Abstract
Besides its role in glycolysis, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) initiates a cell death cascade1–9. Diverse apoptotic stimuli activate inducible nitric oxide synthase (iNOS) or neuronal NOS (nNOS), with the generated nitric oxide (NO) S-nitrosylating GAPDH, abolishing its catalytic activity and conferring on it the ability to bind to Siah1, an E3-ubiquitin-ligase with a nuclear localization signal (NLS). The GAPDH–Siah1 protein complex, in turn, translocates to the nucleus and mediates cell death; these processes are blocked by procedures that interfere with GAPDH–Siah1 binding. Nuclear events induced by GAPDH to kill cells have been obscure. Here we show that nuclear GAPDH is acetylated at Lys 160 by the acetyltransferase p300/CREB binding protein (CBP) through direct protein interaction, which in turn stimulates the acetylation and catalytic activity of p300/CBP. Consequently, downstream targets of p300/CBP, such as p53 (refs 10–15), are activated and cause cell death. A dominant-negative mutant GAPDH with the substitution of Lys 160 to Arg (GAPDH-K160R) prevents activation of p300/CBP, blocks induction of apoptotic genes and decreases cell death. Our findings reveal a pathway in which NO-induced nuclear GAPDH mediates cell death through p300/CBP.
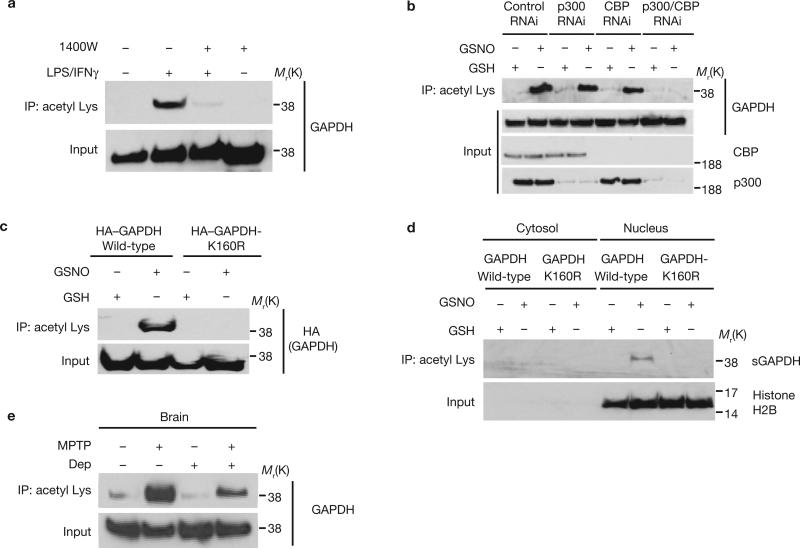
p300 and its closely related homologue CBP are the most prominent nuclear protein acetyltransferases16,17. In macrophage RAW264.7 cells activated with lipopolysaccharide (LPS) and interferon-γ (IFNγ), the active components of endotoxin, GAPDH translocates to the nucleus in an NO-dependent manner1. Under the same conditions, we observed acetylation of GAPDH, which was abolished by treatment with N-(3-(aminomethyl)benzyl)acetamidine (1400W), a selective iNOS inhibitor (Fig. 1a). To ascertain whether p300 and/or CBP are physiologically responsible for GAPDH acetylation in intact cells, we depleted p300 and CBP by RNA interference (RNAi). Depletion of either protein decreased GAPDH acetylation, which was abolished following depletion of both CBP and p300 (Fig. 1b). We also observed acetylation of GAPDH by p300 in vitro (Supplementary Information, Fig. S1a). Moreover, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometric analysis of nuclear GAPDH in HEK293 cells following apoptotic stress revealed acetylation at Lys 160 (Supplementary Information, Fig. S1b, c). To confirm the site of acetylation in intact cells, we transfected HEK293 cells with GAPDH or a mutant GAPDH (GAPDH-K160R). We observed acetylation of wild-type but not the K160R mutant GAPDH (Fig. 1c). We previously reported that nuclear GAPDH is sulphonated1. In HEK293 cells, sulphonated GAPDH in the nucleus was acetylated (Fig. 1d). The K160R mutation selectively abolished acetylation, but did not alter GAPDH catalytic activity, S-nitrosylation or nuclear translocation (Supplementary Information, Fig. S1d, e, f).
Figure 1.
GAPDH is acetylated in the nucleus at Lys 160 following NO stimulation. (a) GAPDH acetylation in RAW264.7 cells treated with LPS/IFNγ for 16 h is abolished by the iNOS inhibitor 1400W. Cell lysates were immunoprecipitated with an anti-acetyl Lys antibody, and the immunoprecipitates were analysed by western blotting with an anti-GAPDH antibody. (b) Both p300 and CBP contribute to acetylation of GAPDH in U2OS cells treated with the NO donor, GSNO. Depletion of p300 or CBP by RNAi leads to diminished acetylation of GAPDH. (c) GAPDH mutation at Lys 160 abolishes its acetylation in the presence of 200 μM GSNO in HEK293 cells. (d) Acetylation of sulphonated GAPDH (sGAPDH) is observed only in the nucleus where it requires intact Lys 160. Cytosolic or nuclear fractions of HEK293 cells were immnoprecipiated with an anti-acetyl Lys antibody, and the immunoprecipitates were analysed by western blotting with a specific anti-sulphonated-GAPDH antibody. (e), R-(−)-Deprenyl (Dep) inhibits the acetylation of GAPDH in brains of mice treated with MPTP.
We previously reported activation of GAPDH–Siah signalling in brains of mice treated with the dopamine neuronal toxin MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine)18. Augmented acetylation of GAPDH occurs in MPTP-treated mouse brains and is reversed by pre-treatment with R-(−)-deprenyl, which selectively blocks S-nitrosylation and nuclear translocation of GAPDH18 (Fig. 1e; Supplementary Information, Fig. S2a). In dopaminergic neuroblastoma SH-SY5Y cells treated with MPP+ (1-methyl-4-phenylpyridium), an active metabolite of MPTP, acetylation of GAPDH was increased, an effect blocked by the NOS inhibitor N(G)-nitro-L-arginine methyl ester (L-NAME) (Supplementary Information, Fig. S2b).
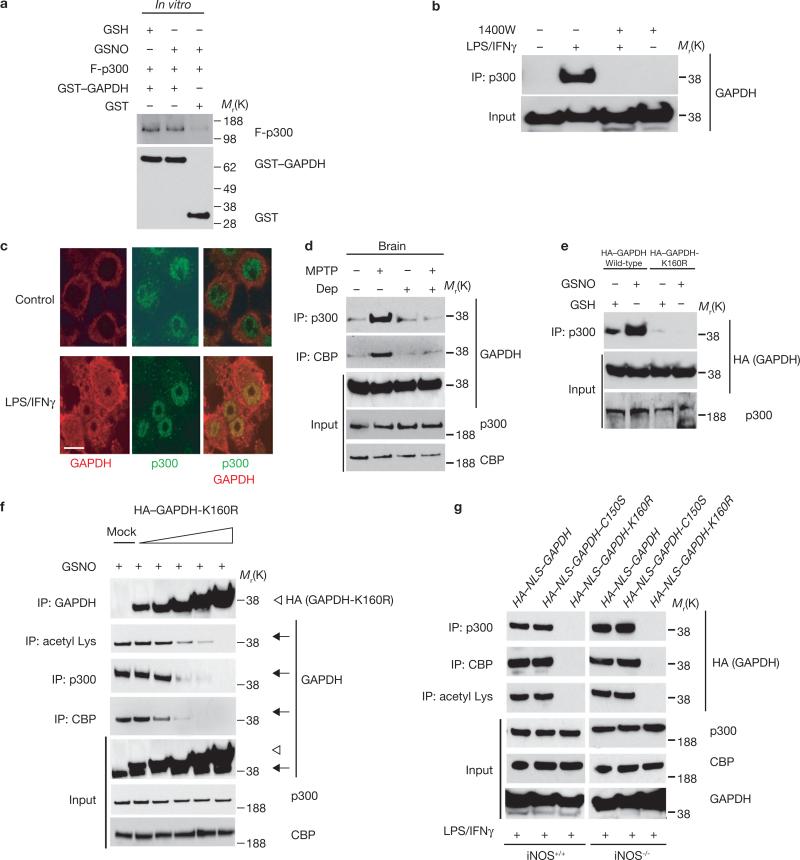
p300 binds directly to many of its substrate targets, such as p53 (refs 10–12). We observed binding of p300 to GAPDH in vitro (Fig. 2a). In intact RAW264.7 cells, LPS/IFNγ treatment elicited GAPDH–p300 binding, which was prevented by inhibition of iNOS (Fig. 2b). GAPDH also bound to CBP in an NO-dependent manner (Supplementary Information, Fig. S2c). In RAW264.7 cells p300 and GAPDH were colocalized in the nuclei in a granular pattern (Fig. 2c, Supplementary Information, Fig. S2d). Interactions between GAPDH and p300/CBP were augmented in brains of mice treated with MPTP; this was reversed by pretreatment with R-(−)-deprenyl (Fig. 2d). Binding of GAPDH to p300/CBP requires intact Lys 160 and is lost in the GAPDH-K160R mutant (Fig. 2e). This suggests that acetylation of GAPDH at Lys 160 is required for binding of GAPDH to p300/CBP, which is consistent with the preserved binding detected when Lys 160 is replaced by Gln (GAPDH-K160Q), which mimics acetylation19 (Supplementary Information, Fig. S2e).
Figure 2.
GAPDH interacts with p300/CBP and GAPDH-K160R acts as a dominant-negative mutant. (a) GAPDH, native or NO-modified, binds similarly in vitro to a fragment of human p300 (F-p300) (amino acids 1135−2414). GST or GST–GAPDH was pre-treated with 50 μM GSH or GSNO for 30 min at 37 °C. F-p300 was added and binding assessed by a GSH-agarose pulldown assay. (b) GAPDH–p300 binding occurs in RAW264.7 cells treated with LPS/IFNγ for 16 h and is abolished by the iNOS inhibitor 1400W (100 μM). Cell lysates were immunoprecipitated with an anti-p300 antibody and the immunoprecipitates were analysed by western blotting with an anti-GAPDH antibody. (c) GAPDH and p300 are colocalized in the nucleus of RAW264.7 cells after exposure to LPS/IFNγ. Cells were stained with immunofluorescent anti-GAPDH and anti-p300 antibodies (green, p300; red, GAPDH). Scale bar, 10 μm. (d) R-(−)-Deprenyl (Dep) inhibits the enhancement of GAPDH–p300/CBP binding in mouse brain elicited by MPTP treatment. (e) K160R mutation of GAPDH abolishes GAPDH–p300 interactions in HEK293 cells. (f) GAPDH-K160R expression prevents acetylation of endogenous GAPDH and its binding to p300/CBP in a concentration-dependent manner, suggesting that GAPDH-K160R functions as a dominant-negative mutant. Forty-eight hours after transfection with 0.1, 0.5, 2, 3, or 4.5 μg of HA–GAPDH-K160R, HEK293 cells were treated with 200 μM GSNO for 24 h. Cell lysates were immunoprecipitated with anti-GAPDH, anti-p300, anti-CBP or anti-acetyl Lys antibody and the immunoprecipitates were analysed by western blotting with anti-HA or anti-GAPDH antibodies. Arrows indicate endogenous GAPDH and arrowheads indicate exogenous HA–GAPDH-K160R. (g) Nuclear localization of GAPDH augments its binding to p300/CBP and acetylation at Lys 160. Peritoneal macrophages from wild-type and iNOS knockout mice were transfected with various GAPDH constructs. Cell lysates were immunoprecipitated with anti-p300, anti-CBP or anti-acetyl Lys antibody and the immunoprecipitates were analysed by western blotting with anti-HA antibody. Both HA–NLS–GAPDH and HA–NLS–GAPDH–C150S, but not HA–NLS–GAPDH-K160R, augmented GAPDH–p300/CBP binding, as well as GAPDH acetylation in both iNOS knockout and wild-type cells.
GAPDH-K160R seems to function as a dominant-negative mutant, as it blocked endogenous GAPDH acetylation and its binding to p300/CBP in HEK293 cells treated with the NO donor S-nitroso-glutathione (GSNO; Fig. 2f). Similarly, in activated peritoneal macrophages treated with LPS/IFNγ, the GAPDH-K160R construct prevented acetylation of GAPDH and binding of GAPDH to p300/CBP (Fig. 2g).
To determine the role of GAPDH nuclear translocation in p300/CBP interactions, we examined the disposition of GAPDH with a fused nuclear localization signal (NLS–GAPDH). In LPS/IFNγ-treated macrophages NLS–GAPDH bound to p300/CBP even in iNOS-deleted cells or following mutation at Cys 150, the site of S-nitrosylation/sulphonation (Fig. 2g). Thus, NO does not seem to influence GAPDH–p300/CBP interaction directly but functions principally to facilitate GAPDH nuclear translocation.
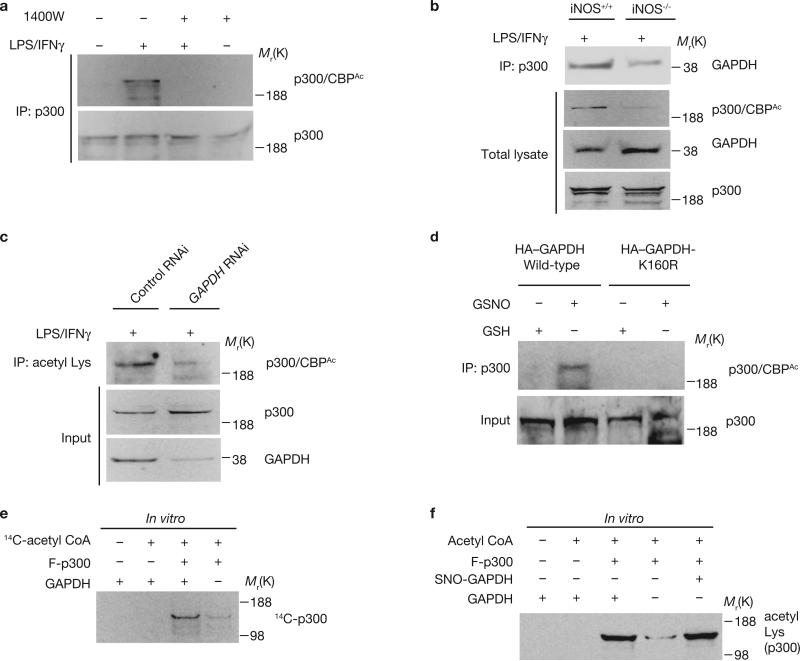
Auto-acetylation of p300 augments its acetyltransferase activity20. We examined the possibility that NO–GAPDH signalling regulates p300/CBP auto-acetylation. In RAW264.7 cells, LPS/IFNγ treatment enhanced p300 acetylation in an NO-dependent manner (Fig. 3a). In peritoneal macrophages, p300 acetylation and GAPDH–p300 binding were diminished in preparations from iNOS knockout mice (Fig. 3b). Acetylation of p300/CBP is dependent on GAPDH, as it was eliminated in RAW264.7 cells depleted of GAPDH by RNAi (Fig. 3c). Acetylation of p300 was also lost in cells overexpressing GAPDH-K160R, confirming a role for this mutant as dominant-negative (Fig. 3d). GAPDH augmented auto-acetylation of p300 in vitro, measured by incorporation of 14C isotope from acetyl CoA to a p300 fragment (Fig. 3e) with no alteration by GSNO (Fig. 3f). This is consistent with the primary role of NO being to convey GAPDH to the nucleus where it comes into contact with p300, facilitating auto-acetylation.
Figure 3.
GAPDH increases the catalytic activity of p300. (a) Increased acetylation of p300 occurs in RAW264.7 cells treated with LPS/IFNγ for 16 h; this was abolished by the iNOS inhibitor 1400W (100 μM). Cell lysates were immunoprecipitated with an anti-p300 antibody, and the immunoprecipitates were analysed by western blotting with an acetylation-specific p300/CBP antibody. (b) Genetic deletion of iNOS abolished the increased acetylation of p300/CBP and diminishes p300–GAPDH binding in peritoneal macrophages treated with LPS/IFNγ for 16 h. (c) Increased acetylation of p300 in RAW264.7 cells treated with LPS/IFNγ is abolished with depletion of GAPDH by RNAi. (d) GAPDH-K160R dominant-negative mutant diminished the increased acetylation of p300 elicited by GSNO in HEK293 cells. Forty-eight hours after transfection with wild-type HA–GAPDH or HA–GAPDH-K160R construct, cells were treated with 200 μM GSH or GSNO. (e) GAPDH augments auto-acetylation of a fragment of p300 (F-p300) in vitro. Auto-acetylation of F-p300 was assessed with 14C-acetyl-CoA in the presence or absence of purified GAPDH. (f) GAPDH, native or NO-modified, augments auto-acetylation of a fragment of p300 (F-p300) in vitro. Auto-acetylation of p300 was analysed by western blotting with anti-acetyl Lys antibody. SNO-GAPDH, S-nitrosylated GAPDH.
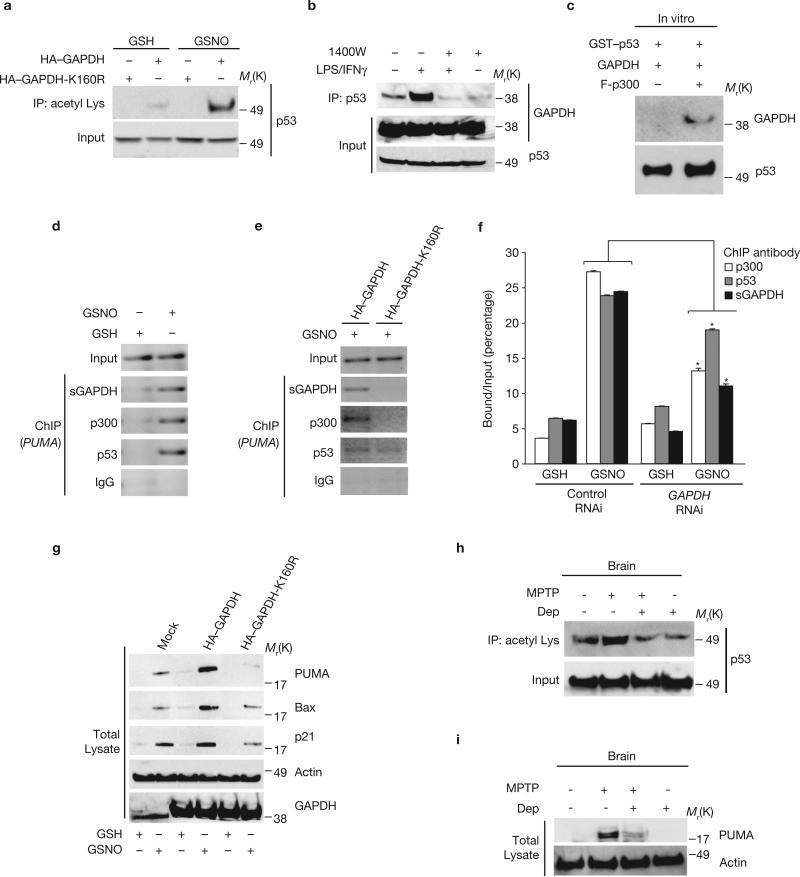
Physiological activation of p300/CBP by GAPDH should lead to acetylation of downstream targets. p300/CBP activates several genes, including some associated with apoptosis, of which p53 is particularly well characterized10–12. Moreover, a major role for p53 in cell death induced by oxidative stress is well established21–23. GSNO treatment of GAPDH-transfected HEK293 cells enhanced p53 acetylation (Fig. 4a). This process may reflect binding of GAPDH to p300/CBP, as it is abolished in the GAPDH-K160R mutant. Acetylation of p53requires NO, being prevented by treatment with 1400W (Supplementary Information, Fig. S2f), and is dependent on GAPDH, being lost following GAPDH depletion by RNAi (Supplementary Information, Fig. S2g). These influences suggest protein interactions between GAPDH, p300 and p53 in intact cells, consistent with the NO-dependent binding of GAPDH and p53 (Fig. 4b). Moreover, in the absence of p300, GAPDH does not bind to p53 in vitro (Fig. 4c). In contrast to GAPDH, p53 does not stimulate acetylation of p300 (Supplementary Information, Fig. S2h).
Figure 4.
GAPDH–p300 activates downstream targets, such as p53 and PUMA. (a) p53 is acetylated in GSNO-treated HEK293 cells; this is abolished by expression of GAPDH-K160R. Cell lysates were immunoprecipitated with anti-acetyl Lys antibody and the immunoprecipitates were analysed by western blotting with an anti-p53 antibody. (b) GAPDH–p53 binding was augmented by treatment with LPS/IFNγ for 16 h in RAW 264.7 cells. This was blocked by 1400W (100 μM). Cell lysates were immunoprecipitated with an anti-p53 antibody and the immunoprecipitates were analysed by western blotting with an anti-GAPDH antibody. (c) GAPDH–p300–p53 forms a complex in vitro, with p300 required for the interaction. Binding was examined by GSH-agarose pulldown assay. (d) Formation of a p53–p300-sulphonated GAPDH (sGAPDH) complex at the PUMA promoter region in U2OS cells treated with GSNO, assayed by ChIP assay. Cells were treated with 200 μM GSH or GSNO for 24 h. (e) Formation of p53–p300–sGAPDH complex at the PUMA promoter region in U2OS cells with GSNO was blocked by expression of GAPDH-K160R. Forty-eight hours after transfection with HA–GAPDH or HA–GAPDH-K160R, cells were treated with 200 μM GSNO for 24 h. (f) RNAi depletion of GAPDH diminishes the formation of the p53–p300– sGAPDH complex at the PUMA promoter in U2OS cells. Forty-eight hours after transfection with control or GAPDH siRNA, cells were treated with 200 μM GSH or GSNO for 24 h. *P < 0.001, n = 3, mean ± s.e.m., one-way ANOVA. (g) Overexpression of GAPDH, but not of GAPDH-K160R, in U2OS cells increases PUMA, BAX and p21 protein levels in the presence of GSNO. Forty-eight hours after transfection with mock, HA–GAPDH or HA–GAPDH-K160R constructs, cells were treated with 200 μM GSH or GSNO. (h) Acetylation of p53 was increased in brains of mice treated with MPTP, which was decreased by pre-treatment with R-(−)-deprenyl (Dep). (i) PUMA levels were increased in brains of mice treated with MPTP, effects prevented by pre-treatment with R-(−)-deprenyl (Dep).
p53 induces apoptosis mainly by transactivating downstream genes among which PUMA is particularly prominent13–15. To determine whether the GAPDH–p300–p53 cascade acts through such genes, we used a chromatin immunoprecipitation (ChIP) procedure24 to determine whether these proteins form a complex with the PUMA promoter. In U2OS cells, which express endogenous wild-type p53, we observed association of sulphonated GAPDH (the nuclear form of GAPDH), p300 and p53 at the PUMA promoter (Fig. 4d). This complex is dependent on the GAPDH–p300 interaction. Thus, GAPDH-K160R abolished association of endogenous sulphonated GAPDH and p300 with the PUMA promoter (Fig. 4e; Supplementary Information, Fig. S3a). Depletion of GAPDH by RNAi substantially reduced the interaction of sulphonated GAPDH, p300 and p53 with the PUMA promoter (Fig. 4f). Consistent with the ChIP results, augmentation of PUMA protein levels in the presence of LPS/IFNγ was blocked by GAPDH-K160R (Supplementary Information Fig. S3b, c). The induction of PUMA is prevented by either genetic depletion of iNOS or the iNOS inhibitor 1400W, indicating that it is dependent on NO (Supplementary Information, Fig. S3c–e). The influence of NO on PUMA, similarly to its effect on p300/CBP (Fig. 2g), reflects enhanced nuclear translocation of GAPDH. Thus, NLS–GAPDH induces PUMA similarly in both wild-type and iNOS-deleted macrophages. Moreover, NLS–GAPDH with the Cys 150 mutation can induce PUMA (Supplementary Information, Fig. S3c).
Other p53 targets, such as Bax and p21, are also induced by overexpression of wild-type GAPDH and blocked by expression of GAPDH-K160R or RNAi to GAPDH (Fig. 4g; Supplementary Information, Fig. S3f, g). Brains of mice treated with MPTP, in which the GAPDH–p300 cascade is activated (Figs 1e, 2d), show augmented p53 acetylation and PUMA induction (Fig. 4h, i). Consistent with the notion that p53/PUMA is regulated by the GAPDH–p300 cascade, pretreatment with R-(−)-deprenyl in the mice blocked augmented p53 acetylation and PUMA induction (Fig. 4h, i).
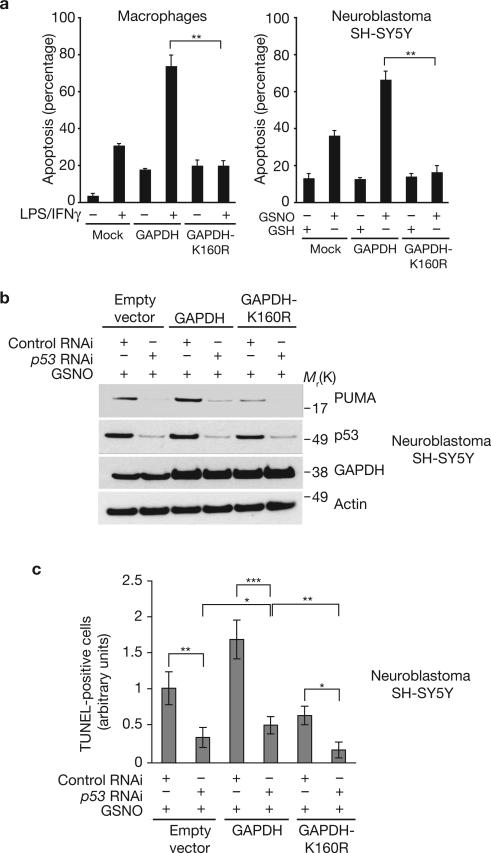
To determine whether cell death associated with GAPDH involves the p300 cascade, we transfected peritoneal macrophages with GAPDH or GAPDH-K160R (Fig. 5a). Overexpression of wild-type GAPDH doubled cell death in the presence of LPS/IFNγ; this was blocked by expression of GAPDH-K160R. Similar results were seen in the dopaminergic neuroblastoma SH-SY5Y cells (Fig. 5a). The principal role for NO is to facilitate nuclear translocation of GAPDH, as p300/CBP binding and PUMA expression (Fig. 2g; Supplementary Information, Fig.S3c), as well as LPS/IFNγ-induced cell death (Supplementary Information, Fig. S4a) are maintained when NLS–GAPDH is overexpressed in iNOS-deleted macrophages. This GAPDH cascade is mediated by p53 and p300/CBP, because genetic depletion of p53 or RNAi depletion of p300/CBP decreased apoptosis (Supplementary Information, Fig. S4b, c). The GAPDH–p300 cascade also mediates etoposide-triggered apoptosis, as expression of GAPDH-K160R inhibited etoposide-initiated cell death (Supplementary Information, Fig. S4d, e). To further assess a role for p53 in cell death mediated by the GAPDH–p300–CBP pathway, we depleted p53 by RNAi in SH-SY5Y cells stably overexpressing GAPDH or GAPDH-K160R. In these cells we examined levels of PUMA (Fig. 5b) and apoptotic cell death monitored by TUNEL assay (Fig. 5c). GSNO treatment substantially augmented binding of p300/CBP to GAPDH, levels of PUMA, as well as cell death, which was greatly diminished in cells expressing GAPDH-K160R (Fig. 5b; Supplementary Information, Fig. S4f, g). Depletion of p53 virtually eliminated the augmentation of PUMA and cell death. This supports the notion that cell death elicited by this signalling cascade is associated with p53 and its downstream target PUMA.
Figure 5.
Influence of GAPDH and GAPDH-K160R on cell death. (a) Increased apoptosis in peritoneal macrophages with LPS/IFNγ or in dopaminergic neuroblastoma SH-SY5Y cells with GSNO is blocked by GAPDH-K160R. Forty-eight hours after transfection with either mock, HA–GAPDH or HA–GAPDH-K160R, peritoneal macrophages isolated from wild-type mice were treated with LPS/IFNγ for 16 h. SH-SY5Y cells were treated with 200 μM GSH or GSNO for 24 h, after transfection with mock, HA–GAPDH- or HA–GAPDH-K160R. Apoptosis was assessed by counting condensed nuclei in transfected cells. **P < 0.001, n = 3, mean ± s.e.m., one-way ANOVA. (b) PUMA levels are increased in SH-SY5Y cells stably overexpressing GAPDH but decreased in cells overexpressing GAPDH-K160R, in comparision with cells with the empty vector, in the presence of 200 μM GSNO for 24 h. Depletion of endogenous p53 by RNAi significantly decreased the levels of PUMA. (c) Knockdown of p53 expression in SH-SY5Y cells stably overexpressing wild-type GAPDH or GAPDH-K160R in the presence of 200 μM GSNO for 24 h leads to decrease apoptosis, suggesting a major role of p53 in NO–GAPDH–p300/CBP death cascade. Cell death was measured by TUNEL assay. *P < 0.01, **P < 0.001, ***P < 0.0001, n = 4, mean ± s.e.m., one-way ANOVA.
In summary, nuclear S-nitrosylated/sulphonated GAPDH is acetylated by p300/CBP and, in turn, stimulates these proteins to acetylate and activate downstream targets, such as p53 (Supplementary Information, Fig. S4h). This cascade seems to mediate the NO-triggered GAPDH cell death pathway. Evidence for this conclusion includes the anti-apoptotic action of the GAPDH-K160R dominant-negative mutant that selectively blocks GAPDH–p300/CBP signalling. p300/CBP acetylates nuclear GAPDH which enhances the ability of GAPDH to stimulate auto-acetylation of p300/CBP. This suggests a feed-forward activation cycle wherein p300/CBP acetylates and augments the ability of GAPDH to stimulate acetylation of p300/CBP, which further acetylates GAPDH. This cascade is consistent with previous reports that overproduction of NO results in p53-mediated transactivation25, and that loss of p53 function by inactivating mutations abrogates NO-induced apoptosis in human lymphoblastoid cells26.
Recently Green and collaborators27 reported that GAPDH diminishes caspase-independent cell death by increasing expression of Atg12, a protein involved in autophagy, and by augmenting autophagy of damaged mitochondria. It is unclear how the caspase-independent cytotoxicity of the HeLa, Jurkat and murine embryonic fibroblast cells used by Green and associates relates to cytotoxicity in the six types of cells we used. In their study the glycolytic activity of GAPDH was important for its role in caspase-independent death, whereas the NO–GAPDH–p300/CBP cascade required a catalytically-inactive S-nitrosylated GAPDH.
The NO-triggered GAPDH death pathway acts through downstream genes including p53 (ref. 28). Besides their role in p53 regulation, p300 and CBP are co-activators for a large number of genes, which may also mediate apoptosis induced by GAPDH–p300/CBP. Furthermore, separate interactions of GAPDH with p300 and CBP have different consequences, and synergy of p300 and CBP is also possible29–31. Alterations of p300/CBP have been linked to a broad spectrum of diseases ranging from the immunodeficiency Rubinstein Taybi syndrome to neurological conditions, such as Huntington's disease32–34. We have detected a role for the GAPDH–p300/CBP cascade in the MPTP-induced model of Parkinson's disease18. This model affords a specific example in which therapeutic interventions, such as drugs that selectively prevent binding of components of the GAPDH–p300/CBP complex, may be developed.
METHODS
Reagents and animals
Unless otherwise noted, chemicals were purchased from Sigma. Anti-rabbit and anti-mouse IgG agarose were purchased from eBiosciences. 1400W was purchased from Alexis. p300 fragment was purchased from ProteinOne. Two methods for RNAi were used in this study: synthesized small oligonucleotides for siRNA were from Dharmacon RNA Technologies (for GAPDH). The vector system for shRNA (for CBP and p300) is as described previously1 The following sequences for silencing gene expressions were chosen, according to previous publications1,35. GAPDH, 5′-CGGGAAGCTCACTGGCATG-3′; p300, 5′-GTTCAAACGCCGAGTCTTCTT-3′; CBP, 5′-TCAACTCCTGTGTCGTCTTTT-3′; and control 5′-TTCTCCGAACGTGTCACGTTT-3′. Antibodies were obtained from the following companies: anti-GAPDH monoclonal antibody (Biogenesis); anti-PUMA, anti-histone H2B and anti-phospho-histone H2B (Ser 14) antibodies (Upstate); anti-HA antibody (Covance); anti-actin antibody (Chemicon); anti-CBP antibody, anti-p53 antibody, anti-Bax antibody, anti-p300 antibody and normal IgG (SantaCruz); anti-acetyl-CBP/p300 and anti-acetyl-lysine antibodies (Cell Signaling). The polyclonal antibodies against GAPDH and sulphonated-GAPDH were prepared as described1. Animal models treated with MPTP were prepared and processed as published18.
Mass spectrometry
HEK293 cells were treated with staurosporine (1 μM) for 24 h. Cells were lysed in RIPA-A (0.3% Triton X-100, 50 mM Tris pH 7.4 and 1 mM EDTA) with rotation at 4 °C for 30 min. Cell lysates were centrifuged at 14,000 g for 10 min. GAPDH was extracted from the pellet with RIPA-B (1% Triton X-100, 1% SDS, 50 mM Tris-Cl, pH 7.4, 500 mM NaCl and 1 mM EDTA), affinity-precipitated with an anti-GAPDH antibody, subjected to SDS–PAGE under non-reducing conditions and visualized by colloidal Coomassie staining. Gel-purified GAPDH was tryptic-digested and analysed by MALDI-TOF mass spectrometry as described1.
Protein interaction assays
GST-tagged proteins were prepared according to the manufacturer's recommendations (Pharmacia Biotech) and purified through glutathione–sepharose (Amersham Biosciences)1. Co-immunoprecipitation studies used anti-acetyl Lys (1:200 dilution) or anti-p300 (1:200 dilution) or anti-CBP (1:200) antibody following the published protocol1.
Cell culture
RAW264.7, SH-SY5Y, HEK293, HCT116, and U2OS cells were maintained in Dullbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 2 mM l-glutamine at 37 °C with a 5% CO2 atmosphere in a humidified incubator. Peritoneal macrophages were prepared by intraperitoneal injection of 1.5 ml of 3% sterile thioglycolate medium as described1.
For RNAi experiments, media for RAW264.7 cells or U2OS was supplemented with 10 mM pyruvate and cells were transfected with 167 nM GAPDH or control siRNA using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. Twenty-four hours after transfection, RAW264.7 cells were incubated in the presence of LPS/IFNγ (LPS 1 μg ml−1 and IFNγ 10 ng ml−1) for an additional 24 h. Immunofluorescent staining of cells for confocal microscopy was carried out as described previously1. For transient transfection of expression constructs, we used Polyfect reagent (Invitrogen) for HEK293 cells and Lipofectamine 2000 for SH-SY5Y cells, U2OS cells and peritoneal macrophages.
SH-SY5Y cells stably overexpressing wild-type GAPDH or GAPDH-K160R were generated by transfecting cells with a construct of pcDNA-GAPDH or GAPDH-K160R in the presence of neomycin (600 μg ml−1). The selected cell lines were further maintained in medium containing neomycin (200 μg ml−1; ref. 36). Silencing of p53 in these stable cell lines was by using MISSION shRNA lentiviral particles (Sigma), according to the manufacturer's protocol.
S-nitrosylation biotin switch assay and glycolytic assay for GAPDH
These assays were performed as described1.
Quantification of colocalization
The correlation coefficient for colocalization of GAPDH and p300 was determined using the colocalization function of the Zeiss LSM software, restricting the analysis to cell nuclei. The reported values represent the cumulative average of five cells from each of three separate plates.
In vitro acetylation assay
In 30 μl of reaction buffer (20 mM Tris-Cl, pH 8.0, 20% glycerol, 100 mM KCl, 1 mM DTT and 0.2 mM EDTA), 20 μM of acetyl-CoA or 1 μl of 14C-acetyl-CoA (60 mCi mmol−1), and 100 ng of a p300 fragment were added. After incubation at 30 °C for 1 h, the reaction was stopped by the addition of 10 μl of SDS sample buffer. The samples were subjected to SDS–PAGE gels and western blotting or autoradiography.
Chromatin immnoprecipitation (ChIP) assay
This was performed as described previously24. In brief, intact cells were treated with 2 mM disuccinimidyl glutarate (Pierce) to crosslink protein complexes, then were treated with formaldehyde to link protein to DNA covalently. Cells were lysed, the nucleoprotein complexes were sonicated and the crosslinked DNA–protein complexes were enriched by immuno-precipitation. The retrieved complexes were then analysed by PCR amplification to detect and quantify specific DNA targets. PUMA promoter elements were detected by PCR using the forward primer, 5′-CTGTGGCCTTGTGTCTGTGAGTAC-3′, and the reverse primer, 5′-CCTAGCCCAAGGCAAGGAGGAC-3′ (ref. 37). For real-time PCR we used Brilliant SYBR green master mix (Stratagene) according to the manufacturer's protocol.
RT–PCR analysis
Total RNA was extracted from cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription as well as PCR procedures used Superscript One Step RT–PCR with Platinum Taq (Invitrogen) for p21, Bax, PUMA and actin genes. The sequences are as follows: p21 forward, 5′-CTGGAGACTCTCAGGGTCGAAA-3′; p21 reverse, 5′-GATTAGGGCTTCCTCTTGGAGAA-3′; PUMA forward, 5′-AGAGGGAGGAGTCTGGGAGTG-3′; PUMA reverse; 5′-GCAGCGCATAT ACAGTATCTTACAGG-3′; Bax forward, 5′-TGCAGAGGATGATGCTGAC-3′; Bax reverse, 5′-CCTGTGACCTGAAGGAGG-3′
Apoptosis detection
Peritoneal macrophages and SH-SY5Y cells were transfected with HA-tagged wild-type GAPDH or GAPDH-K160R and treated with LPS/IFNγ or GSNO, respectively. Cells were permeabilized for 15 min in 0.1% Triton X-100 in PBS and incubated with a monoclonal anti-HA antibody followed by a secondary antibody conjugated with FITC (green). Cells were co-stained with the DNA-binding dye, 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI). Nuclear morphology of transfected cells stained green was assessed by visualization of DAPI-stained cells with a Zeiss fluorescence microscope. The proportion of condensed nucleus was used as a semi-quantitative indicator of apoptosis, for example, apoptosis (percentage).
HEK 293 cells were transfected with HA–GAPDH or HA–GAPDH-K160R separately and treated with 200 μM GSNO or 100 μM etoposide for 16 h in the presence of 10% FBS. Apoptosis, indicated by externalization of phosphatidylserine from the inner to the outer leaflet of the plasma membrane, was monitored by measurement of annexin V-FITC binding cells (Roche Biosciences) with flow cytometry (FACScalibur, Becton Dickinson)27.
Apoptosis was also quantified in peritoneal macrophages from wild-type or iNOS knockout mice, wild-type or p53 knockout HCT116 and U2OS cells using an enzyme immunoassay (Roche Applied Science, cat no. 11544675001) based on a combination of antibodies that recognize histones and DNA, as described previously38. Briefly, primary macrophages were transfected with various GAPDH constructs and treated with LPS/IFNγ for 16 h. HCT116 and U2OS cells were treated with GSNO for 16 h. Cells were lysed in buffer (supplied by the manufacturer) and an aliquot of the cytosolic fraction was used to quantify proteins using standard methods. Cytoplasmic fractions containing fragmented DNA were transferred to streptavidin-coated microtitre plates that had been incubated with a biotinylated monoclonal anti-histone antibody. The amount of fragmented DNA bound to the anti-histone antibody was evaluated by a peroxidase-conjugated monoclonal anti-DNA antibody using ABTS (2,2′-azino-di(3 ethylbezthiazoline sulfonic acid)) as a substrate at 405 nm. Absorbance values were normalized by the values obtained from control cell lysates and is expressed as arbitrary units.
We also used the levels of phosphorylated histone H2B at Ser 14 in western blotting, as another marker associated with apoptosis.
Cell death of SH-SY5Y cells stably expressing either GAPDH or GAPDH-K160R was assayed 16 h after exposure to GSH or GSNO (200 μM). Numbers of total and apoptotic cells were determined by counting nuclei stained with 100 ng ml−1 DAPI and TUNEL, respectively. Cells were fixed in 4% paraformaldehyde and then stained using the In situ cell death detection kit (Roche) according to protocols provided by the manufacturer. The cells were examined under a confocal microscope (Zeiss) and cell death was evaluated as the ratio of TUNEL-positive cells to total cells.
Statistical analysis
P values were calculated by one-way ANOVA by using MINITAB 13 (Minitab, State College, PA).
Supplementary Material
Figure S1 GAPDH is acetylated at its K160 residue, and effects of K160R on GAPDH's glycolytic activity, S-nitrosylation, and nuclear translocation. a, GAPDH is acetylated by p300 in vitro. b, Putative acetylation of GAPDH at K160 in HEK293 cells treated with an apoptotic inducer staurosporine, shown by mass spectrometry. Typically trypsin does not cleave after acetylated lysines but in some instances a minor proportion is cleaved, presumably accounting for our findings1. c, Original spectrum of mass spectrometry of acetylated GAPDH, unmodified GAPDH and sulfonated GAPDH. d, The K160R mutation that abolishes acetylation does not alter GAPDH catalytic activity in U2OS cells. Forty-eight h after transfection with HA-GAPDH-wt or HA-GAPDH-K160R, GAPDH was assayed in cell lysates. e, S-nitrosylation of GAPDH-K160R in HEK293 cells. Forty-eight h after transfection with GAPDH-wt or GAPDH-K160R, cells were treated with 200 μM GSNO for 1 h. Cell lysates were subjected to the biotin switch assay. f, Nuclear translocation of GAPDH-wt and GAPDH-K160R in HEK293 cells. Forty-eight h after transfection with HA-GAPDH-wt or HA-GAPDH-K160R, cells were treated with 200 μM GSNO for 24 h and stained with an anti-HA antibody. Red, GAPDH; blue, DAPI (nucleus). Scale bar, 10 μm.
Figure S2 Effects of MPTP or MPP+ on acetylation of GAPDH, binding of p300/CBP to GAPDH, and effects of GAPDH on acetylation of p53 and p300. a, Quantitative assessment of GAPDH acetylation in brains from mice treated with MPTP in Fig. 1e. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. b, Acetylation of GAPDH is increased in SH-SY5Y cells treated with 5 μM MPP+ for 16 h, which is reversed by a NOS inhibitor L-NAME. Cell lysates were immunoprecipitated with an anti-acetyl lysine antibody, and the immunoprecipitates analyzed by Western blotting with an anti-GAPDH antibody. c, Wild-type GAPDH, but not GAPDH-K160R, binds to CBP in HEK293 cells treated with GSNO. d, Co-localization of p300 and GAPDH in the nucleus in control or LPS-IFNγ-treated RAW264.7 cells was assessed by correlation coefficient analysis. e, HA-GAPDH-K160Q (a pseudo-acetylation mutant), but not HA-GAPDH-K160R (acetylation-null mutant), binds to p300 in HEK293 cells exposed for 16 h to 200 μM sodium nitroprusside (SNP). f, Acetylation of p53 is increased in RAW264.7 cells treated with LPS-IFNγ for 16 h, which is blocked by 100 μM 1400W. g, Depletion of GAPDH by RNAi decreases acetylation of p53 in RAW 264.7 cells treated with LPS-IFNγ. h, GAPDH augments auto-acetylation of a fragment of p300 (F-p300) in vitro. Auto-acetylation of F-p300 (100 ng) with GAPDH (1 μg) or p53 (1 μg) was assessed by Western blotting with an anti-acetyl lysine antibody.
Figure S3 Influences of GAPDH on PUMA mRNA and protein levels. a, Interaction of sulfonated GAPDH (sGAPDH) at the PUMA promoter region is abolished by expression of GAPDH-DN in U2OS cells treated with GSNO. Cell lysates were used for ChIP assay with an sGAPDH antibody. b, Protein levels of PUMA are increased by expression of GAPDH, effects blocked by GAPDH-DN, in RAW264.7 cells treated with LPS-IFNγ for 16 h. c, Transfection of HA-NLS-GAPDH, HA-NLS-GAPDH-C150S, or HA-NLS-GAPDH-DN in peritoneal macrophages of wt or iNOS knockout mice. Both NLS-GAPDH and NLS-GAPDH-C150S elicit increased expression of PUMA, regardless of iNOS disposition, after treatment with LPS-IFNγ. d, e, Expression of PUMA mRNA and protein levels are increased in the presence of LPS-IFNγ, which is blocked by 1400W, in RAW 264.7 cells. f, Expression of PUMA, Bax, and p21 is increased in GSNO-treated U2OS cells expressing wild-type GAPDH, which is blocked by expression of GAPDH-DN. g, GAPDH augments induction of PUMA, Bax, and p21 in U2OS cells. Forty-eight h after transfection with shRNA to GAPDH, or control (CTL), transfected cells were treated with 200 μM GSNO for 24 h.
Figure S4 Acetylation and activation of CBP/p300 in cells stably overexpressing wild-type GAPDH, but not in cells with GAPDH-DN, and influences of GAPDH and GAPDH-DN on apoptosis. a, Nuclear localization of GAPDH augments apoptosis in peritoneal macrophages from iNOS knockout mice. Peritoneal macrophages from wt and iNOS knockout mice were transfected with HA-NLS-GAPDH, HA-NLS-GAPDH-C150S, or HA-NLS-GAPDH-DN. Forty-eight h after transfection, cells were treated with LPS-IFN-γ for 16 h. Apoptosis was assessed with cell death ELISA. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. b, DNA fragmentation in HCT116 cells (wild-type or p53 null) was monitored by ELISA after transfection with GAPDH and GAPDH-DN in the presence of 200 μM GSNO for 24 h. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. c, DNA fragmentation was measured by ELISA in U2OS cells after depletion of p300 or CBP during over-expression of GAPDH with 200 μM GSNO. n=3, mean + S.E.M., *p<0.01; **p<0.001, one-way ANOVA. d, FACS analysis of apoptosis with annexin V indicates that the death triggered by GSNO or etoposide is blocked by GAPDH-DN. Forty-eight h after transfection with GAPDH-wt or GAPDH-DN, HEK293 cells were treated with either 200 μM GSNO or 100 μM etoposide. Cells were stained with annexin V-FITC to identify Annexin V-positive cells (M2 region, x axis). e, Forty-eight h after transfection with GAPDH-wt or GAPDH-DN, cells were treated with either 200 μM GSNO or 100 μM etoposide for 24 h. Apoptosis elicited by GSNO or etoposide is blocked by GAPDH-DN, which is assessed by another apoptotic marker, phosphorylation of histone H2B at serine-14 (pH2B)2. f, In SHSY5Y cells stably overexpressing wild-type GAPDH exposed to 200 μM GSNO for 24 h, autoacetylation of CBP/p300 or GAPDH and GAPDH-CBP/p300 protein interactions are significantly increased, in comparison with cells stably overexpressing GAPDH-DN. g, Cell death in SH-SY5Y cells stably over-expressing GAPDH is increased compared to SH-SY5Y cells stably expressing GAPDH-DN. Cells were treated with 200 μM GSH or GSNO for 24 h. Cell death was measured by TUNEL assay. n = 4, mean + S.E.M, *p< 0.01, **p<0.001, one-way ANOVA. h, Schematic diagram of NO-GAPDH-p300/CBP cell death signaling. NO triggers S-nitrosylation/sulfonation of GAPDH, augments its binding to Siah, translocating the GAPDH-Siah protein complex to the nucleus. GAPDH is acetylated by p300/CBP at K160R. In turn, nuclear acetylated GAPDH augments acetyltransferase activity of p300/CBP. Acetylated p300 acetylates its substrates, such as p53, inducing downstream apoptotic genes and cell death.
Figure S5 Full scans of key Western blot data.
Supplementary References
1. Wang, D., Thompson, P., Cole, P.A. & Cotter, R.J. Structural analysis of a highly acetylated protein using a curved-field reflectron mass spectrometer. Proteomics 5, 2288−2296 (2005).
2. Cheung, W.L. et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507−517 (2003).
ACKNOWLEDGEMENTS
This work was supported by USPHS grants MH-069853 (A.S.); DA-00266, Research Scientist Award DA-00074 (S.H.S); NS-48206, NS-38377, DA-00226 (T.M.D, V.L.D) and grants from Stanley, NARSAD and S-R foundations (A.S.). We thank Yukiko L. Lema for preparing the figures and organizing the manuscript. We appreciate technical assistance provided by A. Kamiya.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 2.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 3.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim. Biophys. Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 5.Ishitani R, et al. Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J. Neurochem. 1996;66:928–935. doi: 10.1046/j.1471-4159.1996.66030928.x. [DOI] [PubMed] [Google Scholar]
- 6.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl Acad. Sci. USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlile GW, et al. Reduced apoptosis after nerve growth factor and serum withdrawal: conversion of tetrameric glyceraldehyde-3-phosphate dehydrogenase to a dimer. Mol. Pharmacol. 2000;57:2–12. [PubMed] [Google Scholar]
- 8.Chen RW, et al. Involvement of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and p53 in neuronal apoptosis: evidence that GAPDH is upregulated by p53. J. Neurosci. 1999;19:9654–9662. doi: 10.1523/JNEUROSCI.19-21-09654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldmeier PC, Boulton AA, Cools AR, Kato AC, Tatton WG. Neurorescuing effects of the GAPDH ligand CGP 3466B. J. Neural. Transm. Suppl. 2000;60:197–214. doi: 10.1007/978-3-7091-6301-6_13. [DOI] [PubMed] [Google Scholar]
- 10.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 11.Avantaggiati ML, et al. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 12.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 13.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 14.Villunger A, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 16.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 17.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 18.Hara MR, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc. Natl Acad. Sci. USA. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-κB transcription factor complex by acetylation. J. Mol. Med. 2003;81:549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 20.Thompson PR, et al. Regulation of the p300 HAT domain via a novel activation loop. Nature Struct. Mol. Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 21.Messmer UK, Ankarcrona M, Nicotera P, Brune B. p53 expression in nitric oxide-induced apoptosis. FEBS Lett. 1994;355:23–26. doi: 10.1016/0014-5793(94)01161-3. [DOI] [PubMed] [Google Scholar]
- 22.Messmer UK, Brune B. Nitric oxide-induced apoptosis: p53-dependent and p53-independent signalling pathways. Biochem. J. 1996;319(Pt 1):299–305. doi: 10.1042/bj3190299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Y, et al. Involvement of p85 in p53-dependent apoptotic response to oxidative stress. Nature. 1998;391:707–710. doi: 10.1038/35648. [DOI] [PubMed] [Google Scholar]
- 24.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 25.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nature Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 26.Li CQ, et al. Apoptotic signaling pathways induced by nitric oxide in human lymphoblastoid cells expressing wild-type or mutant p53. Cancer Res. 2004;64:3022–3029. doi: 10.1158/0008-5472.can-03-1880. [DOI] [PubMed] [Google Scholar]
- 27.Colell A, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 28.Giordano A, Avantaggiati ML. p300 and CBP: partners for life and death. J. Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Yao TP, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki H, et al. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 31.Kung AL, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 32.Oike Y, et al. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- 33.Giles RH, Peters DJ, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 34.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 35.Turnell AS, et al. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 36.Dhakshinamoorthy S, et al. Protein/DNA arrays identify nitric oxide-regulated cis-element and trans-factor activities some of which govern neuroblastoma cell viability. Nucleic Acids Res. 2007;35:5439–5451. doi: 10.1093/nar/gkm594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koutsodontis G, Vasilaki E, Chou WC, Papakosta P, Kardassis D. Physical and functional interactions between members of the tumour suppressor p53 and the Sp families of transcription factors: importance for the regulation of genes involved in cell-cycle arrest and apoptosis. Biochem. J. 2005;389:443–455. doi: 10.1042/BJ20041980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frade JM, Rodriguez-Tebar A, Barde YA. Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature. 1996;383:166–168. doi: 10.1038/383166a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 GAPDH is acetylated at its K160 residue, and effects of K160R on GAPDH's glycolytic activity, S-nitrosylation, and nuclear translocation. a, GAPDH is acetylated by p300 in vitro. b, Putative acetylation of GAPDH at K160 in HEK293 cells treated with an apoptotic inducer staurosporine, shown by mass spectrometry. Typically trypsin does not cleave after acetylated lysines but in some instances a minor proportion is cleaved, presumably accounting for our findings1. c, Original spectrum of mass spectrometry of acetylated GAPDH, unmodified GAPDH and sulfonated GAPDH. d, The K160R mutation that abolishes acetylation does not alter GAPDH catalytic activity in U2OS cells. Forty-eight h after transfection with HA-GAPDH-wt or HA-GAPDH-K160R, GAPDH was assayed in cell lysates. e, S-nitrosylation of GAPDH-K160R in HEK293 cells. Forty-eight h after transfection with GAPDH-wt or GAPDH-K160R, cells were treated with 200 μM GSNO for 1 h. Cell lysates were subjected to the biotin switch assay. f, Nuclear translocation of GAPDH-wt and GAPDH-K160R in HEK293 cells. Forty-eight h after transfection with HA-GAPDH-wt or HA-GAPDH-K160R, cells were treated with 200 μM GSNO for 24 h and stained with an anti-HA antibody. Red, GAPDH; blue, DAPI (nucleus). Scale bar, 10 μm.
Figure S2 Effects of MPTP or MPP+ on acetylation of GAPDH, binding of p300/CBP to GAPDH, and effects of GAPDH on acetylation of p53 and p300. a, Quantitative assessment of GAPDH acetylation in brains from mice treated with MPTP in Fig. 1e. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. b, Acetylation of GAPDH is increased in SH-SY5Y cells treated with 5 μM MPP+ for 16 h, which is reversed by a NOS inhibitor L-NAME. Cell lysates were immunoprecipitated with an anti-acetyl lysine antibody, and the immunoprecipitates analyzed by Western blotting with an anti-GAPDH antibody. c, Wild-type GAPDH, but not GAPDH-K160R, binds to CBP in HEK293 cells treated with GSNO. d, Co-localization of p300 and GAPDH in the nucleus in control or LPS-IFNγ-treated RAW264.7 cells was assessed by correlation coefficient analysis. e, HA-GAPDH-K160Q (a pseudo-acetylation mutant), but not HA-GAPDH-K160R (acetylation-null mutant), binds to p300 in HEK293 cells exposed for 16 h to 200 μM sodium nitroprusside (SNP). f, Acetylation of p53 is increased in RAW264.7 cells treated with LPS-IFNγ for 16 h, which is blocked by 100 μM 1400W. g, Depletion of GAPDH by RNAi decreases acetylation of p53 in RAW 264.7 cells treated with LPS-IFNγ. h, GAPDH augments auto-acetylation of a fragment of p300 (F-p300) in vitro. Auto-acetylation of F-p300 (100 ng) with GAPDH (1 μg) or p53 (1 μg) was assessed by Western blotting with an anti-acetyl lysine antibody.
Figure S3 Influences of GAPDH on PUMA mRNA and protein levels. a, Interaction of sulfonated GAPDH (sGAPDH) at the PUMA promoter region is abolished by expression of GAPDH-DN in U2OS cells treated with GSNO. Cell lysates were used for ChIP assay with an sGAPDH antibody. b, Protein levels of PUMA are increased by expression of GAPDH, effects blocked by GAPDH-DN, in RAW264.7 cells treated with LPS-IFNγ for 16 h. c, Transfection of HA-NLS-GAPDH, HA-NLS-GAPDH-C150S, or HA-NLS-GAPDH-DN in peritoneal macrophages of wt or iNOS knockout mice. Both NLS-GAPDH and NLS-GAPDH-C150S elicit increased expression of PUMA, regardless of iNOS disposition, after treatment with LPS-IFNγ. d, e, Expression of PUMA mRNA and protein levels are increased in the presence of LPS-IFNγ, which is blocked by 1400W, in RAW 264.7 cells. f, Expression of PUMA, Bax, and p21 is increased in GSNO-treated U2OS cells expressing wild-type GAPDH, which is blocked by expression of GAPDH-DN. g, GAPDH augments induction of PUMA, Bax, and p21 in U2OS cells. Forty-eight h after transfection with shRNA to GAPDH, or control (CTL), transfected cells were treated with 200 μM GSNO for 24 h.
Figure S4 Acetylation and activation of CBP/p300 in cells stably overexpressing wild-type GAPDH, but not in cells with GAPDH-DN, and influences of GAPDH and GAPDH-DN on apoptosis. a, Nuclear localization of GAPDH augments apoptosis in peritoneal macrophages from iNOS knockout mice. Peritoneal macrophages from wt and iNOS knockout mice were transfected with HA-NLS-GAPDH, HA-NLS-GAPDH-C150S, or HA-NLS-GAPDH-DN. Forty-eight h after transfection, cells were treated with LPS-IFN-γ for 16 h. Apoptosis was assessed with cell death ELISA. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. b, DNA fragmentation in HCT116 cells (wild-type or p53 null) was monitored by ELISA after transfection with GAPDH and GAPDH-DN in the presence of 200 μM GSNO for 24 h. n=3, mean + S.E.M., **p<0.001, one-way ANOVA. c, DNA fragmentation was measured by ELISA in U2OS cells after depletion of p300 or CBP during over-expression of GAPDH with 200 μM GSNO. n=3, mean + S.E.M., *p<0.01; **p<0.001, one-way ANOVA. d, FACS analysis of apoptosis with annexin V indicates that the death triggered by GSNO or etoposide is blocked by GAPDH-DN. Forty-eight h after transfection with GAPDH-wt or GAPDH-DN, HEK293 cells were treated with either 200 μM GSNO or 100 μM etoposide. Cells were stained with annexin V-FITC to identify Annexin V-positive cells (M2 region, x axis). e, Forty-eight h after transfection with GAPDH-wt or GAPDH-DN, cells were treated with either 200 μM GSNO or 100 μM etoposide for 24 h. Apoptosis elicited by GSNO or etoposide is blocked by GAPDH-DN, which is assessed by another apoptotic marker, phosphorylation of histone H2B at serine-14 (pH2B)2. f, In SHSY5Y cells stably overexpressing wild-type GAPDH exposed to 200 μM GSNO for 24 h, autoacetylation of CBP/p300 or GAPDH and GAPDH-CBP/p300 protein interactions are significantly increased, in comparison with cells stably overexpressing GAPDH-DN. g, Cell death in SH-SY5Y cells stably over-expressing GAPDH is increased compared to SH-SY5Y cells stably expressing GAPDH-DN. Cells were treated with 200 μM GSH or GSNO for 24 h. Cell death was measured by TUNEL assay. n = 4, mean + S.E.M, *p< 0.01, **p<0.001, one-way ANOVA. h, Schematic diagram of NO-GAPDH-p300/CBP cell death signaling. NO triggers S-nitrosylation/sulfonation of GAPDH, augments its binding to Siah, translocating the GAPDH-Siah protein complex to the nucleus. GAPDH is acetylated by p300/CBP at K160R. In turn, nuclear acetylated GAPDH augments acetyltransferase activity of p300/CBP. Acetylated p300 acetylates its substrates, such as p53, inducing downstream apoptotic genes and cell death.
Figure S5 Full scans of key Western blot data.
Supplementary References
1. Wang, D., Thompson, P., Cole, P.A. & Cotter, R.J. Structural analysis of a highly acetylated protein using a curved-field reflectron mass spectrometer. Proteomics 5, 2288−2296 (2005).
2. Cheung, W.L. et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507−517 (2003).