Abstract
cAMP regulates cellular functions primarily by activating PKA. The involvement of PKAs in various signaling pathways occurring simultaneously in different cellular compartments necessitates stringent spatial and temporal regulation. This specificity is largely achieved by binding of PKA to protein scaffolds, whereby a distinct group of proteins called A kinase anchoring proteins (AKAPs) play a dominant role. AKAPs are a diverse family of proteins that all bind via a small PKA binding domain to the regulatory subunits of PKA. The binding affinities between PKA and several AKAPs can be different for different isoforms of the regulatory subunits of PKA. Here we employ a combination of affinity chromatography and mass spectrometry-based quantitative proteomics to investigate specificity in PKA-AKAP interactions. Three different immobilized cAMP analogs were used to enrich for PKA and its interacting proteins from several systems; HEK293 and RCC10 cells and rat lung and testis tissues. Stable isotope labeling was used to confidently identify and differentially quantify target proteins and their preferential binding affinity for the three different cAMP analogs. We were able to enrich all four isoforms of the regulatory subunits of PKA and concomitantly identify more than 10 AKAPs. A selective enrichment of the PKA RI isoforms could be achieved; which allowed us to unravel which AKAPs bind preferentially to the RI or RII regulatory domains of PKA. Of the twelve AKAPs detected, seven preferentially bound to RII, whereas the remaining five displayed at least dual specificity with a potential preference for RI. For some of these AKAPs our data provide the first insights into their specificity.
cAMP is an ubiquitous second messenger that transduces signals from a variety of hormones, neurotransmitters, and inflammatory mediators to regulate a large number of key cellular processes. cAMP can influence cell growth, differentiation, and movement as well as regulating specialized actions unique to specific cell types. The principal target of cAMP is cAMP-dependent protein kinase (PKA)1. Several other proteins such as cyclic nucleotide gated ion channels (1), phosphodiesterases (PDE) (2), and guanine nucleotide exchange factors (Epac) (3) bind cAMP. Interestingly, localized pools of cAMP regulate defined physiological events. It appears that for such events a supramolecular complex is required that comprises of the appropriate effector system together with signal termination enzymes such as PDEs and phosphatases that are sequestered by scaffolding proteins (4). Some of the best described scaffolding proteins are the so-called A-kinase anchoring proteins (AKAPs), which all bind specifically to the N-terminal dimerization domain of the PKA regulatory domain. The organization of a few of these individual supramolecular complexes containing PKA/AKAPs/PDE etc. has been described (4); numerous more of such complexes are expected to exist.
The regulatory domains of mammalian PKAs exist in several isoforms such as RIα, RIβ, RIIα, and RIIβ, which are all encoded by separate genes. The two major isoforms i.e. RI and RII differ in molecular weight, isoelectric point, amino acid sequence, phosphorylation status, tissue distribution, and sub-cellular localization. RI and RII subunits are known to bind to AKAPs with distinct levels of affinity adding another level of intracellular organization for PKA and also facilitating the diversity of the cAMP-mediated signal transduction pathways. Although the PKA-R isoforms differ in functionality, they share a similar overall organization i.e. a dimerization domain, the catalytic subunits inhibitor region, and two cAMP binding domains. The two cAMP binding domains differ in cAMP binding kinetics and are known as site A and site B, respectively (5). Both sites share considerable sequence identity, as a result of a tandem gene duplication, and have conserved phosphate binding cassettes that can be considered as signature motif for cAMP binding. The relative orientation of these two sites is nonetheless, quite different in RI and RII. Additionally, A and B sites have different binding affinity to cAMP derivatives. Site A has a preference for N6-substituted analogs whereas site B is preferred by C2- and C8-substituted analogs (6).
PKA has been studied extensively (7, 8). One of the important goals therein is to develop different cAMP analogs that can result in specific binding, activation, and/or inhibition for each individual cAMP interaction site of the RI and RII isoforms (9). This can help to decipher in detail specific cyclic nucleotide signaling pathways (10). To fully interpret such pathways, analogs should ideally not cross-activate (or inhibit) with other cAMP-regulated proteins such as the before mentioned PDEs, Epac, cyclic nucleotide gated ion channels, and the cGMP-dependent protein kinase (PKG). Although the latter is mainly activated by cGMP, it also binds to cAMP (11, 12). It has been suggested that cGMP and cAMP can cross-activate their respective kinases (13). This cross-talk between PKA and PKG hampers, to some degree, the study of these proteins individually, as dissecting the individual pathways of PKA and PKG requires specific binders, activators, or inhibitors (9, 14). Compared with PKA, PKG is involved in quite different signaling pathways, such as the well characterized nitric oxide-mediated relaxation of smooth muscle cells (15).
The development of synthetic cAMP and cGMP analogs as tools to unravel specific signal transduction pathways requires the sensitive identification and characterization of their cyclic nucleotide interacting proteins. These proteins are typically relatively low abundant, and therefore specific enrichment techniques are essential to study these so-called cyclic-nucleotide interactomes. In recent years such affinity enrichment techniques have been coupled to sensitive mass spectrometric identification of the enriched proteins, nowadays often referred to as chemical proteomics. Chemical proteomics using small molecules as baits, i.e. messenger molecules, drugs, or metabolites, becomes more and more widely used to selectively isolate target proteins from whole cell lysates enabling the analysis of protein subcomplexes and/or signaling pathways (16, 17).
In the present study we compare the properties of three cAMP analogs immobilized individually on agarose beads, for enrichment, isolation, and detection of cyclic nucleotide interacting proteins and their interaction partners, like AKAPs, directly from a crude lysate of cells and tissue. To quantify differential affinity, we use a common strategy in proteomics, namely stable isotope labeling, whereby we introduce the label via reductive amination (18–20). Most interestingly, a very selective enrichment of PKA RI isoforms can be achieved by using cAMP-agarose beads in which the hydroxyl group at the 2′ position on the ribose was replaced with a methoxyl group. This allows us to distinguish, which AKAPs bind preferentially to the RI or RII isoforms. Therefore, this approach provides an elegant tool to further decipher specific cyclic nucleotide signaling pathways.
EXPERIMENTAL PROCEDURES
Materials—
The cAMP-coupled-agarose beads were purchased from BIOLOG (Bremen, Germany). The amount of immobilized cyclic nucleotide on the beads was ∼6 μmol/ml. Protease inhibitor mixture was from Roche Diagnostics. All other chemicals were purchased from commercial sources and were of analysis grade, unless stated otherwise. Isotope-labeled CD2O formaldehyde (20% solution in D2O) was from Sigma-Aldrich. The rabbit polyclonal PKA RIα, RIIα antibodies, and HRP-S-conjugated secondary antibody were from Santa Cruz Biotechnology Inc. High purity water, obtained from a Milli-Q system (Millipore, Bedford, MA), was used for all experiments.
Tissue and Cell Lysate Preparation—
Lysate and tissue preparation protocols were largely as described previously (17, 21). Briefly, HEK293 cells (1.5 × 106 cells/ml) were lysed by dounce homogenization and were left on ice for 10 min. The soluble fraction was taken after centrifugation at 20,800 × g for 10 min. The yield was ∼8 mg of protein, as determined by Bradford assay. For the tissue sample, ∼800 mg of tissue was pulverized in a custom-made steel mortar, which was pre-cooled with liquid nitrogen. Subsequently, 1 ml of ice-cold lysis buffer (50 mm K2HPO4, pH 7.0, 150 mm NaCl. 0.1% Tween 20, protease inhibitor mixture) was added, and the sample was left at room temperature for 5 min. These treated tissue samples were stored on ice for another 10 min. followed by the same procedure as that for HEK293 cells. The final yield of protein was ∼80 mg.
Pulldown Assay—
Prior to pull down, a 50 μl dry volume of immobilized cAMP beads (∼300 nmol of cAMP) were washed with 1 ml of phosphate-buffered saline buffer (50 mm K2HPO4, 150 mm NaCl). For control, beads blocked by ethanolamine were used in a parallel identical pulldown procedure. Prior to the pulldown assays, tissue and cell lysates were incubated with 10 mm ADP/GDP and incubated for 15 min at 4 °C to reduce nonspecific binding, mainly contributed by ADP- and GDP-binding proteins (17, 21). cAMP-agarose beads were added to the lysate in the volume ratio of 1:100 beads to lysate ratio. The lysate-bead suspension was incubated for 2 h at 4 °C by rotary shaking. The supernatant was collected as unbound fraction. The bead bound fraction was washed in several steps with in total 12 ml of lysis buffer to further reduce nonspecific binding. Then the bound fraction was eluted with 90 μl of 8 m urea for denaturing. After digestion with LysC (Roche Diagnostics) for 4 h at 37 °C, reduction and alkylation of the peptides were performed in 2 m urea, followed by further digestion by trypsin (Roche Diagnostics). The dual protease digestion procedure resulted in the detection of more peptides. Reproducibility of the pulldown experiments was evaluated in the rat testis tissue. The ratios observed in this duplicate experiment were reproducible as they were within an experimental error of 10% (supplemental Table S1).
Stable Isotope Labeling Strategy—
The resulting peptides were desalted, dried in vacuo, and re-suspended in 100 μl of triethylammonium bicarbonate (100 μm). Subsequently, 4 μl of formaldehyde-H2 (4% v/v in water for light labeling) was added, vortexed for 2 min followed by the addition of 4 μl of freshly prepared sodium cyanoborohydride (600 mm). The resultant mixture was vortexed for 5 min at room temperature. 16 μl of ammonium hydroxide (1% in water) was added to consume the excess formaldehyde, and 5% formic acid (in water) was added to acidify the solution. For perdeutero-methylation labeling, formaldehyde-D2 (4% v/v in water) was used. The light H4- and heavy D4-di-methyl-labeled samples were mixed in 1:1 ratio based on the total protein input material for the pull down and dried in vacuum and then re-dissolved in 5% acetonitrile containing 0.1 formic acid, prior to preparation for mass spectrometric analysis.
Protein Identification and Quantification—
For mass spectrometric analysis peptides were injected onto a nano-scale liquid chromatography system. Therefore, an Agilent 1100 series liquid chromatography system was equipped with an 20-mm Aqua C18 (Phenomenex, Torrance, CA) trapping column (packed in-house, i.d., 100 μm; resin, 5 μm) and a 250 mm ReproSil-Pur C18-AQ f analytical column (packed in-house, i.d., 50 μm; resin, 3 μm). Trapping was performed at 5 μl/min solution A (0.6% acetic acid) for 10 min, and elution was achieved with a gradient of 0–32% solution B (80% acetonitrile, 0.6% acetic acid) in 60 min, 32–40% solution B in 5 min, 40–100% solution B in 2 min and 100% B for 2 min leading to a total analysis time of 90 min. The flow rate was passively split from 0.4 ml/min to 100 nl/min when performing the elution analysis. Nanospray was achieved using a distally coated fused silica emitter (New Objective, Cambridge, MA) (outer diameter, 360 μm; i.d., 20 μm, tip i.d., 10 μm) biased to 1.8 kV. The LC system was coupled to a 7 Tesla Finnigan LTQ-FT ICR mass spectrometer (Thermo Electron, Bremen, Germany). Briefly, the mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey full scan MS spectra were acquired from m/z 350 to m/z 1500 in the FT-ICR with a resolution of r = 100,000 at m/z 400 after accumulation to a target value of 2,000,000 in the linear ion trap. The five most intense ions were fragmented in the linear ion trap using collisionally induced dissociation at a target value of 10,000. Spectra were processed with Bioworks 3.3 (Thermo, Bremen, Germany), and the subsequent data analysis was carried out using the Mascot (version 2.1.0) software platform (Matrix Science, London, UK) against the IPI-Rat database version 3.19 (44845 protein sequences; 23003239 residues) allowing 1 missed cleavage, carbamidomethyl (C) as fixed modification, “light” and “heavy” methylation of peptide N termini and lysine residues and oxidation (M) as variable modifications. The peptide tolerance was set to 10 ppm and the MS/MS tolerance to 0.9 Da. Proteins were organized using the scaffold software package. Quantification was performed using MSQuant, which is a modified version of 1.4.2a. Peptide ratios between the monoisotopic peaks of “normal” and “heavy” forms of the peptide were calculated and averaged over consecutive MS cycles for the duration of their respective LC-MS peaks in the total ion chromatogram using only FT-MS scans; heavy and light labeled peptides were found to co-elute. All data presented in this manuscript is available in the pride database under the PRIDE experiment accession numbers (3706–3714) and project title name “Selectivity in enrichment of PKA isoforms and their interactors”.
Immunoblotting—
After separating the affinity purified proteins from HEK293 cells on a 1D SDS-PAGE gel, they were transferred to nitrocellulose membranes. Blocking was performed using a solution of TBS (pH 7.5) containing 5% non-fat milk powder. The membranes were then respectively incubated with the PKA RIα (goat polyclonal antibody, PKA Iα reg (C-14): sc-18800; Santa Cruz Biotechnology) in a dilution ratio of 1:100 and PKA RIIα (rabbit polyclonal antibody, PKA IIα reg (M-20): sc-30666; Santa Cruz Biotechnology) in a ratio of 1:100 for 1 h at room temperature. The membranes were washed for 40 min, changing the TBS buffer every 10 min, and then incubated with an HRP-conjugated secondary antibody (sc-2004 and sc-2768; Santa Cruz Biotechnology) in a dilution ratio of 1:10,000 for 1 h at room temperature. After washing the membrane with TBS buffer, the membrane was subjected to chemiluminescence detection according to the manufacturer's protocol (ECL, Amersham Biosciences).
RESULTS
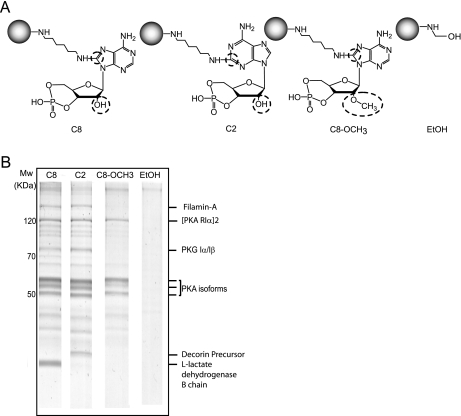
For the affinity purification of cAMP-interacting proteins, various cAMP analogs are available. These cAMP analogs can be immobilized to beads using different coupling positions and different linkers. Here we chose to use agarose beads on which cAMP was immobilized via an aminohexylamino spacer that was attached to either the 8-position of the imidazole ring or 2-position of the pyrimidine ring in the adenine moiety (referred to further on as C8 for the 8-position-linked cAMP analog; and C2 for the 2-position-linked cAMP analog). We took a long chain spacer, as it is expected that the conformational flexibility of the ligand is also increased by increasing the chain length. In addition, we selected 8AHA-2′-O-Me-cAMP-agarose beads (further referred to as C8_OCH3) in which the hydroxyl group at the 2′-position on the ribose was replaced with a methoxyl group. This modified cAMP analog, which is thought to have specific activation characteristics for EPAC1 (22), is linked to the agarose beads via a similar 1,6-diaminohexyl spacer at the C8-position of the adenine ring. Finally, as a negative control we used beads that were blocked by ethanolamine. The chemical structures of the cAMP analogs immobilized on the beads are shown schematically in Fig. 1A.
Fig. 1.
cAMP pull-downs in rat lung tissue using 3 differently immobilized cAMP analogs. A, the chemical structures of the modified immobilized cAMP analogs beads B. SDS-page gels (Coomassie-stained) of rat lung tissue incubated with C8 (lane 1), C2 (lane 2), C8_OCH3 (lane 3), and EtOH control (lane 4) beads. The triplet bands around 50 kDa originate from the PKA-R isoforms, whereas the band at ∼75 kDa is PKG.
Differential Affinity Enrichment of the cAMP Beads Visualized by 1D Gel Analysis—
In the first experiment, we divided rat lung tissue lysate into four equal aliquots. These four samples were individually incubated with the four different, above-mentioned, affinity beads. Following incubation and affinity purification, the enriched proteins were separated using SDS-PAGE. As shown in Fig. 1B, the gel obtained with the control ethanolamine blocked beads (EtOH) did not show any significant protein bands in contrast to protein bands that were pulled down using the C8 beads, C2 beads and C8_OCH3 beads, respectively. These three lanes clearly show the high degree of selectivity obtained using our specific enrichment, comparable with results reported previously (17). Although the three gel lanes show high similarity, there are also striking differences, especially in the molecular weight region where the PKA RI and RII subunits are expected. To confidently identify these differences, we cut each of the four lanes into 20 equal pieces and subjected them to LC-MS analysis. The resulting mass spectra were analyzed using MASCOT and organized using the Scaffold software package. For confident identifications we set a threshold at a minimum of 3 unique peptides with a peptide Mascot score of 30 (p < 0.05). Using these strict settings, we identified 56 proteins in the C8 dataset, 62 proteins for the C2 gel lane, 40 proteins for the C8_OCH3 sample (supplemental Table S2), and no significant proteins in the control lane for the ethanolamine blocked beads. The number of proteins detected is relatively low, but, as illustrated in supplemental Table S2, very specific. A large fraction of proteins are ‘real’ cyclic nucleotide binding proteins or interactors thereof. We attribute this to the fact that we are able to significantly reduce ‘nonspecific’ binding proteins by pre-clearing the lysates with ADP/GDP (17). To confidently determine the relative affinity purification levels of each protein with each of the four types of beads we chose to use isotope labeling by chemical reductive dimethylation labeling (18–20).
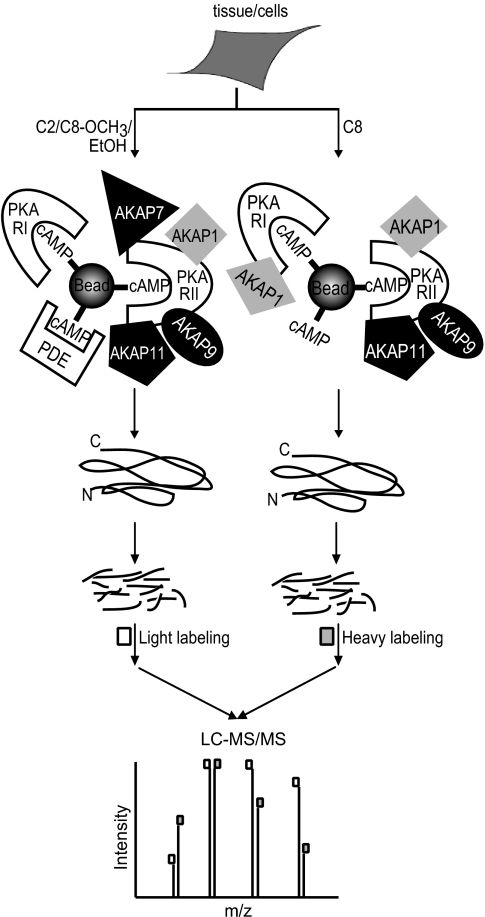
Differential Affinity Enrichment Determined by Stable Isotope Labeling Based Quantitative Proteomics—
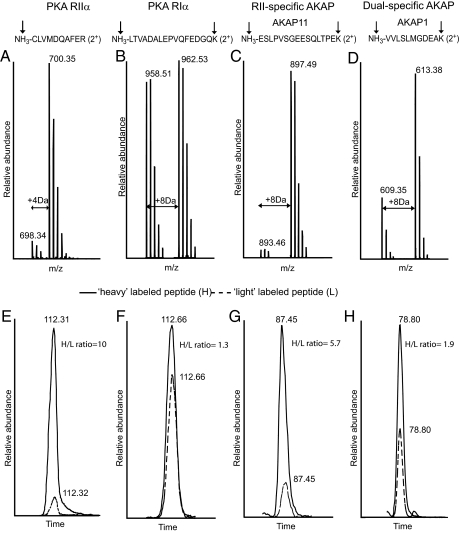
Stable isotope labeling in combination with mass spectrometry is a very elegant approach to quantify differential protein expression (23, 24). To differentially quantify the protein abundance in each pulldown fraction using the four different affinity beads, we designed an experiment as depicted schematically in Fig. 2 and in more detail in supplemental Fig. S1. Following the affinity pull down, we digested all proteins in-solution and chemically labeled the tryptic peptides originating from the pull down with C8 beads with perdeutero-formaldehyde. In individual experiments, the peptides originating from the pull-downs using C2, the C8_OCH3 and the ethanolamine blocked beads were chemically labeled using normal formaldehyde. In dimethyl labeling, formaldehyde reacts with the N terminus and the α-amino group of lysine residues of a peptide, which are subsequently reduced with sodium cyanoborohydride to generate secondary amino groups that are relatively more reactive than their original primary amino groups. Subsequently, each of these species reacts with another formaldehyde moiety to produce a dimethyl substituted tertiary amino group adding 28 (CH2O, “light”) or 32 Da (CD2O, “heavy”) in mass for each N terminus or lysine residue in each peptide (18–20). We mixed the “heavy” labeled sample obtained with the C8 beads, with one of the other three “light” labeled samples. Each set of two labeled samples were mixed in a 1:1 ratio and then analyzed by LC-coupled nanospray LTQ-FT-ICR based mass spectrometry for protein identification and quantification. In Fig. 3, an illustrative example of our quantitative analysis is given, whereby MS spectra (upper panel) and extracted ion chromatograms (lower panel) are depicted in which the “light” (dashed line) and “heavy” (solid line) of individual peptide pairs can be distinguished. Each peptide is detected as a pair with a typical mass difference of 4 Da per amine group, i.e. for the N terminus and each lysine residue present in the peptide. For instance, in the doubly charged peptide of CLVMDVQAFER (Fig. 3A), formaldehyde dimethylates the N terminus generating a m/z difference of 2 which is corresponding to a mass difference of 4 Da between the light- and heavy-labeled peptide. Likewise, Fig. 3B shows that the pair of the doubly charged peptide LTVADALEPVQFEDGQK has an m/z difference of 4Da because of the reaction of formaldehyde with the N terminus and the amino group of the C-terminal lysine. It has been reported that hydrogen-based isotopes can have an isotopic effect during LC separation in which the heavy and light tags show a partial separation which can obviously reduce the accuracy of quantification. However, as shown in Fig. 3, E, F, G, and H, both the light- and heavy-labeled peptide ions co-elute, indicating that such an isotopic effect turns out to be negligible. This is possibly because of deuterium labeling occurring on charged amino residues, which may reduce the differential interaction of hydrogen and deuterium based methyl labels with the RP-HPLC stationary phase (19).
Fig. 2.
Experimental strategy to probe selective enrichment of PKA isoforms and their interacting partners using stable isotope labeling. Following the parallel affinity enrichments using C8 and C2 beads, proteins were digested in-solution with LysC and trypsin, respectively. The tryptic peptides originating from the pull-down using the C8 beads were chemically labeled using CD2O (“heavy-label”) whereas those originating from C2 beads (and also C8_OCH3 and EtOH beads) were labeled with CH2O (“light-label”). Each set of CD2O- and CH2O-labeled samples were mixed in a 1:1 ratio (supplemental Fig. S1) and then analyzed by LC-coupled nanospray LTQ-FT-ICR mass spectrometry for protein identification and quantification.
Fig. 3.
Relative quantitation using stable isotope labeling by reductive dimethylation induces predictive mass shifts. MS spectra (Upper panels, A–D) and extracted ion chromatograms (lower panels, E–H) of peptides originating from proteins in the affinity-enriched HEK293 cell lysate are depicted. The upper panel shows that each detected peptide exists as a pair with a typical 2 Da m/z difference for doubly charged peptides that have no lysine residues whereas a 4 Da m/z difference is observed for doubly charged peptides with one lysine residue. Labeling sites are indicated by the arrows. In the lower panel, the heavy-labeled peptides (peptides which are enriched by C8) are in solid line and light-labeled peptides (peptides which are enriched by C8-OCH3) are in dashed line. Each individual peptide pair is used for the assessment of differential binding affinities.
We performed affinity enrichments with three different immobilized cAMP beads in the lysates of HEK293 cells, RCC10 cells, rat lung and rat testis tissue. We selected these four systems as literature predicted that they may contain quite a diversity of cAMP interacting proteins. A summary of proteins detected, together with the number of identified and quantified peptides, with each of the four beads (i.e. C8, C2, C8_OCH3, EtOH) with different samples is given in supplemental Tables S3A–I. All peptide pairs used for differential quantification are listed in this summary together with their ratios. The protein differential binding ratio was calculated by averaging over all peptide pairs measured for that particular protein. In the remainder of the paper we do not further discuss the results with the ethanolamine blocked beads, as for all proteins discussed below, we were not able to detect them using these beads underscoring the selectivity of our affinity-purifications.
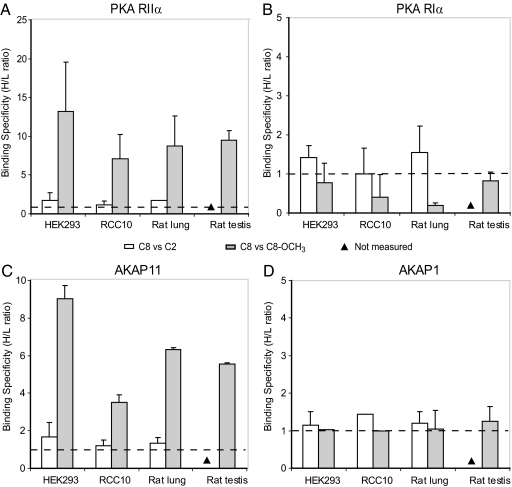
In our pull-downs, we enriched most specifically for the isoforms of the regulatory domain of PKA, represented by the triplet dark bands on the 1D gel of Fig. 1. We detected all four described PKA-R isoforms. Fig. 4A illustrates the differential enrichment of PKA RIIα in the affinity pull-downs with the three beads (i.e. C8, C2, and C8_OCH3). White bars indicate the relative abundance of PKA RIIα pulled down with C8 versus C2 beads, whereas gray bars depict the relative abundance of PKA RIIα recovered with the C8 versus C8_OCH3 beads (Fig. 4). The C8 to C2 ratios are close to 1, irrespective of the origin of the sample (i.e. rat lung tissue, rat testis tissue, HEK293 cells, or RCC10 cells), indicating that both C8 and C2 beads have equal binding specificity towards PKA RIIα. In sharp contrast, the averaged ratios are five and higher for the C8 versus C8_OCH3 pairs, again irrespective of the origin of the sample, indicating that the C8 beads have substantial higher affinity for PKA RIIα than the C8_OCH3 beads. PKA RIIβ displays similar patterns as observed for PKA RIIα as illustrated in supplemental Fig. S2A.
Fig. 4.
Binding specificity patterns of the PKA regulatory isoforms. The ratios were measured in two cell lysates (i.e. HEK293 and RCC10) and two tissue lysates (i.e. rat lung and rat testis tissue) for A (PKA RIIα), B (PKA RIα), and their secondary binding proteins C (AKAP11) and D (AKAP1). The binding specificity ratios of C8 versus C2 are in white, and C8 versus C8_OCH3 are in gray.
Fig. 4B shows that PKA RIα has no substantial binding specificity for either C8 or C2 beads, as revealed by the ratios (∼ 1) (note the different scale of the y axis in Fig. 4B, compared with A). However, in sharp contrast to the results obtained for RII, the binding specificity ratio of C8 to C8_OCH3 is also close to 1, indicating that the C8_OCH3 beads have similar affinity for PKA RIα compared with C8. Similarly, PKA RIβ that we observed in HEK293 and RCC10 exhibits a C8/C8_OCH3 specificity ratio of ∼1 as well, proving that PKA RIβ binds to C8 and C8_OCH3 with similar specificity as well (supplemental Fig. S2B).
In addition to the primary binders of cAMP, we enriched for several secondary binders, primarily many of the expected A kinase anchoring proteins. The enrichment ratios of AKAP11 and AKAP1, which were identified in all four biological systems, are depicted in Fig. 4, C and D, respectively. In Fig. 4C, the measured enrichment patterns of AKAP11 are shown, which are markedly similar to PKA RII shown in Fig. 4A. There is no significant difference between the C8 and C2 beads but lower enrichment when C8_OCH3 beads are used. We attribute this similarity in patterns to the fact that AKAP11 is a PKA RII targeting AKAP. In contrast, AKAP1 exhibited a quite different pattern, as it was found to be equally enriched using the C8, C2, or C8_OCH3 beads (Fig. 4D). This can be rationalized by the ability of AKAP1 to bind equally well to RI and RII meaning that it is a dual specificity AKAP. Other examples are shown in supplemental Fig. S3, which reveals that AKAP7 is RII-specific, and AKAP2 is dual-specific.
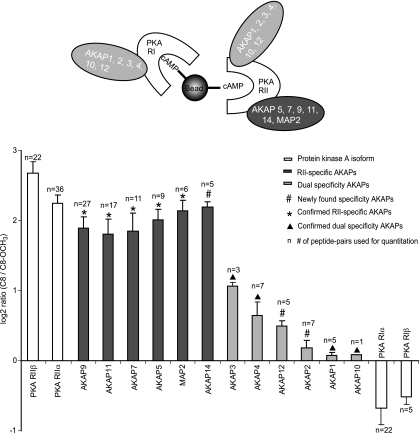
Next to the two AKAPs shown in Fig. 4, C and D, we detected and determined differential binding patterns for a further 10 AKAPs in the four different samples. Fig. 5 summarizes all results for C8 versus C8_OCH3 on a log 2 scale. For each AKAP, the peptides detected in the four samples (HEK293, RCC10, Rat lung tissue and Rat testis tissue) were pooled into one data set and the mean ratios and their standard deviations were calculated. The PKA isoforms, RIIα and RIIβ (white bars on the left) show a log 2 ratio of 3.2 (stdev = 0.12) and 3.9 (stdev = 0.17) whereas RIα and RIβ have similar log 2 ratios of −0.79 (stdev = 0.20) and −0.89 (stdev = 0.16), respectively. Furthermore, 6 AKAPs have similar ratios compared with the RII isoforms:AKAP5, AKAP7, AKAP9, AKAP11, MAP2, and AKAP14 (dark gray bars), indicating they are RII specific AKAPs, in agreement with literature data for AKAP5 (25), AKAP7 (26), AKAP11 (27), and MAP2 (28).
Fig. 5.
Summary of all specificity ratios obtained for each PKA isoform and all detected AKAPs. Overview of averaged binding specificity ratios of all AKAPs affinity purified in the four studied lysates of HEK293, RCC10, rat lung, and rat testis tissue. The protein ratios are calculated by pooling all peptide pairs detected in each tissue for a certain protein. The overall standard deviation was calculated over all spectra in the pool. For comparison, all averaged ratios are depicted on a log 2 scale. The number of spectra, which were used for the average ratios are shown.
On the other hand, AKAP1 (light gray bars) had average ratios of around one indicating that it has similar binding affinities to both RI and RII, so it can be classified as dual specificity AKAP. AKAP10 (d-AKAP2) was also observed as a dual specificity AKAP with a binding pattern similar to AKAP1. Although we only identified one quantifiable peptide of AKAP10 (SIEQDAVNTFTK, supplemental Table S3I), the peptide was identified with high confidence (mascot score of 59, mascot delta score to the next best hit of 47). A manual annotation of the tandem MS spectrum of this peptide is provided in supplemental Fig. S4. Additionally, we performed a BLAST analysis of the peptide sequence against the non-redundant NCBI database to verify the specificity of this peptide referring to AKAP10. The quantitation, was manually inspected in the raw data by extracted ion chromatograms of the heavy and light versions of this peptide. This showed ample intensity to confidently quantitate this peptide (supplemental Fig. S4). In agreement, AKAP1 (25, 29), AKAP10 (30), AKAP3 (31), and AKAP4 (32) have been reported as dual specificity AKAPs previously. In the case of AKAP3 and AKAP4, it seems to be dual specificity with a preferred binding to RII, as the average binding specificity ratios of 1.1 and 0.64, respectively are slightly higher than that of other dual specificity AKAPs in this dataset.
In addition to the enrichment of PKA R isoforms and their binding proteins, we also attained enrichment of PKG. This kinase is known to cross-react with cAMP (supplemental Fig. S2C). Although PKG was identified with good sequence coverage, we were not able to unambiguously differentiate between the known isoforms (Iα or Iβ) because of the lack of unique peptides in the quantitative experiment. In the qualitative experiment in rat lung tissue, detailed in supplemental Table S2, we identified peptides of both PKG I isoforms. In our experiments PKG was pulled down with equal efficiency by C8 and C2 beads with a ratio of 1.2 (HEK293 cell lysate) and 1.6 (rat lung tissue lysate). In sharp contrast, the C8_OCH3 beads exhibited poor levels of PKG enrichment with a ratio of >100 (supplemental Fig. S2C). Therefore, it seems that cAMP modification at the ribose moiety is detrimental for binding to PKG, as recently confirmed in an in vitro binding assay with recombinant PKG (9). Besides PKG, IRAG, a known PKG Iβ binding protein (33), could be detected and displayed a differential enrichment pattern similar to that of PKG (supplemental Table S3I), also with a strikingly lower enrichment using the C8_OCH3 beads, confirming that IRAG is likely enriched as secondary binder to PKG. Furthermore, we enriched and detected PDE2A and PDE10A. PDE2A was found to be less enriched using the C8 beads compared with the C2 beads (data not shown), consistent with earlier reports that an unmodified C8 position in cAMP is essential for phosphodiesterase interactions (17, 34).
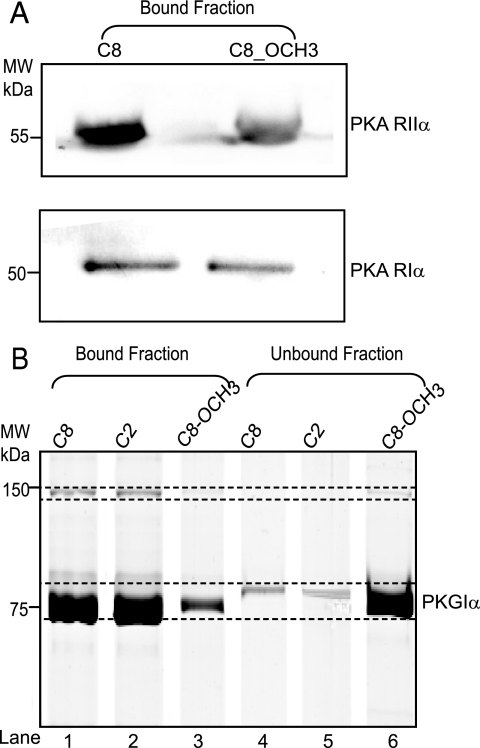
We validated some of the observed differential enrichment characteristics especially for the isoforms RIα and RIIα of PKA (Fig. 6A) by Western blotting with isoform specific antibodies, taking the HEK293 cells as test sample. As shown in Fig. 6A, PKA RII bound to the C8 beads with ∼2.5-fold higher specificity than to the C8_OCH3 beads, in agreement with our chemical proteomics data. Using the RI specific antibodies, Western blotting analysis confirmed that there is no significant selectivity between C8 and C8_OCH3 beads. To confirm the differential enrichment observed for PKG, we performed pull-downs with recombinant PKG (rPKG). The same amount of rPKG was incubated with the three different beads and the amounts of bound and unbound protein were visualized by SDS-PAGE. These results are shown in Fig. 6B, with lane 1, 2, and 3 showing the relative amount of PKG bound to the C8, C2, and C8_OCH3 beads, respectively. The fraction of unbound protein is depicted in the lanes 4, 5, and 6 for C8, C2, and C8_OCH3 beads, respectively. The results clearly confirm that PKG binds to C8 and C2 beads with similar affinities, whereas the binding to the C8_OCH3 beads is significantly less, confirming our chemical proteomics data.
Fig. 6.
In vitro validation of PKA and PKG binding specificities for the three types of cAMP beads. Validation of quantitative chemical proteomics approach. A, Western blot analysis of PKA RIα and PKA RIIα affinity enriched from HEK293 cells by the C8 and C8_OCH3 beads, respectively. B, affinity pull-down assays using recombinant PKG Iα, visualized using SDS gels in which analysis of PKG bound to the bead fractions is shown in lanes 1, 2, and 3. The unbound fractions are depicted in lanes 4, 5, and 6.
Overall these validation experiments on PKA RI & RII and PKG reveal good agreement with the quantitative proteomics experiments, providing confidence to the latter approach for the quantification of the binding ratios of the other proteins.
Background Proteins Can be Bead-specific—
In our affinity pull-downs we also detected some high abundance “background” proteins such as actin, hemoglobin, alpha 2-globulin, l-lactose dehydrogenase B-chain and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). When analyzing the differential binding behavior of these proteins we observed that actin, hemoglobin and alpha 2-globulin were equally abundant no matter which of the four beads (C8, C2, C8_OCH3, and control beads) we used. Remarkably, several “background” proteins revealed also to some extent specific enrichments. For instance, l-lactose dehydrogenase B-chain bound to C8 beads with a relative affinity ratio of 100, compared with C2. GAPDH was enriched by the C8 beads with a relative affinity ratio of 4:1 to both C2 and C8_OCH3. Finally, various ribosomal proteins were found to be preferentially enriched using the C2 beads (supplemental Table S2).
DISCUSSION
To gain further insights into the protein scaffolds involved in cAMP/PKA-directed signaling, such protein complexes are best analyzed and compared in a large set of different cellular and tissue samples. Recently, we introduced methodology to identify at a medium-large scale different cAMP/PKA-linked scaffolds in a qualitative way identifying several AKAPs present in heart tissue (17, 21). For this, we applied specific enrichment of PKA·AKAP complexes using immobilized cAMP. Although these studies gave good insight in the presence of over 14 different AKAPs in heart tissue and also allowed the identification of a novel potential AKAP, it did not provide any detailed information on the PKA-isoform specificity of these AKAPs. A way to address this would be to make use of cAMP analogs that have a specificity for RI over RII or vice versa; however these are poorly recognized thus far (9). In this study we discovered that immobilized C8_OCH3 binds much weaker to the PKA RII than the PKA RI isoforms. By applying a quantitative MS approach, using differential stable isotope labeling, we now show we can evaluate AKAPs for their PKA RI/RII specificity in a variety of tissues, making use of the differential co-enrichment of the AKAPs with either RI or RII.
In summary, the enrichment characteristics of three differently immobilized cAMP analogs were probed in HEK293 cells, RCC10 cells, rat lung, and rat testis tissue. These tissues were chosen as they were expected to contain a wide variety of different AKAPs. Stable isotope dimethyl labeling at the peptide level was applied to quantify the enrichment characteristics on the different beads. For each specifically binding protein an affinity ratio could be calculated based on the comparison of mass spectra peak intensities. We were able to enrich and relatively quantify all four different isoforms of the regulatory subunits of PKA, about a dozen of AKAPs, and several other primary (PKG, PDE) and secondary protein binders of cAMP. Below we will discuss in some more detail the different classes of affinity enriched proteins focusing first on the primary bait proteins, followed by the AKAPs, which are secondary binding proteins.
Primary-binding Proteins—
Our quantitative proteomics data clearly indicated that both RII isoforms (RIIα and RIIβ) bind equally well to C8 and C2 cAMP beads. In contrast, RII seems to have a much lower affinity towards the C8_OCH3 beads (Fig. 4A). The two beads have the same linking position at C8 but in the latter the 2′-OH is methylated.
Detailed structural studies on cAMP-bound PKA RI and RII (35, 36) point out that the cAMP binding domains of RI and RII are highly conserved. Also the residues interacting directly with the 2′-OH seem to be similarly oriented in RI and RII. The invariant Gly (Gly-204/Gly-334 for RIIα and Gly-220/Gly-349 for RIIβ) and Glu (Glu-205/Glu-335 and Glu-221/Glu-350 for RIIα and RIIβ, respectively) bind to the 2′OH of the ribose moiety by forming hydrogen bond. This would point out that the modification at 2′OH ribose moiety (C8_OCH3) likely reduces the binding to RII by disruption of the hydrogen bond. Intriguingly, the same interactions were revealed for RI in which both the Gly (Gly-201/Gly-325 for RIα and RIβ) and the Glu (Glu-202/Glu-326 for RIα and RIβ) residue are conserved. (36). This means that the formation of the hydrogen bond is apparently not solely responsible for diminishing the binding of RII to our C8_OCH3 beads. This observation hints at different interactions in RI and RII that help to stabilize their complex with cAMP and especially with 2′-methoxy-cAMP. We attribute this partly to possible different stacking interactions of cAMP with RI and RII. A sequence alignment between RI and RII reveals that the key hydrophobic residues required for this stacking interaction such as Val (Val-184), Val (Val-186), Val (Val-302), Val (Val-315), Leu (Leu-318), Ile (Ile-327), and Ser (Ser-375) in RI are differently positioned and poorly conserved in RII in which the aligned residues are Ile (Ile-183), Val (Val-185), Ile (Ile-305), Ile (Ile-324), Cys (Cys-327), Leu (Leu-336), and Glu (Glu-384). These variable residues between the two isoforms could influence binding specificity to cAMP. Detailed molecular structures of the two regulatory isoforms bound to the different cAMP analogs would be required to validate the above speculations and possibly point to other differences in the cAMP binding domains of RI and RII.
Another known primary bait protein for the beads is PKG. Our results indicate that PKG binds equally well to C8 and C2; however binding to C8_OCH3 is dramatically reduced (supplemental Fig. S2C). Interestingly, the PKG hydrogen bonding residues: Arg (Arg-177/Arg-403), Gly (Gly-167/Gly-393), and Glu (Glu-168/Glu-394) are conserved with respect to PKA RII. The data on PKG suggest that the ribose 2′OH moiety-related hydrogen bonding is even more crucial for binding of PKG than PKA RII.
Additionally, we enriched for two phosphodiesterases: PDE2A3 in rat lung tissue and PDE10A in rat testis tissue. PDE2A3 was found to bind C2 with an ∼10-fold higher than C8 in binding specificity, indicating that hydrophobic binding between the imidazole ring of cAMP is stronger than that to the pyrimidine ring, both of which are required for cyclic nucleotide discrimination.
Recently the affinities of a range of widely used cyclic nucleotides and derivatives were tested on all subtypes of PKA and PKG, as well as on Epac1 and 6 types of PDE (9). This study by Poppe et al. (9) sheds light on the potential promiscuity of cyclic nucleotide derivatives when applying them as pharmacological tools. The results obtained by Poppe et al. (9) on recombinant proteins in vitro are completely consistent with our findings in vivo. The non-linked version of C8_OCH3 binds to PKA RI with higher affinity than to RII, whereas it does not seem to bind to PKG at all.
Secondary-binding Proteins—
In the cell PKA is localized through interaction of the regulatory domains with the family of distinct but functionally homologous AKAPs. Through an extensive body of work it has now been established that some AKAPs prefer to bind the RII regulatory subunit whereas others have dual specificity for both RI and RII. To some extent the amino acid residues that play a role in whether an AKAP protein binds preferentially RII or displays dual specificity have been defined by extensive mutagenesis studies and biochemical analysis (29, 37–40).
As described above we observed distinct binding specificities for C8 and C8_OCH3 beads, which can be further exploited as a discriminating factor to establish the preference of the secondary AKAP binders for RI or RII. In other words, we can differentiate between RII-specific and dual-specific AKAPs depending on the measured binding specificity ratios between the pull-downs performed with the C8 and C8_OCH3 beads. AKAPs with a ratio significantly similar to RII isoforms were considered RII-specific AKAPs. We detected in total 12 AKAPs, of which 6 could be classified as preferentially binding to the RII regulatory subunit of PKA, whereas the remaining 6 were considered to be dual specificity AKAPs (Fig. 5). In cases where prior literature is available our data confirmed reported specificities of AKAPs providing the validation of our method. However, for a number of AKAPs no information is available about their specificity. Below we discuss for each detected AKAP our findings in the context of literature data.
RII-specific AKAPs—
Our data clearly shows that AKAP5 is an RII-binding protein. AKAP5 was shown to be an RIIβ binding AKAP by deletion experiments and site-directed mutagenesis (41) revealing that several amino acid residues at the C terminus with long aliphatic side chains play a central role. The microtubule-associated protein MAP2 is the first protein shown to co-purify with RII (28, 42). In our experiments, we identified MAP2c in testis tissue and MAP2a/b in HEK293, as based on unique peptide identifiers of these splice isoforms. The binding specificity ratios of these two isoforms clearly confirm that both are RII-specific AKAPs. AKAP11, which we detected in all our cells and tissues, is a clear RII-specific binding AKAP. This protein is known to be expressed abundantly in human testis (43). The peptide Ht31, which is an AKAP-PKA anchoring antagonist, has been successfully used to block RII-AKAP11 interactions, suggesting that AKAP11 may bind RII in a manner similar to other AKAPs (27). AKAP9, which was relatively abundant in HEK293 and RCC10 cells, was also a clear RII-specific AKAP (Fig. 5). Using a yeast two-hybrid screen Feliciello et al. (44) showed that the RII regulatory subunits of PKA bind in vitro Yotiao (AKAP9) with nanomolar affinity (Kd 50–90 nm). Of AKAP7, identified in our experiments (supplemental Fig. 3), at least four splice variants have been reported. Our initial 1D gel analyses indicated that in our systems AKAP7 had a molecular weight of around 40,000 Da indicating that it is likely the γ or δ isoform, which we were unfortunately unable to distinguish, based on the detected peptides. Hundsrucker et al. (26) showed that AKAP7δ is a high affinity RIIα-binding protein with a Kd value of 31 nm. In contrast, yeast two-hybrid analysis and co-immunoprecipitation studies (45) showed that AKAP7γ binds also to the RI subunit. Based on these studies, it seems likely that we mainly enriched for AKAP7δ, being a specific RII-binding AKAP.
Finally we clearly show, for the first time, that AKAP14 preferentially binds to RII. Although relatively little information has been reported about AKAP14, sequence analysis indicates that this protein is closely related to TAKAP-80, which is exclusively expressed in testis (46). In line, we detected this protein only in rat testis.
Dual Specificity AKAPs—
Our data confirm that AKAP1, AKAP10, AKAP2, and AKAP12 have dual specificity. Huang et al. (29) identified d-AKAP1 (AKAP1) as a fragment from a yeast two-hybrid screen based on specific interaction with the RI portion of Ret/ptc2, which includes most of the RI/RII binding domain. The interacting regions on both RI and RII are localized to their N termini. AKAP10 (d-AKAP2) has been well studied and shown to display dual specificity (Kd ∼ 2 and 48 nm for RII and RI, respectively) (47–49), in agreement with our analysis. AKAP2 (AKAP-KL) isoforms contain a 20-residue domain (amino acid 586–605) that binds RII with nanomolar affinity (Kd ∼ 10 nm) and a partial AKAP2 protein isoform (aa 354–741), which includes the putative tethering site that binds both RIIα and RIIβ (50, 51). Although AKAP2 is known to bind RII, our data showed that AKAP2 seems to have dual specificity based on the observed ratio profile (supplemental Fig. 3). AKAP12 (Gravin), a 250 kDa AKAP, was originally identified as a cytoplasmic antigen recognized by myasthenia gravis sera. Our data establish for the first time that AKAP12 is a dual-specific AKAP. Studies in vitro revealed that the residue stretch 1526–1780 of AKAP12 binds RII with nanomolar binding affinity (52).
AKAP3 appears to be expressed specifically in spermatoids and spermatozoa and as expected in our experiments we only detected it in rat testis. AKAP3 has been reported to bind to both RII and RI (31), which is in agreement with our finding. In the sequence of AKAP3, an RII binding domain has been identified (residues 124–141), which is conserved between the human, murine, and bovine AKAP3 (31). In an earlier study, AKAP4 has been reported as an RII-binding protein (53, 54). Nonetheless, our finding is consistent with Vijayaraghavan et al. (31) in which AKAP4 is reported as dual specificity AKAP. Our data indicate that both proteins display dual specificity, however, with a preference to bind to RII because the specificity ratios are slightly different from the other dual specificity AKAPs (Fig. 5). It is possible that some residues required for RII binding are different, although there is a significant homology between the putative RII binding domains of AKAP3 and AKAP4 (i.e. 12 of 18 amino acids are identical). These differences in the core of the binding domain might contribute to the observed different binding specificity.
Conclusion—
We combined a chemical proteomics approach with stable isotope labeling and mass spectrometry to enrich specifically for cAMP-interacting proteins, directly from cell or tissue lysates. cAMP-agarose affinity resins with three different derivatives of cAMP were used to probe their selectivity in the enrichment; C2, C8, and C8_OCH3. Stable isotope labeling facilitated the identification of unspecifically binding background proteins and allowed us to probe the differential binding behavior of relevant proteins to the different analogs directly in tissue. Our data reveal that all PKA regulatory isoforms have a similar affinity for C8 and C2 beads. In contrast, RII isoforms have a significant lower affinity for the C8_OCH3 beads, whereas the RI isoforms seem to be unaffected by this small chemical modification. We employed this effect to monitor the specificity of secondary-binding proteins to the PKA RI and RII isoforms with our LC-MS stable isotope labeling method as read out. In four different input lyastes we detected 12 main AKAP families, of which six were observed to have specificity for the PKA RII isoform, whereas the other six displayed dual specificity for both RI and RII. Besides confirming previously reported PKA-AKAP specificities, the specificity of other PKA-AKAP complexes was also elucidated. We have observed for the first time that AKAP14 has RII specificity whereas AKAP2 and AKAP 12 are dual-specific AKAPs (# in Fig. 5). In conclusion, our chemical proteomics screen sheds new light on nature's complex diversity in signaling mechanisms, as each AKAP, and each isoform thereof, may direct different PKA isoforms to different compartments of the cell, providing the basis of the complex multifunctional platform of this single kinase.
In general, our data reveal that chemical biology approaches, combined with stable isotope labeling and mass spectrometry, provide comprehensive ways to study the interplay between small molecules and target proteins which can be applied to complex biological systems, such as total cell or mammalian tissue lysates.
Supplementary Material
Footnotes
Published, MCP Papers in Press, December 31, 2008, DOI 10.1074/mcp.M800226-MCP200
The abbreviations used are: PKA, cAMP-dependent protein kinase; AKAP, A kinase anchoring protein; PDE, phosphodiesterases; Epac, guanine nucleotide exchange protein directly activated by cAMP; PKG, cGMP-dependent protein kinase; LC, liquid chromatography; TBS, Tris-buffered saline; C2, 2-(6-aminohexlylamino)adenosine-3′, 5′-cyclic monophosphate, immobilized on agarose; C8, 8-(6-aminohexylamino)adenosine-3′, 5′-cyclic monophosphate, immobilized on agarose; C8_OCH3, 8-(6-aminohexylamino)-2′-O-methyladenosine-3′, 5′-cyclic monophosphate, immobilized on agarose; i.d., inner diameter; FT-MS, Fourier transform mass spectrometry; LC-MS, liquid chromatography mass spectrometry; 1D, one-dimensional.
This work was supported by the Netherlands Proteomics Centre.
The on-line version of this article (available at http://www.mcp.org) contains supplemental material.
REFERENCES
- 1.Bradley, J., Reisert, J., and Frings, S. ( 2005) Regulation of cyclic nucleotide-gated channels. Curr. Opin. Neurobiol. 15, 343–349 [DOI] [PubMed] [Google Scholar]
- 2.McConnachie, G., Langeberg, L. K., and Scott, J. D. ( 2006) AKAP signaling complexes: getting to the heart of the matter. Trends Mol. Med. 12, 317–323 [DOI] [PubMed] [Google Scholar]
- 3.de Rooij, J., Zwartkruis, F. J., Verheijen, M. H., Cool, R. H., Nijman, S. M., Wittinghofer, A., and Bos, J. L. ( 1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 4.Wong, W., and Scott, J. D. ( 2004) AKAP signaling complexes: focal points in space and time. Nat. Rev. Mol. Cell. Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 5.Ogreid, D., and Doskeland, S. O. ( 1980) Protein kinase II has two distinct binding sites for cyclic AMP, only one of which is detectable by the conventional membrane-filtration method. FEBS Lett. 121, 340–344 [DOI] [PubMed] [Google Scholar]
- 6.Ogreid, D., Ekanger, R., Suva, R. H., Miller, J. P., and Doskeland, S. O. ( 1989) Comparison of the two classes of binding sites (A and B) of type I and type II cyclic-AMP-dependent protein kinases by using cyclic nucleotide analogs. Eur. J. Biochem. 181, 19–31 [DOI] [PubMed] [Google Scholar]
- 7.Taylor, S. S., Kim, C., Vigil, D., Haste, N. M., Yang, J., Wu, J., and Anand, G. S. ( 2005) Dynamics of signaling by PKA. Biochim. Biophys. Acta 1754, 25–37 [DOI] [PubMed] [Google Scholar]
- 8.Scholten, A., Aye, T. T., and Heck, A. J. ( 2008) A multi-angular mass spectrometric view at cyclic nucleotide dependent protein kinases: in vivo characterization and structure/function relationships. Mass Spectrom. Rev. 27, 331–353 [DOI] [PubMed] [Google Scholar]
- 9.Poppe, H., Rybalkin, S. D., Rehmann, H., Hinds, T. R., Tang, X. B., Christensen, A. E., Schwede, F., Genieser, H. G., Bos, J. L., Doskeland, S. O., Beavo, J. A., and Butt, E. ( 2008) Cyclic nucleotide analogs as probes of signaling pathways. Nat. Methods 5, 277–278 [DOI] [PubMed] [Google Scholar]
- 10.Schwede, F., Maronde, E., Genieser, H., and Jastorff, B. ( 2000) Cyclic nucleotide analogs as biochemical tools and prospective drugs. Pharmacol. Ther. 87, 199–226 [DOI] [PubMed] [Google Scholar]
- 11.Landgraf, W., Hullin, R., Gobel, C., and Hofmann, F. ( 1986) Phosphorylation of cGMP-dependent protein kinase increases the affinity for cyclic AMP. Eur. J. Biochem. 154, 113–117 [DOI] [PubMed] [Google Scholar]
- 12.Pohler, D., Butt, E., Meissner, J., Muller, S., Lohse, M., Walter, U., Lohmann, S. M., and Jarchau, T. ( 1995) Expression, purification, and characterization of the cGMP-dependent protein kinases I beta and II using the baculovirus system. FEBS Lett. 374, 419–425 [DOI] [PubMed] [Google Scholar]
- 13.Pelligrino, D. A., and Wang, Q. ( 1998) Cyclic nucleotide crosstalk and the regulation of cerebral vasodilation. Prog. Neurobiol. 56, 1–18 [DOI] [PubMed] [Google Scholar]
- 14.Dostman, W. R. ( 2003) Inhibitors of cyclic nucleotide-dependent protein kinases in Handbook of cellular signaling (Bradshow, R. A. and Dennis, E. A., eds) 2nd Ed., pp. 487–493, Academic Press, Washington DC
- 15.Lincoln, T. M., Dey, N., and Sellak, H. ( 2001) Invited review: cGMP-dependent protein kinase signaling mechanisms in smooth muscle: from the regulation of tone to gene expression. J. Appl. Physiol. 91, 1421–1430 [DOI] [PubMed] [Google Scholar]
- 16.Kim, E., and Park, J. M. ( 2003) Identification of novel target proteins of cyclic GMP signaling pathways using chemical proteomics. J. Biochem. Mol. Biol. 36, 299–304 [DOI] [PubMed] [Google Scholar]
- 17.Scholten, A., Poh, M. K., van Veen, T. A., van Breukelen, B., Vos, M. A., and Heck, A. J. ( 2006) Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1-interacting protein as a genuine and highly abundant AKAP. J. Proteome Res. 5, 1435–1447 [DOI] [PubMed] [Google Scholar]
- 18.Boersema, P. J., Aye, T. T., Veen, T. A. B. van, Heck, A. J. R., and Mohammed, S. ( 2008) Triplex protein quantification based on stable isotope labelling by peptide dimethylation applied to cell and tissue lysates. Proteomics 8, 4624–4632 [DOI] [PubMed] [Google Scholar]
- 19.Hsu, J. L., Huang, S. Y., Chow, N. H., and Chen, S. H. ( 2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 20.Raijmakers, R., Berkers, C. R., de Jong, A., Ovaa, H., Heck, A. J., and Mohammed, S. ( 2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholten, A., van Veen, T. A., Vos, M. A., and Heck, A. J. ( 2007) Diversity of cAMP-dependent protein kinase isoforms and their anchoring proteins in mouse ventricular tissue. J. Proteome Res. 6, 1705–1717 [DOI] [PubMed] [Google Scholar]
- 22.Christensen, A. E., Selheim, F., de Rooij, J., Dremier, S., Schwede, F., Dao, K. K., Martinez, A., Maenhaut, C., Bos, J. L., Genieser, H. G., and Doskeland, S. O. ( 2003) cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 278, 35394–35402 [DOI] [PubMed] [Google Scholar]
- 23.Bantscheff, M., Schirle, M., Sweetman, G., Rick, J., and Kuster, B. ( 2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 24.Heck, A. J., and Krijgsveld, J. ( 2004) Mass spectrometry-based quantitative proteomics. Expert Rev. Proteomics 1, 317–326 [DOI] [PubMed] [Google Scholar]
- 25.Herberg, F. W., Maleszka, A., Eide, T., Vossebein, L., and Tasken, K. ( 2000) Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 298, 329–339 [DOI] [PubMed] [Google Scholar]
- 26.Hundsrucker, C., Krause, G., Beyermann, M., Prinz, A., Zimmermann, B., Diekmann, O., Lorenz, D., Stefan, E., Nedvetsky, P., Dathe, M., Christian, F., McSorley, T., Krause, E., McConnachie, G., Herberg, F. W., Scott, J. D., Rosenthal, W., and Klussmann, E. ( 2006) High-affinity AKAP7delta-protein kinase A interaction yields novel protein kinase A-anchoring disruptor peptides. Biochem. J. 396, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lester, L. B., Coghlan, V. M., Nauert, B., and Scott, J. D. ( 1996) Cloning and characterization of a novel A-kinase anchoring protein. AKAP 220, association with testicular peroxisomes. J. Biol. Chem. 271, 9460–9465 [DOI] [PubMed] [Google Scholar]
- 28.Rubino, H. M., Dammerman, M., Shafit-Zagardo, B., and Erlichman, J. ( 1989) Localization and characterization of the binding site for the regulatory subunit of type II cAMP-dependent protein kinase on MAP2. Neuron 3, 631–638 [DOI] [PubMed] [Google Scholar]
- 29.Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. ( 1997) Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J. Biol. Chem. 272, 8057–8064 [DOI] [PubMed] [Google Scholar]
- 30.Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. ( 1997) D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. U. S. A. 94, 11184–11189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijayaraghavan, S., Liberty, G. A., Mohan, J., Winfrey, V. P., Olson, G. E., and Carr, D. W. ( 1999) Isolation and molecular characterization of AKAP110, a novel, sperm-specific protein kinase A-anchoring protein. Mol. Endocrinol. 13, 705–717 [DOI] [PubMed] [Google Scholar]
- 32.Miki, K., and Eddy, E. M. ( 1998) Identification of tethering domains for protein kinase A type Iα regulatory subunits on sperm fibrous sheath protein FSC1. J. Biol. Chem. 273, 34384–34390 [DOI] [PubMed] [Google Scholar]
- 33.Ammendola, A., Geiselhoringer, A., Hofmann, F., and Schlossmann, J. ( 2001) Molecular determinants of the interaction between the inositol 1,4,5-trisphosphate receptor-associated cGMP kinase substrate (IRAG) and cGMP kinase Iβ. J. Biol. Chem. 276, 24153–24159 [DOI] [PubMed] [Google Scholar]
- 34.Xu, R. X., Rocque, W. J., Lambert, M. H., Vanderwall, D. E., Luther, M. A., and Nolte, R. T. ( 2004) Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J. Mol. Biol. 337, 355–365 [DOI] [PubMed] [Google Scholar]
- 35.Diller, T. C., Madhusudan, Xuong, N. H., and Taylor, S. S. ( 2001) Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure 9, 73–82 [DOI] [PubMed] [Google Scholar]
- 36.Kim, C., Xuong, N. H., and Taylor, S. S. ( 2005) Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science 307, 690–696 [DOI] [PubMed] [Google Scholar]
- 37.Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D., Bishop, S. M., Acott, T. S., Brennan, R. G., and Scott, J. D. ( 1991) Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266, 14188–14192 [PubMed] [Google Scholar]
- 38.Newlon, M. G., Roy, M., Morikis, D., Hausken, Z. E., Coghlan, V., Scott, J. D., and Jennings, P. A. ( 1999) The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 6, 222–227 [DOI] [PubMed] [Google Scholar]
- 39.Miki, K., and Eddy, E. M. ( 1999) Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J. Biol. Chem. 274, 29057–29062 [DOI] [PubMed] [Google Scholar]
- 40.Banky, P., Newlon, M. G., Roy, M., Garrod, S., Taylor, S. S., and Jennings, P. A. ( 2000) Isoform-specific differences between the type Ialpha and IIalpha cyclic AMP-dependent protein kinase anchoring domains revealed by solution NMR. J. Biol. Chem. 275, 35146–35152 [DOI] [PubMed] [Google Scholar]
- 41.Hausken, Z. E., Dell’Acqua, M. L., Coghlan, V. M., and Scott, J. D. ( 1996) Mutational analysis of the A-kinase anchoring protein (AKAP)-binding site on RII. Classification Of side chain determinants for anchoring and isoform selective association with AKAPs. J. Biol. Chem. 271, 29016–29022 [DOI] [PubMed] [Google Scholar]
- 42.Obar, R. A., Dingus, J., Bayley, H., and Vallee, R. B. ( 1989) The RII subunit of cAMP-dependent protein kinase binds to a common amino-terminal domain in microtubule-associated proteins 2A, 2B, and 2C. Neuron 3, 639–645 [DOI] [PubMed] [Google Scholar]
- 43.Reinton, N., Collas, P., Haugen, T. B., Skalhegg, B. S., Hansson, V., Jahnsen, T., and Tasken, K. ( 2000) Localization of a novel human A-kinase-anchoring protein, hAKAP220, during spermatogenesis. Dev. Biol. 223, 194–204 [DOI] [PubMed] [Google Scholar]
- 44.Feliciello, A., Cardone, L., Garbi, C., Ginsberg, M. D., Varrone, S., Rubin, C. S., Avvedimento, E. V., and Gottesman, M. E. ( 1999) Yotiao protein, a ligand for the NMDA receptor, binds and targets cAMP-dependent protein kinase II(1). FEBS Lett. 464, 174–178 [DOI] [PubMed] [Google Scholar]
- 45.Brown, R. L., August, S. L., Williams, C. J., and Moss, S. B. ( 2003) AKAP7gamma is a nuclear RI-binding AKAP. Biochem. Biophys. Res. Commun. 306, 394–401 [DOI] [PubMed] [Google Scholar]
- 46.Mei, X., Singh, I. S., Erlichman, J., and Orr, G. A. ( 1997) Cloning and characterization of a testis-specific, developmentally regulated A-kinase-anchoring protein (TAKAP-80) present on the fibrous sheath of rat sperm. Eur. J. Biochem. 246, 425–432 [DOI] [PubMed] [Google Scholar]
- 47.Burns-Hamuro, L. L., Hamuro, Y., Kim, J. S., Sigala, P., Fayos, R., Stranz, D. D., Jennings, P. A., Taylor, S. S., and Woods, V. L., Jr. ( 2005) Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 14, 2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burns, L. L., Canaves, J. M., Pennypacker, J. K., Blumenthal, D. K., and Taylor, S. S. ( 2003) Isoform specific differences in binding of a dual-specificity A-kinase anchoring protein to type I and type II regulatory subunits of PKA. Biochemistry 42, 5754–5763 [DOI] [PubMed] [Google Scholar]
- 49.Kinderman, F. S., Kim, C., von Daake, S., Ma, Y., Pham, B. Q., Spraggon, G., Xuong, N. H., Jennings, P. A., and Taylor, S. S. ( 2006) A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong, F., Feldmesser, M., Casadevall, A., and Rubin, C. S. ( 1998) Molecular characterization of a cDNA that encodes six isoforms of a novel murine A kinase anchor protein. J. Biol. Chem. 273, 6533–6541 [DOI] [PubMed] [Google Scholar]
- 51.Glantz, S. B., Li, Y., and Rubin, C. S. ( 1993) Characterization of distinct tethering and intracellular targeting domains in AKAP75, a protein that links cAMP-dependent protein kinase II beta to the cytoskeleton. J. Biol. Chem. 268, 12796–12804 [PubMed] [Google Scholar]
- 52.Nauert, J. B., Klauck, T. M., Langeberg, L. K., and Scott, J. D. ( 1997) Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr. Biol. 7, 52–62 [DOI] [PubMed] [Google Scholar]
- 53.Carrera, A., Gerton, G. L., and Moss, S. B. ( 1994) The major fibrous sheath polypeptide of mouse sperm: structural and functional similarities to the A-kinase anchoring proteins. Dev. Biol. 165, 272–284 [DOI] [PubMed] [Google Scholar]
- 54.Visconti, P. E., Johnson, L. R., Oyaski, M., Fornes, M., Moss, S. B., Gerton, G. L., and Kopf, G. S. ( 1997) Regulation, localization, and anchoring of protein kinase A subunits during mouse sperm capacitation. Dev. Biol. 192, 351–363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.