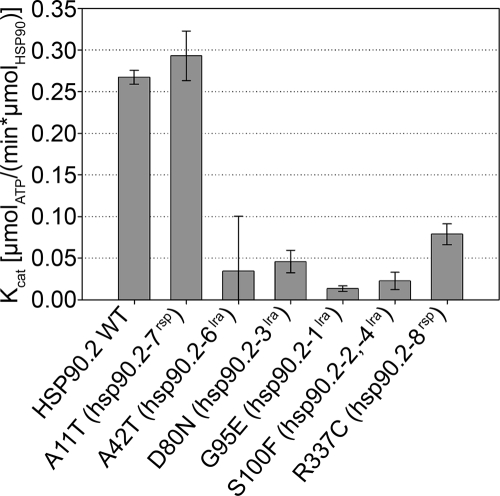
Nearly all of the hsp90.2–8lra alleles are missense changes in amino acids that contact bound nucleotide in the crystal structure, and D80N (hsp90.2–3lra; D79N in ScHSP90) loses ATP hydrolysis. Hence, it was unsurprising that these proteins lacked ATPase activity (Fig. 5). R337C (hsp90.2–8rsp) expressed only very weak ATPase activity (≈2-fold above negative control). Surprisingly, A11T (hsp90.2–7rsp), maintained full ATPase activity, but with a ≈5-fold increase in the observed KM [wild type = 0.04 μM ± 0.01; A11T (hsp90.2–7rsp) = 0.20 μM ± 0.05]. However, plant cytosolic ATP concentrations were ≈3 mM (23), suggesting that the change in KM is probably not relevant to the phenotype exhibited by the mutant. Addition of RAR1 and/or SGT1b to these assays did not alter ATPase activity; this negative result may merely mean that we lack other required conditions and/or components for in vitro reconstruction.
Fig. 5.
HSP90 ATPase activity does not predict hsp90.2 mutant phenotype. In vitro ATPase activity of HSP90.2 and mutant variants with a range of ATP concentrations was used to determine the Kcat. HSP90 concentration was 5 μM, and ATP concentrations ranged between 0 and 1.2 mM (see Materials and Methods).
N-Terminal Dimerization Is Retained in hsp90.2rsp Proteins.
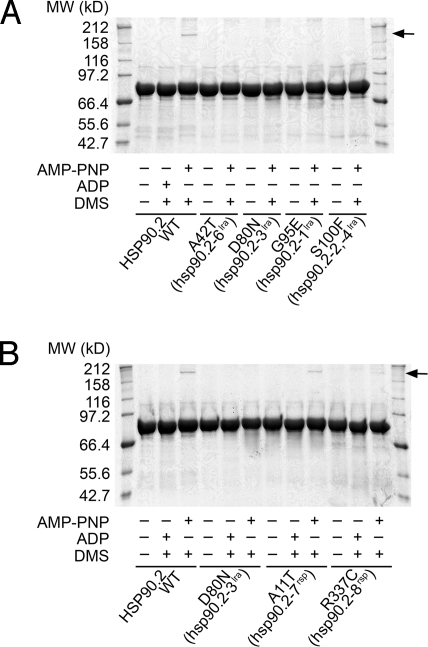
Yeast HSP90 functions as a dimer formed via separate N-terminal and C-terminal dimerization domains. In yeast, N-terminal dimerization is mediated by a short-N-terminal stretch of each monomer (Fig. S4) and requires ATP binding (24). Using the same assay, we found that dimerization of full-length Arabidopsis HSP90.2 depended on the presence of the nonhydrolyzable ATP analogue, adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP). We did not observe HSP90.2 dimers in the absence of AMP-PNP or the presence of ADP. We also tested purified lra and rsp HSP90 mutant proteins for dimerization. The lra mutant variants were unable to dimerize in the presence of any tested nucleotide (Fig. 6A) The rsp mutant variants could dimerize, A11T (hsp90.2–7rsp) more than R337C (hsp90.2–8rsp), but both less than wild type (Fig. 6B). Addition of RAR1 and/or SGT1b to these assays did not alter dimerization activity under these conditions. Hence, ATP binding is required for Arabidopsis HSP90 dimerization.
Fig. 6.
HSP90.2rsp mutant proteins retain dimerization capability. Chemical cross-linking of wild-type and mutant forms of HSP90.2 in the presence of ADP or the nonhydrolysable ATP analog AMP-PNP. All variants of HSP90.2 are unable to dimerize in the presence of ADP. However, although no lra mutant variants (A) are able to dimerize even in the presence of AMP-PNP, both rsp mutant variants (B) can dimerize in the presence of AMP-PNP. The experiment was performed with an HSP90 concentration of 0.25 mg/mL, 15 molar equivalents of DMS, and 10 mM nucleotide.
The proportion of HSP90.2 that we observed in the dimerized form was low compared with the results reported for yeast HSP90 (24). It is unclear whether this was caused by our buffer conditions or it is an intrinsic property of Arabidopsis HSP90.2. However, this lower proportion of dimer was not caused by our cross-linking conditions, because increasing the concentration of cross linker ≈10-fold did not result in an increased proportion of dimerized HSP90.2.
In yeast, an ATP-independent C-terminal domain is sufficient (defined using N-terminal truncations), but not necessary (defined using C-terminal truncations), for HSP90 dimerization (25). Given our dimerization results and the positions of the rsp mutations on HSP90.2, we wanted to make sure that we were assaying the ATP dependent N-terminal activity in our assay. We purified HSP90.2 containing a short C-terminal truncation, known to abolish ATP-independent dimerization in yeast HSP90 (see Materials and Methods and ref. 24). This protein was unable to dimerize, even in the presence of AMP-PNP (Fig. S5). We thus conclude that the C-terminal dimerization domain of Arabidopsis HSP90.2 is necessary, but not sufficient (e.g., as in the cases where the N-terminal domain is mutated), for dimerization of Arabidopsis HSP90.2 and that the dimerization we measured was caused by the N-terminal domain.
Discussion
We performed 2 genetic screens to identify components affecting RPM1-mediated disease resistance in Arabidopsis. We demonstrate here that 2 specific hsp90.2 mutations suppress rar1 and restore NB-LRR protein accumulation, and hence, function. These hsp90.2rsp alleles demonstrate that HSP90.2 plays a broader role in disease resistance in Arabidopsis than previously considered (8). The hsp90.2rsp alleles are unique in 3 ways: (i) The particular mutations, A11T and R337C, have not been identified in any genetic screen, although the residues are strictly conserved across all eukaryotic species. (ii) These mutations translate into HSP90 proteins that bypass the requirement for a cochaperone. (iii) Most importantly, these mutations result in HSP90 alleles that result in a recovery of client protein accumulation and function (Table S1). The particular features of the hsp90.2rsp alleles, together with emerging structural analyses of HSP90 and its cochaperones, allow us to examine HSP90 function and propose an explicit mechanism for the function of the RAR1 cochaperone in NB-LRR protein stabilization.
hsp90.2–7rsp is recessive and encodes an A11T change. hsp90.2–8rsp is weakly semidominant and encodes a R337C change. Both suppress all known rar1 phenotypes to similar degrees. For example, both partially suppress rar1 for RPS5 function and fully suppress rar1 for RPM1 and RPS2 function, as measured by restoration of HR and pathogen growth restriction. They also suppress the rar1-enhanced disease susceptibility phenotype. Most importantly, both restore accumulation of RPM1-myc in rar1. Neither rsp allele has any discernible phenotype in the presence of RAR1. Neither expresses enhanced RPM1 activity in the presence of RAR1. Both provide some level of HSP90.2 function, at least as it pertains to viability in the context of a decrease in overall HSP90 levels. We used these 2 hsp90.2rsp alleles and the 4 previously-identified hsp90lra alleles (8) in a variety of tests designed to address how they might differentially influence 3 properties of HSP90: interaction with RAR1 and SGT1b, ATPase activity, and HSP90 dimerization. The rsp mutant proteins have different properties in these assays, as noted in Results, although both can dimerize to differing degrees.
The rsp alleles allowed us to examine the relationship between HSP90 ATPase activity and HSP90 function. It has long been assumed that ATPase activity is required for HSP90 function (26, 27). The data presented here argue against this concept in 2 ways. First, R337C (hsp90.2–8rsp) restores NB-LRR accumulation in rar1, yet the R337C mutation exhibits a nearly full loss of ATPase activity. Further, R337C is a more efficient suppressor of rar1 than A11T based on its semidominance, yet it nevertheless has lower ATPase activity than A11T. Hence, at least in the absence of RAR1, high ATPase activity is not required for NB-LRR accumulation. Second, D80N (hsp90.2–3lra), which is unable to bind ATP, does provide some function, to the extent that the hsp90.2–3lra hsp90.1KO double mutant was viable, and expresses no novel phenotype. In fact, given the nearly lethal phenotype observed with a half-dose of HSP90.2 in the absence of HSP90.1 the D80N mutation must exhibit more than half the activity of wild-type HSP90.2. But we leave open the possibility of an entirely different explanation for the phenotype of the lra alleles than simple loss of activity. Together, these 2 lines of evidence suggest that HSP90 ATPase activity can be separated from HSP90 function as it pertains to the modulation of NB-LRR function.
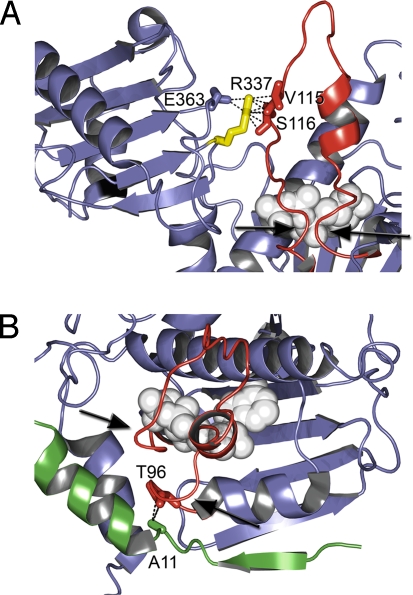
Fig. 7 presents close-up views of part of the X-ray structure of nucleotide-bound yeast HSP90 (28). Because HSP90 is so highly conserved across kingdoms, the Arabidopsis sequence threads onto this sequence with high confidence. The 2 HSP90 monomers in this structure are held together via an N-terminal clasp (and at the C terminus, although that is not relevant here). Each monomer of HSP90 contains a lid segment, hinged at residues G95 and G122 (Fig. 7), that swings through nearly 180° from its open position in the ADP-bound form of HSP90, to a closed ATP-bound conformation. This movement locks in the ATP molecule and places the catalytic arginine (371 in Arabidopsis; 380 in yeast) in position for interaction with the γ-phosphate of ATP. This movement also facilitates formation of the N-terminal dimerization clasp (28).
Fig. 7.
rsp mutations affect residues in the lid region of HSP90 in the closed conformation. Ribbon structures of yeast HSP90 (Protein Data Bank ID code 2CG9) bound to ATP (light gray). This lid (red) is hinged at G95 and G122 and swings 180° to fold over the nucleotide-binding pocket (yeast G94 and G121). (A) R337 (yellow) coordinates interaction of the central client-binding domain (purple, left) with the flexible lid (red) by interacting with V115 and S116 in the lid region and E363 within the middle domain of HSP90 (yeast R346, V114, S115, and E372). (B) A11 (yeast A10) from 1 monomer (green) interacts directly with T96 (yeast T95; red side chain) within the hinge of the other monomer. Black arrows indicate the locations of the hinges of the lid.
The structures of nucleotide-bound HSP90 suggest a mechanism for rar1 suppression by the rsp mutant proteins and present a clear prediction for RAR1 function in NB-LRR accumulation. We postulate that A11T and R337C act to favor the transition between the ADP- and ATP-bound conformations of HSP90.2, a transition characterized by the open–close cycling of the hinged lid. HSP90 has an 5-fold higher affinity for ADP over ATP (20). Thus, favoring the transition state, perhaps counterintuitively, favors the ATP bound conformation. The HSP90rsp proteins demonstrate that lid conformation is critical for client stabilization. Our ATP hydrolysis results suggest that ATP hydrolysis per se is irrelevant to HSP90 activity. Instead, the conformation of the N-terminal domain is important. However, it is likely that ATP hydrolysis is part of a regulatory mechanism allowing for control of the relative time spent in either conformation.
In the nucleotide-bound HSP90 structure (Fig. 7A), V114 and S115 from the closed lid (Fig. 7A in red) contact R337 from the same monomer. In R337C (hsp90.2–8rsp), these interactions are very likely to be destabilized, favoring a lid-open conformation, consistent with an inability to continue efficient nucleotide cycling. As shown in Fig. 7B, A11T lies within the N-terminal strand of HSP90. This strand switches from an intramolecular interaction in the ADP-bound, lid-open form of HSP90 to an intermolecular interaction with the opposing subunit of the HSP90 dimer in the ATP-bound, lid-closed form (Fig. S4). This intermolecular interaction should act to maintain and/or strengthen N-terminal dimerization. A11 contacts T96 near the base of the hinge on the opposing monomer. Consequently, the A11T mutation would be expected to both decrease the binding of the N-terminal strand to the opposing monomer and alter the stability of the lid-closed conformation. This conformational effect would decrease, but not abolish N-terminal dimerization, as we observed in Fig. 6.
We propose that destabilization of the lid-closed conformation by R337C is responsible for the diminution of dimerization, loss of interaction with RAR1 (and SGT1b), and nearly full loss of ATPase activity. The loss of RAR1 interaction with this presumably misregulated “floppy lid” does not have ill effects for the function of this HSP90 allele because it is, in essence, blind to RAR1 presence or absence. Hence, the R337C mechanism of action defines normal RAR1 function, namely, enhancing the cycling of the HSP90.2 lid, N-terminal dimerization cycling and client accumulation. This proposal is consistent with A11T, where we observed interaction with RAR1 (and SGT1b), normal nucleotide hydrolysis, some dimerization, and a recessive phenotype (meaning that it is a less efficient rar1 suppressor than the semidominant R337C). We propose that A11T is also able to bypass normal RAR1 function via its less efficient ability to maintain a lid-closed conformation.
Our model is consistent with recent observations studying the kinetics of N-terminal dimerization using a single molecule FRET-based HSP90 folding assay. In these in vitro experiments, 2 conformational states between the lid-open and lid-closed conformations were studied (29, 30). These intermediate conformations were shown to be rate-limiting steps in the ATPase reaction cycle (30). This work also demonstrated that the yeast cochaperone AHA1 is able to enhance the rate of ATP hydrolysis by bypassing an intermediate conformational state (lid closure) that follows ATP binding and precedes N-terminal dimerization, in favor of a lid-closed, ATP-bound, dimerized, prehydrolysis state. Our interpretation of the HSP90.2 rsp proteins above is consistent with this model.
Based on this model, we expect RAR1 (and by analogy our rsp alleles) to disfavor progression of the HSP90 cycle presented by Hessling et al. (30) at different points before the lid-closed, N-terminal dimerized intermediate they define as I2. R337C dimerizes very poorly compared with wild type, is likely to act by loosening the HSP90 lid, and is the stronger of the 2 rsp alleles. We infer that it diminishes the ATP-bound to the lid-closed I1 intermediate transition proposed by Hessling et al. (30). A11T dimerizes only slight less efficiently than wild type, is likely to act by disrupting N-terminal clasp formation, and expresses less than wild-type weak ATP hydrolytic efficiency. We infer that this allele is unable to transition efficiently beyond the ATP-bound, lid-closed I2 intermediate. Given these inferences, and the fact that A11T retains interaction with RAR1 whereas R337C loses it, we suggest that RAR1 binds to HSP90.2 at, and potentially after, the I1 conformation. Furthermore, we suggest that RAR1 acts to slow the progression of the HSP90 conformational cycle. These transition states can be reached in the absence of nucleotide (29). Hence, our data further suggest that overall HSP90 function and consequent effects on NB-LRR function are more coupled to the HSP90 conformational state than to ATP hydrolysis per se.
The sum of our data and recent single-molecule folding studies (29, 30) suggest that NB-LRR proteins are destabilized in rar1 because the HSP90 lid-open–lid-close cycle cannot be properly regulated, consistent with a model in which the balanced activities of RAR1, SGT1, and other cochaperones acting with HSP90 determine steady-state NB-LRR protein accumulation and signaling competence (7).
Materials and Methods
Plant Lines.
Transgenic Arabidopsis ecotype Columbia (Col-0; line a11) containing estradiol-inducible avrRpm1 has been described (9). For the double RPM1 screen, we used line a11 plant with an additional transgenic, myc-epitope-tagged copy of RPM1 introgressed (13). For the rar1 suppressor screen we used the originally-isolated rar1–21 mutant identified in line a11 (13). For pathology and double mutant analysis, we used rar1–21, rar1–28, or hsp90.2–3lra lines with the estradiol-inducible avrRpm1 removed by backcrossing to Col-0 and subsequent PCR-based marker-assisted breeding (8, 13). Mutant lines used (all in Col-0 unless noted) were rpm1–3 (31), rps2–101c (32), rps5–2 (14), ecotype Ws-0 as an RPP4 mutant control (18), and hsp90.2–5KO (8). We constructed double mutants of hsp90.2 alleles and rar1–21 by identifying F2s with a recombination event placing these linked mutations in cis. These plants were selfed, and resultant F3 individuals were further selected with PCR-based markers. hsp90.1KO was produced by selecting a homozygous insertion in the SALK T-DNA insertion line 075596 [previously referred to as hsp90.1–2; ref. 5] that was identified by molecular analysis of a segregating pool. The insertion site was confirmed by sequencing of the T-DNA-specific product.
Pathogen Strains, Inoculation, and Growth Quantification.
Pto DC3000 derivatives containing pVSP61 (empty vector), avrPphB, avrRpm1 or avrRpt2 have been described (33). Plant inoculations and bacterial growth assays were performed as described (11). Results for all bacterial growth assays represent 3 replicates with error bars representing +/− the standard deviation, a 95% confidence interval. All assays were performed independently a minimum of 3 times with similar results. High concentrations of Pto DC3000 (avrRpm1) (OD600 = 0.1, 5 × 107 cfu/mL) were syringe-infiltrated into leaves of 4- to 5-week-old plants to induce HR. Ion leakage assays were carried out as described (17).
Hyaloperonospora arabidopsidis (Hpa) propagation and inoculation were performed as described (34). Ten-day-old cotyledons of plants were inoculated with the asexual spores of Hpa isolate Emwa1. Asexual sporangiophores were counted 7 days postinoculation on at least 40 cotyledons for each genotype. Trypan blue staining for cell death and the Hp structures has been described (35). Pictures of trypan blue-stained leaves were taken with a light microscope (Nikon Eclipse).
Identification and Map-Based Cloning of Mutations in HSP90.2.
The double RPM1 screen was performed as described (9). The rar1 suppressor screen was performed by using a spray inoculation method in which 2-week old plants were sprayed with a 10 mM MgCl2 suspension containing Pto DC3000 (avrPphB) at a concentration of OD600 = 0.05 (2.5 × 107 cfu/mL) with 0.02% silwet L-77, covered with a clear lid for 4 h, and assessed for chlorosis and other symptoms of bacterial infection 4–6 days later.
Standard genetic crosses and analyses of F1 and F2 progeny were used. From the rar1 suppressor screen, rough mapping was preformed by crossing rsp rar1–21 mutants and the Landsberg erecta rar1–10 mutant (12). F2 plants were tested for rsp rar1–21-like resistance responses by spray inoculation as described above. Resistant F2 individuals were allowed to self and confirmed in the F3 generation. DNA from the F2 individuals was used in PCR amplification of known PCR-based molecular markers (www.arabidopsis.org) to obtain approximate mapping positions. Independent rough mapping of the 2 mutants showed linkage to the same interval. This interval was refined by using molecular markers we developed. We used 423 resistant F2 individuals to define a 4.5-Mb interval on the bottom arm of chromosome V containing HSP90.2 that is known as a regulator of RPM1 stability. By sequencing HSP90.2 in the originally-isolated double mutant, a G/A transition at position 31 (nucleotide positions relative to the translation start site of the published sequence of HSP90.2; At5g56030) was identified in hsp90.2–7rsp. The other mutant, rsp2, also contains a mutation (C1423T giving rise to R337C) in HSP90.2.
Yeast 2-Hybrid Analysis.
HSP90.2 and mutant derivatives were cloned into pJG4–5 by using the EcoRI and XhoI restriction sites and site-directed mutagenesis via overlap extension. RAR1, SGT1a, and SGT1b were cloned into a Gateway-compatible version of pEG202, pEG202gw (gift of Hiro Kaminaka, Tottori University, Tottori, Japan; ref. 7). Interactions were analyzed in yeast strain EGY48. Normal function of the SGT1a construct was shown by testing its interaction with RAR1 in the pJG4–5gw vector. Assays were performed with a plate reader (Tecan) as described (36). Protein levels were analyzed as described (7).
Protein Blot.
For detection of RPM1-myc levels in plants, we introgressed a transgene expressing RPM1-myc from the native RPM1 promoter as described (13). Protein extraction and immunodetection from plant tissue were carried out as described (8).
Production of Recombinant Proteins.
HSP90.2, mutant variants, and a 110-aa C-terminal truncation were cloned into pGEX-6p1 as described above and transformed into RIL codon plus cells (Strategene). Cells were grown in 2× yeast extract tryptone (YT) to an OD of ≈0.4 at 37 °C, and then the temperature was decreased to 22 °C for 45 min, and cells were induced for 3 h with 1 mM IPTG. Cell pellets were resuspended in buffer A [20 mM Tris (pH 8.0), 300 mM NaCl, and 1 Complete EDTA-Free″ protease inhibitor tablet (Roche)]. After resuspension, cells were lysed by using an Avestin Emulsiflex-C5. The lysates were cleared by centrifugation for 45 min at 15,000 rpm in an SS-34 rotor. The cleared lysates were run on a 5-mL High Trap glutathione column (GE Healthcare) and washed with 10 column volumes of buffer A. The protein was eluted with 5 column volumes of buffer A with 20 mM glutathione. PreScission protease (50 units/mL; GE Healthcare) was then added to the sample, and the protein was cleaved overnight at 4 °C while being dialyzed into buffer B [20 mM sodium phosphate (pH 6.5), 150 mM NaCl, 2 mM DTT]. The next morning, the protein was loaded onto a HiLoad 16/10 Q Sepharose High Performance anion exchange column equilibrated in buffer B and eluted with a 150- to 600-mM linear gradient. Fractions were analyzed for purity by SDS/PAGE, and clean fractions were pooled and dialyzed into buffer C (40 mM Hepes, 150 mM KCl, and 5 mM MgCl2 at pH 7.5).
Biochemical Methods.
Circular dichroism experiments were performed on a Pistar-180 circular dichroism/fluorescence spectrophotometer (Applied Photophysics). Samples at ≈20 μM were placed in a 0.1-cm cuvette, and scans were taken from 195 to 260 nm with 1-nm increments and 30,000 repetitions per increment.
ATP hydrolysis assays were performed as described (27, 37). Briefly, 2.5 μM purified HSP90 was incubated with 0.4 mM phosphoenol pyruvate, 0.25 mM NADH, and 1% PK/LDH enzyme mix (Sigma). Proteins were incubated with multiple concentrations of ATP between 0 and 1.2 mM. Experiments were performed in duplicate with a control containing 0.5 mM radicicol to measure HSP90-specific activity. Experiments were performed in 200-μL reactions in a plate reader (GENios; Tecan).
Cross-linking experiments were performed as described (24). Purified HSP90 (0.25 mg/mL) was incubated for 2 h with 10 mM ADP or AMP-PNP, after which a 15 molar excess of dimethyl suberimidate dihydrochloride (DMS) was added for an additional 2-h incubation. Reactions were stopped by addition of SDS/PAGE loading buffer and loading on an 8% gel.
Supplementary Material
Acknowledgments.
We thank Prof. David Toft and Dr. Sara Felts (Mayo Clinic College of Medicine, Rochester, MN) for advice and protein samples to optimize the ATPase assays; Prof. John Sondek (University of North Carolina) for use of equipment and helpful discussions; Ryan Chao, Jonny Chen, Anna Newton, Allison Osborne, David Rybnicek, and Linda Yang for help with plant maintenance and molecular genotyping; and all current and former members of J.L.D.'s laboratory. T-DNA insertion lines were provided by Joe Ecker (Salk Institute, La Jolla, CA) via the Arabidopsis Biological Resource Center. Terry Law and Petra Epple, for helpful discussions and technical advice. This work was supported by National Science Foundation Arabidopsis 2010 Project Grant IOB-0520003 (to J.L.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904877106/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 3.Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.da Silva Correia J, Miranda Y, Leonard N, Ulevitch R. SGT1 is essential for Nod1 activation. Proc Natl Acad Sci USA. 2007;104:6764–6769. doi: 10.1073/pnas.0610926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boter M, et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt BF, III, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 8.Hubert DA, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tornero P, Chao R, Luthin W, Goff S, Dangl JL. Large-scale structure–function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2002;14:435–450. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tornero P, Dangl JL. A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- 12.Muskett PR, et al. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14:979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tornero P, et al. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieri S, et al. RAR1 positively controls steady-state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JH, et al. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JDG. Arabidopsis RPP4 is a member the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signaling components. Plant J. 2002;29:439–451. doi: 10.1046/j.0960-7412.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- 19.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 21.Kawasaki T, et al. A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J. 2005;44:258–270. doi: 10.1111/j.1365-313X.2005.02525.x. [DOI] [PubMed] [Google Scholar]
- 22.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 23.Stitt M, Lilley RM, Heldt HW. Adenine nucleotide levels in the cytosol, chloroplasts, and mitochondria of wheat leaf protoplasts. Plant Physiol. 1982;70:971–977. doi: 10.1104/pp.70.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prodromou C, et al. The ATPase cycle of Hsp90 drives a molecular “clamp” via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52, and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 30.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 31.Grant MR, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 32.Mindrinos M, Katagiri F, Yu G-L, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 33.Ritter C, Dangl JL. Interference between 2 specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell. 1996;8:251–257. doi: 10.1105/tpc.8.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt BF, III, et al. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell. 2002;2:807–817. doi: 10.1016/s1534-5807(02)00174-0. [DOI] [PubMed] [Google Scholar]
- 35.Koch E, Slusarenko AJ. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serebriiskii IG, Toby GG, Golemis EA. Streamlined yeast colorimetric reporter activity assays using scanners and plate readers. BioTechniques. 2000;29:278–279. 282–284, 286–288. doi: 10.2144/00292st03. [DOI] [PubMed] [Google Scholar]
- 37.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.