Abstract
KIR3DL1 shows extensive polymorphism, and its variation has functional significance in terms of cell-surface expression levels and inhibitory capacity. We characterized nine KIR3DL1 alleles (*022, *028, *029, *033, *035, *051, *052, *053, and *054), four of which were identified for the first time in this study, and compared them to known alleles in phylogenetic analysis. Blood was available from eight individuals with these alleles, and cell-surface expression on NK cells could be determined for six of them using the KIR3DL1-specific Ab DX9. Four of the alleles were expressed at clearly detectable levels, and two others showed exceptionally low levels of expression. Site-directed mutagenesis demonstrated that single amino acid changes can result in either diminished or enhanced DX9 staining compared with the respective related KIR3DL1 allotypes. These results raise the possibility that KIR3DL1 evolution maintains variation in KIR3DL1 cell-surface expression levels, potentially due to the effect of such variation on functional capacity.
Killer cell Ig-like receptors (KIR)5 are a diverse family of cell-surface glycoproteins that are expressed on NK cells and a subpopulation of CD8+ T lymphocytes with an activated or memory phenotype (1, 2). KIR trigger inhibitory or activating signals upon binding to their MHC class I ligands, thereby regulating cell cytotoxicity and cytokine secretion (3). Like their MHC ligands, KIR are also polymorphic (4): 14 expressed KIR genes and 2 pseudogenes have been identified in humans so far. KIR haplotypes exhibit variability in the number and type of genes present and in allelic polymorphism of individual genes, resulting in the extensive genetic polymorphism characterizing this complex (5). The KIR genes show high sequence similarity, which facilitates nonallelic homologous recombination (6). Indeed, the propensity for nonallelic homologous recombination likely explains the expansion and contraction of the KIR complex (7, 8). Two basic groups of KIR haplotypes have been defined based on their gene content (5): group A haplotypes have a fixed organization of seven genes, which are mostly inhibitory in nature, and two pseudogenes, while the group B haplotypes have variable numbers of KIR genes, many with an activating function.
The KIR3DL1 gene is unique in terms of its diversity and expression patterns (9). It is the only KIR locus that encodes either inhibitory (KIR3DL1) or activating (KIR3DS1) allotypes. KIR3DL1 can be on both A and B haplotypes, while KIR3DS1 is only on haplotype B. The inhibitory KIR3DL1 allotypes have specificity for HLA-B (and sometimes HLA-A) molecules with the Bw4 serological motif (10, 11). Although the ligands for the KIR3DS1 allotype have not been defined by binding studies, both population and disease association data suggest that it may interact with HLA-B Bw4 molecules with isoleucine at position 80 (Bw4–80I) (12, 13).
Polymorphisms in the KIR3DL1 gene have phenotypic consequences. Staining with the KIR3DL1-specific mAb DX9 revealed that KIR3DL1 subtypes are expressed at different levels (high, low, and null) on the NK cell surface (9), a phenomenon due at least in part to variation in the extracellular domains (14). Because DX9 blocks the interaction between KIR3DL1 and Bw4, enabling NK cells to kill Bw4+ targets (15), the quantity of KIR3DL1 on the cell surface could affect the inhibitory capacity of cells expressing these molecules (16, 17). KIR3DL1 polymorphism also has direct influence on the recognition of Bw4 molecules with specific peptides sitting in their binding groove (10). The KIR proximal promoter region has recently been shown to possess bi-directional transcription activity (18, 19), and the relative strength of the competing promoters may affect the frequency of NK cells expressing a given KIR within a given individual (S. K. Anderson, manuscript in preparation). Also, the proximal promoter of KIR3DL1 is polymorphic at transcription factor binding sites known to affect promoter activity (19). KIR promoter polymorphisms could therefore affect either expression level of different KIR3DL1 allotypes or the frequency at which they are expressed on NK cells within an individual. Ultimately, the functional consequences of KIR3DL1 genetic variants could affect susceptibility or resistance to infections and other diseases.
In this study we characterize expression patterns of several recently discovered KIR3DL1 alleles, including four that were identified for the first time. Flow cytometry analysis of a subset of the corresponding KIR3DL1 allotypes emphasizes the importance of single amino acid changes in altering levels of cell-surface expression detected by DX9 mAb in part as a consequence of intracellular retention, and shows that independently generated mutations with converging functional consequences are maintained in the human population.
Materials and Methods
Subjects
Subjects with novel KIR3DL1 alleles were identified from HIV-1-infected individuals and healthy volunteers. The ethnic backgrounds of the samples were Caucasian (74.3%), African-American (13.4%), Hispanic (6.5%), and others (5.8%). The study was approved by the local Institutional Review Boards of the participating institutions, and all individuals gave informed consent for participation in this study.
KIR genotyping and KIR3DL1 subtyping
Genomic DNA was typed for presence or absence of KIR3DL1 and KIR3DS1 by PCR with sequence-specific primers as previously described (20). Where available, RNA was extracted from PBMCs with TRIzol (Invitrogen), and first strand cDNA was synthesized using oligo(dT) and SuperScript II reverse transcriptase (Invitrogen). The promoter region and cDNA were amplified using gene-specific primers, and the products were sequenced using the Big Dye terminator kit (Applied Biosystems). Due to the unavailability of PBMCs from some donors, genomic DNA was sequenced using gene specific primers. Overlapping fragments spanning each pair of adjacent exons from exon 1 to exon 9 were generated by PCR amplification with primers designed to recognize conserved or gene-specific sequences of the KIR3DL1 exons. All primer sequences are available on request.
Sequence analysis
All of the KIR3DL1 alleles were aligned using the desktop sequence assembly and analysis program Sequencher 4.6 (http://www.genecodes.com). A neighbor-joining tree of the KIR3DL1 and KIR3DS1 alleles was constructed using the distance method (21) with the software PAUP version 4 (22).
Site-directed mutagenesis of KIR3DL1*005, *00101, and *01502
Plasmids containing KIR3DL1*005, *00101, or *01502 (10) were mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Position 115 of KIR3DL1*005 was changed from G to C using primers 5′-ggcccagcgc tgtgctgcctcgaggagg-3′ and 5′-cctcctcgaggcagcacagcgctgggcc-3′, resulting in a V18L amino acid change. Position 740 of KIR3DL1*00101 was changed from G to A using primers 5′-ccttgtcctgtagctcccagagctcctatgac-3′ and 5′-gtcataggagctctgggagctacaggacaagg-3′, resulting in an R226Q change. Position 286 of KIR3DL1*01502 was changed from C to T using primers 5′-gggaactacacatgttggggttcacacccacactcc-3′ and 5′-ggagtgtgggtgtgaaccccaacatgtgtagttccc-3′, and position 496 was changed from C to A using primers 5′-ctctaaggacccctcaagcctcgttggacagatcc-3′ and 5′-ggatctgtccaacgaggcttgaggggtccttagag-3′, resulting in R75W and R145S amino acid changes, respectively. Also, position 829 of KIR3DL1*01502 was changed from C to A using primers 5′-ggtcaacagaacattcaaggcagatttccctctgg-3′ and 5′-ccagagggaaatctgccttgaatgttctgttgacc-3′, making a Q256K change. The full coding sequence of the resulting plasmids KIR3DL1*005V18L (*053), KIR3DL1*00101R226Q (*052), KIR3DL1*01502R75W+R145S (*028), and KIR3DL1*01502Q256K (*029) were sequenced to confirm the mutations.
Analysis of KIR3DL1 expression on HEK293 cell lines or peripheral blood NK cells
HEK293T cells were transfected as previously described (23) using plasmids containing full-length KIR3DL1*005, *01502, *00101, *005V18L, *00101R226Q, 01502R75W+R145S, or KIR3DL1*01502Q256K. PBMCs were isolated from EDTA-treated blood using Histopaque density gradient centrifugation. PBMCs derived from HIV+ individuals were cryopreserved the day of collection and then thawed just before flow cytometric analysis. Expression of KIR3DL1 on transfected cells or PBMCs was analyzed by mAb staining using PE-conjugated anti-KIR3DL1 mAb clone DX9 (Bio-Legend) or clone Z27 (Beckman Coulter) as described (23), followed by flow cytometry. All experiments included staining with isotype control Abs to permit accurate positioning of electronic gates for analysis of KIR3DL1 expression.
Structural analysis
Homology modeling of KIR3DL1 extracellular domain was conducted using the Swiss PDB modeler (24) “first approach” mode based on the best sequence homology. The D0 domain was modeled on the crystal structure of human platelet glycoprotein VI, the most homlogous Ig domain with known structural coordinates, while the D1 and D2 domains were modeled on the KIR2DL1 template.
Analysis of retained KIR3DL1 by intracellular staining
Intracellular retention of KIR3DL1 allotypes was assessed by comparing relative KIR expression detected by cell-surface staining with that of permeabilized cells. Because neither DX9 nor Z27 recognize KIR3DL1 in permeabilized, fixed cells (data not shown), the leader sequences of KIR3DL1 alleles *01502, *005, *053, and *028 were replaced with that of human CD8 fused to the FLAG epitope. HEK293T cells were then transfected with FLAG-tagged versions of specific KIR3DL1 alleles. One set of transfectants was stained for cell-surface KIR3DL1 and another was fixed and permeabilized using Cytofix/Cyotoperm solution (BD Pharmingen) to determine total KIR3DL1 expression. Cells were stained with FITC-conjugated anti-FLAG mAb (Sigma-Aldrich) or control FITC-conjugated mAb, and analyzed on a BD FACSort flow cytometer (BD Biosciences). The surface and total levels of KIR3DL1 expression are expressed as percentage of the appropriate reference allotype (*01502 vs *028 and *005 vs *053). Data from three independent experiments were analyzed and a p value of <0.05 by Student’s t test was considered significant.
Results
Identification of novel KIR3DL1 alleles
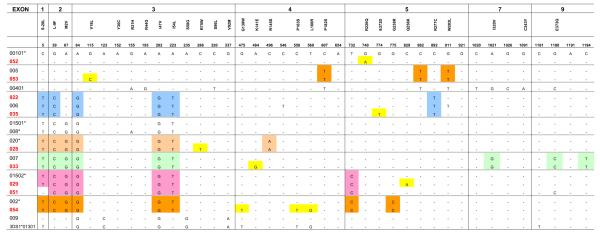
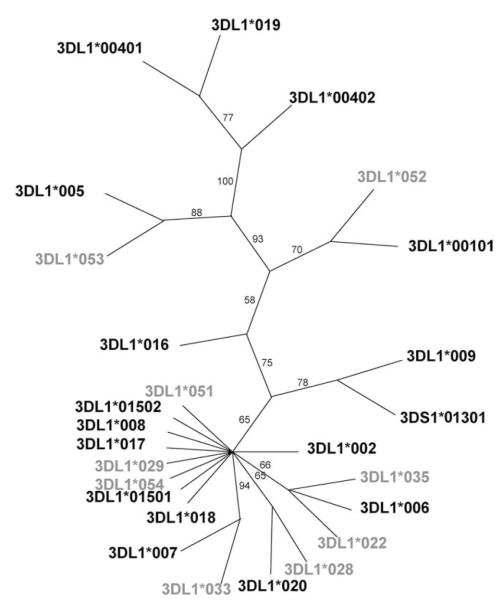
We identified four novel KIR3DL1 alleles in this study, and their names were officially assigned by the KIR Nomenclature Committee (25) as *051 (EF472680), *052 (EF472681), *053 (EF472671, EF472677), and *054 (EF472673, EF472679). Additionally, we confirmed the sequences of several recently reported alleles (*022, *028, *029, *033, *035) (26, 27). These nine alleles were compared with the existing members of the KIR3DL1 family (Fig. 1). Phylogenetic analysis illustrated the relationship between the previously identified KIR3DL1 alleles and newly described alleles (Fig. 2). Although it was not possible to determine the precise frequencies of our four novel alleles, each was present at a frequency below 10% based on KIR3DL1 subtyping data derived from a previous study involving 685 European Americans and 136 African-Americans (28). KIR3DL1 alleles *028, *029, *033, *052, and *053 appear to be the result of single base pair substitutions with respect to previously characterized KIR3DL1 alleles, whereas *022, *035, and *051 possess more than one point mutation, generally in two exons (Fig. 1). KIR3DL1*054 is identical to *002 in all exons except exon 4, where it is identical to KIR3DS1.
FIGURE 1.
Comparison of DNA sequences of the novel KIR3DL1 alleles with other known KIR3DL1 alleles and KIR3DS1. New and recently described 3DL1 alleles included in this study are shown in red whereas known alleles are in black. Unique substitutions identified are highlighted in yellow. Other common substitutions shared by closely related alleles are shown in different shades of color. Hyphen indicates sequence identity with 3DL1*00101.
FIGURE 2.
Phylogenetic analysis of the KIR3DL1 alleles. The neighbor joining tree of full-length coding sequences (4) shows the relationship between the different KIR3DL1/S1 alleles. Confidence of groupings was estimated by 1000 bootstrap replicates and its percentage is shown beside each branch. The alleles discussed in this study are shown in gray.
Differential expression levels of KIR3DL1 alleles
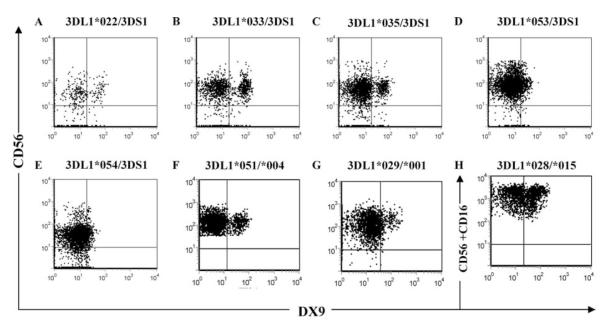
We next sought to assess expression of these KIR3DL1 allotypes on peripheral blood NK cells. PBMCs from HIV+ patients carrying KIR3DL1*022, *028, *029, *033, *035, *051, *053, and *054 alleles were available to assess NK cell-surface expression using DX9. In each case, quadrant gating was based on isotype staining for that specific donor (data not shown). Five of the eight alleles (KIR3DL1*022, *033, *035, *053, and *054) were identified in subjects with KIR3DS1 on the opposite haplotype, and one (*051) was observed in a KIR3DL1*004-positive donor. Because the DX9 mAb does not recognize KIR3DS1 (23, 29–31), staining with this mAb was specific for these KIR3DL1 allotypes and showed that each was expressed on the NK cell surface, although at differing levels of intensity (Fig. 3, A—E). KIR3DL1 allotypes that stained dimly with DX9 also stained dimly with Z27 (data not shown). The expression profile from the donor with KIR3DL1*051/*004 shows that KIR3DL1*051 is also definitely expressed on the cell surface (Fig. 3F), because KIR3DL1*004 is known to be retained within the cell (14). KIR3DL1*029 and *028 were identified in donors with KIR3DL1*001 and *015 on the opposite haplotype, respectively (Fig. 3, G and H). Because KIR3DL1*001 and *015 stain brightly with DX9 mAb, it was not possible to determine whether *029 and *028 are expressed at a high level (similar to *001 and *015), not expressed at all, or are not recognized by the DX9 mAb. We do not think that the relatively low detection of KIR3DL1 allotypes in patient samples reflects an effect of HIV infection but rather is due to the cryopreservation of these samples and conditions required for data acquisition from infected individuals (E. Yamada and D. W. McVicar, unpublished results).
FIGURE 3.
Novel KIR3DL1 alleles are differentially expressed on NK cells. PBMCs from HIV-infected individuals were stained with anti-CD3, anti-CD16, anti-CD56, and anti-KIR3DL1 clone DX9 mAbs. KIR3DL1 expression was analyzed on CD3-CD16+ CD56+/- NK cells. Dot plots show DX9 vs CD56 staining of CD3-CD16+ gated NK cells from donors with KIR3DL1*022/3DS1 (A), KIR3DL1*033/3DS1 (B), KIR3DL1*035/3DS1 (C), KIR3DL1*053/3DS1 (D), KIR3DL1*054/3DS1 (E), KIR3DL1*051/*004 (F),andKIR3DL1*029/*001 (G), or DX9 vs CD56 + CD16 staining profiles of CD3-CD16+CD56+ gated cells from a donor with KIR3DL1*028/*015 (H).
Low expression of KIR3DL1*053 on NK cells from a normal donor
HIV infection has been shown to modulate the expression of various NK receptors (32). Expression of KIR3DL1*053 and *054, which were first identified in HIV-infected patients, appears to be considerably lower than that of other known allotypes (Fig. 3, D and E). KIR3DL1*053 differs from *005 by a single amino acid in the first Ig-like domain. To address whether the low expression of KIR3DL1*053 (Fig. 3D) is due to viral infection, we compared expression levels of *053 and *005 on NK cells from normal donors, each of whom also carried the highly expressed *008 allotype. The expression level of KIR3DL1*008 was determined by staining cells from a KIR3DL1*004/*008 donor (Fig. 4A). As expected, the donor with KIR3DL1*005/*008 showed a population of NK cells staining brightly (cells expressing *008 alone, and those expressing both *008 and *005), and a second population with a lower level of DX9 staining (those with *005 only) (Fig. 4B). The KIR3DL1*053/*008-positive donor also showed a bright staining population of NK cells (cells expressing *008 alone, and both *008 and *053), but the second population, representing only *053, stained poorly with both DX9 (Fig. 4C) and Z27 (not shown), even when compared with the low expression of *005. Importantly, staining of these NK cells with various concentrations of DX9 indicated that the differences observed were not due to variable mAb affinities, but rather reflected the abundance of KIR3DL1 molecules on the cell surface (data not shown) (17). Thus, the expression level of KIR3DL1*053 is even lower than that of conventionally “low” KIR3DL1 allotypes.
FIGURE 4.
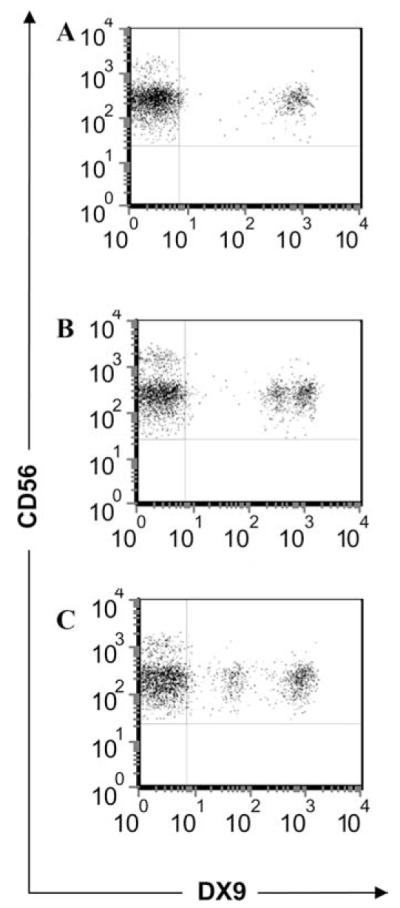
Expression of KIR3DL1*053 on NK cells is lower than that of *005. PBMCs from individuals with KIR3DL1*004/*008 (A), *005/*008 (B), and *053/*008 (C) were stained with anti-CD3, anti-CD56, and anti-KIR3DL1 DX9 mAbs, and expression of KIR3DL1 was analyzed on CD3-CD56+ NK cells. Dot plots show DX9 vs CD56 staining of NK cells.
Variation in the promoter regions of the KIR3DL1 alleles does not account for differential expression patterns
The proximal promoter region 300 bp upstream of the KIR3DL1 start codon (18) was sequenced to explore whether the expression pattern of the alleles could be due to variation in this region. Six polymorphic sites were identified, three of which (-173, -65, -26) disrupt consensus transcription factor binding sites (STAT/YY1, E2F, and Sp1, respectively) (Table I). Position -9 was polymorphic (A/G) across *022 alleles of different donors, but this position is not contained within a known transcription factor binding site. KIR3DL1*005 and *053 were identical in the promoter region, indicating that the difference in expression levels between the corresponding allotypes is not likely to be due to the 5′ up-stream promoter region.
Table I.
Polymorphism in the KIR3DL1 and KIR3DS1 promoter regiona
| KIR Alleles | -173 STAT/YY1 | -65 E2F | -29 | -26 Sp1 | -23 | -9 |
|---|---|---|---|---|---|---|
| 00101 | T | A | C | A | C | G |
| 052 | —b | — | — | — | — | — |
| 005 | — | G | — | — | — | — |
| 053 | — | G | — | — | — | — |
| 00401/02 | — | — | — | — | — | — |
| 022 | — | — | — | G | T | A/G |
| 006 | — | — | T | G | T | — |
| 035 | — | — | — | G | T | — |
| 01501 | — | — | — | G | — | — |
| 008 | — | — | — | G | — | — |
| 020 | — | — | — | G | — | — |
| 028 | — | — | — | G | — | — |
| 007 | — | — | — | G | — | — |
| 033 | — | — | — | G | — | — |
| 01502 | — | — | — | G | — | — |
| 029 | — | — | — | G | — | — |
| 051 | — | — | — | G | — | — |
| 002 | — | — | — | G | — | — |
| 054 | — | — | — | G | — | — |
| 009 | — | — | — | — | — | — |
| 3DS1*01301 | C | — | — | — | — | — |
New and recently described alleles included in this study are shown in bold.
—, Sequence identity with 3DLI*00101.
A single amino acid change affects KIR3DL1 expression
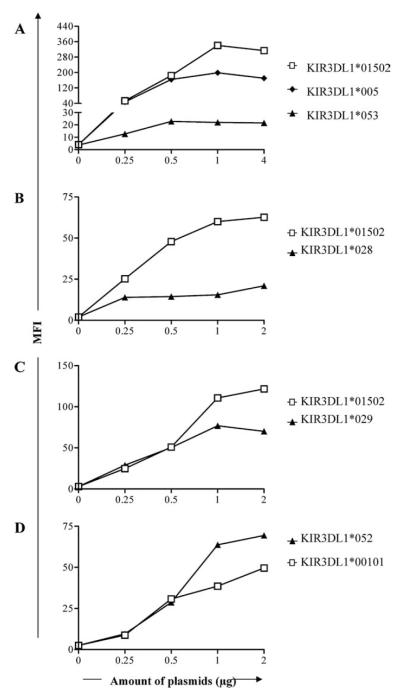
The extracellular domain of KIR3DL1*053 differs from KIR3DL1*005 by a single nucleotide in exon 3 resulting in a V to L substitution at amino acid residue 18. To determine whether the low expression level of *053 is due to this change, we introduced a V to L mutation at amino acid 18 of KIR3DL1*005, creating KIR3DL1*053 (*005V18L). HEK293T cells were transfected with various concentrations of plasmids carrying the “high” KIR3DL1*01502, the “low” *005, or *053 (*005V18L) allele, and cell-surface expression was determined by staining with DX9. Cells transfected with plasmids carrying KIR3DL1*01502, *005, and *053 (*005V18L) were saturated when >0.5 μg plasmids were used. The expression level of KIR3DL1*053 determined by mean fluorescence intensity (MFI) was exceptionally low compared with that of KIR3DL1*005 and *01502 (Fig. 5A). Because the staining of transfected cells became saturated at the same points, differences observed in the percentage of DX9+ cells were due to variability in their expression levels. This result indicates that the leucine at amino acid 18 is responsible for the low expression of KIR3DL1*053.
FIGURE 5.
A single amino acid change alters KIR3DL1 expression. HEK293T cells transfected with indicated concentrations of plasmids carrying KIR3DL1*005, *01502, or *053 (*005V18L)(A), *01502 or *028 (*015R75W+R145S)(B), *01502 or *029 (*015Q256K)(C), and *00101 or *052 (001R266Q)(D) were stained with DX9. The results show the MFI of DX9.
The expression of KIR3DL1*028, *029, and *052 on PBMCs could not be determined due to the unavailability of suitable donors. Therefore, plasmids carrying these alleles were made by mutating nucleotides of the most closely related KIR3DL1 allele. The extracellular domains of KIR3DL1*028, *029, and *052 differ from KIR3DL1*01502 or KIR3DL1*001 by one or two amino acids. Introduction of mutations resulting in an R to W at amino acid residue 75 and an R to S at amino acid residue 145 in KIR3DL1*01502 created KIR3DL1*028 (*015R75W+R145S). The resulting allotype was identical to KIR3DL1*028 at the amino acid sequence level. A single nucleotide change in exon 5 of KIR3DL1*01502 resulted in a Q to K substitution at amino acid 256, creating KIR3DL1*029 (*015Q256K). Finally, an R to Q substitution was introduced at amino acid 226 of KIR3DL1*00101 to create KIR3DL1*052 (*001R226Q). These plasmids were transfected into HEK293T cells and expression was detected by staining with DX9.
Both KIR3DL1*028 and *029 were expressed on the cell surface, and the expression levels were lower than KIR3DL1*01502 (Fig. 5, B and C). KIR3DL1*052 was also expressed on the cell surface, and interestingly the apparent expression level was higher than that of KIR3DL1*00101 (a “high” allotype) (Fig. 5D). Taken together, these data confirm the ability of even a single amino acid substitution in the extracellular region of KIR3DL1 to alter its conformation and suggest the existence of many KIR3DL1 allotypes with wide variation in DX9 reactivity and/or cell-surface expression.
Intracellular retention of KIR3DL1*028 and *053
Because alterations within the D0 domain of KIR3DL1*004 are known to affect the expression of this allotype by causing intracellular retention (14), we asked whether KIR3DL1*028 or *053 might be partially retained within the cytoplasm. KIR3DL1*028 differs from *01502 by two residues: W75 in the D0 domain and S145 of the D1 domain. The serine residue at position 145 of *028 is known to have little effect on expression, as it is shared with KIR3DL1*020, a highly expressed allotype. Thus, we compared retention of KIR3DL1*028 to *01502 (Fig. 6A). In parallel, we compared expression of KIR3DL1*005 with *053, as these allotypes differ by only one residue at position 75 of D0 (Fig. 6B). Consistent with the data shown in Fig. 5, A and B, detection of surface expression of *053 and *028 by anti-FLAG Ab was lower than that of their reference allotypes, with *053 showing a greater effect than *028 (Fig. 6, left columns). As anticipated, analysis of fixed and permeabilized cells with anti-FLAG significantly reduced these differences (Fig. 6, right columns). These results indicate that low expression levels of KIR3DL1*028 and *053 on the cell surface are partially due to intracellular retention of these molecules.
FIGURE 6.
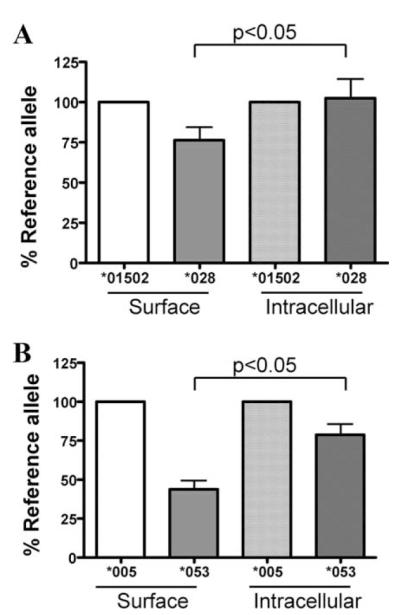
Intracellular retention of KIR3DL1*028 (A) and *053 (B). HEK293T cells were transfected with equal amounts of the indicated FLAG-tagged KIR3DL1 cDNAs. Twenty-four hours after transfection, cells were harvested and surface KIR3DL1 expression was assessed using FITC-conjugated anti-FLAG Ab (left two columns). Parallel samples were fixed, permeabilized, and stained for total KIR3DL1 with the same Ab (right two columns). Staining of each novel allotype (darker columns) was normalized to expression of its reference allotype (lighter columns).
Discussion
Allelic diversity within the KIR genes is thought to be generated by point mutation, gene conversion, and recombination (33, 34), but the relative contribution of these mechanisms varies across the KIR genes. At least two inhibitory KIR genes, 3DL1 and 3DL2, are known to have high allelic polymorphism as a result of point mutation and recombination (9, 33). The KIR3DL1 alleles studied herein, *022, *028, *029, *033, *035, *051, *052, and *053, appear to be the result of point mutations in one or more exons with respect to previously characterized KIR3DL1 alleles (Fig. 1). The generation of the chimeric KIR3DL1*054 allele is most easily explained by a gene conversion event involving ancestral *002 and KIR3DS1 alleles. This patchwork pattern of sequence similarity between a novel allele and two other alleles/genes has also been described for KIR2DL1*004 (2DL1v), which is similar to KIR2DL1 except for exons 5 and 6 where it is identical to KIR2DS1 (33); KIR2DS2*005 (2DS2v2), which shows sequence homology to KIR2DS2 in the extracellular regions and to KIR2DS3 in the transmembrane and cytoplasmic domains (34); and KIR3DL1*009, which is identical to KIR3DS1 in exons 2 and 3, but has a KIR3DL1 background otherwise.
We have been able to investigate for the first time herein the expression levels of the novel KIR3DL1 alleles as well as several recently described, but uncharacterized KIR3DL1 alleles. Previous data indicate that KIR3DL1 allotypes can be grouped into high (*001, *002, *008, *015, and *020), low (*005, *006, and *007), or no (*004) cell-surface expression. However, even within these groups, variations in expression levels are detected (9, 17). These differential expression patterns of KIR3DL1 allotypes have been attributed to minor differences in their coding sequences (9, 14). The expression levels of inhibitory KIR receptors can dramatically affect NK cell function by potently inhibiting NK cell lysis of MHC class I+ healthy cells and therefore unleash more potent killing when their ligands are down-regulated (35).
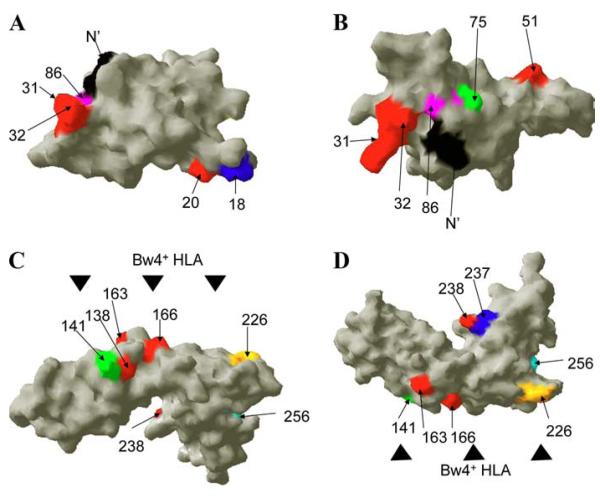
Increasing evidence suggests that the D0 domain plays an important role in KIR3DL1/S1 function (14, 27, 36). Although the D0 domain is not predicted to directly participate in HLA binding, residues 50 and 51 have been shown to modulate the interaction between KIR3DL1 and HLA-B (36). KIR3DL1*028 and *053 differ from *020 (high expressing allotype) and *005 (low expressing allotype), respectively, by one amino acid in D0. Therefore, it is conceivable that these changes may affect the strength of interaction between these allotypes and HLA molecules. Additionally, previous data indicate that leucine at amino acid 86 disrupts the WSAPS motif found in the D0 domain of all other KIR. The disruption of the analogous region of the erythropoietin receptor prevents proper folding and leads to retention of the receptor within the cell (37). Similarly, mutation in this motif participates in the intracellular retention of KIR3DL1*004 (14), confirming that residues in D0 can also dramatically regulate KIR3DL1 expression. Interestingly, the R75W change, which is unique to *028, is also in close proximity to position 86 (Fig. 7B), and *028 is expressed on the cell surface at a much lower level than the high expressing *01502 allotype. In accordance with this notion we find that both *053 and *028 are retained within transfectants, demonstrating that the low expression of KIR3DL1*028 and *053 is due, at least in part, to retention in the cytoplasm in a manner similar to *004 (Fig. 6). Furthermore, Norman et al. recently reported five residues within the D0 domain that are under strong positive selection (amino acids 5, 20, 31, 32, and 51) (27). The novel substitution found in *053 is in close proximity to residue 20, suggesting that the alteration in expression of *053 might be due to perturbation of this otherwise conserved region of D0 (Fig. 7A). Regardless, our data on these new alleles support the contention that residues clustered in key positions of D0 play critical roles in KIR3DL1 expression and therefore biology.
FIGURE 7.
Several amino acid changes found in the novel and recently discovered KIR3DL1 allotypes are located at positions that potentially result in functional differences. Two views of models of the D0 domain of KIR3DL1 are shown (A and B). The model is based on the crystal structure of human platelet glycoprotein VI. The N terminus amino acid residue (amino acid 7) is shown in black. Positively selected residues (amino acids 20, 31, 32, and 51) reported by Norman et al. (27) are shown in red. Residue 86 unique to KIR3DL1*004 is shown in pink, while residues 18 and 75 that are unique to KIR3DL1*053 and *028 are shown in blue (A) and green (B), respectively. Two views of models of the D1 and D2 domains are shown (C and D). The model was based on the crystal structure of KIR2DL1. Black triangles indicate a predicted HLA-interaction face. Residues in red (amino acids 138, 163, 166, and 238) are unique to KIR3DL1*054. Colored residues (amino acids 141, 226, 237, and 256) represent unique amino acids identified in KIR3DL1*033, *035, *029, and *052.
In contrast to *028 and *053, KIR3DL1 alleles *022, *029, *033, *035, *052, and *054 differ from well-defined KIR3DL1 alleles by amino acids in the D1 and D2 domains, suggesting that they may alter binding characteristics and/or expression. Residues 138, 141, 163, 166, and 226 of KIR3DL1 are all predicted to be within the HLA-contacting loops (Fig. 7, C and D) and, accordingly, some of these residues are also strongly positively selected in evolution (27). Residues 237 and 256, unique to KIR3DL1*035 and *029, respectively, are well away from these predicted HLA contact sites (Fig. 7, C and D), suggesting that as with changes in D0, these positions primarily alter the expression patterns of KIR3DL1. In contrast, KIR3DL1*033 differs from *007 by one amino acid at residue 141. Because these allotypes are comparably expressed on the cell surface, it will be interesting to determine whether this change in an HLA-contacting residue affects ligand binding.
KIR3DL1*052 and *054 have acquired mutations that may alter both recognition of HLA ligand and their expression. In the first case, a single amino acid change in the D2 domain of KIR3DL1*052, relative to *001, resulted in a very high expression level of this novel allotype (Fig. 5D). This substitution in *052 is also in the HLA contacting face and occurs within the RSSYDM motif of the D2 domain that is otherwise highly conserved in KIR3DL1 and KIR3DS1 allotypes (38). Similarly, KIR3DL1*054 is identical to *002 (a high expressing allotype) except in exon 4 (D1 domain), where it is identical to KIR3DS1. Based on staining with Z27, which recognizes both inhibitory and activating allotypes of the KIR3DL1/S1 locus, KIR3DS1 shows low Z27 binding levels relative to most KIR3DL1 allotypes (31). Therefore, the low expression pattern of KIR3DL1*054 is likely due to the variation in the D1 domain that it shares with KIR3DS1. Additionally, amino acid residues 138, 163, and 166 unique to KIR3DL1*054 and KIR3DS1 are part of the predicted MHC contact site and they may be sites of Ab binding. Thus, although it is likely that KIR3DL1*054 and KIR3DS1 share some unique ligand binding characteristics, alteration of Ab binding sites on or near the HLA contact face may be responsible for the apparent low expression of these allotypes.
Taken together, our data support and extend the contention that the large number of KIR3DL1 alleles arose by point mutation to provide a wide spectrum of Bw4+ ligand recognition within the NK compartment. In some cases complexity is achieved by altering the apparent levels of KIR on the cell surface and in others specific mutations within predicted sites of HLA contact suggest diverse ligand binding characteristics.
Altogether, each of these cases involves a distinct and independently derived genetic variant, and in each case the cell-surface expression was altered relative to the closely related allele from which the respective allele was presumably derived. These data hint at the possibility of convergent evolution of KIR3DL1 phenotype variation, where the maintained variants represent a spectrum in terms of cell-surface expression levels. They also build upon recent functional and disease association studies that highlight the biological significance of KIR3DL1 variation (17, 28).
Acknowledgments
We thank Jim Gattis for helping with the structure modeling and Laura Quigley and Geraldine O’Conner for assistance with the analysis of intracellular staining.
Footnotes
This work was supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-C0-12400. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. A.B. was supported by National Institutes of Health Grant DA13324.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
- KIR
- killer cell Ig-like receptor
- MFI
- mean fluorescence intensity
Disclosures The authors have no financial conflicts of interest.
References
- 1.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 2.Ugolini S, Arpin C, Anfossi N, Walzer T, Cambiaggi A, Forster R, Lipp M, Toes RE, Melief CJ, Marvel J, Vivier E. Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat. Immunol. 2001;2:430–435. doi: 10.1038/87740. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD: the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 6.Carrington M, Cullen M. Justified chauvinism: advances in defining meiotic recombination through sperm typing. Trends Genet. 2004;20:196–205. doi: 10.1016/j.tig.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: Expansion of the KIR locus by unequal crossing over. J. Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 10.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, et al. Cutting edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J. Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 11.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 13.Single RM, Martin MP, Gao X, Meyer D, Yeoger M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 14.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 15.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 1994;180:537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 17.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J. Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 19.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 20.Martin MP, Carrington M. KIR locus polymorphisms: genotyping and disease association analysis. In: Ewbank J, Vivier E, editors. Methods in Molecular Biology: Innate Immunity. Humana Press; Totowa, NJ: 2007. pp. 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Swofford DL. Paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- 23.Pascal V, Yamada E, Martin MP, Alter G, Altfeld M, Metcalf JA, Baseler MW, Adelsberger JW, Carrington M, Anderson SK, McVicar DW. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J. Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 24.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Hum. Immunol. 2003;64:648–654. doi: 10.1016/s0198-8859(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 26.Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics. 2006;58:474–480. doi: 10.1007/s00251-006-0108-3. [DOI] [PubMed] [Google Scholar]
- 27.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 28.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EB, Schupert WL, Phair J, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 32.Fauci AS, Mavilio D, Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat. Rev. Immunol. 2005;5:835–843. doi: 10.1038/nri1711. [DOI] [PubMed] [Google Scholar]
- 33.Shilling HG, Lienert-Weidenbach K, Valiante NM, Uhrberg M, Parham P. Evidence for recombination as a mechanism for KIR diversification. Immunogenetics. 1998;48:413–416. doi: 10.1007/s002510050453. [DOI] [PubMed] [Google Scholar]
- 34.Rajalingam R, Gardiner CM, Canavez F, Vilches C, Parham P. Identification of seventeen novel KIR variants: fourteen of them from two non-Caucasian donors. Tissue Antigens. 2001;57:22–31. doi: 10.1034/j.1399-0039.2001.057001022.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 36.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J. Exp. Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilton DJ, Watowich SS, Katz L, Lodish HF. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J. Biol. Chem. 1996;271:4699–4708. doi: 10.1074/jbc.271.9.4699. [DOI] [PubMed] [Google Scholar]
- 38.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol. Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]