Abstract
A key problem in computational proteomics is distinguishing between correct and false peptide identifications. We argue that evaluating the error rates of peptide identifications is not unlike computing generating functions in combinatorics. We show that the generating functions and their derivatives (spectral energy and spectral probability) represent new features of tandem mass spectra that, similarly to Δ-scores, significantly improve peptide identifications. Furthermore, the spectral probability provides a rigorous solution to the problem of computing statistical significance of spectral identifications. The spectral energy/probability approach improves the sensitivity-specificity trade-off of existing MS/MS search tools, addresses the notoriously difficult problem of “one-hit-wonders” in mass spectrometry, and often eliminates the need for decoy database searches. We therefore argue that the generating function approach has the potential to increase the number of peptide identifications in MS/MS searches.
Introduction
Tandem mass spectrometry (MS/MS) has become the leading high-throughput technology for protein identification. These experiments often generate millions of spectra, and interpreting them leads to challenging statistical problems (see Nesvizhskii et al., 2007 [1] and Kall et al., 2008 [2] for recent reviews). One of the major problems in tandem mass spectrometry is the lack of theoretical (as opposed to empirical) estimates of statistical significance of peptide identifications. Indeed, the Proteomics Publication Guidelines [3, 4] recommend searching in decoy databases to determine the statistical significance of peptide identifications (this is in contrast to genomics searches that do not employ decoy databases). We argue that if the error rates reported by existing MS/MS software tools were reliable (as in the case of genomics searches), the search in decoy databases would not be necessary. The major difference here is that MS/MS searches are currently based on empirical database-dependent estimates of error rates (often represented by Poisson, Gaussian, hypergeometric, or other approximations of tails of score distributions [5, 6, 7]) as opposed to the analytically derived and database-independent error rates in genomics tools like BLAST [8]. Although the target-decoy search strategy is currently viewed as the best way to distinguish between the correct and false identifications [9, 10, 11, 12, 13, 14], this valuable approach has certain shortcomings. While the shortcomings of such strategies are well recognized in genomics (see [15]), they are often overlooked in proteomics. Also, decoy databases take a toll on every lab engaged in MS/MS searches effectively doubling the search time. We argue that using decoy databases is an acknowledgment of our inability to solve the following problem:
Spectrum Matching Problem
Given a spectrum S and a score threshold T for a spectrum-peptide scoring function, find the probability that a random peptide matches the spectrum S with score equal to or larger than T.
The Spectrum Matching Problem was first posed by Fenyo and Beavis, 2003 [16] (see also [17]).1 They acknowledged that the theoretical solution of this problem is unknown and suggested a heuristic approach to its solution based on approximating the tail of the score distribution. Solving the Spectrum Matching Problem is equivalent to computing the False Positive Rates (FPR) of spectral matches. FPR is a property of an individual spectrum as opposed to the False Discovery Rate (FDR), the property of multiple spectra (proportion of incorrect identifications among all identifications judged correct).2
Search in a decoy database looks like an attractive approach for approximating the solution of the Spectrum Matching Problem as , where m is the number of matches between the spectrum and the decoy database of size n (with scores equal to or larger than the threshold T). However, for an individual spectrum, the number of matches for typical n is usually zero thus making this approach problematic (decoy and target databases usually have the same size). To obtain reliable FPR for an individual spectrum, one can increase n (e.g., making giant decoy databases 1000 times larger than target databases). Since this is impractical, some existing approaches bundle all spectra with the same score to evaluate the FDR of all spectra in the bundle and to use FDR as a surrogate for FPR (see [1]). Figure 1 in the Supplement 1 illustrates that spectra with the same score may have vastly different FPRs thus implying that careful analysis of all peaks in the spectrum (rather than the scores alone) may be necessary to compute the database matching statistics for individual spectra.
Assigning the same FPR to all identifications with identical scores [18, 19, 20] is a dangerous oversimplification since the scoring functions of existing MS/MS tools are not based on rigorous probabilistic models and are often inaccurate (see Supplement 1). Recognizing this problem, Fenyo and Beavis, 2003 [16] pioneered computing FPR for an individual spectrum as an empirical solution of the Spectrum Matching Problem.3 They constructed the empirical score distribution of low-scoring (erroneous) peptide identifications and extrapolated it to evaluate the FPR of high-scoring peptide identifications in the tail of the distribution. Such approaches are not free of shortcomings: Waterman and Vingron, 1994 [15] wrote: “Theory is needed because simulations rarely cover the extreme tails of a distribution.” criticizing similar approaches in genomics. In another paper criticizing such empirical approaches, Nagarajan et al., 2005 [21] demonstrated that all existing motif finding tools are statistically flawed and can be off by orders of magnitude in computing P-values. This flaw remained uncovered for 15 years and affected 1000s of studies. Needless to say, the mass spectrometry community is not immune to similar flaws suggesting that re-examination of existing approaches to estimation of statistical significance in MS/MS searches is timely. In this paper, we demonstrate that the analysis of statistical significance in various MS/MS tools is often unreliable (see Supplement 1).
We further argue that use of decoy databases is not free from shortcomings. The intuition behind using a decoy database is to estimate the number of spectra that match the database by chance. If a spectrum S has probability p(S) of matching a random database, then a decoy database is simply a time-consuming way to evaluate Σp(S) over all spectra in the dataset (this sum represents the expected number of hits in the decoy database) but not a good way to estimate individual probabilities p(S). The generating function approach, in difference from the decoy database approach, accurately computes probabilities p(S) for the individual spectra, an important advantage for addressing the problem of “one-hit-wonders” in MS/MS searches. An ideal approach to evaluating the statistical significance of MS/MS searches would be to use a database containing all possible peptides up to a certain length, and use the number of identifications in this database to evaluate the error rate. However, the time required to search this database renders this approach infeasible. Below we show that it is nevertheless possible to compute the precise number of the identified peptides in this huge database thus computing the solution of the Spectrum Matching Problem exactly rather than empirically. This illustrates the advantages of (fast) analysis of scores over the huge database of all peptides as compared to (slow) analysis of scores over the much smaller decoy databases.
Solving the Spectrum Matching Problem is not unlike computing the generating function in combinatorics [22, 23]. Given a spectrum S and a score X, define E(S, X) as the number of peptides (among all possible peptides) that match the spectrum S with score X. To evaluate FPRs one has to compute E(S, X) for every spectrum S and every score X (more precisely, the sum of probabilities of all peptides contributing to E(S, X)). Figure 1(b) illustrates the notion of the generating function in the simple case when the score X of a match between a spectrum and a peptide is defined as the number of peaks in the spectrum explained as b or y ions. Figure 1(c) shows the generating function for a more advanced scoring described below. We show how to compute E(S, X) and to use it for improving the sensitivity-specificity trade-off of various database search tools. We further introduce the notion of spectral energy (Figure 1) that represents the difference between the best de novo spectral interpretation and the best database spectral interpretation. We show that while the Energy-score (in difference from the Δ-score) was ignored in MS/MS searches so far, it greatly improves the separation between the correct and false identifications. Finally, we introduce the notion of spectral probability (the total probability of all peptides with scores exceeding a threshold) that further improves the separation between the correct and false identifications (Figure 1).
Figure 1. Illustration of the generating function.
(a) A spectrum S of peptide GAIDKAEEIR (top 43 peaks after removal of low-intensity peaks). (b) The number of peptides (E(S, X)) that explain X b/y peaks in this spectrum. For example, there are 360 peptides with 13 b/y ions explained (E(S, 16) = 360), 12940 peptides with 12 b/y ions explained, and so on. The score of the top-scoring database peptide GAIDKAEEIR is 11, the optimal score among all possible peptides is 13 (such as for the peptide QP MGAEAELR), thus Energy-score is 2. The second top-scoring peptide in the database (DQELLSEIR) has score 5, therefore Δ-score is 6. For simplicity, a peak that explains both a b-ion and a y-ion in a particular peptide is counted as explaining two b and y peaks. (c) The (uniformly weighted) generating function of the same spectrum. The table shows the number of peptides with score X, the overall probability of peptides with score X and the total probability of all peptides with scores equal to or larger than X (spectral probability). The peptides QIDKAEEIR and QIDGAAEEIR represent better spectral interpretations (score 64) than the correct peptide GAIDKAEEIR identified by InsPecT (score 60) resulting in Energy-score 4. There are 24 optimal de novo reconstructions that are all derived from QIDKAEEIR and QIDGAAEEIR via I/L and Q/K substitutions (16 for QIDKAEEIR and 8 for QIDGAAEEIR). The total probability of these 24 peptides is 16 · 20−9 + 8 · 20−10 = 3.20 · 10−11. The second best peptide in the database (IRSIESQLR) has score 27, therefore Δ-score is 33.
While this paper is limited to identifications of non-modified peptides, the generating function approach can be extended to modified peptides as well (see the Discussion section). Our MS-GF software for computing generating function/spectral energy/spectral probability of tandem mass spectra is available as open source from http://www.cs.ucsd.edu/users/ppevzner/ software.html.
Methods
The generating function
To introduce the notion of the generating function of tandem mass spectra, we use the analogy with the classical Ising model of ferromagnetism, one of the pillars of statistical mechanics [24]. The model consists of n magnetic spins such that each spin can be in two states (up and down). This results in 2n possible states each with its own energy defined by the elementary interactions between neighboring spins on the lattice. The partition function represents the key technique for analyzing the Ising model and is defined as Σall states π e−Energy(π) (in this paper we ignore the “temperature” parameter of the Ising model).4
Interpreting a spectrum S with a peptide P is not unlike choosing a state in the Ising model. Instead of 2n states of magnetic spins, there are 20n possible interpretations of the spectrum S by peptides of length n. Each of these interpretations has its own “energy” given by the score of the match between spectrum S and peptide P. The goal is to compute the partition (generating) function of the spectrum S and to apply it for analyzing statistics of the MS/MS searches rather than the statistics of the Ising model. While, the generating function of tandem mass spectra involves 20n terms, we show below how to efficiently compute it. For the sake of simplicity, we first introduce the notion of generating function for boolean spectra that ignore intensities, charges, inaccuracies in peak positions, and C-terminal ions. While the boolean spectra are impractical, they proved to be useful as a stepping stone for introducing simple scoring/algorithms and later generalizing them to real spectra and more complex algorithms (see [25, 26, 27]). Later, we will illustrate how to define the generating function for real spectra.
We represent a boolean spectrum S with parent mass k as 0-1 vector s1 … sk, where si = 1 if there is a peak at mass i in the spectrum, and si = 0, otherwise. This representation assumes that the spectra are discretized and all masses are integers (Figure 2). For example, for ion-trap spectra this can be approximated by multiplying all masses by 10 and taking integer parts (see Kim et al., 2008 [28] for details). The match score between spectra s1 … sk and is defined as .
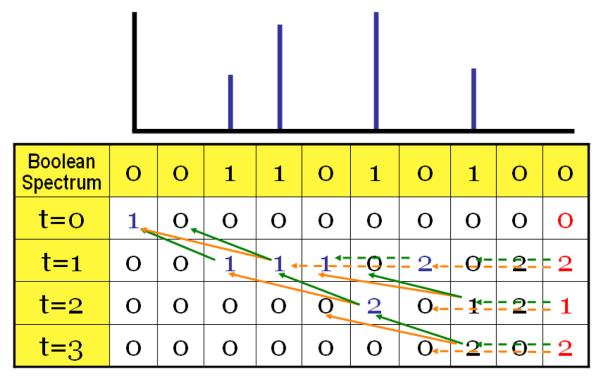
Figure 2. Illustration of the dynamic programming algorithm for computing the generating function.
The MS-GF dynamic programming algorithm is illustrated with the help of a simplified amino acid model (only two amino acids A and B with masses 2 and 3 Daltons respectively) and a simplified discretized spectrum (only 4 peaks at 2,3, 5, and 7 Da). The scoring function used for this illustration is the number of matching prefix ions. The spectrum is converted into its boolean representation 011010100 with 1s at positions 2,3,5, and 7 (extra zero in the beginning is added to represent the variable x(0, t)). The vertical axis in the dynamic programming table represents scores (t). The value in each cell of the matrix represents the number of peptide reconstructions that explain the initial part of the spectrum till that position with the corresponding score. The first cell in the matrix (0,0) is initialized with 1, and the matrix is filled progressively from left to right and top to bottom. The value of each cell is computed as the sum of the values of previously filled cells which are 2 (green arrow) or 3 (orange arrows) columns before the cell under consideration. If there is a peak at the current position of the spectrum, sum is taken over the cells in the previous row, otherwise in the same row. In this example, the maximum achievable score (t) is 3, which can be obtained by two peptide reconstructions. The sequences of these reconstructions can be obtained by backtracking, as indicated by the arrows, and are found to be ABAA and BAAA. We also see that there are 2 reconstructions with score 1 and 1 reconstruction with a score of 2.
Given a peptide P = p1 … pn, we define its theoretical spectrum Spectrum(P) as a 0-1 spectrum s1 … sk with (n − 1) 1s, such that si = 1 iff i is the mass of the peptide p1 … pi. The score (denoted as Score(P, S)) between a peptide P and a spectrum S (with the same parent mass) is defined as the match score between spectra Spectrum(P) and S. For convenience, we assume that Score(P, S) = −∞ if peptide P and spectrum S have different parent masses. Let SCORE = SCORE(S) = maxall peptides P Score(P, S) be the maximum value of Score(P, S) among all possible peptides P. SCORE can be estimated using de novo peptide sequencing algorithms [29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40]. We define energy of a peptide-spectrum pair as Energy(P, S) = SCORE − Score(P, S) and define the generating function of the spectrum S as Σtx(t) · e−t, where x(t) is the number of peptides with energy t.5
Given the probabilities of individual amino acids (e.g., computed empirically from a set of protein sequences), we define the probability prob(P) of a peptide P = a1 … am as the product of probabilities of its amino acids . We will also consider the weighted generating function: Σall peptides P prop(P) · e−Energy(P,S) = Σt y(t) · e−t, where y(t) is the overall probability of all peptides with energy t.
Computing the generating function for boolean spectra
Given a spectrum S, we introduce a variable x(i, t) equal to the number of peptides of mass i that have t peaks in common with spectrum S, i.e, the number of peptides P such that Score(P, Si) = t (Si stands for “i-prefix” s1 … si of the spectrum S). In the case S has a peak at position i (si = 1), the variable x(i, t) can be computed as follows (|a| denotes the mass of an amino acid a):
Otherwise (si = 0):
Below we provide an equivalent and more compact representation of these recurrencies:
We initialize x(0, 0) = 1, x(0, t) = 0 for t > 0, and assume that x(i, t) = 0 for negative i. The maximum value SCORE of Score(P, S) among all possible peptides P is simply the maximum value of t with non-zero x(k, t). See Figure 2.
The recurrence for computing the weighted generating function is very similar. In this case the variable y(i, t) equals to the overall probability of peptides of mass i that have t peaks in common with spectrum S. The variable y(i, t) is initialized in the same way as x(i, t)6 and is computed using the following recurrence:
The above algorithm for computing the generating function has complexity O(|S|·|SCORE|·Mult · PeptideLength · A), where A = 20 is the number of amino acids, PeptideLength is the maximum length of a peptide with the mass equal to |S|, and Mult is the multiplication co-efficient that was applied to all masses in the spectrum to satisfy the assumption that they are integers (typically, Mult = 10 for ion-traps). In practice, it requires 0.1-0.2 seconds to compute the generating function on a desktop machine with 2.16 Ghz Intel processor.
Computing the generating function for real spectra
MS-GF transforms tandem mass spectra into its integer-valued scored version s1 … sk (rather than boolean spectra) using the probabilistic model similar to [30, 32, 40]. This transformation takes into account peptide length, peak intensities, neutral losses, dependencies between ion types, noise, etc. Most de novo and database search algorithms use such representation (explicitly or implicitly) by assigning intensity-dependent scores to peaks, further adjusting for imprecisions in mass-measurements, and applying dot-product for scoring spectra against peptides. However, these scores are typically attached to the positions of peaks in the spectrum s1 … sk and will not enable a computation of the generating function in the low-accuracy setting with accuracy threshold δ. However, as long as we redefine the spectrum s1 … sk as with , the generating function (in case of imprecise mass measurements) can be easily computed as described below.
The score Score(P, S) between a peptide P and a spectrum S (with the same parent mass) is defined as the dot-product between the theoretical spectrum Spectrum(P ) and S (now S is defined as an arbitrary integer-valued vector and Spectrum(P ) is defined to allow for both N-terminal and C-terminal ions as in [27]). Let SCORE be the maximum value of Score(P, S) and Energy(P, S) = SCORE − Score(P, S). Given a spectrum S, we define x(i, t) as the number of peptides of mass i with score t, i.e, the number of peptides P such that Score(P, Si) = t. The variable x(i, t) can be computed as in the case of boolean spectra.
We emphasize that MS-GF can handle scored spectra generated by any MS/MS tool with additive scoring functions. The scoring function chosen in this paper can be viewed as a variation of Sherenga and PepNovo [30, 42] with improved analysis of peak intensities and doubly charged ions (the details are described in [28]). Some MS/MS analysis tools (e.g., SEQUEST or tools using sequence-specific peak intensities [43, 44, 45]) have non-additive scoring components and thus cannot be modeled by this generating function framework. However, MS-GF still can be used to re-score their results (Supplement 4 illustrates how such additive re-scoring improves non-additive SEQUEST scoring).
Let 𝒜 be a peptide identification algorithm that accepts a peptide P as an interpretation of a spectrum S as long as the peptide-spectrum score Score(P, S) is larger or equal to the threshold T. Given the allowed (integer) parent mass error ∊, the weighted generating function allows one to compute the overall probability of peptides with scores equal to or larger than T (spectral probability) as
For example, the spectral probability Prob60(S) = 2.76 · 10−10 represents the total probability of all 306 peptides with scores larger or equal to the score of the correct peptide in Figure 1(c). The probability that the algorithm 𝒜 identifies the spectrum S in a random database of size n is computed as 1 − (1 − ProbT(S))n. Since the parameter T is usually chosen in such a way that ProbT(S) is much smaller than , one can assume that 1 − (1 − ProbT(S))n ≈ ProbT(S) · n. If a user attempts to identify peptides with a fixed F P R in a database of size n (e.g., F P R = 0.01 is commonly used in MS/MS searches), then the parameter T is chosen in such a way that . The corresponding value of T can be derived from the generating function (see the last column in Figure 1(c)).
Results
Datasets
The Shewanella oneidensis MR-1 dataset used here (14.5 million spectra) and peptide identifications based on this dataset are described in [46]. 28,377 unmodified peptides were identified in this dataset by InsPecT with an error rate of 5% (1% spectrum-level error rate) as measured using a decoy database [20].
Due to its large size, searching the entire Shewanella dataset with tools like SEQUEST is rather time-consuming. To make it easier to benchmark our approach against other tools and to summarize the results, we constructed two smaller datasets (geared to peptides of length 10) that are used in this study. The Supplement 4 describes benchmarking for other peptide lengths (the results are similar).
Shewanella-1784: From 28,377 peptides identified in Shewanella oneidensis MR-1, we selected all doubly-charged tryptic peptides of length 10. It resulted in 1745 and 39 peptides identified in the target and decoy databases (2.2% error rate). For each of these 1745 + 39 = 1784 peptides, we retained one spectrum (chosen randomly if the peptide is identified from multiple spectra) to construct the final dataset of 1784 spectra.
Shewanella-50000: From all 14.5 million Shewanella spectra, we randomly selected 50,000 doubly-charged spectra with parent masses ranging from 1100 to 1200 Da (these spectra typically correspond to peptides of length ≈ 10 aa). Each spectrum in this dataset was searched against all Shewanel la proteins (1.47 million of amino acids) and against the randomized decoy database (of same size) with SEQUEST (TurboSEQUEST v.27, rev. 12), InsPecT (20060907), and X!Tandem (2007.01.01.2), as well as analyzed with MS-GF and PeptideProphet (v3.0).
Using generating functions to estimate the statistical significance of peptide identifications
We found that the error rates reported by existing database search tools do not provide accurate estimates of the statistical significance of individual peptide identifications (they are often off by an order of magnitude) while the error rates evaluated by MS-GF are very accurate (see Supplements 1 and 2).
To evaluate whether MS-GF accurately estimates the number of hits in decoy database (thus eliminating the need for the decoy database search) we conducted the following experiment. For each spectrum in the Shewanella-50000 dataset, we generated top-scoring peptides whose total probability sums up to the parameter SpectralProbability. A spectrum is considered identified in a database if any of the generated reconstructions is present in the database. We varied the value of SpectralP robability, and computed the number of spectra that were identified in the Shewanella database and the decoy database of the same size. Table 1 shows the distribution of these numbers, compares them against SpectralProbability · n · 50000 (the expected number of matches in the database of size n) and shows that the number of matches in the decoy database is very close to the expected number of matches computed by MS-GF.
Table 1.
Number of spectra in Shewanella-50000 dataset that are identified in the Shewanella database (Column 2) and the decoy database (Column 3) by top peptide reconstructions with probability SpectralProbability. Column 4 provides the expected number of spectra that will match the decoy database given SpectralProbability, as computed by MS-GF without actually doing the search.
| SpectralProbability | # Correct IDs (in target DB) |
# False IDs (in decoy DB) |
# False IDs (predicted by MS-GF) |
|---|---|---|---|
| 2e-9 | 8314 | 161 | 146 |
| 1e-9 | 7721 | 76 | 75 |
| 8e-10 | 7525 | 60 | 59 |
| 6e-10 | 7272 | 44 | 44 |
| 5e-10 | 7115 | 34 | 37 |
| 4e-10 | 6937 | 28 | 29 |
| 2e-10 | 6333 | 15 | 15 |
| 1e-10 | 5755 | 6 | 7 |
| 1e-11 | 3820 | 0 | 0.7 |
Figures 3(a,b) show the distributions of InsPecT and X!Tandem scores for the peptides identified in Shewanella-1784 dataset against the target and decoy database. Advanced peptide identification tools are expected to have similar score distributions in target and decoy databases (otherwise, the difference between the distributions can be used to better separate the correct and false identifications). For InsPecT, the distributions in the target and decoy databases are similar, with Kolmogorov-Smirnov (KS) distance of 0.28, indicating that InsPecT scoring cannot further differentiate between the correct and the false identifications. In case of X!Tandem E-value, there is some separation between the distributions in target and decoy database, however the distributions still have a large overlap and it is unclear what additional features can separate the correct and false identifications.
Figure 3. Separation between correct and incorrect identifications.
Distribution of (a) InsPecT MQScore and (b) X!Tandem E-Value, for the peptides identified in Shewanella-1784 dataset against Shewanella and decoy databases. X-axes show the database search scores, and Y-axes show the fraction of identifications with that score. The Kolmogorov-Smirnov (KS) distance between the two distributions is 0.28 for InsPecT scores and 0.58 for X!Tandem scores. (c) Distribution of Energy(P, S) for the same dataset (the KS distance is 0.77). (d) Distribution of −log10(SpectralP robability) (the KS distance is 0.78). Spectral P robability of the pair (P, S) is defined as the sum of probabilities of all peptides whose score is larger or equal to the score Score(P, S) of the match between peptide P and spectrum S.
Figure 3(c) shows the distribution of Energy(P, S) for identifications from Shewanella-1784 dataset and demonstrates that spectral energy provides an excellent separation between the correct and false identifications. In particular, Energy = 0 for a significant portion of correct identifications (in these cases, the identified peptide also represents an optimal de novo reconstruction). The false identifications, on the other hand, have no identifications with Energy = 0. Moreover, the separation in Figure 3(c) indicates that the Energy is complementary to many other parameters used for scoring spectra (recall that InsPecT scoring combines seven parameters but still does not attain the separation power of Energy). Figure 4 further shows the joint distribution of SCORE and Energy and provides an intuitive explanation why the generating function approach improves the sensitivity/specificity ratio of existing MS/MS search tools. Note that the target and decoy identifications are well separated in 2-D, with low SCORE and Energy for the target database and high SCORE and Energy for the decoy database.
Figure 4. Joint distribution of SCORE and Energy.
The distribution is plotted for the identifications in the Shewanella-1784 dataset, for the peptides identified in the Shewanella database and the decoy database. The blue dots (decoy database) are laid over the red dots (Shewanella database), so that all decoy database identifications are visible.
Let Score(P, S) be the match score of a peptide P and a spectrum S. We denote the spectral probability ProbScore(P,S)(S) of the peptide-spectrum pair (P, S) as the sum of probabilities of all peptides with match scores larger or equal to Score(P, S) (when compared to S). Figure 3(d) shows the distribution of the spectral probability (as computed by MS-GF) for correct and false peptide identifications. This parameter also provides excellent separation between the correct and false identifications, with false identifications typically having much larger spectral probabilities ProbScore(P,S)(S). This is in agreement with Figure 3(c), further confirming that most identifications on the decoy database, in spite of their high scores, actually represent poor (sub-optimal) de novo solutions, and could be distinguished from correct solutions using MS-GF.
Generating functions improve the sensitivity-specificity trade-off of MS/MS database searches
Generating functions can be used to re-score the identifications obtained by various database search tools and to improve the sensitivity-specificity trade-off. We illustrate this result using Shewanella-50000 dataset searched against the target Shewanella database and the decoy database using X!Tandem [47] (see Supplement 3 for similar analysis using SEQUEST and PeptideProphet). The existing database search tools use two types of scores that we refer to as raw and combined scores. Raw scores (used for scanning databases) are defined by a spectrum and a peptide alone without any reference to the scores of other peptides encountered in the database search. The database-dependent combined scores integrate raw scores with other information like Δ-score of the second best peptide match (like in SEQUEST), or the distribution of scores of all peptides in the database (like in X!Tandem). We emphasize that the generating function (and the spectral probability) represents the raw score since it does not use any additional information about other peptides in the database. Below we show that the spectral probability improves on previously proposed raw scores and even outperforms the combined scores of the existing database search tools.
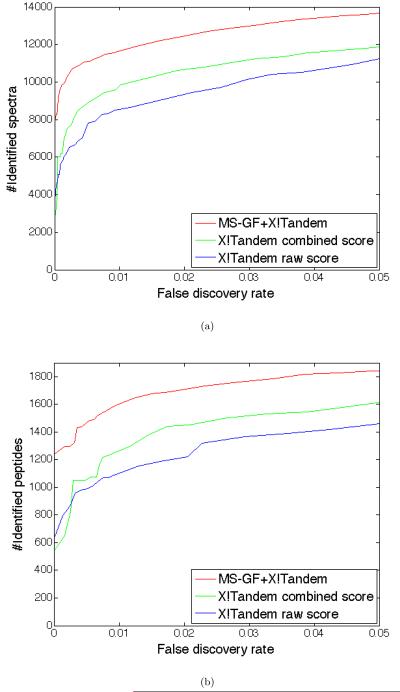
For each spectrum in the Shewanella-50000 dataset, three different scores are used for analyzing the peptide identifications and constructing ROC curves: (i) X!Tandem raw score used for scanning the database, (ii) X!Tandem combined score (E-value) that integrates the raw score with the distribution of the scores for all peptides in the database, and (iii) spectral probability as reported by MS-GF for the X!Tandem identification. For each score, a varying cutoff is used, and the number of spectra that have an identification with scores above the cutoff in the Shewanella database and the corresponding error rate (ratio of the number of identifications on a decoy database of the same size and the number of identifications in the target database) are plotted in Figure 5(a).
Figure 5. Sensitivity-specificity trade-offs.
(a) Comparison of MS-GF with X!Tandem. The number of spectra identified in the Shewanella database and the corresponding error rate. Three scores are compared (from top to bottom): (i) MS-GF+X!Tandem: FPR as reported by MS-GF for the X!Tandem identifications, (ii) X!Tandem combined score: X!Tandem E-value that uses the raw score as well as the distribution of scores of all peptides for the given spectrum and (iii) X!Tandem raw score: X!Tandem hypergeometric score. (b) Similar to (a), but counting the number of unique peptides identified in the Shewanella and the decoy database instead of the number of identified spectra.
The Supplementary Table 5 lists all identified spectra (in both target and decoy databases) along with their X!Tandem and MS-GF scores. For example, the minimum X!Tandem E-value is 3.8·10−14 for target database (Etarget) and 2.9·10−5 for decoy database (Edecoy). The Supplementary table 5 illustrates that X!Tandem identifies 2689 spectra in the target database with E-values below Edecoy (X!Tandem identifies these spectra with virtually zero FDR). MS-GF simply rescores and ranks the same spectra using spectral probabilities instead of X!Tandem E-values. For example, the highest scoring spectrum identified by X!Tandem is ranked only as 234-th by MS-GF (peptide ADAVVIAAGGFAK), while the peptide ranked as 1-st by MS-GF is only ranked as 3445-th by X!Tandem (peptide ALGGASGGFTSGK). The Supplementary Table 5 reveals that the minimum spectral probability is 4.88 · 10−17 for target database (SpectralProbabilitytarget) and 1.31·10−10 for decoy database (SpectralProbabilitydecoy). There are 7887 spectra in the target database with spectral probabilities below SpectralProbabilitydecoy (MS-GF identifies these spectra with virtually zero FDR). It represent nearly three-fold increase compared to 2689 spectra identified by X!Tandem with virtually zero FDR. There is a similar increase with respect to the number of peptide identified with virtually zero FDR (1243 for MS-GF versus 493 for X!Tandem)
MS-GF results in significantly higher number of identifications in the Shewanella database for a given error rate (number of identifications on the decoy database) when compared to the raw X!Tandem scores. Similarly, it significantly improves on SEQUEST and PeptideProphet (see Supplement 3). Figure 5(b) shows similar curves for the number of unique peptides instead of the number of spectra. For 5% error rate, X!Tandem raw/combined score identifies 1449/1613 peptides, while MS-GF identifies 1837 peptides. The advantage of MS-GF is particularly pronounced for extremely accurate identifications. For example, for 0.3% error rate (very few false identifications) MS-GF identified 1326 peptides while X!Tandem identified 943/1050 peptides with raw/combined scores. Such extremely accurate identifications are important for a notoriously difficult problem of identifying proteins based on a single peptide hit (one-hit-wonders). Indeed a single peptide hit with the error rate 0.3% may be more reliable than two peptide hits with the error rate 3% each [48, 49, 50, 44]. The fact that MS-GF has better sensitivity-specificity than even the combined X!Tandem score is surprising since MS-GF has no access to the valuable information about other peptides in the database that is incorporated into the combined X!Tandem score. We therefore argue that the spectral probability represents a valuable addition to the various “raw” scores proposed for MS/MS searches so far.
We remark that the MS-GF+X!Tandem curve in Figure 5 was constructed using the information about matches in the decoy database. The superior performance of MS-GF+X!Tandem over X!Tandem raises a question whether a database search based on MS-GF (i.e., using SpectralProbability as a score) would be better off on its own (without using matches identified by X!Tandem). In other words, we are interested in how a database search with MS-GF scoring would fare in comparison with other database search tools. Figure 6 illustrates that MS-GF alone (without using X!Tandem identifications) performs better than X!Tandem. For each spectrum in the Shewanella-50000 dataset, we generated the top-scoring peptides whose probabilities sum up to the parameter SpectralProbability. A spectrum is considered identified in a database if any of the generated reconstructions is present in the database. We varied the value of SpectralProbability, and computed the number of spectra that were identified in the Shewanella database and the decoy database of the same size. This essentially mimics the database search with the spectral probability as the scoring function computed by MS-GF. Figure 6 provides a comparison between the number of identifications made by MS-GF and X!Tandem. Despite the fact that X!Tandem combined score utilizes information that MS-GF does not have access to, MS-GF outperforms X!Tandem. In addition, MS-GF, accurately estimates the number of hits in decoy database thus eliminating the need for the decoy database search altogether (see Supplement 2). This observation illustrates that computing scores over all possible peptides is better than observing scores over the relatively small decoy database.
Figure 6. Performance of MS-GF vs. X!Tandem.
The plots show the number of spectra identified in the Shewanella database and the corresponding error rate. (a) The spectral identifications in the Shewanella and decoy databases are divided into three groups, depending on whether the peptide endpoints are consistent with trypsin cleavage specificity: tryptic (both endpoints consistent), semi-tryptic (only one endpoint consistent) and non-tryptic (both endpoint inconsistent). The partition into these three groups illustrates MS-GF generates more tryptic peptides than the total number of peptides generated by X!Tandem.(b) Same as (a), but based on the number of unique peptides identified in each database (instead of the number of spectra). As expected, the number of peptides with both non-tryptic endpoints is very small.
Interpreting the “one-hit-wonders” is a difficult problem that often amounts to manual validations. The subjective nature of such inferences have resulted in the Proteomics Publication Guidelines to virtually discard single-hit protein identifications. In a large scale study, this inevitably results in the loss of large amounts of valuable information. For example, there are 402 proteins with single peptide hits in Shewanella oneidensis MR-1 [46] as opposed to 1992 proteins with multiple hits (over 20% of the expressed proteome).7 While we estimated that nearly 75% of these “one-hit-wonders” are correct identifications (as discussed in [46, 51]), no means were available to objectively separate them from the false identifications. Below we show how MS-GF (that provides a superior separation between correct and incorrect peptide identifications for low error rates) can be used for reliable identification of the single-hit proteins.
We computed SpectralProbability for the peptides identified in the decoy database in [46]8. The lowest value of SpectralP robability among all these decoy identifications is 1.55 × 10−8. Similarly, SpectralP robability was computed for the peptides from the single-hit proteins and the spectral probability for 345 of them was lower than 1.55 × 10−8. These 345 peptides represent better identifications than every identification in the decoy database, and the corresponding proteins must be considered reliably identified with virtually zero empirical error rate.9 We further remark that many single-hit-wonders with SpectralP robability below 1.55 × 10−8 are actually more statistically significant than some proteins with multiple peptide hits but larger SpectralP robability values (see [48, 49, 50, 44] for combining peptide significance scores into protein significance scores).
Discussion
While the previous approaches to evaluating the statistical significance of spectral identifications greatly improved the state of the art in peptide identification, they have not yet eliminated the decoy databases and empirical approximations from MS/MS searches. PeptideProphet [5] combines multiple scores into a single discriminant score, and fits its observed distribution to a mixture model comprising of a gaussian distribution for correct identifications and a gamma distribution for incorrect identifications. Sadygov and Yates, 2003 [6] argue that the frequencies of matches between fragment ions predicted from a random peptide and an experimental spectrum follow a hypergeometric distribution that is used to compute the probability that a peptide identification is correct. On the other hand, OMSSA tools [7] consider the same to be a Poisson distribution and accordingly compute the statistical significance of peptide identifications. These studies were taken further by Wan et al., 2006 [52] who realized the importance of generating some random peptides for estimating the statistical significance of the individual spectra (see also [53]) but stopped short of proposing a technique for analyzing all peptides. In an earlier work, Bafna and Edwards, 2003 [33] proposed an algorithm for generating suboptimal de novo reconstructions and suggested to use their score distribution for validating the optimal de novo reconstruction.
While the approaches [5, 6, 7, 16] are very valuable, neither of them rigorously solves the Spectral Matching Problem for individual spectra: instead they compute the error rates based on approximate fitting the empirical distributions to a standard distribution that may not carefully reflect the specifics of an individual spectrum. Moreover, they assume the same null hypothesis for all spectra in the sample, the assumption that may not be adequate for mass spectrometry searches. Our approach does not assume any “null hypothesis” or “noise model” for spectra generation as in [16]. Also, it does not assume any particular approximation for the tail of the score distribution. Instead, it rigorously solves the Spectrum Matching Problem, the same problem the existing approaches attempt to solve via decoy databases and various approximations.
MS-GF allows one to accurately estimate the statistical significance of individual spectral interpretations. As described above, MS-GF can be used either to complement the decoy searches or on its own. The former case illustrates the synergy between the decoy database and the generating function approaches in cases when the generating function framework can only be applied to the results of the decoy database searches10. The generating function approach can be further used to generate a list of all peptides whose score exceeds a threshold and match these peptides in the protein database, thus enabling a hybrid approach to peptide identification [54, 55, 28].
While the generating function described here evaluates the statistical significance over the set of all unmodified peptides, it can be extended to analyze modified peptides in both restricted and blind [56, 57, 25] modes. The former case amounts to adding “modification edges” of fixed length while the latter case amounts to adding modification edges of arbitrary length to the amino acid graph. The dynamic programming in the resulting graph should take into account the maximum allowed number of modifications per peptide.
Supplementary Material
Acknowledgments
We thank Richard Smith for providing the MS/MS datasets, Seungjin Na for help with some computational experiments, and Vineet Bafna, Nuno Bandeira, Alexey Nesvizhskii, and Stephen Tanner for many valuable comments. This work was supported by National Institutes of Health Grant NIGMS 1-R01-RR16522 and by Howard Hughes Medical Institute Professor Award.
Footnotes
The Spectrum Matching Problem assumes a certain probabilistic distribution on the set of all peptides and computes the total probability of all peptides P with score(P, S) > T.
Different papers on statistics of MS/MS searches often use inconsistent terminology. The solution of the Spectral Matching Problem provides E-values (the expected number of peptides with the scores equal to or larger than the observed score) or can be used for computing p-values in the hypothesis testing framework. To avoid a confusion, we follow the terminology from the recent review [1] and use the term FPR (and the related term Spectral Probability defined below) in the remainder of this paper.
The approach in [16] is particularly attractive since it can be implemented without decoy databases.
Partition functions in statistical mechanics represent a special class of generating functions and we use them below only to illustrate this notion in application to tandem mass spectra.
This expression represents the exponential generating function [22] of the vector x = (x(0), x(1), …). Similarly to many applications of generating functions outside physics, we follow Herbert Wilf's interpretation of generating functions (“a clothesline on which we hang up a sequence of numbers” as defined in [23]) rather than using it as a model of a physical process. As some other applications of generating functions in bioinformatics [41], we do not analyze the analytical behavior of the MS/MS generating functions in this paper.
We initialize x(0, 0) = 1 since the “empty” peptide is the only peptide with mass 0 that has 0 peaks in common with the spectrum S. We initialize y(0, 0) = 1 since the probability of the empty peptide is defined as 1.
For typical bacterial MS/MS projects, the percentage of one-hit-wonders is closer to 30% (see [51]). The percentage is somewhat smaller for the unusually large Shewanella dataset.
1417 peptides were identified in the decoy database as compared to 28,377 peptides identified in the Shewanella database as described in [46]. From 1417 peptides we selected the Charge-2 and unmodified peptides for this analysis, giving 1065 peptides.
See [28] for detection of sequencing errors and programmed frameshifts using a similar approach.
This is particularly relevant for estimating the error rates of protein identifications, re-scoring of complex non-additive scoring functions, or projects that can tolerate higher error rates (MS-GF in the database search mode becomes rather slow when high error rates are acceptable)
References
- 1.Nesvizhskii A, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nature Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 2.Kall L, Storey J, Maccoss M, Noble W. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 3.Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The Need for Guidelines in Publication of Peptide and Protein Identification Data: Working Group On Publication Guidelines For Peptide And Protein Identification Data. Mol Cell Proteomics. 2004;3:531. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw R, Burlingame A, Carr S, Aebersold R. Reporting Protein Identification Data: The next Generation of Guidelines. Mol Cell Proteomics. 2006;5:787–8. doi: 10.1074/mcp.E600005-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Keller A, Nesvizhskii A, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 6.Sadygov R, Yates J. A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence databases. Anal Chem. 2003;75:3792–3798. doi: 10.1021/ac034157w. [DOI] [PubMed] [Google Scholar]
- 7.Geer L, Markey S, Kowalak J, Wagner L, Xu M, Maynard D, Yang X, Shi W, Bryant S. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Elias J, Gygi S. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 10.Fenyo D, Phinney B, Beavis R. Determining the overall merit of protein identification data sets: rho-diagrams and rho-scores. J Proteome Res. 2007;6:1997–2004. doi: 10.1021/pr070025y. [DOI] [PubMed] [Google Scholar]
- 11.Higdon R, Hogan J, Belle G, Kolker E. Randomized sequence databases for tandem mass spectrometry Peptide and protein identification. OMICS. 2005;9:364–79. doi: 10.1089/omi.2005.9.364. [DOI] [PubMed] [Google Scholar]
- 12.Higgs R, Knierman M, Freeman A, Gelbert L, Patil S, Hale J. Estimating the statistical significance of peptide identifications from shotgun proteomics experiments. J Proteome Res. 2007;6:1758–1767. doi: 10.1021/pr0605320. [DOI] [PubMed] [Google Scholar]
- 13.Beausoleil S, Jedrychowski M, Schwartz D, Elias J, Villen J, Li J, Cohn M, Cantley L, Gygi S. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proceedings of the National Academy of Sciences. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W, Liu T, Monroe M, Strittmatter E, Jacobs J, Kangas L, Petritis K, Camp D, Smith R. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 15.Waterman M, Vingron M. Rapid and Accurate Estimates of Statistical Significance for Sequence Data Base Searches. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4625–4628. doi: 10.1073/pnas.91.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenyo D, Beavis R. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson J, Chait B, Fenyo D. A statistical basis for testing the significance of mass spectrometric protein identification results. Anal. Chem. 2000;72:999–1005. doi: 10.1021/ac990792j. [DOI] [PubMed] [Google Scholar]
- 18.Eng J, McCormack A, Yates J. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. Journal Of The American Society For Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Perkins D, Pappin D, Creasy D, Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Tanner S, Shu H, Frank A, Wang L, Zandi E, Mumby M, Pevzner P, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 21.Nagarajan N, Jones N, Keich U. Computing the P-value of the information content from an alignment of multiple sequences. Bioinformatics. 2005;21:i311–i318. doi: 10.1093/bioinformatics/bti1044. [DOI] [PubMed] [Google Scholar]
- 22.Graham R, Knuth D, Patashnik O. Concrete mathematics: a foundation for computer science. Addison-Wesley Longman Publishing Co., Inc.; Boston, MA, USA: 1989. [Google Scholar]
- 23.Wilf H. Generatingfunctionology. Academic Press; Boston, MA: 1994. [Google Scholar]
- 24.Pathria R. Statistical Mechanics. 2nd edition Butterworth-Heinemann; Oxford: 1996. [Google Scholar]
- 25.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner P. Identification of post-translational modifications via blind search of mass-spectra. Nature Biotechnology. 2005;23:1562–2567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 26.Bandeira N, Tsur D, Frank A, Pevzner P. Protein Identification via Spectral Network Analysis. Proceedings of the National Academy of Sciences. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandeira N, Olson J, Mann M, Pevzner P. De Novo Peptide Sequencing via MultiStage Mass Spectrometry. Bioinformatics. doi: 10.1093/bioinformatics/btn184. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Gupta N, Bandeira N, Pevzner P. Spectral dictionaries: Integrating de novo peptide sequencing with database search of tandem mass spectra. doi: 10.1074/mcp.M800103-MCP200. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor J, Johnson R. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal Chem. 2001;73:2594–2604. doi: 10.1021/ac001196o. [DOI] [PubMed] [Google Scholar]
- 30.Dancík V, Addona T, Clauser K, Vath J, Pevzner P. De novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 1999;6:327–342. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- 31.Chen T, Kao M, Tepel M, Rush J, Church G. A dynamic programming approach to de novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 2001;8:325–337. doi: 10.1089/10665270152530872. [DOI] [PubMed] [Google Scholar]
- 32.Frank A, Pevzner P. PepNovo: De Novo Peptide Sequencing via Probabilistic Network Modeling. Analytical Chemistry. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 33.Bafna V, Edwards N. On de-novo interpretation of tandem mass spectra for peptide identification; Proceedings of the Seventh Annual International Conference on Computational Molecular Biology; 2003. pp. 9–18. [Google Scholar]
- 34.Lu B, Chen T. A suboptimal algorithm for de novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 2003;10:1–12. doi: 10.1089/106652703763255633. [DOI] [PubMed] [Google Scholar]
- 35.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 36.Bern M, Goldberg D. De novo analysis of peptide tandem mass spectra by spectral graph partitioning. Journal of Computational Biology. 2006;13:364–78. doi: 10.1089/cmb.2006.13.364. [DOI] [PubMed] [Google Scholar]
- 37.Fischer B, Roth V, Roos F, Grossmann J, Baginsky S, Widmayer P, Gruissem W, Buhmann J. NovoHMM: A Hidden Markov Model for de Novo Peptide Sequencing. Anal. Chem. 2005;77:7265–7273. doi: 10.1021/ac0508853. [DOI] [PubMed] [Google Scholar]
- 38.Grossmann J, Roos F, Cieliebak M, Liptak Z, Mathis L, Muller M, Gruissem W, Baginsky S. AUDENS: a tool for automated peptide de novo sequencing. J. Proteome Res. 2005;4:1768–1774. doi: 10.1021/pr050070a. [DOI] [PubMed] [Google Scholar]
- 39.Dimaggio P, Jr, Floudas C. De Novo Peptide Identification via Tandem Mass Spectrometry and Integer Linear Optimization. Anal Chem. 2007;79:1433–1446. doi: 10.1021/ac0618425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo L, Dutta D, Wan Y. Msnovo: a dynamic programming algorithm for de novo peptide sequencing via tandem mass spectrometry. Anal. Chem. 2007;79:4870–4878. doi: 10.1021/ac070039n. [DOI] [PubMed] [Google Scholar]
- 41.Dewey T. A sequence alignment algorithm with an arbitrary gap penalty function. J Comput Biol. 2001;8:177–90. doi: 10.1089/106652701300312931. [DOI] [PubMed] [Google Scholar]
- 42.Frank A, Tanner S, Bafna V, Pevzner P. Peptide sequence tags for fast database search in mass-spectrometry. J. Proteome Res. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- 43.Tabb D, Fernando C, Chambers M. Myrimatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 45.Shilov I, Seymour S, Patel A, Loboda A, Tang W, Keating S, Hunter C, Nuwaysir L, Schaeffer D. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Gupta N, Tanner S, Jaitly N, Adkins J, Lipton M, Edwards R, Romine M, Osterman A, Bafna V, Smith R, Pevzner P. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craig R, Beavis R. TANDEM: matching proteins with tandem mass-spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 48.Nesvizhskii A, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 49.Tabb D, McDonald W, Yates J. Dtaselect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Chambers M, Tabb D. Proteomic Parsimony through Bipartite Graph Analysis Improves Accuracy and Transparency. J. Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta N, Benhamida J, Bhargava D, Goodman E, Kain I, Nguyen N, Ollikainen N, Rodriguez J, Wang J, Lipton M, Romine M, Bafna V, Smith R, Pevzner P. Comparative Proteogenomics: Combining Mass Spectrometry and Comparative Genomics to Analyze Multiple Genomes. Genome Res. doi: 10.1101/gr.074344.107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan Y, Yang A, Chen T. PepHMM: A Hidden Markov Model Based Scoring Function for Mass Spectrometry Database Search. Anal Chem. 2006;78:432–7. doi: 10.1021/ac051319a. [DOI] [PubMed] [Google Scholar]
- 53.Venable J, Yates J., 3rd Impact of ion trap tandem mass spectra variability on the identification of peptides. Anal Chem. 2004;76:2928–37. doi: 10.1021/ac0348219. [DOI] [PubMed] [Google Scholar]
- 54.Alves G, Yu Y. Robust accurate identification of peptides (RAId): deciphering MS2 data using a structured library search with de novo based statistics. Bioinformatics. 2005;21:3726–3732. doi: 10.1093/bioinformatics/bti620. [DOI] [PubMed] [Google Scholar]
- 55.Frank A, Savitski M, Nielsen M, Zubarev R, Pevzner P. De novo Peptide sequencing and identification with precision mass spectrometry. J Proteome Res. 2007;6:114–23. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen B, Davey S, Ham A, Liebler D. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J Proteome Res. 2005;4:358–68. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 57.Searle B, Dasari S, Turner M, Reddy A, Choi D, Wilmarth P, McCormack A, David L, Nagalla S. High-throughput identification of proteins and unanticipated sequence modifications using a mass-based alignment algorithm for MS/MS de novo sequencing results. Anal. Chem. 2004;76:2220–2230. doi: 10.1021/ac035258x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.