Abstract
The human Y chromosome shows frequent structural variants, some of which are selectively neutral, while others cause impaired fertility due to the loss of spermatogenic genes. The large-scale use of multiple Y-chromosomal microsatellites in forensic and population genetic studies can reveal such variants, through the absence or duplication of specific markers in haplotypes. We describe Y chromosomes in apparently normal males carrying null and duplicated alleles at the microsatellite DYS448, which lies in the proximal part of the azoospermia factor c (AZFc) region, important in spermatogenesis, and made up of “ampliconic” repeats that act as substrates for nonallelic homologous recombination (NAHR). Physical mapping in 26 DYS448 deletion chromosomes reveals that only three cases belong to a previously described class, representing independent occurrences of an~1.5-Mb deletion mediated by recombination between the b1 and b3 repeat units. The remainder belong to five novel classes; none appears to be mediated through homologous recombination, and all remove some genes, but are likely to be compatible with normal fertility. A combination of deletion analysis with binary-marker and microsatellite haplotyping shows that the 26 deletions represent nine independent events. Nine DYS448 duplication chromosomes can be explained by four independent events. Some lineages have risen to high frequency in particular populations, in particular a deletion within haplogroup (hg) C*(xC3a,C3c) found in 18 Asian males. The nonrandom phylogenetic distribution of duplication and deletion events suggests possible structural predisposition to such mutations in hgs C and G. Hum Mutat 29(10), 1171–1180, 2008.
Keywords: Y chromosome, AZFc, microsatellite, deletion, duplication
INTRODUCTION
The human Y chromosome is remarkable for its high level of structural variability. Cytogenetic and molecular studies have shown that many structural variants exist within human populations, including deletions [Jobling et al., 1996, 2007; Repping et al., 2003, 2006], duplications [Bosch and Jobling, 2003; Jobling et al., 1996; Repping et al., 2006], and inversions [Affara et al., 1986; Bernstein et al., 1986; Page, 1986; Repping et al., 2006; Verma et al., 1982]. While many variants appear to be selectively neutral, some are certainly pathogenic, causing failure of spermatogenesis [Vogt et al., 1996], through loss of azoospermia factor (AZF) loci. Others may have subtle effects on fertility through variation in the copy number of spermatogenic genes, though this is difficult to demonstrate conclusively, and remains controversial [McElreavey et al., 2006]. Underlying the structural variability of the Y chromosome is a high rate of mutation through nonallelic homologous recombination (NAHR) between highly similar paralogous sequences, of which the Y bears a particularly large proportion [Skaletsky et al., 2003]. These paralogs also act as substrates for frequent gene conversion events [Bosch et al., 2004; Rozen et al., 2003].
As well as its importance in male infertility, the Y chromosome is a powerful marker for studying issues in human evolutionary genetics. The strength of this haplotypic system [Jobling and Tyler-Smith, 2003] comes from combining a robust and well-resolved phylogeny based on slow-mutating binary markers such as single-nucleotide polymorphisms (SNPs) [Underhill et al., 2000; Y Chromosome Consortium, 2002], with fine-scale haplo-typing based on rapidly mutating multiallelic markers, principally multiple microsatellites. The total number of informative microsatellites available is over 200 [Kayser et al., 2004], and some population studies analyze more than 50 of these [King et al., 2007; Xue et al., 2005].
When multiple microsatellites distributed throughout the chromosome are used in population studies, in effect they also act as a surveillance system for Y-chromosomal structural variants. When a particular microsatellite lies in a deleted region, its absence from a haplotype clearly signals the deletion (a “null” allele). The case of a duplication is less straightforward, and this could lead to underascertainment: if both copies of the duplicated microsatellite have the same number of repeat units, they are indistinguishable by length, though quantitative analysis could demonstrate that two copies are present. If the two copies have different repeat numbers due to microsatellite mutation, then the “duplicated” allele can clearly be seen as two peaks in an electropherogram, though it needs to be distinguished from possible mosaicism following somatic mutation [Clayton et al., 2004].
Microsatellite haplotype anomalies have already been used to identify a number of different Y-chromosomal rearrangements. Previously, we have used duplicated [Bosch and Jobling, 2003] and null alleles [King et al., 2005] of several clustered microsatellites to diagnose duplications and deletions of the AZFa region on Yq; the counting of multiple alleles and semiquantitative analysis of the microsatellite DYS464 [Butler and Schoske, 2005] reveals copy number variation of parts of the AZFc region, and the absence of all copies of the same marker indicates pathogenic AZFc deletions [King et al., 2005]; finally, absence of the microsatellite DYS458 signals recurrent and apparently nonpathogenic deletions on Yp that encompass the Amelogenin Y gene [Jobling et al., 2007].
Here, we describe Y chromosomes in apparently normal males carrying null and duplicated alleles at the hexanucleotide-repeat microsatellite DYS448. In the Y-chromosomal reference sequence [Skaletsky et al., 2003] this microsatellite lies within “u2” (Fig. 1a), a~170-kb segment in the proximal part of the 4.5-Mb region on Yq that includes AZFc [Kuroda-Kawaguchi et al., 2001]. This region is mostly composed of large “ampliconic” repeat units [Skaletsky et al., 2003], many arranged as palindromic sequences, which may act as substrates for illegitimate recombination events that cause chromosomal rearrangements.
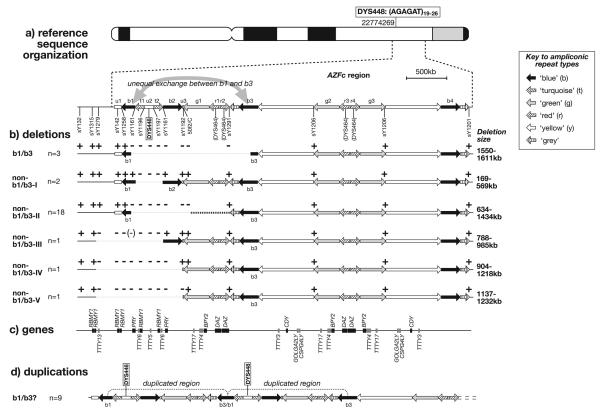
FIGURE 1.
Deletions and duplications of DYS448 caused by rearrangements within the proximal AZFc region. a: Reference sequence organization around DYS448, showing position of the microsatellite on an idiogram of the Y chromosome (with genome position of start of the marker given according to build 36.1 of the reference assembly), and below, a schematic view of the region around DYS448, including large ampliconic repeat units shown as arrows (nomenclature in key after Kuroda-Kawaguchi et al. [2001]). Paralogous repeat copies are indicated by shading or hatching, as indicated in the key to the right. STSs used in mapping are shown below, and the gray curved arrow indicates putative sites of unequal exchange between the b1 and b3 repeat units. The positions of the multi-locus microsatellite DYS464 are also shown. b: Mapping of presence (+) or absence (−) of STSs in DYS448 deletion chromosomes, and proposed extents of deletions in the six different classes. The duplicated nature of sY1161 means that this marker is uninformative in the non-b1/b3 class III deletion (indicated as“−” in parentheses). c: Genes in the region, based on the reference sequence [Skaletsky et al., 2003]. Names above the line are protein-coding genes, and names below are nontranslated genes (in gray). d: Putative b1/b3-mediated arrangement of region in DYS448 duplication chromosomes.
We use deletion mapping, as well as haplotyping with binary markers and multiple microsatellites, to explore the molecular basis for the underlying rearrangements, which reveal novel deletions and duplications affecting gene copy number and illustrate the complexity and variability of the organization of this important and dynamic region of the Y chromosome. Some rearrangements have come to high frequency in particular populations, and the phylogenetic distributions of independent duplication and deletion events suggest that some branches of the Y phylogeny might bear structures that predispose to, or protect against, rearrangement mutations.
MATERIALS AND METHODS
DNA Samples
Most DNA samples were from collections of the authors, and were obtained with appropriate informed consent. Some samples form part of sets described previously [Jobling et al., 1996; Parkin et al., 2006, 2007; Roewer et al., 2007]. The 684 male samples from the Centre d'Etude du Polymorphisme Humain–Human Genome Diversity Project (HGDP-CEPH) panel [Cann et al., 2002] were also included. Some samples were subjected to whole genome amplification [Dean et al., 2002] using the Genomiphi kit (GE Healthcare, Amersham, United Kingdom) before analysis. Two of the deletion chromosomes (448del1 [m38] and 448del6 [m252]) were previously described in a set carrying deletions of the marker 50f2/C [Jobling et al., 1996].
Identification of DYS448 Deletions and Duplications
Deletions and duplications were ascertained using a published multiplex incorporating DYS448 [Butler et al., 2002] or the commercial forensic kit Y-filer (Applied Biosystems, Warrington, United Kingdom), and were confirmed by repeated typing. DYS448 was considered to be duplicated when its two peaks in an electropherogram were of approximately equal height and area.
Deletion Mapping
Y-specific sequence-tagged sites (STSs) around DYS448, with primer sequences available from the literature [Jobling et al., 1998; Repping et al., 2003; Skaletsky et al., 2003; Tilford et al., 2001; Vollrath et al., 1992], were amplified by PCR and analyzed by agarose gel electrophoresis. An STS was considered to be deleted when reproducibly absent in the presence of a larger independent Y-specific control amplicon coamplified in the same PCR reaction [Jobling et al., 2007]. The PCR system in most cases was as described [Jobling et al., 2007], and cycling conditions were as follows: 33 cycles of (94°C for 30 s, 60°C for 30 s, and 70°C for 30 s). The marker 50f2/C (DYS7C) was typed using a previously described assay which generates a small (196-bp) test amplicon and a larger control amplicon from Yp (minisatellite MSY1) using a single primer pair.
Y Chromosome Haplotyping
Binary markers were typed in a hierarchical fashion, using either the SNaPshot minisequencing protocol (Applied Biosystems) on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems), or primer extension on the Sequenom mass spectrometry system (Sequenom, San Diego, CA). Amplification and extension primers were based on ones published previously [Bosch et al., 2006; Hurles et al., 2005; Paracchini et al., 2002], with additional primers based on published sequences [Y Chromosome Consortium, 2002]. The binary markers define haplogroups that are represented in a maximum parsimony tree [Jobling and Tyler-Smith, 2003; Y Chromosome Consortium, 2002].
A total of 24 Y-specific microsatellites (DYS19, DYS385a/b, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS426, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, YCAIIa/b, and Y-GATA-H4.1) were typed in two multiplexes [Butler et al., 2002; Parkin et al., 2006]. PCR products were resolved on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems), and analyzed using GeneMapper software (Applied Biosystems). Allele nomenclature was as described [Parkin et al., 2006], and compatible with International Society for Forensic Genetics (ISFG) recommendations [Gusmão et al., 2006].
Estimation of DYS464 Copy Number
DYS464 was amplified using published primers [Redd et al., 2002], the forward primer being 5′-labeled with the dye 6-FAM. PCR reactions were set up as described [Butler et al., 2002], with an annealing temperature of 60°C, and analyzed by capillary electrophoresis. A total of five DNA samples from the YCC collection [Y Chromosome Consortium, 2002] for which published electropherograms were available [Redd et al., 2002] were used as controls.
Microsatellite Network Construction
A weighted median-joining network [Bandelt et al., 1999] was constructed from microsatellite haplotypes using Network 4.0 (www.fluxus-engineering.com/sharenet.htm). The network assumes a single-step microsatellite mutational model, allows nodes (median vectors) to be unoccupied by sampled haplotypes, and represents the union of the most parsimonious trees. Of the total 24 microsatellites analyzed we omitted DYS448 itself, and also YCAIIa, YCAIIb, and DYS385a, DYS385b because of allele assignment problems, leaving 19 microsatellites. Two chromosomes (448del2 and 448del3) carried duplications at DYS19 and DYS461, respectively (Supplementary Table S1; available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat); to allow these loci to be included in the network, we considered only the smaller allele in each case. Weighting [Qamar et al., 2002] was used to remove some reticulations (closed structures) within networks by taking into account the range of different mutation rates of the markers, reflected indirectly by their allele length variances among all chromosomes included in the network.
TMRCA Estimation
Time-to-most-recent-common-ancestor (TMRCA) of clusters within networks, together with standard deviation, was estimated using the rho statistic within the Network program. This measure represents the mean number of mutations between a root haplotype and every other haplotype in the cluster, counted from the network itself. Root haplotypes were chosen to be as close as possible to the center of a cluster, and to be comprised of more than one haplotype. In the case of haplogroup (hg) C3*(xC3a,C3c), choice of alternative roots made little difference to the TMRCA. Rho is related to time in generations (t) by ρ=μt, where μ is the per-haplotype per-generation mutation rate, calculated from the per-microsatellite per-generation mutation rate of 2.0 × 10−3 [Gusmão et al., 2005]. Generation time was taken to be 31 years [Fenner, 2005].
Relative Frequency of Deletions and Duplications in Different Haplogroups
To test whether rearrangements involving DYS448 were randomly distributed across the Y phylogeny, we combined all of our data (on Central Asian, Bhutanese, Nepalese, French, English, and CEPH-HGDP samples; see Supplementary Table S2) into haplogroup designations at the “letter” clade level (A–R) for compatibility. We then carried out Fisher's exact tests to ask whether rearrangements were over- or underrepresented in particular haplogroups given their relative frequencies, using a false discovery rate correction for multiple testing [Benjamini and Hochberg, 1995].
RESULTS
Physical Characterization of DYS448 Deletions and Duplications
While the majority of Y chromosomes carry a single DYS448 allele, we observed that some carry null alleles; in principle, this can result from small-scale primer-site mutations, but the analysis described below confirms locus deletion. We also found that some chromosomes carry two alleles of different lengths, but of equal peak heights in electropherograms produced by capillary electrophoresis, suggesting a genuine allele duplication. From our population studies (a total of 3,303 chromosomes; our unpublished data) [Parkin et al., 2006, 2007; Roewer et al., 2007] we assembled a collection of 26 deletions and nine duplications for further molecular analysis (Table 1).
TABLE 1.
DYS448 Deletion and Duplication Chromosomes†
| Name | Population | Class | Offspring? | Haplogroup | DYS448 | DYS464 alleles |
|---|---|---|---|---|---|---|
| 448del1 (m38) | China (Chengdu) | b1/b3 | Y | O3e | Null | 13–16 |
| 448del2 | Russia (Kalmyk) | b1/b3 | n.k. | C3c | Null | 13 |
| 448del3 | Nepal (Tamang) | b1/b3 | n.k. | C* | Null | 15–18 |
| 448del4 | Tajikistan (Urgut) | I | Y | E3b | Null | 17–18 |
| 448del5 | Nepal (Tharu) | I | n.k. | O2 | Null | 14–15–17 |
| 448del6 (m252) | Mongolia (Khalkh) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–18 |
| 448del7 | Kyrgyzstan (Andijan) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del8 | Kyrgyzstan (Andijan) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del9 | Kyrgyzstan (Andijan) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del10 | Kyrgyzstan (Andijan) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del11 | Kyrgyzstan (Jankatalab) | II | Y | C3*(xC3a,C3c) | Null | 12–13–19 |
| 448del12 | Kyrgyzstan (Doboloo) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del13 | Kyrgyzstan (Doboloo) | II | Y | C3*(xC3a,C3c) | Null | 13–18 |
| 448del14 | Kyrgyzstan (Doboloo) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del15 | Kyrgyzstan (Doboloo) | II | Y | C3*(xC3a,C3c) | Null | 13–18 |
| 448del16 | Tajikistan (Kamangaron) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del17 | Tajikistan (Agalic) | II | Y | C3*(xC3a,C3c) | Null | 12–13–17 |
| 448del18 | Russia (Kalmyk) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–14–18 |
| 448del19 | Russia (Kalmyk) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–14–18 |
| 448del20 | Russia (Kalmyk) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–14–18 |
| 448del21 | Russia (Kalmyk) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–14–18 |
| 448del22 | Russia (Kalmyk) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–18 |
| 448del23 (HGDP1296) | China (Han) | II | n.k. | C3*(xC3a,C3c) | Null | 12–13–19 |
| 448del24 | Egypt (Qena) | III | n.k. | G | Null | 12–13–14 |
| 448del25 (HGDP1179) | China (Yizu) | IV | n.k. | O2 | Null | 14–15–16–18 |
| 448del26 | Nepal (Magar) | V | n.k. | D* | Null | 15–17 |
| 448dup1 | Tajikistan (Richtan) | – | Y | G | 20–22 | 13–14–15 |
| 448dup2 | Tajikistan (Richtan) | – | Y | G | 20–22 | 13–14–16 |
| 448dup3 | Tajikistan (Richtan) | – | Y | G | 20–21 | 13–14–17 |
| 448dup4 | Tajikistan (Richtan) | – | Y | G | 20–22 | 13–14–16 |
| 448dup5 | Nepal (Magar) | – | n.k. | O3e | 20–21 | 13–15–16 |
| 448dup6 (HGDP997) | Brazil (Karitiana) | – | n.k. | Q(x Q3a) | 19–20 | 14–16–19 |
| 448dup7 (HGDP617) | Israel (Bedouin) | – | n.k. | E1 | 19–20 | 15–17 |
| 448dup8 (HGDP1200) | Senegal (Mandenka) | – | n.k. | E1 | 19–20 | 15–17–18 |
| 448dup9 (HGDP1284) | Senegal (Mandenka) | – | n.k. | E1 | 19–20 | 16–18 |
Designations m 38 and m 252 are from Jobling et al. [1996]. HGDP numbers are those given by HGDP-CEPH (www.cephb.fr/HGDP-CEPH-Panel). Information on whether or not individuals have fathered children is indicated under “Offspring?”
Y, yes; n.k., not known.
The location of DYS448 within an ampliconic repeat region immediately suggests a candidate mechanism for deletions and duplications (assuming that the starting structure has the same organization as the reference sequence): NAHR causing unequal exchange between the repeat units b1 and b3 (Fig. 1a). Three b1/b3-mediated deletions have been reported before in one fertile and two infertile men [Hucklenbroich et al., 2005; Repping et al., 2003].
To investigate this possibility, we undertook deletion mapping in chromosomes carrying null alleles, using STSs around DYS448 following an established approach [Repping et al., 2003]. This mapping reveals six physically distinct deletion classes, and, surprisingly, supports the b1/b3 NAHR mechanism in only 3 out of 26 cases: in these chromosomes the adjacent set of six STSs, sY1161, sY1196, sY1197, sY1192, 50f2/C, and sY1291, are absent, while flanking STSs sY1258 (proximal) and sY1206 (distal) are present (Fig. 1b).
Among the remaining 23 deletion chromosomes, STS deletion mapping reveals five physically distinct non-b1/b3 deletion classes (Fig. 1b; Table 1). The smallest deletions (non-b1/b3-class I) are carried by two chromosomes, and lack only the two STSs flanking DYS448. A total of 18 chromosomes (non-b1/b3-class II), like the b1/b3 deletions, have a proximal breakpoint within the b1 repeat, but a distal breakpoint proximal to b3, between STSs 50f2/C and sY1291. The remaining three classes of non-b1/b3-mediated deletions are all found as singletons, and all share a proximal breakpoint outside the AZFc region (between sY1315 and sY1279), but have three different distal breakpoints. Sparse and uneven spacing of informative STSs means that the physical size of the deletions is uncertain (Fig. 1b), but even given this uncertainty all of the non-b1/b3 deletion classes must have smaller deletions than the b1/b3 class (1,550–1,611 kb). Importantly, all of the five non-b1/b3 deletion classes cannot be mediated by NAHR between ampliconic repeats, at least on the basis of a starting structure resembling the reference sequence organization. They may result from nonhomologous processes, though verification of this would require the sequencing of breakpoints.
In the case of the DYS448 duplication chromosomes, breakpoint positions cannot be determined without the use of techniques such as quantitative PCR or fluorescence in situ hybridization (FISH) analysis, and unfortunately the duplication DNA samples are unsuitable for these methods. While b1/b3-mediated duplication may be the most parsimonious mechanism to explain DYS448 duplications (Fig. 1d), it is abundantly clear from the complexity of the different deletion classes we have observed that other duplication mechanisms are also possible.
Haplotype Analysis
Haplotype analysis provides a means to ask whether rearrangements that are indistinguishable by physical mapping are likely to be identical by descent, or identical by state; i.e., recurrent. If a rearrangement is mediated by NAHR between substantial repeat sequences, then we expect it to recur; in contrast, if a rearrangement is not NAHR-mediated it is more likely to be due to a sporadic event, in which case a set of chromosomes that shares it will probably have common ancestry.
We first used binary polymorphisms to determine the haplogroups of deleted and duplicated chromosomes (Table 1; Fig. 2a), reasoning that if two deletion or duplication chromosomes belong to different haplogroups in the Y-chromosomal binary-marker phylogeny, then they must have occurred independently. We also determined 23-locus microsatellite haplotypes (Supplementary Table S1), to provide an additional estimator of the relatedness between the chromosomes carrying rearrangements. Membership of the same haplogroup, and possession of similar microsatellite haplotypes, can indicate that a rearrangement carried by a set of chromosomes is identical by descent.
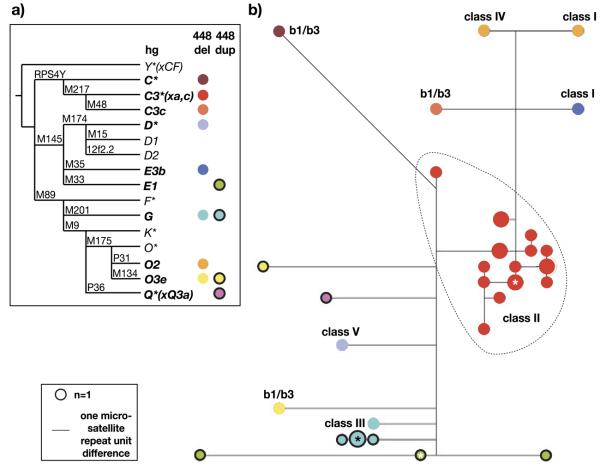
FIGURE 2.
Haplogroups andY-STR haplotypes of DYS448 deletion and duplication chromosomes. a: Binary marker phylogeny of the Y chromosome, with haplogroups (hg) [Y Chromosome Consortium, 2002] containing DYS448 deletion (“448del”) and duplication (“448dup”) chromosomes indicated by colored circles. Binary markers (e.g., M175) used to define haplogroups are given above each branch. C3*(xC3a,C3c) is abbreviated as C3*(xa,c). b: Weighted median joining network [Bandelt et al., 1999] representing the molecular relationships between the 19-locusY-STR haplotypes of 27 deletion and nine duplication chromosomes. Circles represent haplotypes, with area proportional to frequency and colored according to haplogroup as in (a). Lines between circles represent microsatellite mutational steps, and asterisks indicate root nodes used for dating purposes.
The three b1/b3 deletion chromosomes belong to three different haplogroups (O3e, C*, C3c; Fig. 2a), thus providing clear evidence of independent recurrent mutation, and supporting NAHR as the mutational mechanism. In contrast, the 18 non-b1/b3-class II chromosomes all belong to hg C3*(xC3a,C3c), with all but one of their microsatellite haplotypes forming a tight cluster in a median joining network (Fig. 2b), strongly suggesting a common deletion origin for this group. Use of the rho statistic within Network yields a TMRCA for this group of 2,900±766 years. Of the remaining four classes of deletion chromosomes, three (classes III, IV, and V) are represented by singletons, in haplogroups (hgs) G, O2, and D*; the pair of chromosomes in class I belong to two different haplogroups, O2 and E3b, indicating that they have independent origins.
The nine duplication chromosomes belong to four haplogroups (Fig. 2a), again indicating recurrence of DYS448 duplication. A total of four lie in hg G, and form a tight cluster in the network (Fig. 2b) clearly suggesting identity-by-descent of the duplication; they have a TMRCA of 408±289 years. A total of three lie in hg E1, suggesting a common origin, although their diverged microsatellite haplotypes indicate ancient divergence (TMRCA of 5,984±1,246 years); and the remaining two independent cases belong to haplogroups O3e1* and Q*(xQ3a).
In summary, the 26 deletion chromosomes can be explained by nine independent deletion events, of which three are probably mediated by b1/b3 recombination; the nine duplication chromosomes can be explained by four independent events.
Indirect Estimation of DAZ Gene Copy Number Using a Multilocus Microsatellite, DYS464
The testis-specific deleted in azoospermia (DAZ) genes [Reijo et al., 1995] play an essential role in both primordial germ-cell development and in the differentiation and maturation of sperm [Yen, 2004]. In one of the non-b1/b3 deletion classes (class II), the distal breakpoints lie between the STSs 50f2/C and sY1291. This interval contains the so-called “red” repeats r1 and r2 (Fig. 1a), including two copies of DAZ (Fig. 1c)—present in four copies within the reference sequence. To address the issue of DAZ copy number variation in these 18 class II DYS448 deletion chromosomes, we exploited the presence of another microsatellite, DYS464, that also lies within the “red” repeats (Fig. 1a). Previous work [Berger et al., 2003; Butler and Schoske, 2003, 2005; Redd et al., 2002] has shown that most Y chromosomes give DYS464 allelic patterns compatible with the presence of four copies. In electropherograms, this can present as four peaks if all four copies of the microsatellite carry different repeat numbers, or fewer peaks if some copies carry the same repeat number. In some studies [Butler and Schoske, 2003, 2005] the ratio of peak heights or areas has been used to infer copy number; for example, apparent ratios of 2:1:1, or 1:3 being interpreted as four copies. Our experiments suggest that, in the absence of control samples in which copy number is known for certain, such semiquantitative inferences are risky. Here, we therefore rely on the less ambiguous evidence of the number of alleles distinguishable by length, thereby determining the minimum number of DYS464 copies.
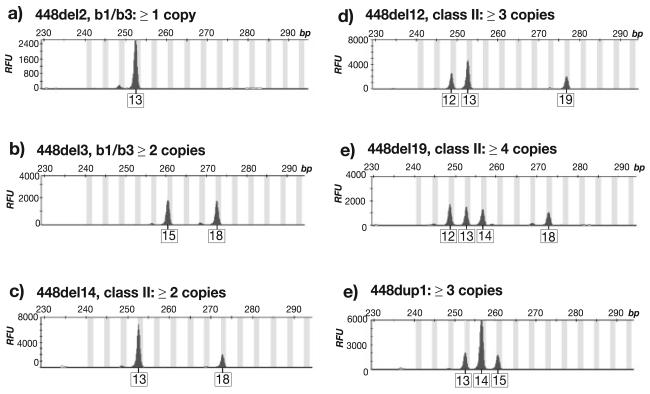
We analyzed the allelic patterns for DYS464 for the deletion chromosomes; examples are shown in Fig. 3. In non-b1/b3 class I, III, IV, and V deletion chromosomes we do not expect the deletions to have affected “red” repeat copy number, so they should each carry four copies. In agreement with this expectation, chromosomes in classes I, III, and V present two or three peaks, and the class IV chromosome presents four. In b1/b3 deletion chromosomes we expect two copies: the chromosomes in this class present only one or two DYS464 peaks (Fig. 3a and b), which is also consistent with our expectations. We now turn to the chromosomes in which copy number is in question. The 18 non-b1/b3 class II chromosomes all present either two, three or four peaks (Fig. 3c, d and e), compatible with their carrying four copies. This strongly suggests that the deletion that all these chromosomes share by descent does not encompass the r1 and r2 repeats, and that DAZ gene copy number is “normal”, at a probable four copies.
FIGURE 3.
Examples of electropherograms showing variation in the number of alleles of microsatellite DYS464. a–f: different numbers of DYS464 alleles seen in deletion and duplication chromosomes. Note the differences in peak heights, e.g., (c), which could be regarded as a 3:1 ratio. Ratios are not reliable enough to predict relative copy number (see text). Vertical gray bars indicate allele “bins”, defined in GeneMapper (Applied Biosystems). Allele designations, boxed below each peak, are in repeat number. RFU, relative fluorescent units.
We can also address the issue of whether or not the duplications encompassing DYS448 include the r1 and r2 repeats, which would lead to a DAZ gene copy number of six—this would be the case if the duplications were b1/b3-mediated. We analyzed allelic patterns for DYS464 for the nine duplication chromosomes. All show only two or three peaks (e.g., Fig. 3f), so this provides no evidence to support a “red” repeat copy number of greater than four. However, it is important to note that the greater the number of copies of DYS464, the more likely it is that some copies will be identical in allele length, and therefore indistinguishable in our analysis. Power to diagnose duplications involving the “red” repeats is therefore limited.
Effects of Deletions on Gene Content
The proximal AZFc region contains many other genes in addition to DAZ, and the different deletion or duplication classes are expected to affect the copy numbers of different subsets of these. Here we consider only the deletions, because their breakpoints are better characterized. All five of the novel deletion classes are smaller in extent that the b1/b3 class. Based on the reference sequence, and depending on the breakpoint positions within the b1 and b3 repeat units, b1/b3 deletions remove six or seven of the many testis-specific protein-coding genes and five of the nontranslated transcription units in the region (Fig. 1c). Uncertainty regarding the precise positions of the breakpoints in the other deletion classes leads to corresponding uncertainty in the number of deleted genes, and these are summarized in Table 2.
TABLE 2.
Protein-Coding and Nontranslated Genes Removed by the Smallest and Largest Extents of the Six Physical Deletion Classes
| Smallest extent |
Largest extent |
|||
|---|---|---|---|---|
| Deletion class |
Protein-coding genes deleted | Nontranslated genes deleted |
Protein-coding genes deleted | Untranslated genes deleted |
| b1/b3 |
DAZ × 2; RBMY1× 2; PRY; BPY2; |
TTTY4;TTTY5;TTTY6 × 2 TTTY17; |
DAZ × 2; RBMY1× 2; PRY × 2; BPY2 |
TTTY4;TTTY5;TTTY6 × 2; TTTY17 |
| Class I | – | TTTY5 | RBMY1 × 2 | TTTY5;TTTY6 × 2 |
| Class II | RBMY1 × 2; PRY | TTTY5;TTTY6 × 2 |
DAZ × 2; RBMY1 × 2; PRY × 2; BPY2 |
TTTY4;TTTY5;TTTY6 × 2; TTTY17 |
| Class III | RBMY1 × 3; PRY | TTTY5; TTTY6 | RBMY1 × 6; PRY | TTTY5; TTTY6 × 2; TTTY13 |
| Class IV | RBMY1 × 4; PRY | TTTY5;TTTY6 × 2 | RBMY1 × 6; PRY × 2 | TTTY5; TTTY6 × 2; TTTY13 |
| Class V | RBMY1 × 4; PRY × 2 | TTTY5;TTTY6 × 2 | RBMY1 × 6; PRY × 2 | TTTY5; TTTY6 x 2; TTTY13 |
Mutation Rate of b1/b3 Deletions
Three of the deletion events correspond to independent occurrences of the~1.5-Mb b1/b3 deletion originally described [Repping et al., 2003]. In principle, the mutation rate of b1/b3-mediated deletion can be calculated given an estimate of the number of generations encompassed within the phylogeny relating all sampled chromosomes [Bosch et al., 2004; Repping et al., 2006]. Such an estimate, 52,000 generations, has been calculated for a set of 47 chromosomes representing the major clades of the Y chromosome tree [Repping et al., 2006]. We analyzed 3,303 chromosomes among which all the clades were also represented, and found three independent b1/b3 deletion events. Time elapsed within the phylogeny relating the surveyed chromosomes is difficult to estimate; although our sample size is large, many chromosomes are closely related to each other. A minimum bound must be 52,000 generations, and a maximum bound of 10 times higher seems reasonable, which yields an approximate range of mutation rates of ~5 × 10−5 to ~5 × 10−6 per generation. This rate is similar to that of another NAHR-mediated Y-chromosomal deletion, encompassing AZFa, which has been determined in sperm DNA as 2.16 × 10−5 [Turner et al., 2007].
Population and Haplogroup Distribution of Rearrangements
Most of the rearrangement chromosomes we have identified derive from Asian populations. To ask is this reflected a true over-representation, or sampling bias, we expanded our sample size by including other published data. The inclusion of DYS448 in a commercial forensic multiplex (Y-filer; Applied Biosystems) means that population data are accumulating in the forensic literature, and reporting of “null” and duplicated alleles appears to be thorough (though haplogroup data are not available). We extracted population data from 25 such studies, comprising 6,614 chromosomes, and containing 15 DYS448 deletions and five duplications (Supplementary Table S2). This brings the total sample size to 9,916, containing 41 deletion chromosomes and 14 duplication chromosomes. Deletion chromosomes are strikingly over-represented in Asia, at 0.7% (n=5,776), compared to another large sample, Europe, at only 0.03% (n=2,650). For duplication chromosomes, we see 0.8% in Africa (n=898), compared to 0.1% or less elsewhere. This prevalence of duplications or deletions in certain regions could reflect the presence of founder lineages such as those represented by the duplications observed within haplogroups E1 in Africa and G in Asia, and the class II deletions within hg C3*(xC3a,C3c).
However, DYS448 rearrangements are not only over-represented in terms of the number of chromosomes, but also in the number of independent mutation events that underlie them. It is possible that some haplogroups carry a structural predisposition to, or protection against, rearrangements in the proximal AZFc region. To test this, we considered the population samples for which haplogroup frequency data were available (Supplementary Table S2; P.B., D.C.-S., E.J.P., unpublished observations), allowing comparisons of the frequencies of DYS448 deletion/duplication events in 2,995 chromosomes, belonging to 18 different haplogroups. A total of three haplogroups showed significant over- or underrepresentation of rearrangement events. We observe no DYS448 deletion/duplication events among 783 haplogroup R chromosomes, which is significantly fewer than expected by chance (Fisher's exact test: P=0.027), suggesting that this lineage could be structurally protected against them. The reference sequence, which is the only basis we currently have to propose putative mechanisms for deletions and duplications, belongs to this haplogroup—reinforcing the difficulty of interpreting structural variation on this most dynamic of human chromosomes. In contrast, 60 haplogroup G chromosomes include two rearrangement events (P=0.027) and 97 haplogroup C chromosomes include three events (P=0.007). These significant overrepresentations of DYS448 rearrangement events suggest that sequence structures in these lineages could predispose them to mutation. Associations to haplogroups remain significant after correction for multiple testing (n=3; threshold=0.0416) using the false discovery rate [Benjamini and Hochberg, 1995].
DISCUSSION
Using a combination of deletion mapping and haplotyping, we have ascertained and characterized nine independent deletion events that encompass the microsatellite DYS448, and extend into the proximal part of the AZFc region. The nine events correspond to six physically distinct deletion classes, attesting to the complexity of this region of the Y chromosome, and cautioning against simple interpretations of deletion data in terms of probable NAHR without careful mapping.
These rearrangements are by no means the only examples of polymorphic AZFc structures in apparently normal males. Previous work identified recurrent deletions and duplications involving the marker 50f2/C [Jobling et al., 1996], which lies within the u3 element, ~520 kb distal to DYS448 (Fig. 1a). Inclusion of 50f2/C in our deletion mapping allows clarification of the relationships between these sets of chromosomes. Some chromosomes lack both markers: the b1/b3 and non-b1/b3 class II DYS448 deletion chromosomes correspond to the previously defined 50f2/C deletion classes 2L (e.g., 448del1 [m38]) and 10L (448del6 [m252]). However, typing of DYS448 (data not shown) demonstrates that all other 50f2/C-deletion classes actually possess a DYS448 allele. In these cases loss of 50f2/C cannot be caused by b1/b3 deletion; most have been previously shown to carry different rearrangements, such as a g1/g3-mediated deletion following a b2/b3-mediated inversion [Fernandes et al., 2004].
Considering the smallest extents of all DYS448 deletion classes except class V, the repeated nature of the genes in the proximal AZFc region means that deletion results in a reduction in copy number, rather than a complete elimination, of any gene family. However, larger extents of three deletion classes, and any extent of the class V deletion, would remove both copies of the PRY gene. Reduction of gene copy number within this region, including the loss of two copies of DAZ, may predispose carriers to spermatogenic failure. Sperm count data are unavailable for the individuals we have studied, but for many cases we know whether or not they have fathered offspring (Table 1). This information shows that 448del1, who carries a b1/b3 deletion, and 448del4, who carries a class I deletion, are both fertile. The discovery of a set of chromosomes (class II) that share a deletion by descent and have a TMRCA of 2,900±766 years, shows that the deletion they carry is certainly compatible with male fertility, and this is supported by direct information demonstrating fertility in 12 out of 18 cases. Since this deletion class is relatively common among Central Asians, a directed study of any subtle effects on sperm count, and more detailed physical mapping, could be undertaken in a newly recruited cohort from this region. Class III, IV, and V deletions are present as singletons, and there is no information about fertility, so it remains possible that spermatogenesis is impaired in these cases. Nonetheless, there is no evidence from our study that deletions of parts of the proximal AZFc region are deleterious.
There is less reason to suppose that duplications might affect sperm count, unless gene dosage is critical, which seems improbable. Duplication chromosomes are underascertained, since their discovery relies on a length mutation having occurred at one copy of the duplicated microsatellite of interest; the probability of this depends on the mutation rate of the microsatellite, and the number of generations that the chromosome has persisted in the population [Bosch and Jobling, 2003]. Given the mutation rate of microsatellites (typically ~2×10−3 per locus per generation [Gusmão et al., 2005]), any duplication chromosome carrying two different-length alleles is likely to have been present in the population for a number of generations, and is therefore at least compatible with male fertility [Bosch and Jobling, 2003]. The four duplications within hg G, and the three within hg E1, are identical by descent, and provide further evidence that these rearrangements persist through the generations, and probably represent neutral copy-number variants. Direct information on fertility for the hg G duplication supports this.
Rearrangements such as those we describe here appear to owe their heterogeneous population frequencies to two major factors. First, having once occurred, a neutral rearrangement can be successfully propagated in a particular population due to its social organization. The overrepresentation of particular haplotype clusters in Asia is already well established: two distinct examples within hg C3—the “Manchu” [Xue et al., 2005] and “Khan” [Zerjal et al., 2003] haplotypes—have been ascribed to social selection in the history of dynastic lineages. Cultural transmission of fertility [Heyer et al., 2005] has ensured that these lineages persist at high frequencies today. Second, haplogroups in general are highly geographically differentiated, and some haplogroups appear to be either predisposed to, or protected against, rearrangements.
The advantage of using microsatellites to detect deletions and duplications is the very large number of chromosomes that can be surveyed, allowing the detection of rare rearrangements. As the number of markers [Kayser et al., 2004] and the size of databases [Willuweit et al., 2007] increases, it seems likely that most regions of the chromosome will effectively have been surveyed in most populations, allowing an unbiased assessment of structural polymorphism on this highly variable chromosome.
Supplementary Material
ACKNOWLEDGMENTS
We thank all DNA donors, Sjoerd Repping for information about the phenotype of the b1/b3 deletion male WHT3453, and Matt Hurles for primers. M.A.J. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (grant number 057559); P.B., G.R.B., and C.T.S. were supported by the Wellcome Trust. E.J.P. and D.R.C.S. were supported by the Arts and Humanities Research Council and the EC Sixth Framework Programme under Contract no. ERAS-CT-2003-980409, within the framework of the European Science Foundation EUROCORES programme “The Origin of Man, Language and Languages.” P.dK. was supported by the Netherlands Organization for Scientific Research (NWO) project 231-70-001 within the same EUROCORES programme. I.N. and M.S. were supported by the Max Planck Society.
Grant sponsor: Max Planck Society; Grant sponsor:WellcomeTrust; Grant number: 057559; Grant sponsor: Arts and Humanities Research Council and the EC Sixth Framework Programme; Grant number: ERAS-CT-2003-980409; Grant sponsor: Netherlands Organization for Scientific Research (NWO); Grant number: 231-70-001.
Footnotes
The Supplementary Material referred to in this article can be accessed at http://www.interscience.wiley.com/jpages/1059-7794/suppmat.
REFERENCES
- Affara NA, Ferguson-Smith MA, Tolmie J, Kwok K, Mitchell M, Jamieson D, Cooke A, Florentin L. Variable transfer of Y-specific sequences in XX males. Nucleic Acids Res. 1986;14:5375–5387. doi: 10.1093/nar/14.13.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Berger B, Niederstatter H, Brandstatter A, Parson W. Molecular characterization and Austrian Caucasian population data of the multi-copy Y-chromosomal STR DYS464. Forensic Sci Int. 2003;137:221–230. doi: 10.1016/j.forsciint.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Bernstein R, Wadee A, Rosendorff J, Wessels A, Jenkins T. Inverted Y chromosome polymorphism in the Gujerati Muslim Indian population of South Africa. Hum Genet. 1986;74:223–229. doi: 10.1007/BF00282538. [DOI] [PubMed] [Google Scholar]
- Bosch E, Jobling MA. Duplications of the AZFa region of the human Y chromosome are mediated by homologous recombination between HERVs and are compatible with male fertility. Hum Mol Genet. 2003;12:341–347. doi: 10.1093/hmg/ddg031. [DOI] [PubMed] [Google Scholar]
- Bosch E, Hurles ME, Navarro A, Jobling MA. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, González-Neira A, Flaiz C, Mateu E, Scheil H-G, Huckenbeck W, Efremovska L, Mikerezi I, Xirotiris N, Grasa C, Schmidt H, Comas D. Male and female lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Butler JM, Schoske R, Vallone PM, Kline MC, Redd AJ, Hammer MF. A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci Int. 2002;129:10–24. doi: 10.1016/s0379-0738(02)00195-0. [DOI] [PubMed] [Google Scholar]
- Butler JM, Schoske R. Forensic value of the multicopy Y-STR marker DYS464. In: Doutremepuich C, Morling N, editors. Progress in Forensic Genetics, vol 10. Proceedings of the 20th International International Society of Forensic Genetics (ISFG) Congress, Arcachon, France, 9–13 September 2003; International Congress Series (ICS) 1261; Amsterdam: Elsevier; 2003. pp. 278–280. [Google Scholar]
- Butler JM, Schoske R. U.S. population data for the multi-copy Y-STR locus DYS464. J Forensic Sci. 2005;50:975–977. [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu JY, Carcassi C, Contu L, Du RF, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang XY, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian YP, Shu QF, Xu JJ, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Clayton TM, Guest JL, Urquhart AJ, Gill PD. A genetic basis for anomalous band patterns encountered during DNA STR profiling. J Forensic Sci. 2004;49:1207–1214. [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang LH, Wu XH, Faruqi AF, Bray-Ward P, Sun ZY, Zong QL, Du YF, Du J, Driscoll M, Song WM, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am J Hum Genet. 2004;74:180–187. doi: 10.1086/381132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmão L, Sanchez-Diz P, Calafell F, Martin P, Alonso CA, Alvarez-Fernandez F, Alves C, Borjas-Fajardo L, Bozzo WR, Bravo ML, Builes JJ, Capilla J, Carvalho M, Castillo C, Catanesi CI, Corach D, Di Lonardo AM, Espinheira R, Fagundes de Carvalho E, Farfan MJ, Figueiredo HP, Gomes I, Lojo MM, Marino M, Pinheiro MF, Pontes ML, Prieto V, Ramos-Luis E, Riancho JA, Souza Goes AC, Santapa OA, Sumita DR, Vallejo G, Vidal Rioja L, Vide MC, Vieira da Silva CI, Whittle MR, Zabala W, Zarrabeitia MT, Alonso A, Carracedo A, Amorim A. Mutation rates at Y chromosome specific microsatellites. Hum Mutat. 2005;26:520–528. doi: 10.1002/humu.20254. [DOI] [PubMed] [Google Scholar]
- Gusmão L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int. 2006;157:187–197. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Heyer E, Sibert A, Austerlitz F. Cultural transmission of fitness: genes take the fast lane. Trends Genet. 2005;21:234–239. doi: 10.1016/j.tig.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20:191–197. doi: 10.1093/humrep/deh558. [DOI] [PubMed] [Google Scholar]
- Hurles ME, Sykes BC, Jobling MA, Forster P. The dual origin of the Malagasy in island southeast Asia and East Africa: evidence from maternal and paternal lineages. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C. Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet. 1996;5:1767–1775. doi: 10.1093/hmg/5.11.1767. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Bouzekri N, Taylor PG. Hypervariable digital DNA codes for human paternal lineages: MVR-PCR at the Y-specific minisatellite, MSY1 (DYF155S1) Hum Mol Genet. 1998;7:643–653. doi: 10.1093/hmg/7.4.643. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Lo IC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, Chang YM, Kraaijenbrink T, Henke J, Guanti G, McKeown B, van Oorschot RA, Mitchell RJ, de Knijff P, Tyler-Smith C, Parkin EJ. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet. 2007;16:307–316. doi: 10.1093/hmg/ddl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Kittler R, Erler A, Hedman M, Lee AC, Mohyuddin A, Mehdi SQ, Rosser Z, Stoneking M, Jobling MA, Sajantila A, Tyler-Smith C. A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet. 2004;74:1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Bosch E, Adams SM, Parkin EJ, Rosser ZH, Jobling MA. Inadvertent diagnosis of male infertility through genealogical DNA testing. J Med Genet. 2005;42:366–368. doi: 10.1136/jmg.2004.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Parkin EJ, Swinfield G, Cruciani F, Scozzari R, Rosa A, Lim SK, Xue Y, Tyler-Smith C, Jobling MA. Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur J Hum Genet. 2007;15:288–293. doi: 10.1038/sj.ejhg.5201771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- McElreavey K, Ravel C, Chantot-Bastaraud S, Siffroi JP. Y chromosome variants and male reproductive function. Int J Androl. 2006;29:298–303. doi: 10.1111/j.1365-2605.2005.00637.x. [DOI] [PubMed] [Google Scholar]
- Page DC. Sex reversal: deletion mapping the male-determining function of the human Y chromosome. Cold Spring Harb Symp Quant Biol. 1986;51:229–235. doi: 10.1101/sqb.1986.051.01.028. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res. 2002;30:e27. doi: 10.1093/nar/30.6.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin EJ, Kraayenbrink T, van Driem GL, Tshering K, de Knijff P, Jobling MA. 26-locus Y-STR typing in a Bhutanese population sample. Forensic Sci Int. 2006;161:1–7. doi: 10.1016/j.forsciint.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Parkin EJ, Kraayenbrink T, Opgenort JR, van Driem GL, Tuladhar NM, de Knijff P, Jobling MA. Diversity of 26-locus Y-STR haplotypes in a Nepalese population sample: isolation and drift in the Himalayas. Forensic Sci Int. 2007;166:176–181. doi: 10.1016/j.forsciint.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AJ, Agellon AB, Kearney VA, Contreras VA, Karafet T, Park H, de Knijff P, Butler JM, Hammer MF. Forensic value of 14 novel STRs on the human Y chromosome. Forensic Sci Int. 2002;130:97–111. doi: 10.1016/s0379-0738(02)00347-x. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page DC. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- Roewer L, Krüger C, Willuweit S, Nagy M, Rodig H, Kokshunova L, Rothamel T, Kravchenko S, Jobling MA, Stoneking M, Nasidze I. Y-chromosomal STR haplotypes in Kalmyk population samples. Forensic Sci Int. 2007;173:204–209. doi: 10.1016/j.forsciint.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of massive palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova R, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome: a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Tilford CA, Kuroda-Kawaguchi T, Skaletsky H, Rozen S, Brown LG, Rosenberg M, McPherson JD, Wylie K, Sekhon M, Kucaba TA, Waterston RH, Page DC. A physical map of the human Y chromosome. Nature. 2001;409:943–945. doi: 10.1038/35057170. [DOI] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2007;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi Q, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Verma RS, Rodriguez J, Dosik H. The clinical significance of pericentric inversion of the human Y chromosome: a rare “third” type of heteromorphism. J Hered. 1982;73:236–238. doi: 10.1093/oxfordjournals.jhered.a109627. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre HM, Castel A, Nieschlag E, Weidner W, Gröne H-J, Jung A, Engel W, Haidl G. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page DC. The human Y chromosome - a 43-interval map based on naturally-occurring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- Willuweit S, Roewer L, on behalf of the International Forensic Y Chromosome User Group Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, Xu J, Du R, Fu S, Li P, Yang H, Tyler-Smith C. Recent spread of a Y-chromosomal lineage in northern China and Mongolia. Am J Hum Genet. 2005;77:1112–1116. doi: 10.1086/498583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PH. Putative biological functions of the DAZ family. Int J Androl. 2004;27:125–129. doi: 10.1111/j.1365-2605.2004.00469.x. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.