SUMMARY
Serum response factor (SRF) is a prototypic transcription factor that mediates stimulus-dependent gene expression. Here, we show that SRF mediates NGF signaling, axonal growth, branching, and target innervation by embryonic DRG sensory neurons. Conditional deletion of the murine SRF gene in DRGs results in no deficits in neuronal viability or differentiation but causes defects in extension and arborization of peripheral axonal projections in the target field in vivo, similar to the target innervation defects observed in mice lacking NGF. Moreover, SRF is both necessary and sufficient for NGF-dependent axonal outgrowth in vitro, and NGF regulates SRF-dependent gene expression and axonal outgrowth through activation of both MEK/ERK and MAL signaling pathways. These findings show that SRF is a major effector of both MEK/ERK and MAL signaling by NGF and that SRF is a key mediator of NGF-dependent target innervation by embryonic sensory neurons.
INTRODUCTION
Sensory neurons of the dorsal root ganglion (DRG) undergo complex yet stereotypic changes during embryonic development. In the mouse embryo, migrating neural crest precursors coalesce into DRGs beginning around E9.5 (White et al., 1996). Shortly thereafter, DRG neurons elaborate peripheral and central axonal projections that innervate distinct peripheral targets, such as skin and muscle, and synapse with neurons in the spinal cord, respectively. Despite this generalized projection pattern, DRG neurons are greatly diversified with respect to both morphological and physiological properties. For example, most large-diameter DRG neurons express the neurotrophin receptor TrkC, project myelinated, proprioceptive axons to muscle spindles, and are dependent on the neurotrophin NT-3 for their development (Snider and Silos-Santiago, 1996). In contrast, the majority of DRG neurons possess small-diameter soma, express the neurotrophin receptor TrkA during embryonic development, and project unmyelinated or thinly myelinated fibers to innervate the epidermis (Marmigere and Ernfors, 2007). These cutaneous axons are nociceptors and thermoceptors, and depend on target-derived nerve growth factor (NGF) during development.
NGF is a prototypic neurotrophic growth factor that controls many aspects of cutaneous sensory neuronal development by promoting survival, maturation, and final target innervation. In particular, the establishment of cutaneous innervation involves a carefully orchestrated series of steps, including axonal outgrowth and extension, branching, defasciculation, and penetration of the epidermis. NGF is clearly implicated in several of these steps: indeed, cultured DRG neurons extend axons when exposed to NGF in vitro (Levi-Montalcini and Cohen, 1956). Moreover, NGF is expressed in skin, and newborn mice lacking NGF have impaired branching and epidermal innervation in vivo (Patel et al., 2000).
How does target-derived NGF support DRG axon extension, branching, and epidermal innervation? In vitro studies show that the mitogen-activated protein kinase kinase/extracellular regulated kinase (MEK/ERK) and phosphatidylinositol-3-kinase/Akt (PI3K/AKT) signaling pathways mediate NGF-induced sensory axon growth. The PI3K pathway regulates local assembly of the axonal cytoskeleton via glycogen synthase kinase 3β(GSK3β) and multiple microtubule-binding proteins (Zhou et al., 2004). The MEK/ERK pathway phosphorylates numerous effector proteins and is thought to control axon outgrowth via local signaling mechanisms at the growth cone (Markus et al., 2002). More recently, analyses of mouse mutants lacking particular MEK/ERK pathway intermediates show that signaling through this pathway is crucial for branching and extension of cutaneous sensory axons in the target field in vivo (Zhong et al., 2007).
Despite an abundance of evidence implicating MEK/ERK and PI3K signaling pathways in NGF-dependent axonal growth, the relative contributions of local NGF signaling within the growth cone and retrograde NGF signaling to the cell body and to nuclear effectors such as transcription factors are unclear. Indeed, the identification of transcription factors and downstream target genes that mediate NGF-dependent axonal extension, branching, and target field innervation is currently a major challenge.
The transcription factor serum response factor (SRF) is a key mediator of stimulus-dependent transcription of immediate-early genes (IEGs) in cell lines (Treisman, 1987). SRF is a member of the MADS-box transcription factor family, which binds to a consensus sequence CC(A/T)6GG known as the CArG box (Rivera et al., 1990). SRF is constitutively bound to CArG boxes found in the promoters of numerous cytoskeletal and immediate-early genes, including c-fos, c-jun, Egr-1, Vcl, actin, and SRF itself (Miano, 2003). Although the CArG box is sufficient to mediate SRF-dependent transcription, many growth factor-responsive IEG promoter regions contain one or more serum response elements (SREs), typified by the combination of a CArG box with an adjacent ETS site (Miano, 2003). Growth factors and other stimuli activate SRF-dependent transcription by recruiting cofactors to SRF or to adjacent cis regulatory elements. Although many have been identified, two major families of SRF cofactors have been extensively studied: the ETS-domain-containing ternary complex factor (TCF) family, which includes Elk-1, SAP-1, SAP-2/Net, and the myocardin family, which includes MAL/MKL1/MRTF-A, MKL2/MRTF-B, and myocardin. The combinatorial effect of SRF and the binding of SRF cofactors to transcriptional regulatory sites dictates the specific repertoire of genes induced by different stimuli. In support of this model, MEK/ERK signaling leads to robust phosphorylation of TCFs promoting their binding to the ETS site on the SRE and SRE-mediated transcription (Marais et al., 1993). MEK/ERK-TCF-SRF transcription controls expression of a number of growth-promoting genes. In contrast, activation of Rho GTPases triggers SRF-dependent transcription through a mechanism involving the SRF coactivator, MAL. In its latent form, MAL is sequestered in the cytosol by binding to monomeric G-actin. Activation of local actin treadmilling by Rho GTPases, and the consequent accumulation of F-actin, leads to a commensurate depletion of G-actin and the translocation of MAL to the nucleus (Miralles et al., 2003). Nuclear accumulation of MAL promotes the formation of the MAL-SRF complex and expression of genes involved in reorganization of the cytoskeleton (Miano et al., 2007).
Despite a large body of in vitro work, the in vivo contribution of SRF to growth factor signaling and gene expression during nervous system development is relatively unknown. Targeted deletion of the murine SRF gene reveals that SRF is critical for early mesodermal differentiation; SRF−/− mice die in utero as early as E6.5, precluding the use of these mice in studying nervous system development (Arsenian et al., 1998). Recently, the availability of SRF conditional mutant mouse lines has facilitated the study of the in vivo function of this transcription factor (Ramanan et al., 2005; Wiebel et al., 2002). It is now known that deletion of SRF in the developing nervous system results in impaired migration of neurons in the rostral migratory stream, (Alberti et al., 2005), whereas deletion of SRF in the perinatal hippocampus results in deficits in formation of the mossy fiber pathway (Knoll et al., 2006). In the adult hippocampus, loss of SRF results in mice with impaired induction of IEGs and defects in plasticity (Ramanan et al., 2005). Despite these advances in understanding the role of SRF in the central nervous system, the in vivo contributions of SRF to neurotrophic growth factor signaling, gene expression, and neuronal development remain largely undiscovered.
Here, we show that SRF is a critical mediator of NGF signaling, axonal growth, branching, and epidermal innervation by embryonic DRG sensory neurons. Moreover, SRF is both necessary and sufficient for NGF-dependent axonal outgrowth in vitro, and NGF regulates SRF-dependent axonal outgrowth through activation of both MEK/ERK and MAL signaling pathways. Finally, we demonstrate that NGF is essential for the expression of several SRF-dependent cytoskeletal genes in embryonic DRG neurons in vivo. Together, our findings suggest that SRF is a major effector of both MEK/ERK and MAL signaling by NGF and that activation of SRF target gene expression mediates NGF-dependent cutaneous innervation by embryonic sensory neurons.
RESULTS
Expression of SRF in DRG Neurons Is Regulated by NGF
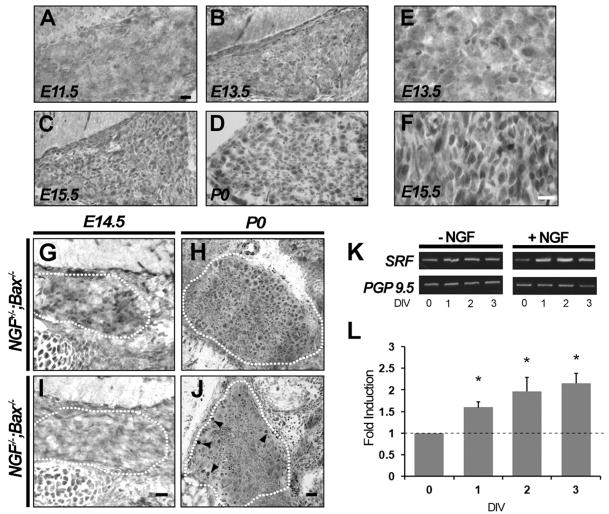
Although SRF is widely expressed in the adult mouse, whether it is expressed in neurons of the peripheral nervous system during development is unknown. To address this, DRGs of embryonic mice at E11.5, E13.5, E15.5, and P0 were immunostained using an antibody against SRF. At E11.5, SRF protein was barely detectable in DRGs, but by E13.5 neuronal nuclei were immunolabeled with anti-SRF (Figures 1A and 1B). In these cells, SRF levels increased progressively, with a peak level of expression observed between E13.5 and E15.5 (Figures 1B–1F). We confirmed that the antibody is specific to SRF, as virtually all neuronal staining was lost in mice with a targeted deletion of SRF (Figures 2A and 2B). Interestingly, the times at which SRF expression peaks in DRG neurons coincide with times at which these sensory neurons encounter target-derived neurotrophins, such as NGF. NGF, in particular, signaling through its receptor TrkA, is essential for the expression of numerous genes that may contribute to survival, differentiation, and axonal outgrowth of cutaneous sensory neurons. Therefore, we next asked whether NGF promotes expression of SRF in vivo.
Figure 1. NGF Regulates Expression of SRF in Developing DRG Neurons.
(A–D) The developmental expression of SRF in DRG neurons at E11.5, E13.5, E15.5, and at P0 is shown. SRF protein was barely detectable in DRGs at E11.5, but by E13.5 neuronal nuclei were clearly immunolabeled with SRF. Scale bar, 20 μm.
(E and F) High magnification of DRG neurons at E13.5 and E15.5 shows that peak increase in SRF expression occurs between these times. After P0, no further increase in the expression of SRF in DRG neurons was observed (data not shown). Similar results were found in each of four to six sections from two or three animals. Scale bar, 20 μm.
(G–J) Expression of SRF in NGF+/−;Bax−/− and NGF−/−;Bax−/− DRGs at E14.5 (G and I) and at P0 (H and J). Note that at E14.5 SRF expression was readily observed in DRG neurons in NGF+/−;Bax−/− control mice, although in NGF−/−;Bax−/− animals SRF immunostaining was virtually undetectable. In contrast, SRF expression was detectable in DRG neurons from NGF−/−;Bax−/− mice at P0, albeit at a significantly reduced intensity compared to littermate controls. Interestingly, large-diameter neurons in the periphery of NGF−/−;Bax−/− DRGs showed unperturbed SRF staining (arrowheads). Scale bar, 20 μm.
(K) RT-PCR analysis of SRF expression in cultured DRG neurons grown for 1–3 days in vitro (DIV) in the presence or absence of NGF. DRG neurons were protected from apoptosis in the absence of NGF by supplementation of culture medium with pan-caspase inhibitor BAF. Expression of the panneuronal marker PGP9.5 serves as a control.
(L) Real-time PCR analysis show that expression of SRF is induced over several days in cultured DRG neurons by NGF. Compared to control unstimulated cultures, NGF stimulation induced expression of SRF 1.63 ± 0.13, 1.96 ± 0.32, and 2.144 ± 0.22 fold at 1, 2, and 3 DIV, respectively. Shown are means ± SEM (n = 3, *p < 0.05 using two-way ANOVA with Mann-Whitney U test post hoc analysis).
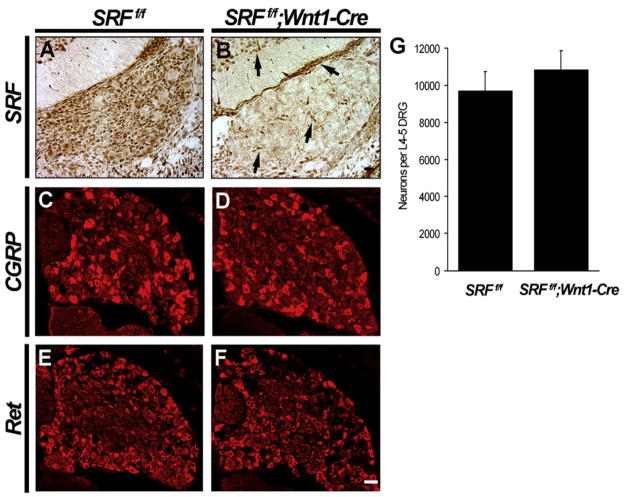
Figure 2. SRF Is Dispensable for Survival and Maturation of DRG Neurons In Vivo.
(A and B) Loss of SRF expression in DRG sensory neurons in SRFf/f;Wnt1-Cre mice at E15.5. Note that the deletion of SRF is limited only to neural crest derived cells in the DRG and not seen in fibroblasts and spinal interneurons (black arrows) (n = 3). Scale bar, 20 μm.
(C–F) CGRP and Ret immunostaining in SRFf/f and SRFf/f;Wnt1-Cre animals at E17.5 reveal that the maturation of peptidergic (C and D) and nonpeptidergic (E and F) neurons are unaffected in the absence of SRF (n = 3). Scale bar, 40 μm.
(G) SRF is dispensable for neuronal viability. Cell counts were obtained by Nissl staining and whole-cell counting by scoring neuronal profiles from L4–L5 DRGs by taking embryonic cross-sections at the boundaries of the renal calyces at E17.5. Shown are means ± SEM of neuronal counts for six L4–L5 ganglia.
Although the majority of DRG neurons die of apoptosis by E14.5 in mice lacking NGF, the concomitant deletion of the proapoptotic gene Bax rescues these neurons and affords us the unique ability to study the survival-independent functions of NGF. Thus, NGF−/−;Bax−/− mice are valuable for understanding how NGF controls gene expression in peripheral neurons (Luo et al., 2007). Immunohistochemistry using the anti-SRF antibody in DRGs from NGF−/−;Bax−/− and NGF+/−;Bax−/− littermate control mice revealed that the level of SRF was significantly reduced in the absence of NGF signaling at both E14.5 and P0 (Figures 1G–1J). A few larger-diameter neuronal nuclei confined to the periphery of the DRG showed unperturbed expression of SRF in NGF−/−;Bax−/− mice at P0 (arrowheads, Figure 1J). This finding can be explained by the NGF independence of large-diameter proprioceptive neurons. To address whether NGF signaling is indeed sufficient to increase SRF expression, DRG neurons from E13.5 mouse embryos were cultured and exposed to NGF in vitro. In keeping with the in vivo findings and previous work showing stimulus-dependent expression of SRF (Misra et al., 1991), we observed a 2.1-fold increase in expression of SRF by real-time PCR in DRG neurons exposed to NGF compared to untreated controls (Figures 1K and 1L). These findings indicate that expression of SRF in developing DRG neurons is regulated by NGF.
SRF Conditional Mutant Mice
We next sought to determine whether SRF-dependent gene expression contributes to development of sensory neurons. Because of the early lethality of SRF−/− embryos, mice harboring a LoxP-based conditional SRF allele (SRFf/f) (Ramanan et al., 2005) were crossed to mice carrying a Wnt1-Cre transgene (Danielian et al., 1998), which directs expression of Cre recombinase in premigratory neural crest cells, including all progenitors of DRG neurons (Figure S1A). Mice heterozygous for both the floxed SRF allele and the Wnt1-Cre transgene were viable and fertile and exhibited no obvious deficits. In contrast, intercrosses of SRFf/f and SRFf/+;Wnt1-Cre mice failed to generate progeny in typical Mendelian ratios (Table S1). Analysis of SRFf/f;Wnt1-Cre embryos revealed that mid-to-late gestational lethality in these mice may be attributable to defects in patterning of the embryonic vasculature, as evidenced by mispatterned blood vessels in the head and neck region as well as other tissues derived from the cranial and cardiac neural crest (unpublished data). Nevertheless, the coalescence of peripheral ganglia derived from the trunk neural crest was unaffected in SRFf/f; Wnt1-Cre mice. The trigeminal ganglia, dorsal root ganglia, and the paravertebral sympathetic ganglia of the trunk all formed normally and were grossly intact.
SRF Is Dispensable for DRG Neuronal Viability and Differentiation
Despite the apparent integrity of DRGs in SRF mutant mice, we sought to ascertain whether loss of SRF disrupted NGF signaling and caused DRG neurons to undergo apoptosis. To ask whether SRF is required for NGF-dependent survival, we performed cresyl violet staining of DRGs from SRFf/f and SRFf/f;Wnt1-Cre DRGs at E17.5. By this age, most naturally occurring cell death in DRGs is complete, and, therefore, we could reliably assess the extent to which SRF is necessary for survival of these neurons. We found that SRF mutant mice had a normal complement of DRG neurons, indicating that SRF is dispensable for viability of sensory neurons (Figure 2G).
We next asked whether SRF is important for maturation of DRG neurons. During embryonic development, TrkA+ small-diameter DRG neurons further differentiate into two distinct classes of nociceptive neurons. These neuronal subsets are classified as nonpeptidergic and peptidergic; each of these neuronal subsets expresses a unique repertoire of genes and elaborates axonal projections in distinct patterns (Marmigere and Ernfors, 2007). In order to ascertain whether SRF influences differentiation of these neuronal classes, we performed a series of immunohistochemistry and in situ hybridization analyses using antibodies and probes against many proteins and genes that are normally expressed in these sensory neurons (Figures 2C–2F and see Table S2). Surprisingly, no obvious defects in expression of either peptidergic or nonpeptidergic markers were observed in the SRF mutants. These findings indicate that DRG neurons undergo phenotypic differentiation normally in the absence of SRF.
SRF Is Required for Branching and Extension of Peripheral but Not Central Projections of DRG Sensory Neurons In Vivo
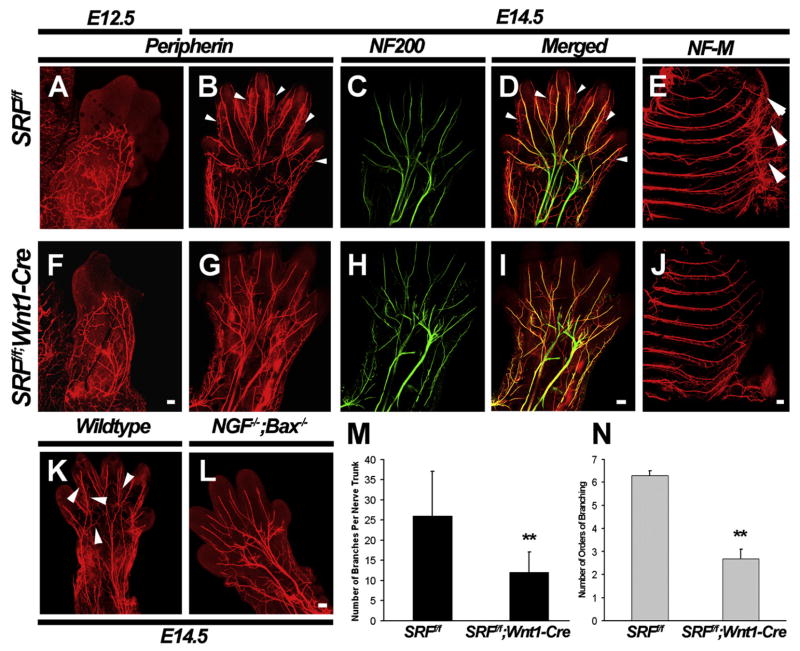
Because of the requirement of NGF in the extension, branching, and epidermal innervation of TrkA+ DRG neurons (Patel et al., 2000), we next investigated whether SRF is required for axonal extension or branching in vivo. For these analyses, a whole-mount immunofluorescent staining assay using antibodies directed against peripherin was developed. This assay enables a detailed assessment of all peripheral projections of small-diameter DRG sensory neurons. Whole-mount peripherin immunolabeling of E12.5 embryos revealed that axons of DRG neurons were beginning to innervate the limb buds at this time. However, little to no difference in the initial extension of these axons was discernible between SRFf/f and SRFf/f;Wnt1-Cre mutant embryos (Figures 3A and 3F). By E14.5, the main trunks of sensory axons innervating the limbs in wild-type animals are seen innervating the tips of the embryonic digits. These main axonal bundles continue to arborize into more superficial cutaneous fibers that spread throughout the surface of the limb (arrowheads, Figures 3B and 3D). In SRFf/f;Wnt1-Cre mutant embryos, in contrast, the terminal extension and arborization of cutaneous sensory axons were severely affected, although the main nerve trunks and their principal branches remained intact (arrowheads, Figures 3G and 3I). In particular, the main trunks did not extend as far into the digits and failed to elaborate as many superficial branches as seen in control embryos. In fact, the number of peripherin+ branches per nerve trunk and the number of times each main nerve trunk underwent subsequent branching events (orders of branching) were both substantially reduced in SRFf/f;Wnt1-Cre mutants compared to littermate controls (Figures 3M and 3N). Similar defects were observed in the intercostal nerves (Figures 3E and 3J). We also determined the extent to which the SRF mutant mice phenocopy those found in NGF−/−;Bax−/− mice. Surprisingly, the majority of axons were able to reach the target field normally in NGF−/−;Bax−/− mice; axonal extension and branching defects were limited to the final phase of target innervation (Figures 3K and 3L), similar to those observed in SRFf/f;Wnt1-Cre mice (Figure 3G). Interestingly, the central spinal projections of small-diameter neurons were seen innervating the dorsal horn of the spinal cord in SRF mutant and control mouse lines (Figures S2C–S2F), similar to that seen in mice lacking NGF (Patel et al., 2000 and data not shown). These findings suggest that SRF mediates NGF-dependent branching and extension of peripheral but not central axonal projections of DRG sensory neurons.
Figure 3. SRF Is Required for Peripheral Target Innervation by Embryonic DRG Neurons In Vivo.
(A and F) Whole-mount peripherin immunostaining of limb buds from SRFf/f and SRFf/f;Wnt1-Cre embryos at E12.5 show little to no defect in axonal branching and extension in peripheral projections of DRG neurons in vivo. Scale bar, 20 μm.
(B–D and G–I) A comparison of whole-mount peripherin and neurofilament-200 (NF200) double immunostaining of E14.5 SRFf/f and SRFf/f;Wnt1-Cre embryos reveals that peripheral projections to the limbs are impaired in the absence of SRF. These defects are, however, only limited to small-diameter peripherin+ neurons that are dependent on NGF. Large-diameter myelinated axons visualized by NF200 staining are unaffected in SRF mutant mice at this age. Scale bar, 100 μm.
(E and J) A similar analysis by immunostaining for Neurofilament-M (NF-M) demonstrates that SRF mutant mice have terminal intercostal nerve fibers that are substantially reduced in branching and extension, compared to littermate controls. Scale bar, 100 μm.
(K and L) Peripherin immunohistochemistry in limbs from wild-type and NGF−/−;Bax−/− embryos at E14.5 show similar defects to those seen in SRF mutant embryos. Scale bar, 100 μm.
(M and N) Quantitation of branching defects in SRFf/f;Wnt1-Cre mice at E14.5. Quantitation was performed by using two parameters. The number of branches per nerve trunk was quantitated by counting the total number of branch points observed along each major nerve trunk entering the forelimb. The number of orders of branching was analyzed by counting the number of times each major nerve trunk underwent subsequent branching events. Shown are means ± SEM (n = 3, for SRFf/f and SRFf/f;Wnt1-Cre embryos for each age, n = 2 for NGF−/−;Bax−/− and Bax−/−) (**p < 0.01 using Student’s t test).
The Function of SRF in Axon Extension and Branching Is Cell Autonomous
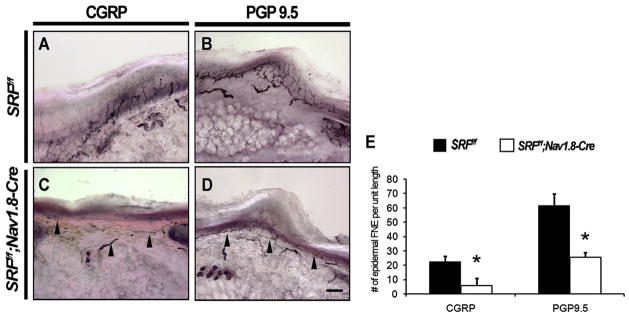
Wnt1-Cre-mediated excision of SRF results in loss of SRF in nonneuronal derivatives of the neural crest, including Schwann cells, which may contribute to the axonal defects observed in SRFf/f;Wnt1-Cre mice. To ask whether SRF functions cell autonomously within sensory neurons to control peripheral axonal projections, SRFf/f mice were crossed to a mouse line in which Cre recombinase has been knocked into the Nav1.8 locus. The Nav1.8-Cre mouse line is useful for excision of floxed genes exclusively in NGF-responsive small-diameter sensory neurons (Nassar et al., 2004). Unlike SRFf/f;Wnt1-Cre mutant mice, SRFf/f;Nav1.8-Cre mutant mice were viable, fertile, and exhibited no obvious morphological or behavioral abnormalities. To assess whether DRG small-diameter neuron-specific deletion of SRF affects axonal extension and skin innervation, we performed immunostaining for the peptidergic neuronal marker CGRP and the panneuronal marker PGP9.5 in glabrous skin sections from the hindlimbs of SRFf/f control and SRFf/f;Nav1.8-Cre mice at P30. We found that SRFf/f;Nav1.8-Cre mutant mice had few PGP9.5+ and CGRP+ axonal termini innervating the epidermis compared to SRFf/f littermate controls (Figure 4). A dramatic reduction of skin innervation in SRFf/f;Nav1.8-Cre mice is also evident at P0, suggesting that the adult phenotypes are due to a persistence of embryonic target innervation defects and not due to postnatal loss of axons from the epidermis (Figures S3E and S3F). Other neuronal populations that do not express Nav1.8-Cre were unaffected in SRFf/f;Nav1.8-Cre mice; neither TH+ sympathetic fibers nor Neurofilament 200+ large-diameter myelinated fibers were impaired in the mutants (Figures S3A–S3D). Together, these observations show that SRF functions cell autonomously, within sensory neurons, to mediate axonal extension, branching, and epidermal innervation.
Figure 4. Target Innervation Defects in Mice Lacking SRF Are Cell Autonomous.
(A–D) Cell-autonomous target innervation defects in mice lacking SRF were analyzed by examining innervation to the hindlimb footpad in SRFf/f and SRFf/f; Nav1.8-Cre mice at P30. A significant loss of epidermal innervation was observed in SRFf/f;Nav1.8-Cre mice by immunostaining for CGRP to visualize peptidergic projections (A and C), as well as by immunostaining with the panneuronal marker PGP9.5 (B and D). Scale bar, 60 μm.
(E) Quantification of the number of CGRP+ and PGP9.5+ free nerve endings (FNE) crossing the dermal-epidermal border. Shown are the means ± SEM of eight to ten sections from six animals. Unit length is 500 μm of glabrous skin. (*p < 0.05 using Student’s t test).
SRF Is Necessary for NGF-Dependent Axonal Outgrowth in DRG Neurons In Vitro
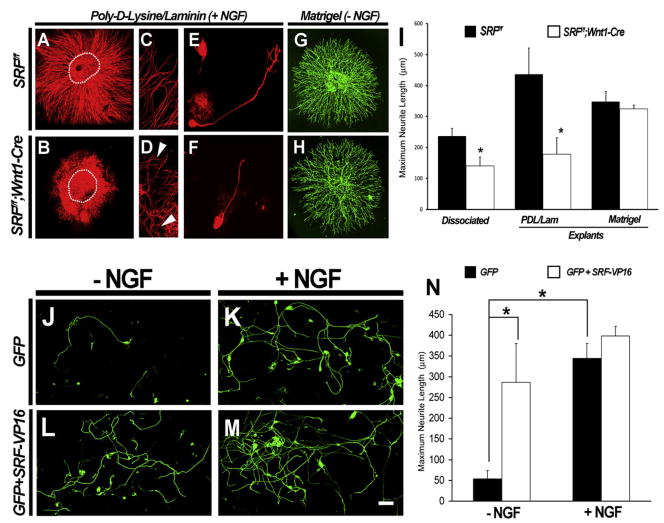
To further characterize the role of SRF in NGF-dependent axon extension and branching, in vitro experiments using dissociated DRG and explant cultures from SRF mutant mice were performed. Embryonic DRG explants from SRFf/f mice robustly project axons that extend from the explant when exposed to NGF (Figures 5A and 5C). In contrast, DRG explants from SRFf/f; Wnt1-Cre mice show fewer and shorter neurites (Figures 5B, 5D, and 5I and Figure S4A). A similar axonal projection deficit was observed in dissociated cultures of DRG neurons from SRFf/f control and SRFf/f;Wnt1-Cre mutant DRGs at E13.5 (Figures 5E, 5F, and 5I). Moreover, differences in axonal outgrowth in control and mutant DRG neurons were not due to differences in neuronal viability because activated caspase-3 immunoreactivity of dissociated DRG neurons was unaffected by the absence of SRF (Figure S5). Thus, SRF is required for NGF-dependent extension of axons from embryonic DRG neurons in vitro.
Figure 5. SRF Is Necessary and Sufficient for NGF-Dependent Axonal Outgrowth In Vitro.
(A–D) Explants from SRFf/f and SRFf/f;Wnt1-Cre mice demonstrate that SRF is necessary for NGF-dependent axonal outgrowth. Arrowheads point to shorter and stunted axons in SRF mutant explants compared to controls (compare [C] with [D]).
(E and F) Dissociated neurons from SRFf/f and SRFf/f;Wnt1-Cre mice cultured on laminin/poly-D-lysine substrate at very low density in the presence of NGF show that neurons from SRF mutant mice respond to NGF with reduced axonal outgrowth.
(G and H) However, when grown on matrigel substrate, both SRFf/f and SRFf/f;Wnt1-Cre mutant DRG explants are able to extend axons to comparable extents, suggesting that SRF is not required for all forms of axonal extension.
(I) Quantification of the data, showing a comparison of axonal outgrowth defects from explants (PDL/laminin and matrigel) and dissociated neurons from SRFf/f and SRFf/f;Wnt1-Cre mice. Shown are means ± SEM (n = 4 for each genotype, n = 3 for explants grown on matrigel, *p < 0.05 using Student’s t test).
(J–N) Transfection of wild-type rat DRG neurons with constructs expressing GFP alone (J and K) or GFP + SRF-VP16 (L and M) demonstrate that constitutively active SRF can promote axonal extension in DRG neurons in the absence of NGF. These data are quantitated in (N). Shown are means ± SEM (n = 4), *p < 0.05 using two-way ANOVA with Mann-Whitney U test post hoc analysis). Scale bar, 30 μm.
Despite the uniformity of the in vivo and in vitro findings, the question of whether SRF regulates the basal machinery responsible for axonal outgrowth remains unanswered. In light of previous work demonstrating SRF as a regulator of the actin cytoskeleton, it is plausible that axonal growth and branching defects seen in the SRF mutants are due to reduced basal expression of cytoskeletal genes such as Actin. We therefore asked whether SRF is required for generalized axonal extension of sensory neurons or whether this transcription factor is strictly required for NGF-dependent axonal extension and branching. Indeed, the in vivo analyses support a case for SRF’s involvement during the NGF-dependent phase of cutaneous sensory neuron axonal outgrowth; the peripheral projections of SRFf/f;Wnt1-Cre and NGF−/−;Bax−/− mutant mice were similarly defective, whereas the central projections of these mutant mice were equally unaffected. To ask whether SRF mediates axonal extension in other populations of sensory neurons, or whether it is required for NGF-independent extension of cutaneous sensory neurons, we cultured DRG explants from SRFf/f and SRFf/f;Wnt1-Cre embryos at E14.5 in the presence of BDNF or NT3, neurotrophins that support distinct populations of DRG neurons, or on matrigel, a substrate that permits outgrowth of cutaneous sensory axons in a neurotrophin-independent manner (Tonge et al., 1997). We observed that loss of SRF modestly affected axonal outgrowth of cultured DRG sensory neurons responsive to NT3, whereas BDNF-mediated axonal outgrowth from SRF-deficient neurons was unaffected (Figure S4B). Remarkably, outgrowth of axons from DRG explants from SRFf/f;Wnt1-Cre mice was virtually identical to outgrowth from SRFf/f control explants after 48 hr on the matrigel substrate (Figures 5G and 5H). These axons are derived from small-diameter cutaneous neurons, as evidenced by immunostaining these explants with TrkA (data not shown). These observations negate the possibility that loss of SRF disrupts the basal machinery for axonogenesis in DRG neurons and suggest that TrkA+ DRG neurons retain the ability to extend axons even in the absence of SRF.
SRF Is Sufficient for Axon Outgrowth in Dissociated DRG Neurons In Vitro
Although our results show that NGF-dependent axonal extension requires a transcriptional component mediated by SRF, they do not address the possibility that SRF-dependent gene expression is sufficient for axonal growth. We therefore tested whether expression of a constitutively active form of SRF (SRF-VP16) (Johansen and Prywes, 1994) is sufficient to promote axonal outgrowth in the absence of NGF. This was achieved through electroporation of dissociated rat DRG neurons in culture with plasmid constructs expressing either GFP alone or GFP and SRF-VP16 in the presence of the pan-caspase inhibitor BAF. In GFP-electroporated cultures, little to no axonal outgrowth was observed in the absence of NGF, whereas robust outgrowth of axons was seen in the presence of NGF (Figures 5J, 5K, and 5N). In striking contrast, DRG cultures electroporated with both GFP and SRF-VP16 showed robust axonal outgrowth even in the absence of NGF (Figures 5L and 5N). Moreover, the addition of NGF to these cultures did not enhance axon outgrowth any further (Figures 5M and 5N). These data, taken together, suggest that activation of SRF-dependent gene expression is both necessary and sufficient for NGF-dependent axonal extension.
SRF Is Not Required for Signaling through the NGF/TrkA Receptor Complex
The similarities in the phenotypes between SRFf/f;Wnt1-Cre and NGF−/−;Bax−/− embryos prompted us to ask whether deletion of SRF leads to a loss of neurotrophin receptors or a reduction of NGF-TrkA signaling. Our earlier observations that loss of SRF affects neither the viability of DRG neurons nor the expression of NGF-dependent genes necessary for nociceptive maturation argue against this possibility. Nonetheless, to further test this idea, we examined the expression of the neurotrophin receptors TrkA, TrkB, TrkC, and p75 in mice lacking SRF by real-time PCR. Expression of none of these genes is affected by the absence of SRF (Figure S6C). Furthermore, immunoblotting for activated effectors of the NGF signaling pathway, pTrkA, pERK1/2, and pAkt, demonstrate that downstream effectors of NGF are properly activated in the absence of SRF (Figure S6A). Finally, localization of the NGF receptors TrkA and p75 to the leading edge of growth cones was indistinguishable in DRG neurons from mutant and littermate control mice (Figure S6B). Thus, loss of SRF does not affect expression, trafficking, or signaling by neurotrophin receptors.
NGF Stimulates SRF-Dependent Transcription through a MAPK-Dependent, MAL-Dependent Pathway
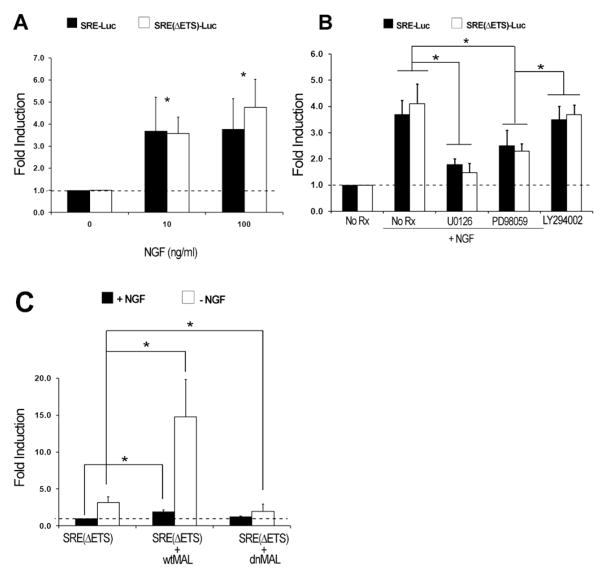
We next considered whether SRF mediates NGF-dependent gene transcription in DRG sensory neurons. Previous work showed that NGF and other stimuli activate immediate-early gene (IEG) transcription in PC12 cells in a manner dependent on the integrity of SREs within IEG promoters (Visvader et al., 1988). Moreover, an isolated SRE is sufficient to mediate growth factor-dependent expression of a heterologous reporter construct in cell lines (Treisman, 1987). To determine whether NGF could indeed stimulate SRF-dependent transcription in DRG neurons, we assessed the activity of an SRE-luciferase reporter gene upon exposure of transfected primary rat DRG neurons to NGF. Following NGF withdrawal from the culture medium, re-exposure to NGF led to a 2.5- to 4.5-fold increase in the activity of the SRE reporter, suggesting that the SRE can indeed mediate NGF-dependent transcription in these neurons (Figure 6A). We next utilized SRE(ΔETS)-luciferase, a reporter construct that contains a multimerized CArG box, which binds to SRF, but no adjacent ETS binding site, which mediates interactions with the TCFs (Kalita et al., 2006). This reporter enables us to test whether TCF activation is a necessary step for NGF-dependent activation of the SRE reporter gene. NGF robustly activated the SRE(ΔETS)-luciferase reporter in embryonic rat DRG neurons, suggesting that NGF activation of SRF-dependent transcription in DRG neurons does not require the association of TCFs to an adjacent ETS binding site (Figure 6A). However, the MEK inhibitors U0126 and PD98059 both strongly repressed transcription of the SRE(ΔETS)-luciferase reporter, as strongly as they repressed the SRE luciferase reporter, showing that MEK/ERK pathway activation was required for NGF signaling to SRF (Figure 6B). Of note, two inhibitors of PI3K/AKT signaling, LY294002 and wortmannin, had little to no effect on NGF-induced transcription of SRE(ΔETS)-luciferase (Figure 6B and data not shown). Together, these findings argue for a NGF-TrkA-MEK/ERK-SRF signaling pathway that mediates SRE-dependent transcription independent of TCFs.
Figure 6. NGF Signals to SRF through Activation of MEK/ERK and MAL Signaling Pathways.
(A) Luciferase reporter assays in primary cultures of embryonic rat DRG neurons using SRE-luciferase and SRE(ΔETS)-luciferase demonstrate that the ETS-binding site for TCF cofactors is dispensable for SRF-dependent transcription in DRG sensory neurons by NGF. Luciferase activity is reported as fold induction, which is the ratio between normalized SRE (or SREΔETS) -firefly luciferase activity and constitutively active thymidine kinase-Renilla luciferase activity of each condition, normalized to the ratio obtained from unstimulated DRG neurons.
(B) Luciferase reporter assays in primary cultures of rat DRG neurons demonstrating that SRF-mediated transcription by NGF is dependent on signaling through MEK/ERK. The pharmacologic MEK inhibitors U0126 and PD98059 were used to inhibit MEK/ERK signaling, while the inhibitor LY294002 was used to inhibit PI3K/AKT signaling.
(C) Luciferase reporter assays in embryonic rat DRG neurons stimulated with NGF using SRE(ΔETS)-luciferase demonstrate the effect of electroporating neurons with wtMAL and dnMAL constructs. Shown are means ± SEM (from at least four independent experiments with each condition performed in four to six replicates) *p < 0.05 using two way-ANOVA with Mann-Whitney U test post hoc analysis.
We next tested the potential involvement of the SRF transcriptional coactivator MAL in NGF-SRF signaling because MAL family members mediate TCF-independent SRF-dependent transcription (Cen et al., 2003). In many cell types, virtually all of the cytoskeletal target genes regulated by SRF are controlled by MAL (Miralles et al., 2003; Selvaraj and Prywes, 2004). In light of these findings, a dominant-negative MAL (dnMAL) construct previously shown to specifically disrupt MAL signaling, but not TCF-dependent signaling (Cen et al., 2003; Selvaraj and Prywes, 2004), was used to ask whether MAL was responsible for the activation of SRF-dependent transcription by NGF. Indeed, DRG neurons expressing dnMAL exhibited greatly reduced SRE(ΔETS)-luciferase reporter activation following exposure to NGF (Figure 6C). Similarly, the use of inhibitors that stabilize G-actin pools preventing MAL nuclear translocation, such as latrunculin B, also greatly diminished reporter activation by NGF (data not shown). Remarkably, neurons expressing full-length MAL (wtMAL) dramatically augmented NGF-induced SRE-reporter activation (Figure 6C). These data suggest that activation of both MEK/ERK and MAL are essential for NGF-dependent SRF/SRE transcription.
NGF Stimulates Neurite Outgrowth in DRG Neurons in a MAP Kinase- and MAL-Dependent Manner
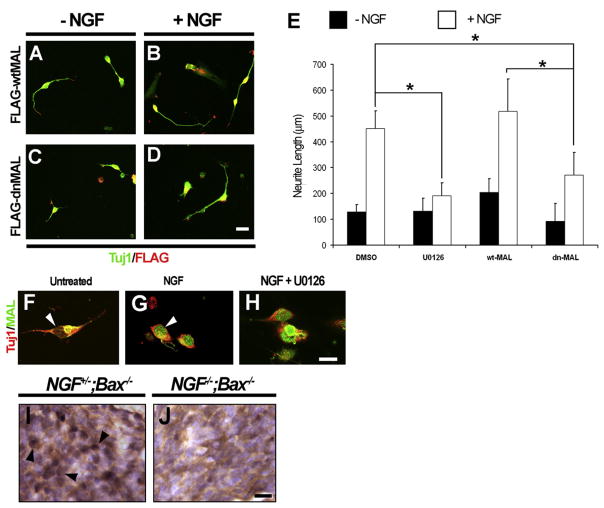
Because SRF-mediated transcription induced by NGF in DRG neurons is dependent on both MEK/ERK and MAL, we investigated whether this signaling dependence translates to a similar requirement of MEK/ERK and MAL signaling in NGF-dependent axonal outgrowth. The requirement of MEK/ERK signaling of NGF-dependent axonal outgrowth of sensory neurons (Markus et al., 2002) was confirmed in experiments using the MEK inhibitor U0126. Compared to DMSO-treated control cultures, embryonic DRG cultures stimulated with NGF in the presence of U0126 yielded substantially shorter axons (Figure 7E). To investigate whether MAL signaling is similarly required for NGF-dependent axon outgrowth, embryonic rat DRG neurons were electroporated with FLAG-tagged full-length MAL (FLAG-wtMAL) or FLAG-tagged dominant-negative MAL (FLAG-dnMAL) and grown in the absence or presence of NGF. Full-length MAL did not affect neurite outgrowth from DRG neurons (Figures 7A, 7B, and 7E). In contrast, FLAG-dnMAL significantly reduced axon outgrowth in DRG neurons treated with NGF (Figures 7C–7E), suggesting that NGF-dependent axon outgrowth requires the activation of MAL.
Figure 7. Axonal Outgrowth Mediated by NGF Is Dependent on Both MEK/ERK and MAL Signaling.
(A–E) NGF-dependent axonal outgrowth in DRG neurons requires both MEK/ERK and MAL signaling. Shown are dissociated cultures of embryonic rat DRG neurons stimulated with or without NGF, electroporated with constructs expressing wtMAL (A and B) or dnMAL (C and D). Cultures were immunostained with antibodies against FLAG (for FLAG-tagged MAL constructs) and the panneuronal marker Tuj1, and axonal measurements were taken to quantify the extent of axonal outgrowth. The MEK inhibitor U0126 was used to assess the effect of MEK/ERK inhibition compared to the inhibition of MAL signaling using dnMAL. These data are quantified in (E). Shown are means ± SEM (n = 3). Scale bar, 25 μm (*p < 0.05, using two-way ANOVA and Mann-Whitney U test post hoc analysis).
(F–H) Dissociated embryonic rat DRG neuronal cultures demonstrating nuclear translocation of MAL following NGF stimulation. (F) After 3 days of serum starvation, MAL immunostaining can be detected in a perinuclear distribution, whereas, (G) after NGF stimulation MAL immunostaining is observed in a predominantly nuclear distribution. (H) Nuclear translocation of MAL following NGF stimulation is not dependent on MEK/ERK signaling, as evidenced by the translocation of MAL even in the presence of MEK inhibitor U0126. The panneuronal marker Tuj1 was used to identify neuronal cells. Scale bar, 20 μm.
(I and J) NGF-dependent nuclear localization of MAL is observed in vivo. Shown are DRG sections from E14.5 NGF+/−;Bax−/−control and NGF−/−;Bax−/− embryos immunostained with anti-MAL antibody and counterstained with hematoxylin. Note that, in the NGF+/−;Bax−/−control animals, MAL staining is visible in numerous neuronal nuclei, whereas MAL is detectable in only a few neuronal nuclei in DRG sections from mice lacking NGF. Scale bar, 10 μm.
Certain stimuli activate MAL-dependent transcription, at least in part, through MAL translocation from the cytosol to the nucleus where it forms a complex with SRF (Miralles et al., 2003). Indeed, activation of Rho GTPases in embryonic cortical neurons and serum stimulation of fibroblasts cause MAL translocation to the nucleus, while, on the other hand, MAL is constitutively retained in the nucleus in adult cortical neurons (Kalita et al., 2006; Miralles et al., 2003; Tabuchi et al., 2005). We therefore tested whether the subcellular distribution of MAL changes in response to NGF stimulation in DRG neurons. When dissociated rat DRG neurons were stimulated with NGF, translocation of MAL from a perinuclear distribution to a predominantly nuclear localization was observed (Figures 7F and 7G). Interestingly, the nuclear translocation of MAL induced by NGF in vitro continued unperturbed in the presence of U0126 (Figure 7H), suggesting that the influence of MEK/ERK signaling on SRF-dependent gene expression and axon outgrowth did not occur through the regulation of MAL trafficking. We were also able to observe NGF dependence for MAL translocation in vivo. DRGs from NGF−/−; Bax−/− mice immunostained for MAL at E14.5 revealed MAL staining in a mostly cytosolic pattern of expression with nuclear sparing (Figure 7J), while, in contrast, MAL immunostaining was seen in many neuronal nuclei in NGF+/−;Bax−/− littermate control mice (arrows, Figure 7I). Therefore, MAL nuclear translocation is dependent on NGF in sensory neurons both in vivo and in vitro and is essential for both NGF-dependent SRE-mediated gene expression and axonal extension.
NGF Regulates the Expression of Several Cytoskeletal Genes through SRF-Dependent Transcription
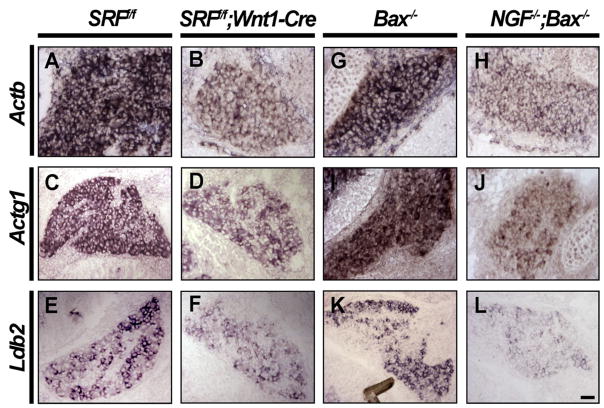
In light of the importance of SRF in NGF-dependent target innervation, we next sought to identify NGF and SRF target genes, some of which are presumed to be important for axon extension, branching, and epidermal innervation. SRF, through MAL signaling in particular, regulates the expression of a large cohort of cytoskeletal genes, including α actin, β actin, γ actin, vinculin, and SRF itself (Selvaraj and Prywes, 2004). It is not known, however, whether SRF is important for the regulation of these genes in sensory neurons or whether NGF increases expression of these SRF-dependent cytoskeletal genes. To address this, expression patterns of a large group of cytoskeletal genes as well as genes whose expression requires NGF (K.M. and D.D.G., unpublished data) were assessed by in situ hybridization on DRG sections from SRFf/f and SRFf/f;Wnt1-Cre mice. At E14.5, DRG neurons lacking SRF express substantially reduced levels of β actin and γ actin (Figures 8A–8D). We also found that expression of the LIM family transcriptional regulator Ldb2, which is found in a highly selective pattern in DRG neurons, was diminished in the absence of SRF by E16.5 (Figures 8E and 8F). Ldb2 is a member of the LIM-domain-binding (CLIM/Ldb) cofactor family that associates with LIM homeodomain proteins, a number of which are implicated in neuronal development. In contrast, expression of several other cytoskeletal genes, including β-III tubulin, was unaffected in SRF mutants (Figure S7).
Figure 8. NGF Controls Expression of Several SRF Target Genes.
(A–D) In situ hybridization using probes against β actin (Actb) and γ actin (Actg1) demonstrate that these cytoskeletal genes are greatly reduced in the absence of SRF in DRGs from E14.5 SRFf/f and SRFf/f;Wnt1-Cre DRGs.
(E and F) SRF controls the expression of a LIM family transcriptional regulator, Ldb2, in E16.5 SRFf/f and SRFf/f;Wnt1-Cre DRGs (n = 3 for each genotype). (G–L) In situ hybridization analyses confirm that these SRF target genes are also regulated by NGF using DRG sections from NGF+/+;Bax−/−and NGF−/−;Bax−/− mice at E14.5 (G–J) and at E16.5 (K and L) (n = 2 for each genotype), Scale bar, 40 μm.
We also determined how loss of NGF influences the expression of these SRF target genes. In DRGs from NGF−/−;Bax−/− mice, expression of β actin and γ actin were greatly compromised compared to Bax−/−littermate controls at E14.5 (Figures 8G–8J). Also, at E16.5, DRGs from NGF−/−;Bax−/− mice showed very low levels of Ldb2 expression compared to Bax−/−control DRGs, suggesting that NGF also controls expression of this transcription factor (Figures 8K and 8L). It is noteworthy that SRF itself is a transcriptional target of SRF, forming a positive-feedback loop to further augment SRF expression (Misra et al., 1991). This positive-feedback loop likely explains our observation that SRF expression increases in an NGF-dependent manner in DRG neurons during embryonic development (Figures 1G–1J). Thus, NGF regulates expression of β actin, γ actin, Ldb2, and probably SRF itself, through an SRF-dependent transcriptional mechanism.
DISCUSSION
We report that SRF is a critical mediator of NGF signaling, gene expression, axonal growth, and target innervation by embryonic DRG sensory neurons. SRF is dispensable for viability and maturation of these neurons but is essential for extension and arborization of their axonal projections in the target field in an NGF-dependent manner. Moreover, SRF is both necessary and sufficient for NGF-dependent axonal outgrowth in vitro, and NGF regulates SRF-dependent axonal outgrowth through activation of both MEK/ERK and MAL signaling pathways. We propose a model in which target-derived NGF retrogradely signals through MEK/ERK and MAL-dependent pathways to promote SRF-dependent transcription in DRG sensory neurons (Figure S8).
NGF Control of Target Innervation Involves an SRF-Dependent Transcriptional Mechanism
The precise requirement of NGF for axon extension and branching of DRG neurons has not been fully understood. Our in vivo findings from NGF−/−;Bax−/− mice implicate a specialized role for NGF, limited to controlling the terminal phase of target innervation in vivo. This suggests that, in the mouse embryo, emerging axons from newly coalesced DRGs reach NGF-rich target fields through NGF-independent mechanisms. The molecules responsible for this initial phase of NGF-independent axonal outgrowth in DRG neurons are unknown. Nevertheless, in vivo, it is clear that distal axon extension, branching, and innervation of the embryonic epidermis are dependent on NGF.
The sequence of events between the initiation of NGF/TrkA signaling and the onset of axonal extension and branching has not been elucidated. A large body of evidence supports a role for MEK/ERK signaling in this process both in vitro and in vivo (Markus et al., 2002; Zhong et al., 2007). Nevertheless, the identity of components downstream of MEK/ERK signaling that mediate NGF-dependent axonal outgrowth and whether local axonal control and retrograde transcriptional mechanisms are both required in this process are unknown. In this study, we implicate SRF as a nuclear target of retrograde MEK/ERK signaling and as a transcriptional mediator of MEK/ERK-dependent target innervation by NGF.
Although several studies implicate the transcription factors CREB and NFAT in axonal outgrowth, the precise roles of these transcription factors in mediating NGF-dependent transcriptional responses in sensory neurons is less clear. First, the onset of phenotypes in both CREB−/−and NFATc2/c3/c4−/− mice occurs considerably earlier than the time at which DRG neurons first become NGF dependent (Graef et al., 2003; Lonze et al., 2002). Second, axonal extension phenotypes in mice lacking these transcription factors are more severe than in mice lacking NGF. Third, these analyses were all performed in null mutant mice, which lack the cell-autonomous control necessary to preclude axonal outgrowth defects secondary to glial cell or target dysfunction. The present findings with SRF support a specific role for this transcription factor in NGF-dependent target innervation. SRF mutant mice show phenotypes that are remarkably similar to innervation defects seen in NGF−/−;Bax−/− mice. Moreover, these phenotypes occur at times when DRG neurons first become NGF dependent and, importantly, occur in a cell-autonomous manner. Our findings argue in favor of a model in which NGF controls target innervation by DRG sensory neurons, at least in part through retrograde MEK/ERK signaling to the nucleus to stimulate SRF-dependent transcription (Figure S8). Future studies will establish the relative contributions of SRF, CREB, and NFAT during sensory neuron axonal development and whether these transcription factors have identical, overlapping, or unique sets of target genes.
An NGF-TrkA-SRF Signaling Pathway Controlling Axon Growth and Branching
Our work implicates signaling through both the MEK/ERK and MAL signaling pathways during SRF-dependent transcription, axonal growth, and target innervation mediated by NGF. These observations, while defining the mechanism of NGF signaling to SRF, raise the question of how these two distinct signaling pathways converge. One attractive model posits that MEK/ERK signaling is able promote MAL phosphorylation, which may be necessary for MAL-dependent transcription. Indeed, in cortical neurons, activation of MEK/ERK signaling by the neurotrophin BDNF leads to phosphorylation of MAL (Kalita et al., 2006).
The prospect of MAL as a key intermediate of NGF to SRF signaling in target innervation is far more compelling. The Rho family GTPases Rac1 and Cdc42 are potent activators of MAL signaling and are also well-characterized effectors of NGF signaling. In fact, Rho GTPases are critical for NGF-mediated axonal extension in neurons (Van Aelst and D’Souza-Schorey, 1997), possibly through the activation of MAL. Numerous studies also report that MAL association with nuclear SRF controls cytoskeletal reorganization by promoting expression of a number of cytoskeletal genes (Selvaraj and Prywes, 2003). Furthermore, in vitro studies using dominant-negative MAL constructs demonstrate that MAL family members are critical for neurite extension (Knoll et al., 2006; Shiota et al., 2006). The ability of MAL to translocate to the nucleus following NGF stimulation also raises the exciting possibility of retrograde transport of MAL from the axon terminus to the nucleus. Because the release of cytosolic MAL is primarily triggered by actin treadmilling, it is conceivable that local Rho GTPase and actin polymerization signals activated in the growth cone and distal axon by NGF/TrkA signaling are conveyed to SRF through the trafficking of MAL from the distal axon to the cell body, a potentially unique mode of retrograde signaling in these neurons.
How Does SRF Exert Control over Target Innervation by NGF?
It is likely that an axon undergoing branching and extension over a short timeframe suffers an acute shortage of cytoskeletal precursors, unless these precursors are replenished. When axons of DRG neurons first encounter NGF, retrograde signaling and SRF-dependent transcription may serve to acutely increase the availability of these cytoskeletal proteins. Indeed, we found that both β actin and γ actin, key cytoskeletal target genes regulated by SRF, are also controlled by NGF. In addition, the LIM domain family cofactor Ldb2, an NGF-dependent SRF target gene, is expressed specifically in DRG neurons during development and has been implicated in axonal outgrowth. Indeed, studies in zebrafish have demonstrated that Ldb proteins are critical for target innervation by peripheral but not central axonal projections of sensory neurons, which is remarkably analogous to our observations in SRF and NGF mutant mice (Becker et al., 2002; Segawa et al., 2001). Together, these data suggest that NGF influences changes in the cytoskeleton through SRF-dependent gene expression both by directly regulating cytoskeletal genes and by regulating the expression of secondary transcriptional events.
It is curious that, despite the reduction in cytoskeletal gene expression, the outgrowth of central projections to spinal laminae are unaffected in the absence of SRF, and the majority of axonal extensions into the limbs and trunk in vivo are unaffected. In vitro, we have shown that extension of axons in matrigel is also SRF independent. These observations imply that either basal expression of cytoskeletal genes in the absence of SRF is sufficient for initial axonal outgrowth or that the mechanism by which SRF controls NGF-dependent target innervation is mediated via expression of target genes that remain to be established. Indeed, the reduction in cytoskeletal gene expression cannot at this time be causally linked to defects in axonal outgrowth or branching.
From a developmental standpoint, the mechanism by which SRF mediates NGF-dependent target innervation illustrates the complexity of neurotrophic factor physiology in the development of cutaneous sensory neurons. NGF/TrkA signaling clearly has pleiotropic effects, but what is astounding is the ability of this single ligand-receptor pair to activate a divergent set of transcription factors, each of which controls a distinct function. Thus, NGF-dependent activation of CREB family members controls the expression of genes that are necessary for growth and survival of DRG neurons (Lonze et al., 2002). In the present study, we show that NGF activates SRF to control axonal branching, extension, and target innervation. Yet, other transcriptional mechanisms that control NGF-dependent expression of Ret, CGRP, and genes that define the peptidergic and nonpeptidergic sensory neuronal subtypes remain undiscovered.
Finally, the potential of SRF-dependent transcription in the postnatal neuron has not escaped our notice. Another scenario where neurons may be required to acutely increase the availability of cytoskeletal precursors is during regeneration following nerve injury. For example, SRF controls expression of a number of IEGs, many of which are also upregulated in DRG neurons following acute nerve injury (Ramanan et al., 2005; Seijffers et al., 2006). Whether SRF is required for the induction of gene expression events that mediate neuronal regeneration is an exciting avenue for future investigation.
EXPERIMENTAL PROCEDURES
Generation of Mice
SRFf/f mice (Ramanan et al., 2005) were mated to either a mouse strain expressing Cre recombinase under control of the Wnt1 promoter (Danielian et al., 1998) to generate SRFf/f;Wnt1-Cre mice or to a mouse strain in which Cre recombinase was inserted into the Nav1.8 locus (Nassar et al., 2004) to generate SRFf/f;Nav1.8-Cre mice. NGF−/−;Bax−/− mice were generated as described (Patel et al., 2000).
Dissociated DRG Neuronal Cultures and DRG Explant Cultures
Dissociated DRG neurons were isolated by enzymatic digestion of whole DRGs from E14.5 C57/BL6 mice or E15–16 Sprague-Dawley rat embryos using a method previously described for the isolation of sympathetic neurons (Kuruvilla et al., 2004). For explant cultures, DRGs were isolated as previously described (Lonze et al., 2002) and plated on coverslips precoated with poly-D-lysine/laminin or matrigel (BD Biosciences) diluted 1:2 in PBS. The culture medium for dissociated and explant studies was Neurobasal medium with B27 supplement (GIBCO), 35 mM glucose, 1 mM L-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin.
Luciferase Reporter Assays
Neurons were electroporated with the appropriate firefly luciferase reporter and renilla luciferase constructs (8:1 ratio) and cultured overnight using Neurobasal-B27-supplemented media with NGF (50 ng/ml). Cells were then serum starved for 48 hr in Neurobasal-B27 medium supplemented with BAF without NGF. NGF and appropriate inhibitors were reintroduced to the culture medium for 24–48 hr. Luciferase assays were performed using a commercial dual-luciferase reporter assay kit following the manufacturer’s instructions (Promega). SRE transcriptional activity was reported by comparing the normalized ratio of firefly luciferase activity to renilla luciferase activity and unstimulated control samples.
Cell Counts
E17.5 animals were sacrificed, and DRG cell counting was performed as described previously (Lonze et al., 2002).
Quantitation of Axonal Outgrowth
Quantitation of axonal outgrowth was performed using low-density (<50,000 cells/well on a 12-well dish) cultures of DRG neurons on coverslips precoated with laminin/poly-D-lysine substrate. Quantitation was performed within 24 hr of seeding, by postfixing cells using 4% paraformaldehyde and following standard immunocytochemistry protocols. At least five images of each coverslip were photographed, and at least 20 axons were quantitated per condition. Axon length measurement and tracing were automated using NeuronJ software (Meijering et al., 2004).
Supplementary Material
Acknowledgments
We thank Drs. Alex Kolodkin, John W. Griffin, Fenquan Zhou, Ravi Misra, and members of the Ginty laboratory for helpful discussion and comments on this manuscript. We thank Michelle Polley, Takako Makita, Wenqin Luo, Ting Guo, and Caitlin Engelhard for reagents, technical support, and valuable comments; Drs. Ron Prywes and Jay Baraban for generously providing plasmid constructs; and Dr. Mohammed Nassar for the generation of Nav1.8-Cre mice. This work was supported by the National Institutes of Health grant NS34814 (D.D.G.), the Silvio Conte Center for Neuroscience Research (D.D.G.), the Medical Research Council, UK (J.N.W.), and the NIH-Medical Scientist Training Program (S.R.W.). D.D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The Supplemental Data for this article, including Experimental Procedures, figures, and tables, can be found online at http://www.neuron.org/cgi/content/full/58/4/532/DC1/.
References
- Alberti S, Krause SM, Kretz O, Philippar U, Lemberger T, Casanova E, Wiebel FF, Schwarz H, Frotscher M, Schutz G, Nordheim A. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci USA. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Ostendorff HP, Bossenz M, Schluter A, Becker CG, Peirano RI, Bach I. Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech Dev. 2002;117:75–85. doi: 10.1016/s0925-4773(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, Prywes R. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Johansen FE, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita K, Kharebava G, Zheng JJ, Hetman M. Role of mega-karyoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J Neurosci. 2006;26:10020–10032. doi: 10.1523/JNEUROSCI.2644-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll B, Kretz O, Fiedler C, Alberti S, Schutz G, Frotscher M, Nord-heim A. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat Neurosci. 2006;9:195–204. doi: 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye H, Ginty DD. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R, Cohen S. In vitro and in vivo effects of a nerve growth-stimulating agent isolated from snake venom. Proc Natl Acad Sci USA. 1956;42:695–699. doi: 10.1073/pnas.42.9.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of non-peptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Misra RP, Rivera VM, Wang JM, Fan PD, Greenberg ME. The serum response factor is extensively modified by phosphorylation following its synthesis in serum-stimulated fibroblasts. Mol Cell Biol. 1991;11:4545–4554. doi: 10.1128/mcb.11.9.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar MA, Stirling LC, Forlani G, Baker MD, Matthews EA, Dickenson AH, Wood JN. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc Natl Acad Sci USA. 2004;101:12706–12711. doi: 10.1073/pnas.0404915101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–357. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schutz G, Linden DJ, Ginty DD. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Sheng M, Greenberg ME. The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev. 1990;4:255–268. doi: 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Allchorne AJ, Woolf CJ. The transcription factor ATF-3 promotes neurite outgrowth. Mol Cell Neurosci. 2006;32:143–154. doi: 10.1016/j.mcn.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota J, Ishikawa M, Sakagami H, Tsuda M, Baraban JM, Tabuchi A. Developmental expression of the SRF co-activator MAL in brain: role in regulating dendritic morphology. J Neurochem. 2006;98:1778–1788. doi: 10.1111/j.1471-4159.2006.03992.x. [DOI] [PubMed] [Google Scholar]
- Snider WD, Silos-Santiago I. Dorsal root ganglion neurons require functional neurotrophin receptors for survival during development. Philos Trans R Soc Lond B Biol Sci. 1996;351:395–403. doi: 10.1098/rstb.1996.0034. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Estevez M, Henderson JA, Marx R, Shiota J, Nakano H, Baraban JM. Nuclear translocation of the SRF co-activator MAL in cortical neurons: role of RhoA signalling. J Neurochem. 2005;94:169–180. doi: 10.1111/j.1471-4159.2005.03179.x. [DOI] [PubMed] [Google Scholar]
- Tonge DA, Golding JP, Edbladh M, Kroon M, Ekstrom PE, Edstrom A. Effects of extracellular matrix components on axonal outgrowth from peripheral nerves of adult animals in vitro. Exp Neurol. 1997;146:81–90. doi: 10.1006/exnr.1997.6498. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Visvader J, Sassone-Corsi P, Verma IM. Two adjacent promoter elements mediate nerve growth factor activation of the c-fos gene and bind distinct nuclear complexes. Proc Natl Acad Sci USA. 1988;85:9474–9478. doi: 10.1073/pnas.85.24.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FA, Silos-Santiago I, Molliver DC, Nishimura M, Phillips H, Barbacid M, Snider WD. Synchronous onset of NGF and TrkA survival dependence in developing dorsal root ganglia. J Neurosci. 1996;16:4662–4672. doi: 10.1523/JNEUROSCI.16-15-04662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebel FF, Rennekampff V, Vintersten K, Nordheim A. Generation of mice carrying conditional knockout alleles for the transcription factor SRF. Genesis. 2002;32:124–126. doi: 10.1002/gene.10049. [DOI] [PubMed] [Google Scholar]
- Zhong J, Li X, McNamee C, Chen AP, Baccarini M, Snider WD. Raf kinase signaling functions in sensory neuron differentiation and axon growth in vivo. Nat Neurosci. 2007;10:598–607. doi: 10.1038/nn1898. [DOI] [PubMed] [Google Scholar]
- Zhou FQ, Zhou J, Dedhar S, Wu YH, Snider WD. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 2004;42:897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.