Abstract
AIM: To characterise expression of known E-cadherin repressors; Snail, Slug and Twist in the development of esophageal adenocarcinoma.
METHODS: E-cadherin, Slug, Snail and Twist mRNA expression in Barrett's metaplasia and esophageal adenocarcinoma specimens was examined by real-time reverse transcription-polymerase chain reaction (RT-PCR). Semi-quantitative immunohistochemistry was used to examine cellular localization and protein levels. The effect of Slug on epithelial mesenchymal transition (EMT) markers was examined by transfection of Slug into an adenocarcinoma line OE33.
RESULTS: Cellular localization of Slug in Barrett’s metaplasia was largely cytoplasmic whilst in adenocarcinoma it was nuclear. Semi-quantitative analysis indicated that Slug was more abundant in adenocarcinoma compared to matched Barrett's metaplastic specimens. Snail and Twist were expressed in adenocarcinoma but were cytoplasmic in location and not induced compared to Barrett's mucosa. These observations were supported by mRNA studies where only Slug mRNA was shown to be over-expressed in adenocarcinoma and inversely correlated to E-cadherin expression. Overexpression of Slug in OE33 mediated E-cadherin repression and induced the mesenchymal markers vimentin and fibronectin.
CONCLUSION: Progression to adenocarcinoma is associated with increased Slug expression and this may represent a mechanism of E-cadherin silencing.
Keywords: Slug, Oesophagus, Cancer, Barrett’s metaplasia, Epithelial-mesenchymal transition
INTRODUCTION
The incidence of esophageal adenocarcinoma is currently rising faster than any other cancer in the Western world. Alarmingly the cause of this increase is largely unknown[1]. The strongest known risk factor for esophageal adenocarcinoma is the condition Barrett’s metaplasia which is characterized by a replacement of the native squamous lined esophagus with a columnar epithelium: a possible consequence of prolonged reflux of gastric contents into the lower esophagus[1,2].
To date, numerous proteins have been implicated in the malignant progression of Barrett’s metaplasia to adenocarcinoma including E-cadherin[3–6]. E-cadherin is a calcium dependent cell adhesion molecule which is essential in the establishment of homotypic adhesion junctions[7]. In nearly all epithelial cancers E-cadherin has been reported to be repressed: an essential step for increased invasiveness and metastasis[8–10], thus this event commonly occurs during late tumourigenesis. In particular in the development of esophageal adenocarcinoma, E-cadherin repression has been reported to be a late event associated with high grade dysplastic Barrett’s metaplasia and adenocarcinoma[4–6].
However, it remains unclear what directs repression of E-cadherin expression in esophageal adenocarcinoma. Unlike in hereditary diffuse gastric cancer E-cadherin is not subject to mutation and neither is there evidence of E-cadherin promoter methylation as is common in colorectal cancers[11,12]. A possible mechanism of E-cadherin silencing, which to date has not been addressed, is transcriptional repression by proteins involved in epithelial mesenchymal transition (EMT) including proteins in the Snail family: Snail and Slug and the transcription factor Twist[13–15].
Several studies have elegantly demonstrated in a variety of cell types that overexpression of Snail and Slug causes an EMT and this is associated with E-cadherin repression[16,17]. Furthermore, it has been demonstrated that these effects are mediated through interaction with specific E-pal elements of the E-cadherin proximal promoter[16,17]. In support of a role for these EMT regulators in E-cadherin repression and carcinogenesis, several studies have shown overexpression in several epithelial cancers including overexpression of Slug in gastric carcinomas[18], Snail in colorectal cancers[19] and Twist in pancreatic cancers[20].
The primary aim of this study was to determine the expression profile of Snail, Slug and Twist in the development of esophageal adenocarcinoma and determine if overexpression of these proteins in an esophageal background is able to mediate a repression in E-cadherin.
MATERIALS AND METHODS
Ethics
This work has been carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association. This study was approved ethically by University Hospital Birmingham Trust (LREC 2002/166). All patients provided informed written consent.
Patient tissue
Esophageal adenocarcinoma resection specimens: Samples of normal squamous esophagus (n = 40) and esophageal adenocarcinoma (n = 40) were collected during surgery. Half of the esophageal adenocarcinoma resection specimens (n = 20) collected also had associated intestinal Barrett’s metaplasia. In addition, samples (n = 20) of long segment (≥ 3 cm) Barrett’s metaplasia, defined as columnar mucosa with intestinal type goblet cells were also collected during endoscopy. All specimens were divided in two, half for RNA extraction and half for immunohistochemistry.
Immunohistochemistry: Immunohistochemistry for E-cadherin was performed using microwave antigen retrieval as previously described with an E-cadherin monoclonal antibody (clone 36, BD Biosciences, Oxford, UK) used at a concentration of 1:300[21]. Immunohistochemistry for Snail, Slug and Twist was performed as follows: Slides were immersed in W-cap buffer (Bio-Optica, Milan, Italy) and cycled in a Pixel antigen retriever (CellPath, Newtown, UK) for 60 min, washed in running water and placed in methanol:hydrogen peroxide (10:1) for 5 min. Sections were then incubated in a primary antibody to either Snail, 1:10, (SNAI1 clone E18, Autogen Bioclear, Wiltshire, UK), Slug, 1:20 (SNAI2 clone G18 Autogen Bioclear, UK) or Twist, 1:50 (clone C17 Autogen Bioclear, UK) in TBS 7.5 × buffer (Bios Europe Ltd, Skelmersdale, UK) at 4°C overnight, washed with TBS and reacted with peroxidase-linked rabbit anti-sheep antibody (Dako, Ely, UK) at a 1:100 dilution in TBS for 1 h. The immunoreactivity was then revealed as above using DAB. Slides were then dipped in hematoxylin, dehydrated and mounted. The slides were scored by a previously described method for (1) intensity of staining (0 = negative, 1 = weak, 2 = moderate, 3 = intense) and (2) percentage of epithelial cells staining (0 = 0%-5%; 1 = 6%-25%; 2 = 26%-50%; 3 = 51%-75%; 4 = 76%-100%); these two scores were multiplied to yield a final staining score[39]. In addition, cellular localization (nuclear, cytoplasmic, cell surface) was assessed. All sections were scored independently by two observers (PJ and CT).
In the series of immunofluorescent experiments following primary antibody incubations, sections were washed extensively and then incubated with either FITC goat anti-mouse or goat anti-rabbit (Jackson Immunoresearch, USA, 1:500) for 1 h. Sections were then washed and incubated in 4’, 6-Diamidino-3-phenylindole dihydrochloride hydrate (DAPI) (1:10 000) for 1 min prior to visualisation. Omission of primary antibody was employed as a negative control. Images were visualized using an Olympus BX40 microscope and digital images taken using a Sensys Photometrics camera (Middlesex, UK). Desksoft SmartCapture 2 software was used for image acquisition (Desksoft, USA).
Real time RT-PCR
Real time RT-PCR reactions were performed as previously described using 18S ribosomal RNA as an internal standard (PE Biosystems, Roche, USA)[21]. Each reaction was performed in triplicate and contained one of the following sets of probes and primers: (1) E-cadherin (probe 5’FAM AAATTCACTCTGCCCAGGACGCGGTAMRA3’), forward primer (5’-GGCGCCACCTCGAGAGA-3’) and reverse primer (5’-TGTCGACCGGTGCAATCTT3’); (2) Slug (probe 5'FAM'CACATACAGTGATTATTTCCC CGTATCTCTATGAGAGTTAMRA3'), forward primer (5’-AAAAGCCAAACTACAGCGAACTG-3’) and reverse primer (5’-AGAATCTCTGCTTGTGGT ATG ACA-3’); (3) Snail (probe 5’FAMTGCAGGACTCTA ATCCAGAGTTTACCTTCCAGCTAMRA3’), forward primer (5’-CCCAATCGGAAGCCTAACTACAG-3’) and reverse primer (5’-CAGGTGGGCCTGGTCGTA-3’), or (4) Twist (probe 5’-FAMACAATGACATCTAGG TCTC-3’), forward primer (5’-GCTCCAGAGTCTCTA GACTGTCCATT-3’) and reverse primer(5’-GGGGCCTGGTCCATGTC-3’).
Western blotting
Western blotting was performed as previously described[21] with antibodies to (1) E-cadherin (1:2 000, clone 36, Autogen Bioclear UK); (2) Fibronectin (1:1000, Clone P1F11 Autogen Bioclear, UK); (3) Vimentin (1:1000, clone C-20, Autogen Bioclear, UK); (4) Cytokeratin 19 (CK-19) (1:2000; Oncogene Research Products USA) and (5) GFP (1:2000, Clone AB290 Abcam, UK). Immunoreactive bands were then subject to densitometry using NIH Image 1.62 software.
Cell culture
Cell lines derived from esophageal adenocarcinoma (OE33[22], SEG-1[23] and oesophageal squamous carcinoma TE-7[24]) were all routinely cultured in Dulbecco’s modified Eagles medium (Gibco, USA) with 10% fetal calf serum supplemented with 1 × 105 Units/L penicillin and 1 g/L streptomycin. Upon reaching 70% confluence cells were lysed into Trizol reagent (Gibco, UK) for mRNA extraction and evaluation of Slug mRNA expression by Real Time PCR.
Transfection studies
Cell line OE33 was transiently transfected with either control empty vector, pMSCV-GFP or full-length human Slug-GFP tagged construct, pMSCV-Slug-GFP[25] or beta-galactosidase plasmid (beta-GAL) using Lipofectamine Plus according to manufacturers instruction. 48 h post transfection cells were either assayed for transfection efficiency using beta-galactosidase assay to determine transfection efficiency, lysed into RIPA buffer for Western blotting or lysed in Trizol for real time RT-PCR studies. Data analysis was only performed in experiments where a minimum of 40% transfection efficiency was achieved as determined by beta-galactosidase assay.
To determine the effect of Slug on the human E-cadherin promoter, an E-cadherin promoter assay was performed as previously described[21]. Briefly, OE33 cells were transiently transfected with full length human slug (pCDNA3-Slug[25]), Renilla luciferase plasmid pRL TK and the wild-type human E-cadherin promoter cloned into the pGL3basic luciferase reporter plasmid (EproWT) a kind gift from Professor Frans van Roy). Controls included empty pCDNA3 and empty pGL3basic vectors. 48 h post transfection cells were assayed for firefly and Renilla luciferase activities using the dual-reporter assay kit Stop ‘N’ Glow (Promega) according to manufacturer’s instruction. Firefly luciferase activity was normalized to Renilla luciferase activity as a transfection control. Promoter activity was expressed as a fold change in relative luciferase units (RLU) compared to that obtained in pGL3basic control transfected cells. Results shown represent the means ± SE of three independent experiments.
Statistical analysis
All data are presented as means ± SE. Statistical significance was calculated using unpaired Student’s t test. To assess the association between Slug and E-cadherin, linear regression analysis was performed on log-transformed mRNA fold change values using log Slug expression as the independent variable and log E-cadherin expression as the dependent variable. The R2 value was determined to demonstrate the extent to which expression of Slug was correlated to E-cadherin. A Bonferroni adjustment was applied to P values attained from multiple comparisons during the analysis of prognostic factors. In univariate analyses significance was accepted at P ≤ 0.05, following Bonferroni adjustment, P ≤ 0.0125 was considered significant. All analyses were performed using SPSS version 10.0 (SPSS Inc, USA).
RESULTS
Immunolocalization of Snail, Slug and Twist in esophageal adenocarcinoma
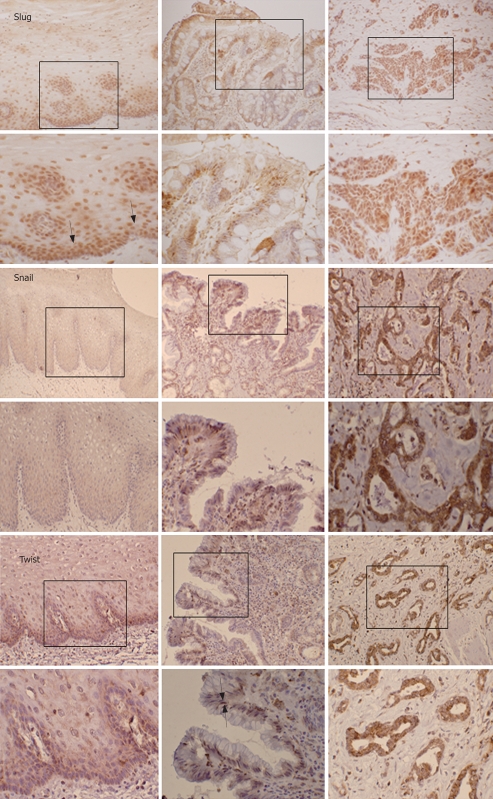
In normal stratified squamous esophagus, there was no detectable expression of Snail and only weak diffuse cytoplasmic immunoreactivity for Twist, while there was strong nuclear immunoreactivity for Slug in basal and supra-basal layers (Figure 1). In Barrett’s metaplasia, Snail and Slug were generally localised to the cytoplasm with staining being patchy and diffuse in nature. In contrast, Twist was largely nuclear in localization (Figure 1). In esophageal adenocarcinoma, all three proteins were abundantly expressed with Snail and Twist being mostly localised in the cytoplasm whilst Slug was almost exclusively nuclear in all adenocarcinoma specimens (n = 20) (Figure 1).
Figure 1.
Immunolocalization of Slug, Snail and Twist in normal esophagus, intestinal type Barrett’s metaplasia and oesophageal adenocarcinoma. Normal squamous esophagus (S), intestinal type Barrett’s metaplasia (BM) and esophageal adenocarcinoma (ADC) were subjected to immunohistochemistry using antibodies to Slug, Snail and Twist. Arrows denote areas of positivity (× 20, × 40).
In addition, all slides were scored and data are shown in Table 1. To determine if Snail, Slug and Twist were over-expressed in adenocarcinoma specimens relative to non-matched Barrett’s metaplastic specimens (BM-ve), statistical analysis was performed and results showed that both Slug and Snail were significantly over-expressed (P < 0.05) in esophageal adenocarcinoma. Twist, however, was not significantly induced. To validate this data, we further performed immunohistochemistry on a further 20 sections of adenocarcinoma with matched Barrett’s metaplastic tissue (BM+ve). When comparing the staining scores of matched Barrett’s metaplastic specimens with adenocarcinoma, only Slug was significantly over-expressed (P < 0.05).
Table 1.
Semi-quantitative analysis of Snail, Slug and Twist in malignant progression of Barrett’s metaplasia to adenocarcinoma (Immunoreactivity score)
Semi-quantitative analysis of immunoreactivity for Snail, Slug and Twist in specimens of squamous esophagus (S); Intestinal type Barrett’s metaplasia without associated adenocarcinoma BM (-ve), intestinal Barrett’s metaplasia with associated adenocarcinoma BM (+ve) and esophageal adenocarcinoma (ADC). The mean staining scores were calculated for each type of mucosa and the mean fold change in protein expression was calculated.
denotes significant (P < 0.05) differences between adenocarcinoma and intestinal type Barrett’s metaplasia without associated adenocarcinoma.
denotes significant (P < 0.05) differences between adenocarcinoma and intestinal type Barrett’s metaplasia with associated adenocarcinoma.
Since there is a growing body of evidence implicating Slug as a repressor of E-cadherin, we further sought to address if Slug overexpression was associated with depressed E-cadherin expression in sections of both Barrett’s metaplasia and adenocarcinoma using dual immunofluorescence with anti-Slug (FITC) and E-cadherin antibodies (Texas Red) (Figure 2). Sections of Barrett’s metaplasia mostly demonstrated preserved cell border E-cadherin immunoreactivity consistent with its role in cell-cell adhesion whilst there was little detectable expression of Slug. Conversely in sections of adenocarcinoma in areas of abundant Slug immunoreactivity, there was almost a complete loss of E-cadherin immunoreactivity with the expression mostly localised to the cytoplasm.
Figure 2.
Co-immunofluoresence of Slug and E-cadherin in Barrett’s metaplasia and esophageal adenocarcinoma. Sections of Barrett's metaplasia (BM) mostly demonstrate preserved cell border E-cadherin (Texas Red) immunoreactivity consistent with its role in cell-cell adhesion. Slug (FITC) immunoreactivity in BM however was minimal and appeared non specific. Conversely in sections of adenocarcinoma (ADC) in areas of abundant Slug immunoreactivity, there was negligible E-cadherin immunoreactivity. DAPI staining was used to highlight nuclei in sections (× 40).
mRNA expression of E-cadherin, Slug, Snail and Twist in Barrett’s metaplasia and esophageal adenocarcinoma specimens
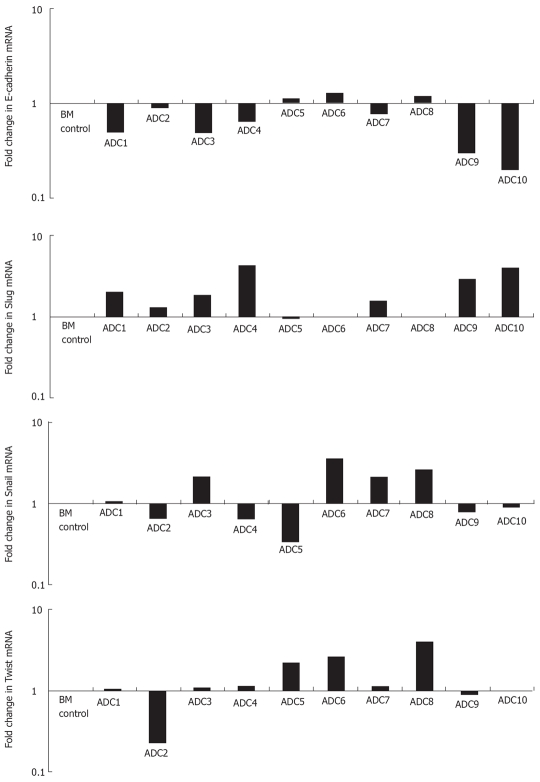
To determine if Slug, Snail, Twist and E-cadherin were modulated at the transcriptional level in esophageal adenocarcinoma, 10 specimens of esophageal adenocarcinoma each with matched intestinal Barrett’s metaplasia were analysed by real-time PCR (Figure 3).
Figure 3.
mRNA expression of E-cadherin, Slug, Snail and Twist in esophageal adenocarcinoma. Real time RT-PCR was employed to examine the expression of E-cadherin, Slug, Snail and Twist in ten esophageal adenocarcinoma specimens (ADC1-ADC10) each compared to Barrett’s metaplastic tissue collected from the same resection specimen. Barrett’s metaplastic tissue control (BM), normalized to 1.0.
E-cadherin mRNA was repressed in 70% of adenocarcinoma specimens whilst Slug, Snail and Twist was over-expressed in 70%, 40% and 50%, respectively. The mean fold decrease for E-cadherin in the ten specimens examined was 0.73 whilst Slug, Snail and Twist had mean fold increases of 2.07, 1.47 and 1.50 respectively. We then sought to determine if the mean fold changes for Slug, Snail and Twist across the ten specimens were significantly different from E-cadherin values. This revealed that only Slug was significantly elevated compared to E-cadherin expression (P < 0.005). Additionally we assessed the association between E-cadherin and these EMT regulators using linear regression analysis. Results of this showed that again only Slug was negatively correlated with E-cadherin expression (R2 0.677, P < 0.03) (Figure 4).
Figure 4.

Correlation of E-cadherin and Slug mRNA expression in esophageal adenocarcinoma specimens. Using linear regression analysis, Slug mRNA expression was demonstrated to be negatively correlated with E-cadherin expression in the ten adenocarcinoma specimens examined (ADC1-10) (R2 0.677, P < 0.03).
Exogenous overexpression of Slug in the cell line OE33 induces an EMT phenotype
Slug mRNA expression was examined in a panel of three esophageal cell lines OE33, SEG1 and TE-7 by real-time PCR and results showed that the cell line OE33 had the lowest expression of Slug mRNA whilst the highest Slug expressing cell line was TE-7. In this regard, the cell line OE33 was chosen for Slug overexpression studies.
The cell line OE33 was transiently transfected with either full length human Slug-GFP vector or the control empty GFP vector. Forty-eight h after transfection, cells were either lysed and processed for mRNA and protein analysis or fixed for immunofluorescence.
mRNA analysis revealed an approximate 6.5 × induction in Slug mRNA in cells transfected with full length human Slug-GFP compared to cells transfected with empty-GFP vector (P < 0.05) (Figure 5A). Exogenous Slug expression was further verified indirectly (due to a lack of a commercially available Slug antibody suitable for Western blotting) by Western blotting for GFP (Figure 5B).
Figure 5.
Overexpression of Slug in the esophageal adenocarcinoma derived cell line OE33. Cell line OE33 was transiently transfected with either full length human Slug-GFP vector (OE33 Slug-GFP) or with the control empty GFP vector (OE33 empty). 24 h post transfection, (A) Slug mRNA and (B) GFP protein expression was determined. CK-19 was used as an epithelial control. aP < 0.05 using Student's t test.
To ascertain whether overexpression of Slug in OE33 cells mediated an EMT phenotype, Western blotting was performed to examine protein expression of three EMT markers E-cadherin, vimentin and fibronectin (Figure 6). Densitometric analysis of Western blots (n = 6) revealed Slug overexpression was associated with a repression in E-cadherin (mean fold decrease of 53% ± 15%, P < 0.05) and increased vimentin (41% ± 11%, P < 0.05) and fibronectin expression (32% ± 12%, P < 0.05). In addition, the cell adhesion and signalling molecule beta-catenin was also examined and shown to be significantly repressed (57% ± 19%, P < 0.05).
Figure 6.
The effect of Slug overexpression on proteins implicated in EMT. Expression of E-cadherin, beta-catenin, fibronectin and vimentin were all examined in Slug over-expressing (OE33 Slug-GFP) and control cells (OE33 empty-GFP). Lysates were normalized using GFP expression.
To address whether Slug mediated E-cadherin protein repression was through a direct effect at the level of the E-cadherin promoter and consequently a transcriptional effect, a luciferase E-cadherin promoter assay and real-time PCR was employed respectively as previously described[21,25]. Overexpression of Slug in OE33 resulted in a significant reduction (35% ± 10%, P < 0.05) in E-cadherin promoter activity (Figure 7A) which was mirrored by decreased E-cadherin mRNA expression (20% ± 5%, P < 0.05) (Figure 7B).
Figure 7.
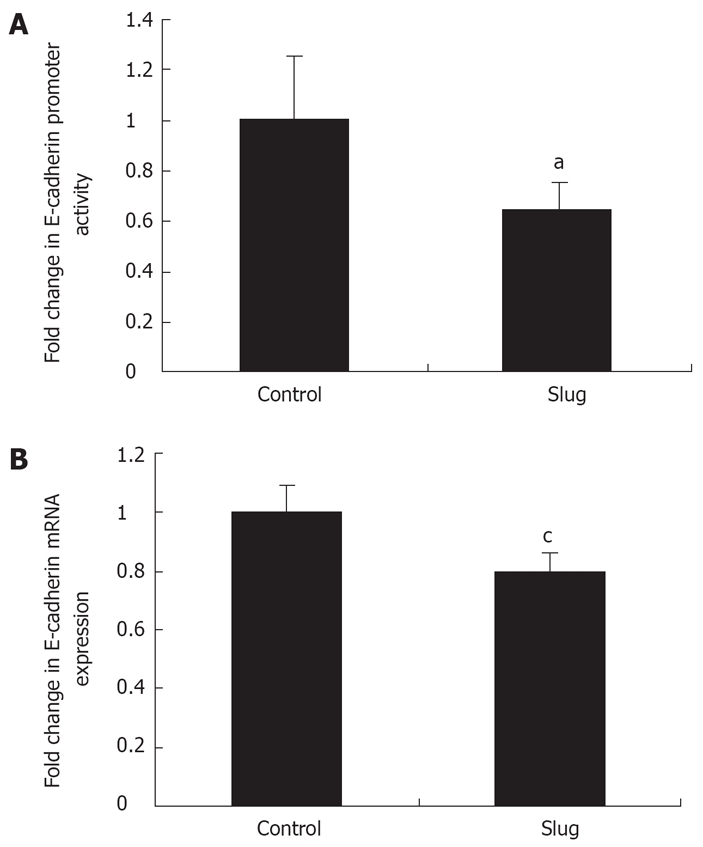
Slug expression causes a decrease in E-cadherin promoter activity and mRNA expression. A: OE33 cells transfected with full length human Slug (Slug) showed decreased E-cadherin promoter activity compared to sham transfected control cells. Promoter activity was expressed as a fold change in relative luciferase units (RLU) compared to control cells. Results represent the means ± SE of three independent experiments. aP < 0.05 using Student’s t test; B: Expression of E-cadherin mRNA in Slug over-expressing cells (Slug) was also repressed in comparison to control cells (control). Relative gene expression was normalized to 1.0 (100%) of controls. Error bars = 2 mean ± SE. cP < 0.05 using Student’s t test. These data are the mean of three independent experiments.
DISCUSSION
Recent reports have highlighted the importance of epithelial mesenchymal regulators including Snail, Slug and Twist in gastrointestinal carcinogenesis[18,19,26–28]. We have demonstrated for the first time a modulation in the expression and localization of these proteins in the malignant progression of normal squamous esophagus to adenocarcinoma.
Twist was only weakly expressed in squamous esophagus consistent with the results of Yuen et al[28]. Their studies reported over 95% of non-neoplastic squamous esophageal samples were either negative or weak for Twist immunoreactivity as assessed by immunohistochemistry. However, in our studies we did observe nuclear Twist staining in Barrett’s metaplasia which in adenocarcinoma had become cytoplasmic in localization. This pattern of cytoplasmic expression for Twist in cancer has also been observed in squamous cell cancers of the esophagus[28]. We were unable to show a difference in Twist expression between matched Barrett’s and adenocarcinoma specimens at both the mRNA and protein level, and neither was its expression associated with prognostic end points such as stage and modal involvement.
In the case of Snail, this was comparably either very weak or nondetectable in stratified squamous oesophagus whilst in both Barrett’s metaplastic tissue and adenocarcinoma Snail was detectable in the cell cytoplasm. Despite moderate expression of Snail in adenocarcinoma specimens, there was no difference in expression between matched adenocarcinoma and Barrett’s specimens at both the protein and mRNA level. A recent study has also reported Snail immunoreactivity in adenocarcinomas of the upper gastrointestinal tract including the esophagus[29]. In their study Snail expression was reported in 11% of esophageal adenocarcinomas examined and no correlation was found between the expression of Snail and tumor grade and stage.
Why both Snail and Twist are localised to the cytoplasm in esophageal adenocarcinoma is unclear though a recent study has suggested that Snail’s localization could in part be determined by post-translational phosphorylation with proteins such as p21-activated kinase 1 (Pak1)[30]. Interestingly, when Pak1 mediated phosphorylation of Snail was inhibited, this not only caused a nuclear to cytoplasmic accumulation but also attenuated its repressor activity[30].
Immunolocalization studies for the transcription factor Slug showed that it was strongly expressed in nuclei of basal and suprabasal esophageal keratinocytes of the stratified squamous epithelium consistent with previous reports in murine studies[31] and other human stratified epithelia including epidermis[25]. Whilst Slug nuclear expression was lost in Barrett’s metaplasia, it was retained in adenocarcinoma and furthermore there appeared to be an overexpression of this protein in adenocarcinoma compared to matched Barrett’s metaplasia at both the mRNA and protein level.
Interestingly Uchikado et al have reported that Slug is also over-expressed in esophageal squamous cell carcinomas (SCC)[27]. Their study showed that the presence of Slug was associated with depth of tumor invasion, lymph node metastasis, stage and lymphatic invasion. Consistent with our study, Slug expression in SCC was significantly correlated with reduced E-cadherin expression.
Thus our data would suggest that of the three EMT regulators examined, Slug was the only transcription factor to show a significant increase at both the mRNA and protein level in the progression from Barrett’s metaplasia to adenocarcinoma. This coupled with nuclear expression in adenocarcinoma and an association with E-cadherin repression suggests functionality in oesophageal tumourigenesis.
The consequence of overexpression of Slug in adenocarcinoma is likely to be complex, as exemplified by the ever-growing list of direct and indirect downstream target genes attributed to be modulated as a consequence of its expression[25,32,33]. As anticipated, many of these targets appear to be involved in cellular survival, proliferation and mesenchymal transition. The latter was exemplified by E-cadherin, a protein commonly silenced in epithelial cancers including esophageal adenocarcinoma[4–6,8–10,17].
To support our hypothesis that Slug might be responsible for the observed repression of E-cadherin in adenocarcinoma, Slug was exogenously over- expressed in an oesophageal cell line OE33. Our results demonstrate that Slug could repress E-cadherin transcription at the level of the E-cadherin promoter. Slug mediated E-cadherin promoter repression has been previously demonstrated in other lineages and this is likely to be mediated through one or more of the E-box elements present within the promoter[17].
To verify the extent of EMT, we further examined the expression of the mesenchymal proteins vimentin and fibronectin[34]. Indeed both proteins were induced as a consequence of Slug expression and consistent with other studies Slug also mediated a repression in beta-catenin, a protein involved in both cell-cell adhesion and Wnt signalling[35].
Abrogating Slug induction may thus represent a potential therapeutic strategy since it appears central to the EMT phenotype observed in esophageal adenocarcinoma. A potential approach might be to silence or block known inducers of Slug. In this regard, the extracellular signals FGF, TGF-beta, and Wnt, which have been reported to be over-expressed and implicated in the pathogenesis of esophageal adenocarcinoma, have been found to induce Slug expression[13,14,36–38]. Thus abrogating the expression of these extracellular signals might represent a mechanism of repressing Slug and potentially mediating a mesenchymal to epithelial transition which is likely to impact on development of disease and ultimately patient survival.
In summary of the EMT regulators examined, Slug appears to be the only transcription factor over-expressed in esophageal carcinogenesis. A downstream consequence of Slug overexpression is likely to be silencing of E-cadherin and ultimately in concert with other signalling molecules induction of invasion and metastasis.
COMMENTS
Background
The incidence of esophageal adenocarcinoma is currently rising faster than any other cancer in the Western world though the cause of this increase is largely unknown. Progression of this disease is associated with a repression in the cell adhesion molecule E-cadherin; a crucial event in invasion and metastasis.
Research frontiers
E-cadherin silencing is a common event in nearly all epithelial malignancies and in several cancers this is either due to mutation, gene deletion or promoter methylation. However, how E-cadherin is silenced in esophageal adenocarcinoma has not been unequivocally addressed. In this study, the authors demonstrate that the overexpression of Slug could be a potential mechanism for mediating E-cadherin repression.
Innovations and breakthroughs
Recent reports have highlighted the importance of epithelial mesenchymal regulators including Snail, Slug and Twist in gastrointestinal carcinogenesis. In particular in esophageal squamous cell cancers, Slug is over-expressed. This is the first study to report that Slug is also over-expressed in esophageal adenocarcinomas. Furthermore, our in vitro studies would suggest that this protein may be the cause of the repression in E-cadherin observed in this cancer.
Applications
By understanding how Slug is induced and by blocking its expression, this study may represent a future strategy for therapeutic intervention in the treatment of patients with esophageal adenocarcinoma.
Terminology
Slug, Snail and Twist are all proteins involved in the process called epithelial mesenchymal transition. This is when epithelial cells lose cell-cell adhesion and behave more like fibroblasts. Such a mechanism is thought to be crucial in invasion and metastasis of cancer. Non-surprisingly, the cell adhesion molecule E-cadherin is repressed during this process.
Peer review
The authors examined the expression of E-cadherin and its repressors; Snail, Slug and Twist in squamous esophagus, Barrett’s metaplasia and esophageal adenocarcinoma. It revealed that Slug was increased in adenocarcinoma and in OE33 cells, the increase of Slug expression was inversely correlated to E-cadherin expression (mRNA and promoter activity) and induced EMT. The results are interesting and may represent a molecular mechanism of esophageal carcinogenesis.
Supported by City Hospital Trust Fund and the University of Birmingham Scientific Project Grant
Peer reviewer: Satoshi Osawa, MD, First Department of Medicine, Hamamatsu University School of Medicine, 1-20-1 Handayama, 431-3192, Japan
S- Editor Li DL L- Editor Ma JY E- Editor Wang HF
References
- 1.Lagergren J. Adenocarcinoma of oesophagus: what exactly is the size of the problem and who is at risk? Gut. 2005;54 Suppl 1:i1–i5. doi: 10.1136/gut.2004.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett's metaplasia. Lancet. 2000;356:2079–2085. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 3.Koppert LB, Wijnhoven BP, van Dekken H, Tilanus HW, Dinjens WN. The molecular biology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:169–190. doi: 10.1002/jso.20359. [DOI] [PubMed] [Google Scholar]
- 4.Bongiorno PF, al-Kasspooles M, Lee SW, Rachwal WJ, Moore JH, Whyte RI, Orringer MB, Beer DG. E-cadherin expression in primary and metastatic thoracic neoplasms and in Barrett's oesophagus. Br J Cancer. 1995;71:166–172. doi: 10.1038/bjc.1995.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swami S, Kumble S, Triadafilopoulos G. E-cadherin expression in gastroesophageal reflux disease, Barrett's esophagus, and esophageal adenocarcinoma: an immunohistochemical and immunoblot study. Am J Gastroenterol. 1995;90:1808–1813. [PubMed] [Google Scholar]
- 6.Bailey T, Biddlestone L, Shepherd N, Barr H, Warner P, Jankowski J. Altered cadherin and catenin complexes in the Barrett's esophagus-dysplasia-adenocarcinoma sequence: correlation with disease progression and dedifferentiation. Am J Pathol. 1998;152:135–144. [PMC free article] [PubMed] [Google Scholar]
- 7.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 8.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198:11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 10.Behrens J. Cadherins and catenins: role in signal transduction and tumor progression. Cancer Metastasis Rev. 1999;18:15–30. doi: 10.1023/a:1006200102166. [DOI] [PubMed] [Google Scholar]
- 11.Wijnhoven BP, de Both NJ, van Dekken H, Tilanus HW, Dinjens WN. E-cadherin gene mutations are rare in adenocarcinomas of the oesophagus. Br J Cancer. 1999;80:1652–1657. doi: 10.1038/sj.bjc.6690577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 13.De Craene B, van Roy F, Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signal. 2005;17:535–547. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 15.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 17.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A The transcription factor Slug represses E-cadherin expression and inducesepithelial to mesenchymal transitions: a comparison with Snail and E47repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 18.Castro Alves C, Rosivatz E, Schott C, Hollweck R, Becker I, Sarbia M, Carneiro F, Becker KF. Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J Pathol. 2007;211:507–515. doi: 10.1002/path.2138. [DOI] [PubMed] [Google Scholar]
- 19.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohuchida K, Mizumoto K, Ohhashi S, Yamaguchi H, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. Twist, a novel oncogene, is upregulated in pancreatic cancer: clinical implication of Twist expression in pancreatic juice. Int J Cancer. 2007;120:1634–1640. doi: 10.1002/ijc.22295. [DOI] [PubMed] [Google Scholar]
- 21.Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55:1449–1460. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockett JC, Larkin K, Darnton SJ, Morris AG, Matthews HR. Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization. Br J Cancer. 1997;75:258–263. doi: 10.1038/bjc.1997.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes SJ, Nambu Y, Soldes OS, Hamstra D, Rehemtulla A, Iannettoni MD, Orringer MB, Beer DG. Fas/APO-1 (CD95) is not translocated to the cell membrane in esophageal adenocarcinoma. Cancer Res. 1997;57:5571–5578. [PubMed] [Google Scholar]
- 24.Boonstra JJ, van der Velden AW, Beerens EC, van Marion R, Morita-Fujimura Y, Matsui Y, Nishihira T, Tselepis C, Hainaut P, Lowe AW, et al. Mistaken identity of widely used esophageal adenocarcinoma cell line TE-7. Cancer Res. 2007;67:7996–8001. doi: 10.1158/0008-5472.CAN-07-2064. [DOI] [PubMed] [Google Scholar]
- 25.Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, Hughes S, Tselepis C, Hotchin NA. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- 26.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, Aikou T. Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1174–1180. [PubMed] [Google Scholar]
- 28.Yuen HF, Chan YP, Wong ML, Kwok WK, Chan KK, Lee PY, Srivastava G, Law SY, Wong YC, Wang X, et al. Upregulation of Twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60:510–514. doi: 10.1136/jcp.2006.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosivatz E, Becker KF, Kremmer E, Schott C, Blechschmidt K, Hofler H, Sarbia M. Expression and nuclear localization of Snail, an E-cadherin repressor, in adenocarcinomas of the upper gastrointestinal tract. Virchows Arch. 2006;448:277–287. doi: 10.1007/s00428-005-0118-9. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 31.Parent AE, Choi C, Caudy K, Gridley T, Kusewitt DF. The developmental transcription factor slug is widely expressed in tissues of adult mice. J Histochem Cytochem. 2004;52:959–965. doi: 10.1369/jhc.4A6277.2004. [DOI] [PubMed] [Google Scholar]
- 32.Bermejo-Rodriguez C, Perez-Caro M, Perez-Mancera PA, Sanchez-Beato M, Piris MA, Sanchez-Garcia I. Mouse cDNA microarray analysis uncovers Slug targets in mouse embryonic fibroblasts. Genomics. 2006;87:113–118. doi: 10.1016/j.ygeno.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 35.Bienz M. beta-Catenin: a pivot between cell adhesion and Wnt signalling. Curr Biol. 2005;15:R64–R67. doi: 10.1016/j.cub.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 36.Soslow RA, Petersen CG, Remotti H, Altorki N. Acidic fibroblast growth factor is expressed sequentially in the progression from Barrett's esophagus to esophageal adenocarcinoma. Dis Esophagus. 2001;14:23–27. doi: 10.1111/j.1442-2050.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- 37.von Rahden BH, Stein HJ, Feith M, Puhringer F, Theisen J, Siewert JR, Sarbia M. Overexpression of TGF-beta1 in esophageal (Barrett’s) adenocarcinoma is associated with advanced stage of disease and poor prognosis. Mol Carcinog. 2006;45:786–794. doi: 10.1002/mc.20259. [DOI] [PubMed] [Google Scholar]
- 38.Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett’s esophagus. Oncogene. 2006;25:3084–3092. doi: 10.1038/sj.onc.1209338. [DOI] [PubMed] [Google Scholar]
- 39.Di Martino E, Wild CP, Rotimi O, Darnton JS, Olliver RJ, Hardie LJ. IGFBP-3 and IGFBP-10 (CYR61) up-regulation during the development of Barrett's oesophagus and associated oesophageal adenocarcinoma: potential biomarkers of disease risk. Biomarkers. 2006;11:547–561. doi: 10.1080/13547500600896791. [DOI] [PubMed] [Google Scholar]