Abstract
Activated macrophages are essential effectors of immunity and a rich source of matrix metalloproteinase-9 (MMP-9; gelatinase B). To search for cellular substrates of the enzyme, we subjected wild-type macrophages and macrophages expressing an autoactivating form of pro-MMP-9 (M9A macrophages) to proteomics analysis. Two-dimensional liquid chromatography together with tandem mass spectrometry identified 467 proteins in medium conditioned by M9A and/or wild-type macrophages. Subtractive proteomics identified 18 candidate MMP-9 substrates. Biochemical studies confirmed that two transmembrane proteins, β2 integrin subunit (CD18) and amyloid protein precursor (APP), were enriched in the medium of M9A macrophages. To identify potential cleavage sites, we synthesized an overlapping library of peptides that spanned 60 residues of the ectodomain and transmembrane domain of β2 integrin. Active MMP-9 cleaved a single peptide, ECVKGPNVAAIVGGT, at residues corresponding to Ala705 and Ile706 of the β2 integrin. Peptides corresponding to this cleavage site were detected by tandem mass spectrometric analysis only in medium from M9A macrophages, strongly supporting the proposal that β2 integrin is shed by autoactivating MMP-9. Our observations indicate that subtractive proteomics in concert with peptide substrate mapping is a powerful approach for identifying proteolytic substrates and suggest that MMP-9 plays previously unsuspected roles in the regulation and shedding of β2 integrin.
Matrix metalloproteinases (MMPs),1 a subfamily of metazincins, are a structurally related group of zinc-dependent proteases (1). They are synthesized in latent form as pro-MMPs, and their prodomain must be removed or modified before they are proteolytically active. Some MMPs are secreted, whereas others are anchored to the cell surface, but their proteolytic activity is thought to be confined locally within the secretory pathway at the cell surface and nearby extracellular space (1–3). Individual MMPs have distinct substrate specificities and act on diverse extracellular and membrane proteins, such as chemokines, cell surface adhesion proteins, and extracellular matrix components. Proteolysis by MMPs plays an important role in a wide variety of normal and pathological processes, such as host defense, inflammation, and tumor progression (1–9).
High levels of MMP-9 (gelatinase B) are expressed by activated macrophages (10), which are key effector cells of both innate and acquired immunity. In addition to having homeostatic functions, MMP-9 secreted by macrophages has been implicated in aneurysm formation, tumor progression, and disruption of atherosclerotic plaques (8, 9, 11, 12). Although the pathogenesis of those processes is generally thought to involve inappropriate degradation of extracellular matrix proteins, it has become increasingly clear that MMPs cleave a number of diverse substrates to mediate their varied functions (3, 13). Because MMP-9 can accumulate on the cell surface (14), it is likely to act on membrane proteins.
To understand the specific roles of individual MMPs in inflammatory and immune responses, it is critical to identify their physiological substrates (3, 15–17). Most studies have focused on identifying substrates by their ability to be cleaved in defined in vitro reactions (18, 19), but this approach is biased in two ways. First, the candidate substrate must be selected a priori. Second, in vitro reactions fail to account for the complexity of the pericellular environment. Another method is to identify sequences in synthetic peptides that MMPs can cleave (20, 21). However, individual MMPs cleave different proteins at a variety of sites rather than at a consensus site. Moreover MMPs often interact with substrates through domains remote from the active site (exosites) (22), and exosites of MMP-2 have been used in a yeast two-hybrid system to trap candidate substrates (23). However, some substrates may bind weakly or not at all to exosites, limiting the utility of this approach for global substrate screening.
An emerging strategy for finding MMP substrates is to conduct an unbiased, global search by coupling gel electrophoresis or liquid chromatography with MS-based protein identification. For example, two-dimensional (2D) gel electrophoresis (24) and derivatization of cysteine-containing peptides with an isotope affinity tag (25) have identified candidate substrates for membrane type-1 MMP (MT1-MMP) in plasma and cultured cells. Quantitative approaches using 2D difference gel electrophoresis have identified potential substrates of MMP-2 and MMP-9 in bronchoalveolar lavage fluid (26) and of MMP-9 and the related metalloproteinases ADAM-10 and ADAM-17 in cancer cells (27, 28). Lectin affinity chromatography detected glycosylated proteins that were selectively enriched in medium from a monocyte cell line expressing ADAM-17 and in phorbol ester-stimulated monocytes (16). Recently iTRAQ (isobaric tags for relative and absolute quantitation) labeling was used to identify substrates of MMP-2 (29). It is important to note, however, that proteases can affect protein abundance by pathways not involving proteolysis. Thus, an important limitation of many of these studies is that they fail to provide evidence that proteins with altered abundance in cells expressing a protease are direct substrates for proteolytic cleavage.
In the current studies, we used subtractive proteomics to identify proteins enriched in the medium of a macrophage cell line. Subtractive proteomics compares two or more proteomes to identify proteins that are specifically enriched or depleted under certain conditions (30, 31). Our biochemical studies confirmed that two integral membrane proteins, amyloid precursor protein (APP) and the β2 integrin subunit (CD18), were shed by macrophages expressing autoactivating MMP-9. We next used a peptide substrate mapping strategy to identify potential MMP-9 cleavage sites in β2 integrin subunit. Targeted MS/MS analysis demonstrated that β2 integrin subunit peptides with the same cleavage site were detected only in the medium of macrophages expressing autoactivating MMP-9, providing strong evidence that β2 integrin is a direct substrate for proteolysis. Our observations indicate that subtractive proteomics in concert with peptide substrate mapping is a robust, high throughput technique for identifying cellular substrates that are proteolytically shed from macrophages.
EXPERIMENTAL PROCEDURES
Cell Culture—
The mouse macrophage cell line RAW264.7 was transduced with high titer retrovirus. Retrovirus was prepared with Phoenix-A packaging cells transfected with the retroviral expression vector pBM-IRES-PURO (32) containing the cDNA for an autoactivating form of mouse MMP-9 (M9A) (33) or with an empty vector control (wild type (WT)) in the first cistron. To select transfected cells, a puromycin resistance gene was inserted into the second cistron. MMP-9 activity in the transduced cell lines was tested by elastin assay as described previously (34).
Macrophage-conditioned Medium—
Transduced macrophages (2 × 107 cells in 150-mm maxiplates) grown to confluence in RPMI 1640 medium supplemented with 10% fetal bovine serum were washed three times with PBS (pH 7.4) and then incubated with 15 ml of serum-free RPMI 1640 medium for 6 or 24 h with or without presence of 50 μm ilomastat (GM6001) (Elastin Products Co., Inc., Owensville, MO). Conditioned medium from the cells was collected, supplemented with 100 μm butylated hydroxytoluene (to inhibit oxidation) and 10 mm EDTA (to inhibit MMP-9 and other metal ion-dependent proteases), and centrifuged for 15 min at 1200 × g. The supernatant was frozen immediately at −80 °C until use.
Protein Isolation—
Macrophage-conditioned medium (15 ml) was supplemented with 0.2% sodium deoxycholate, incubated for 30 min at 4 °C, and adjusted to a final concentration of 20% trichloroacetic acid. After the medium was incubated overnight at 4 °C, it was centrifuged at 15,000 × g for 45 min. The pellet was washed twice with acetone at −20 °C, and the precipitated protein was resolubilized at room temperature in 6 m urea and 50 mm ammonium bicarbonate.
Cell Lysates—
Adherent M9A cells were washed three times with PBS and then incubated on ice for 30 min with lysis buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS supplemented with 1 mm PMSF, 1 mm EDTA, 5 μg/ml aprotinin, and 5 μg/ml leupeptin). The plates were scraped to recover residual cellular material, which was combined with the cell lysates. DNA in the harvested material was sheared by repeated passage through a 25-gauge needle. The lysate was then centrifuged for 15 min at 13,800 × g, and the supernatant was frozen at −80 °C until use. For MS analysis, thawed cell lysates (80 × 106 cells/ml in lysis buffer) were vigorously mixed with an equal volume of lysis buffer. Cellular debris were removed by centrifugation at 15,000 × g for 10 min. Protein concentration was determined using the Bradford assay with albumin as the standard.
Tryptic Digestion—
Proteins were reduced with 10 mm DTT at 37 °C, alkylated with 40 mm iodoacetamide at room temperature in the dark for 45 min, and then digested for 3 h with endo-Lys-C (1:100, w/w; Roche Applied Science). After the protein solution was diluted 1:4 (v/v) with 50 mm ammonium bicarbonate, trypsin (1:50, w/w; Promega) was added, and the sample was digested overnight at 37 °C. For MS analysis, samples were desalted on a C18 solid-phase extraction column (Empore C18HD, 3M Inc.) and eluted with 80% acetonitrile in 0.1% formic acid.
LC-MS—
Peptides were separated with a 2D LC system (Surveyor HPLC, Thermo Fisher) and then introduced into an ion trap mass spectrometer (Deca XP Plus, Thermo Fisher) using ESI (35). For each analysis, the same amount of digested protein was loaded onto the system. In the first dimension, peptides were separated on a strong cation exchange column (BioBasic SCX, 150 × 0.3 mm; Thermo Electron Corp.) with a 12-step salt gradient (0, 10, 20, 30, 40, 50, 60, 70, 100, 200, 500, and 900 mm NH4Cl). In the second dimension, they were separated on a C18 reverse-phase column (Hypersil BioBasic-C18, 5 μm, 100 × 0.3 mm; Thermo Electron Corp.) with a linear gradient of 7–35% acetonitrile in 0.1% formic acid over 90 min. Two reverse-phase columns were alternately loaded to allow simultaneous elution from one column into the mass spectrometer and salt step elution onto the other. MS/MS spectra were acquired in the positive ion mode using data-dependent acquisition with a survey MS scan (m/z 300–1500) followed by MS/MS scans on the four most abundant precursor ions. After two acquisitions of a given precursor ion within 30 s (the average chromatographic peak width), the ion was excluded for 1.2 min.
Targeted LC-MS/MS analyses of candidate MMP-9 substrates were performed on an LTQ linear ion trap instrument (Thermo Fisher) interfaced with a Michrom MS4B HPLC instrument (Michrom Bioresources) containing a nanotrap column (PepTrap, Michrom Bioresources) and a Magic C18AQ analytical column (5 μm, 0.1 × 100 mm; Michrom Bioresources) (35). Tryptic digests (2 μg of protein in 5% acetonitrile, 0.1% formic acid) were loaded onto the trap column and eluted with a linear gradient of 0–37% solvent A (90% acetonitrile, 0.1% formic acid) over 180 min. MS/MS spectra were obtained with a specific ion precursor selection window of 2.5 amu (relative collision energy, 35).
MALDI-TOF and MALDI-TOF/TOF analyses were performed on a 4700 Proteomics Analyzer (Applied Biosystems). MS/MS analysis was performed with a precursor ion resolution of 50 and metastable ion suppression. Samples were desalted with C18 ZipTips (Waters), spotted on the MALDI target plate (2 pmol), dried, and overlaid with matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid in 70% acetonitrile, 0.1% TFA).
Protein Identification—
Proteins were identified with the SEQUEST v2.7 search engine and the UniProt protein database (Swiss-Prot/TrEMBL v.2.0; February 1, 2005) using mouse taxonomy restriction, an enzyme-unrestricted search with a 2.5-amu window for precursor mass, 0.8-amu mass tolerance for fragment ions, fixed cysteine alkylation by iodoacetamide, and variable oxidation of methionine. MS/MS spectra were extracted from original files using Bioworks v3.1 (Thermo Electron) with the default settings. Peptide identifications from SEQUEST database searches were further evaluated using PeptideProphet (36) and compiled into protein identifications using ProteinProphet (37). Only proteins with a ProteinProphet probability of ≥0.96 and ≥2 unique peptides (semitryptic and fully tryptic peptides with PeptideProphet probability of ≥0.90) in two independent analyses were considered as substrate candidates (estimated false discovery rate <5%). For proteins that could not be distinguished from each other because of similar sequence(s), only the isoform assigned the most unique peptides by ProteinProphet was considered in the analysis. Functional analysis of proteins was performed with the UniProt Knowledgebase (v8.0) and Swiss-Prot IDtracker.
Extracted Ion Chromatograms—
Proteins identified as potentially different in relative abundance by spectral counting (38–42) were quantified by extracted ion chromatograms that were manually extracted from MS survey scans (43, 44) of the data-dependent LC-MS/MS data. Extracted ion chromatograms were constructed from the MS data for selected peptides using Xcalibur software (Thermo Electron). Ion chromatograms were reconstructed for the peptide charge state identified by SEQUEST. A wide retention time window in the chromatograms of different samples was examined to ensure that differences in the elution time of peptides did not affect the results. Reconstructed chromatograms for other charge states (up to 3+) were also monitored to ensure that the major charge state of each peptide was used. We also confirmed that each peptide eluted in a single fraction from the strong cation exchange column.
Technical Replicates—
Previous studies have shown that repeated LC-MS/MS analysis of complex biological material yields up to 50% more protein identifications than a single analysis (38, 45). We therefore determined the number of analyses needed to assess the extracellular macrophage proteome by performing replicate analyses on equivalent amounts of total protein from macrophage-conditioned medium. In four independent experiments, a single analysis identified an average of 329 proteins (≥2 unique peptides and a ProteinProphet probability of ≥0.96) in medium from M9A macrophages. Combining two analyses increased the number of protein identifications by 32%. The average number of identified proteins with three and four analyses increased the number by only 13 and 8%, respectively. These observations indicate that duplicate analyses of the same sample give a reasonable estimate of the number of proteins that our 2D LC-MS/MS approach can identify in macrophage medium and that this approach yields reproducible results.
Controlling for Cell Lysis—
Two approaches were used to determine whether the proteins detected in medium might have been released from injured, apoptotic, or necrotic cells. First, the rank order of protein abundance (as assessed by the number of unique peptides detected) in conditioned M9A and WT medium was compared with that in lysates of macrophages expressing autoactivating MMP-9. There was no significant correlation between the medium and lysate samples (ρ = 0.25, Spearman's rank correlation test) even when only the 100 most abundant proteins in the medium were considered (ρ = 0.27). These observations indicate that cellular injury was unlikely to have been a major contributor to the proteins found in macrophage-conditioned medium. The observation that a large number (∼50%) of the proteins detected in medium were undetectable in lysates was consistent with this proposal.
Second, to identify extracellular proteins that were likely derived from injured cells, we devised an empiric relationship, the “medium/lysate ratio,” which we defined as (medium rank/lysate rank)−1. To calibrate the relationship, we calculated the medium/lysate ratio for lactate dehydrogenase (LDH) and aspartate amino acid aminotransferase, two intracellular proteins often used as markers of lysis (46). Lactate dehydrogenase and aspartate amino acid aminotransferase exhibited medium/lysate ratios of 0.9 and 1.2, respectively. Based on this observation, we defined shed and secreted proteins as those with a medium/lysate ratio of >1.5.
Immunoblot Analysis—
Cell lysates and conditioned media were separated by SDS-PAGE under reducing conditions, transferred to PVDF membranes (Immobilon, Millipore, Bedford, MA), and subsequently immunoblotted with specific antibodies before being visualized by chemiluminescence (ECL Plus, Amersham Biosciences). Immunoblot analyses used rabbit antibodies to the N terminus (Sigma) and C terminus (Zymed Laboratories Inc., San Francisco, CA) of β-APP, rat monoclonal anti-β2 integrin (CD18) (BD Pharmingen), and goat anti-murine MMP-9 (R&D Systems, Minneapolis, MN).
Identifying Candidate Proteolytic Cleavage Sites—
An overlapping library of 10 peptides centered on the putative transmembrane domain and adjacent ectodomain region of β2 integrin subunit (residues 651–710) was synthesized (SigmaGenosys). Each peptide had 15 residues with a five-amino acid overlap between peptides. Peptides (28 μg/ml) were individually incubated for 12 h at 37 °C in PBS supplemented with active MMP-9 (250 ng; Calbiochem) in 20 mm phosphate buffer (pH 7.4). At the end of incubation, the sample was desalted using C18 ZipTips (Millipore) and analyzed by MALDI-TOF-MS and LC-MALDI-TOF/TOF-MS/MS.
ELISA—
Conditioned media were analyzed for Aβ1–40 peptide by sandwich ELISA as described previously (47). Rodent Aβ1–40 peptide (Bachem, Inc.) was used as a standard.
Gene Ontology Analysis—
Gene Ontology analysis was performed with DAVID (48) and the BiNGO plug-in module (49) in Cytoscape. Proteins were linked with the current UniProt identification numbers (IDs) and then submitted to analysis (either directly or after conversion to Entrez Gene identifications using the DAVID Gene ID conversion tool). A complete list of UniProt IDs was submitted to the DAVID Gene Functional Annotation Clustering tool. Complete Gene Ontology terms (SP_PIR_KEYWORDS, UP_SEQ_FEATURE, COG_KOG_ONTOLOGY) and medium classification stringency (defined by κ statistics and fuzzy heuristic clustering) were used to classify proteins against the whole mouse genome. Graphical analyses of cellular compartments, biological processes, and molecular functions were performed using Entrez Gene IDs and the BiNGO module of Cytoscape.
Statistical Analysis—
Student's unpaired two-tailed t test and Bolshev's sign test (a Poisson distribution-based non-parametric test) (50) were used to compare the number of unique peptides identified in medium from M9A cells versus WT cells. For all statistical analyses, p < 0.05 was considered significant.
RESULTS
To search for potential substrates of MMP-9, we transduced a mouse macrophage cell line (RAW264.7) with a retrovirus expressing an autoactivating form of MMP-9 (M9A macrophages). Control cells were transduced with an empty vector (WT macrophages). Autoactivating MMP-9 was generated by replacing the glycine residue at position 100 in the prodomain with a leucine residue (33, 51). Using this autoactivating mutant circumvented the need to add exogenous proteases or compounds to activate pro-MMP-9 that could produce nonspecific effects. Because macrophages normally secrete MMP-9, the transduced cells had the mechanisms to accurately deliver the recombinant enzyme to the pericellular compartments in which endogenous MMP-9 proteolysis occurs. Thus, by overexpressing MMP-9 in cells that already express MMP-9, we up-regulated the shedding of authentic substrates, increasing the likelihood of identifying natural substrates. In contrast, exogenous MMP-9 would not be spatially regulated and therefore could act indiscriminately on numerous macrophage proteins.
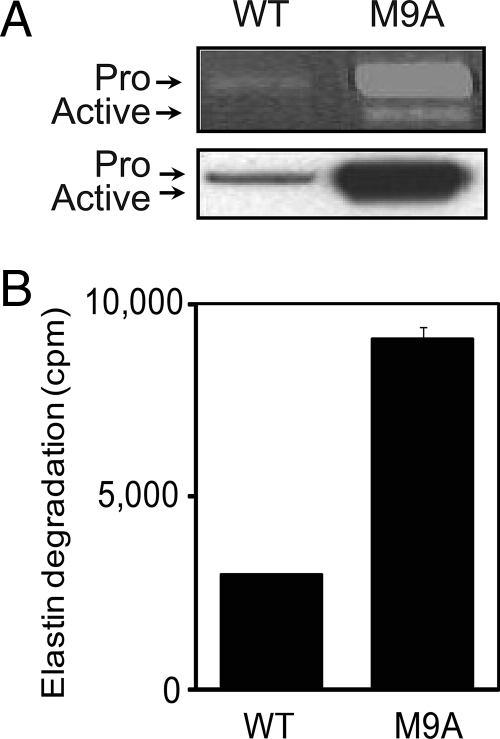
Using gelatin zymography and immunoblotting, we readily detected both pro-MMP-9 and active MMP-9 in conditioned medium from M9A cells. WT macrophages expressed lower levels of pro-MMP-9 and no active MMP-9 (Fig. 1A). Elastin, an established in vitro substrate of MMP-9 (33), was degraded 2.5-fold more rapidly by M9A cells than by WT cells (Fig. 1B). These results indicate that expression of autoactivating MMP-9 modestly increased the level of active enzyme in the medium of M9A cells.
Fig. 1.
Macrophages expressing autoactivating pro-MMP-9 secrete active MMP-9. RAW cells were transduced with retrovirus encoding a puromycin resistance gene (WT) or a puromycin resistance gene plus a gene for autoactivating pro-MMP-9 (M9A). A, equal amounts of protein from cell lysates or conditioned medium were separated by SDS-PAGE and then analyzed by gelatin zymography (upper panel) or immunoblotting with an antibody specific for MMP-9 (lower panel). B, WT and M9A cells were incubated with radiolabeled elastin as described previously (33, 34). At the end of the incubation, the medium was harvested and clarified by centrifugation, and soluble elastin degradation fragments were quantified by scintillation counting (mean ± S.E.). Pro, pro-MMP-9; Active, proteolytically cleaved MMP-9.
LC-MS/MS Identifies a Diverse Array of Proteins in Macrophage-conditioned Medium—
We next searched for transmembrane or membrane-associated proteins whose presence in the medium was enhanced by overexpression of autoactivating MMP-9. Macrophages were incubated for 24 h in serum-free medium, which was then collected and concentrated. A tryptic digest of proteins from the concentrate was analyzed by 2D LC-MS/MS using an ion trap mass spectrometer. Peptides were identified, using strict criteria, by searching MS/MS spectra against the mouse Swiss-Prot/TrEMBL protein database.
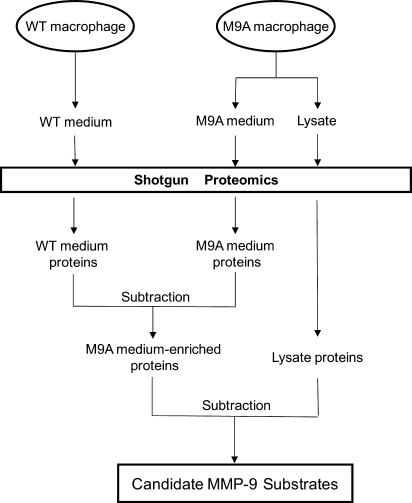
We used a three-tiered strategy to identify shed and secreted proteins that were more abundant in the medium when macrophages overexpressed MMP-9 (Fig. 2). First, we used MS/MS to detect proteins in medium from M9A and WT macrophages. Second, we identified proteins that appeared to be selectively enriched in medium from M9A cells (as judged by the number of unique peptides). Third, we identified proteins that likely were derived from leaky or damaged cells and removed them from the list of candidate substrates.
Fig. 2.
Scheme for identifying candidate MMP-9 substrates. Conditioned medium from WT and M9A cells and lysates from M9A cells were analyzed by 2D LC-MS/MS. Proteins were identified using a peptide probability score of ≥0.90 and a protein probability score of ≥0.96 by the PeptideProphet and ProteinProphet algorithms. We also required that at least two unique peptides be derived from each protein, which had to be detected in at least two of three independent experiments. Proteins potentially derived from leaky or injured cells were removed from the list of proteins in the media. Candidate substrates of MMP-9 were identified using the following criteria: (i) detecting at least 3 times as many unique peptides in medium of M9A cells as in medium of WT cells and (ii) a significant difference in the number of unique peptides as assessed by Student's t test or Bolshev's sign test.
Protein identification was based on the following four criteria: (i) a peptide probability score of ≥0.90 as determined by the PeptideProphet algorithm, (ii) a protein probability score of ≥0.96 as determined by the ProteinProphet algorithm, (iii) detection of at least two unique peptides derived from the protein of interest in a single sample, and (iv) detection of the protein in at least two of three independent experiments.
Our preliminary studies indicated that combining two replicate analyses of macrophage medium gave a reasonable estimate of the number of proteins that 2D LC-MS/MS can identify and that the analyses yielded reproducible results (“Experimental Procedures”). We therefore performed replicate analyses on three independent preparations of serum-free medium incubated with WT or M9A cells for 24 h. With this approach, we detected 467 proteins in conditioned media from WT and/or M9A macrophages (supplemental Table 1). Most of the proteins (∼77%) were present in both samples. However, ∼8% were identified only in medium from M9A cells, and ∼15% were unique to WT medium. Overall >67% of WT proteins and >66% of M9A proteins were found in all three analyses, indicating comparable biological and analytical variability in proteome coverage.
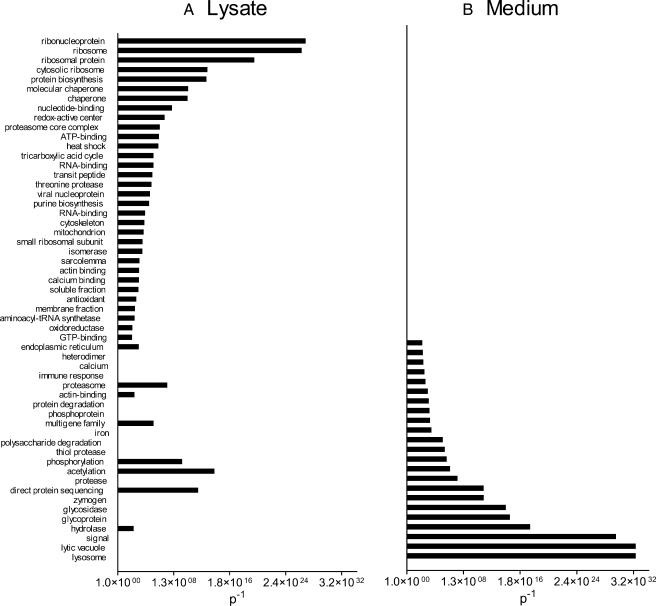
Gene Ontology Analysis Reveals a Distinct Subset of Biological Functions for Shed and Secreted Macrophage Proteins—
We used Gene Ontology annotation (Fig. 3) to organize the proteins found in macrophage-conditioned media and lysates (supplemental Table 2) into functional biologic modules. Proteins in the media, which would represent shed and secreted products, associated strongly with pathways implicated in lysosomal degradation, the secretion pathway (signal peptide-containing proteins), macromolecule degradation, iron metabolism, calcium metabolism, and the immune response (Fig. 3). These modules are consistent with the roles of macrophages in immunity. In contrast, proteins identified in cell lysates segregated into different, non-overlapping functional modules relating to intracellular processes and intermediary metabolism (Fig. 3).
Fig. 3.
Macrophage proteins detected in medium and in lysates represent distinct functional categories. Conditioned medium from M9A and WT cells and lysates from M9A cells were analyzed by 2D LC-MS/MS. Proteins were identified and quantified as described in the legend to Fig. 2. Proteins were ranked by the number of peptides identified, and the medium/lysate ratio was calculated. Functional categories were determined using Gene Ontology analysis for lysate proteins (medium/lysate ratio <0.5) (A) or secreted/shed proteins (medium/lysate ratio >1.5) (B). This approach identified multiple distinct categories of proteins that were significantly overrepresented (categories with p < 0.01; plotted as p−1) in secreted/shed proteins and proteins arising from lysis.
It is worth noting that only 29 of the 467 proteins we detected in medium conditioned by M9A and/or WT macrophages have known or predicted membrane-spanning domains (Table I). Those proteins include the metalloproteases ADAM-8, -10, and -15; several integrins, including α4, αM, and β2; the interleukin 2 receptor; activated leukocyte cell adhesion molecule (CD166/ALCAM); CD44; and syndecan-4. These data suggest that shedding of proteins associated with the plasma membrane accounted for only a fraction of the proteins we detected in medium from M9A macrophages. Interestingly syndecan-4 (52) and CD166 (28) are known substrates of MMP-9.
Table I.
Transmembrane proteins identified in medium conditioned by macrophages
Macrophages were extensively washed to remove serum proteins and then incubated for 24 h in serum-free medium. The medium was harvested and concentrated. Macrophages were lysed with detergent. Desalted tryptic digests of medium or lysate proteins were analyzed by replicate 2D LC-MS/MS analyses. Peptides were identified with SEQUEST and the mouse UniProt protein database using an unrestricted search and a 2.5-amu window for precursor mass, assuming cysteine alkylation and variable oxidation of methionine. Peptide identifications were compiled into protein identifications using ProteinProphet. Protein identification required a ProteinProphet probability ≥0.96 and detection of at least two unique peptides in two of three independent experiments.
| Entry name | Protein name | Total number of unique peptides
|
||
|---|---|---|---|---|
| M9A medium | WT medium | Lysate | ||
| A4_MOUSE | Amyloid-β A4 precursor protein (APP) | 34 | 7 | 0 |
| ADA10_MOUSE | A disintegrin and metallopeptidase domain 10 | 11 | 6 | 0 |
| ADA15_MOUSE | A disintegrin and metallopeptidase domain 15 (metargidin) | 2 | 12 | 0 |
| ADAM8_MOUSE | A disintegrin and metallopeptidase domain 8 | 4 | 6 | 0 |
| APLP2_MOUSE | Amyloid-like protein 2 | 4 | 0 | 0 |
| ATNG_MOUSE | Sodium/potassium-transporting ATPase γ chain | 13 | 8 | 0 |
| CD166_MOUSE | Activated leukocyte cell adhesion molecule | 6 | 3 | 0 |
| CD44_MOUSE | CD44 antigen | 8 | 6 | 0 |
| CSF1R_MOUSE | Colony-stimulating factor 1 receptor | 70 | 60 | 0 |
| CSTN1_MOUSE | Calsyntenin 1 | 16 | 24 | 0 |
| DAG1_MOUSE | Dystroglycan 1 | 9 | 9 | 0 |
| GSLG1_MOUSE | Golgi apparatus protein 1 | 8 | 0 | 0 |
| HA1D_MOUSE | Histocompatibility 2, k1, k region | 7 | 3 | 0 |
| HA1L_MOUSE | Histocompatibility 2, d region locus 1 | 14 | 23 | 0 |
| HG2A_MOUSE | CD74 antigen | 8 | 5 | 0 |
| IGSF8_MOUSE | Immunoglobulin superfamily member 8 (EWI-2, CD316) | 4 | 1 | 0 |
| IL2RG_MOUSE | Interleukin 2 receptor, γ chain | 4 | 6 | 0 |
| ITA4_MOUSE | Integrin α4 | 5 | 10 | 0 |
| ITAM_MOUSE | Integrin αM | 13 | 7 | 1 |
| ITB2_MOUSE | Integrin β2 | 12 | 3 | 1 |
| ITM2B_MOUSE | Integral membrane protein 2b | 2 | 4 | 0 |
| LFNG_MOUSE | β-1,3-N-Acetylglucosaminyltransferase | 9 | 8 | 0 |
| NRP2_MOUSE | Neuropilin 2 | 9 | 6 | 0 |
| RAE1A_MOUSE | Retinoic acid early transcript 1, α | 9 | 0 | 0 |
| SDC4_MOUSE | Syndecan-4 | 5 | 5 | 0 |
| SEM4A_MOUSE | Semaphorin-4a | 7 | 16 | 0 |
| SORL_MOUSE | Sortilin-related receptor | 4 | 9 | 0 |
| ST14_MOUSE | Suppression of tumorigenicity 14 (epithin) | 3 | 9 | 0 |
| VAS1_MOUSE | Vacuolar ATP synthase subunit s1 | 7 | 12 | 0 |
Subtractive Proteomics Identifies Candidate MMP-9 Substrates—
To identify candidate proteins shed specifically by macrophage MMP-9, we used subtractive proteomics (30, 31) to select proteins enriched in medium conditioned by M9A cells. Recent studies strongly suggest that quantifying the number of identified peptides in a shotgun proteomics analysis provides a semiquantitative assessment of relative protein abundance (38, 39, 41, 53, 54). We therefore used the number of unique peptides detected for each protein to quantify the relative abundance of that protein.
Proteins enriched in medium from M9A cells were distinguished by the following criteria: (i) at least 3 times as many unique peptides detected in medium from M9A cells as in medium from WT cells, (ii) subtraction of likely cytosolic proteins (medium/lysate ratio ≤1.5; note that no proteins unique to the M9A cell medium met this criterion), and (iii) a significant difference in the number of unique peptides detected in M9A cells and WT cells as assessed by both Student's t test and Bolshev's sign test. Extracted ion chromatograms (XICs) were used to confirm that peptides derived from candidate proteins were more abundant in medium from M9A macrophages than in medium from WT macrophages (39). XICs were based on precursor ion intensities in the MS survey scan and quantification of peak areas of the peptide ions derived from medium conditioned by WT or M9A macrophages.
Using these criteria, we identified 18 proteins that appeared to be enriched in medium conditioned by M9A cells (Table II). Four of those have a known or predicted transmembrane domain. One protein, retinoic acid early inducible protein 1α (RAET1A), is anchored to the plasma membrane by glycosylphosphatidylinositol. Two of the membrane-associated proteins, APP and the β2 integrin subunit (CD18), are transmembrane proteins implicated in human disease.
Table II.
Candidate MMP-9 substrates identified by subtractive proteomics in macrophage-conditioned medium
Macrophage proteins were identified by MS/MS analysis as described in the legend to Table I. Candidate MMP-9 substrates were identified by requiring (i) at least 3 times as many unique peptides in medium from M9A macrophages as in medium from WT macrophages, (ii) detection of at least two unique peptides in two of three independent analyses, (iii) a medium/lysate ratio of >1.5, and (iv) a significant difference in the number of unique peptides between M9A cells and WT cells as assessed by Student's t test or Bolshev's sign test. Results represent means of three independent experiments. Y, yes; N, no.
| Entry name | Protein name | Average no. of unique peptides
|
Structural annotation
|
Statistical significance
|
|||
|---|---|---|---|---|---|---|---|
| M9A | WT | Signal peptidea | TM domaina | Bolshev's test | t test | ||
| Membrane-associated proteins | |||||||
| GLG1_MOUSE | Golgi apparatus protein 1 precursor (E-selectin ligand 1) (ESL-1) | 2.7 | 0.0 | Y | Y | +b | 0.06 |
| A4_MOUSE | Amyloid-β A4 protein precursor (APP) | 11.3 | 2.3 | Y | Y | + | 0.005 |
| ITGB2_MOUSE | Integrin β2 precursor (CD18) | 4.0 | 1.0 | Y | Y | + | 0.02 |
| IGSF8_MOUSE | Immunoglobulin superfamily member 8, CD316 antigen | 4.0 | 1.0 | Y | Y | + | 0.02 |
| RAE1A_MOUSEc | Retinoic acid early inducible protein 1 α precursor (RAE-1α) | 3.0 | 0.0 | Y | N | + | 0.19 |
| Secreted proteins | |||||||
| HEXB_MOUSE | β-Hexosaminidase β chain precursor | 6.7 | 0.0 | Y | N | + | 0.11 |
| RISC_MOUSE | Retinoid-inducible serine carboxypeptidase precursor | 3.3 | 0.0 | Y | N | + | 0.13 |
| Q61297_MOUSE | Pancreatic α-amylase isozyme | 3.0 | 0.0 | Y | N | + | d |
| ARSB_MOUSE | Arylsulfatase B precursor | 2.7 | 0.0 | Y | N | + | 0.18 |
| MA2B2_MOUSE | Epididymis-specific α-mannosidase precursor | 2.0 | 0.0 | Y | N | + | 0.23 |
| MMP9_MOUSE | Matrix metalloproteinase-9 (MMP-9, gelatinase B) | 70.0 | 2.0 | Y | N | + | 0.00002 |
| Q8BK91_MOUSE | N-Acetylglucosamine-6-sulfatase precursor | 5.7 | 0.7 | ? | N | + | 0.006 |
| CATE_MOUSE | Cathepsin E precursor | 1.7 | 0.3 | Y | N | −b | 0.047 |
| AMYS_MOUSE | α-Amylase, salivary and hepatic precursor | 27.7 | 6.3 | Y | N | + | 0.06 |
| O88325_MOUSE | α-N-Acetylglucosaminidase | 4.3 | 1.0 | Ye | ? | + | 0.38 |
| LICH_MOUSE | Lipase A (cholesteryl esterase) | 6.7 | 1.7 | Y | N | + | 0.31 |
| BGLR_MOUSE | β-Glucuronidase precursor | 14.7 | 4.7 | Y | N | + | 0.21 |
| PRDX4_MOUSE | Peroxiredoxin 4 (Prx-IV) | 8.7 | 2.0 | Yf | N | + | 0.079 |
| PLF4_MOUSE | Platelet factor 4 precursor (PF-4) (CXCL4) | 2.0 | 0.7 | Y | N | − | 0.057 |
| Proteins lacking a signal peptide or membrane-spanning domain | |||||||
| RL6_MOUSE | 60 S ribosomal protein L6 | 2.7 | 0.7 | N | N | − | 0.013 |
Annotation based on UniProt Knowledgebase.
+ or −, passes or fails (Bolshev's sign test at α = 0.05), respectively.
Glycosylphosphatidylinositol-membrane anchored.
No peptide identified in WT cells.
Annotation based on Gene Ontology.
Computational prediction (Phobius and Sig-Pred).
Subtractive proteomics also identified elevated levels of 12 secreted proteins (proteins with known or predicted signal peptides but without transmembrane domains) in medium from M9A cells (Table II), including MMP-9 itself. Platelet factor 4 (PF4), a known substrate of MMP-9 in vitro (18), was also detected, although the difference in levels of unique peptides had borderline significance (p = 0.057, Student's t test).
Because the cells in our model system overexpressed MMP-9 (Fig. 1), pro-MMP-9 was one of the most enriched proteins in medium from M9A macrophages (Table II), being identified by 35 times as many unique peptides as in medium from WT cells. The peptide number for PF4 was 3 times higher in M9A medium than in WT medium, and the enrichment was confirmed by XICs of selected peptides (data not shown). PF4 is generally considered to be a secreted protein that binds with high affinity to cell surface proteoglycans (18). L6, a subunit of the 60S ribosome that lacks a known transmembrane domain or signal peptide, also appeared to be enriched in medium from M9A macrophages. Antimicrobial peptides derived from ribosomes have been identified in the extracellular space (55, 56).
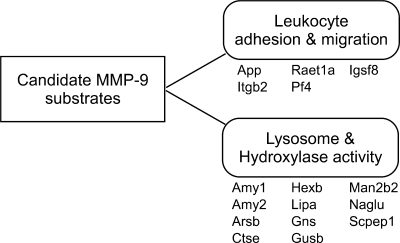
In Gene Ontology analysis of proteins enriched in medium conditioned by M9A cells, increased MMP-9 activity was associated with the presence of proteins involved in leukocyte adhesion and migration, glycosyl bond hydrolysis, and lysosomal functions (Fig. 4). Many of the proteins were hydrolases with amylase, hexosaminidase, or sulfuric ester hydrolase activity being consistent with lysosomal origin (57, 58).
Fig. 4.
Functional analysis of proteins enriched in the medium of M9A macrophages. Conditioned medium from WT and M9A cells was analyzed by 2D LC-MS/MS. Functional categories of proteins enriched in medium from M9A macrophages (Table II) were determined by Gene Ontology analysis. This approach identified two major categories of proteins that were significantly overrepresented (leukocyte adhesion and migration, p = 0.007; lysosome and hydroxylase activity, p = 0.0001 after Benjamini-Hochberg correction) in the medium of macrophages expressing autoactivating MMP-9. Gene name, protein entry ID, protein name (UniProt database) are as follows: App, A4_MOUSE, amyloid protein precursor; Itgb2, ITGB2_MOUSE, integrin β2; Raet1a, RAE1A_MOUSE, retinoic acid early transcript 1, α; Pf4, PLF4_MOUSE, platelet factor 4; Igsf8, IGSF8_MOUSE, immunoglobulin superfamily member 8; Amy1, AMYS_MOUSE, α-amylase, salivary and hepatic; Amy2, Q61297_MOUSE, pancreatic α-amylase isozyme; Arsb, ARSB_MOUSE, arylsulfatase B; Ctse, CATE_MOUSE, cathepsin E; Hexb, HEXB_MOUSE, β-hexosaminidase β chain; Lipa, LICH_MOUSE, lipase A; Gns, Q8BK91_MOUSE, N-acetylglucosamine-6-sulfatase; Gusb, BGLR_MOUSE, β-glucuronidase; Man2b2, MA2B2_MOUSE, epididymis-specific α-mannosidase; Naglu, O88325_MOUSE, α-N-acetylglucosaminidase; Scpep1, RISC_MOUSE, retinoid-inducible serine carboxypeptidase.
Biochemical Confirmation That Overexpression of MMP-9 Induces Shedding of β2 Integrin and Amyloid Protein Precursor—
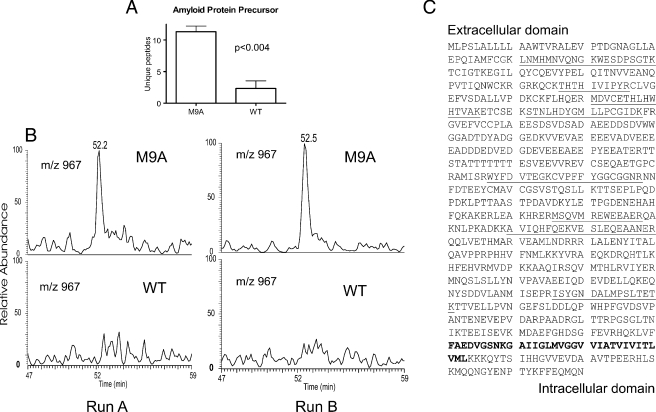
We selected APP and the β2 integrin subunit for detailed biochemical characterization because they are involved in the pathogenesis of human disease. Although MMP-9 was recently implicated in shedding of APP, β2 integrin subunit is not known to be an MMP-9 substrate (59–63). APP is a type I integral membrane protein expressed by macrophages (64); it is proteolytically cleaved to yield amyloid-β peptides. 2D LC-MS/MS identified a mean of 11 unique peptides in three independent preparations of medium exposed to M9A cells for 24 h. In contrast, only three peptides were identified in two of three analyses of WT medium (Fig. 5A). When the incubation period was reduced to 6 h, five unique peptides were identified in medium from M9A macrophages, but none were detected in WT medium. Integrating peak areas of the XIC for multiple different peptide molecular ions yielded similar results as shown in the XICs of peptide STNLHDYGMLLPCGIDK (Fig. 5B; m/z 967, [M + 2H]2+). Only peptides derived from the ectodomain of APP were detected in the medium; this is consistent with proteolytic shedding of APP at the extracellular face of the plasma membrane (Fig. 5C). These observations strongly support the involvement of MMP-9 in a pathway that leads to shedding of APP from M9A macrophages.
Fig. 5.
The ectodomain of APP is shed by macrophages expressing active MMP-9. Conditioned medium from M9A or WT cells was analyzed by 2D LC-MS/MS or immunoblotting. A, the number of unique peptides derived from APP that were detected by MS/MS in conditioned medium incubated with cells for 24 h. Results represent the means and S.D. of three independent experiments. B, XIC for peptide STNLHDYGMLLPCGIDK (m/z 967, [M + 2H]2+) of APP. Results represent three independent experiments. The peptide was detected only in one salt gradient fraction. C, peptides derived from APP identified in medium conditioned by M9A macrophages (underlined). The predicted transmembrane domain is shown in boldface type. Note that only peptides from the extracellular domain were detected.
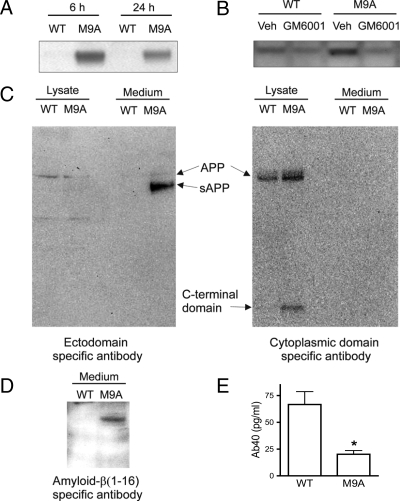
To confirm these observations, we performed immunoblot analysis with antibodies that specifically recognize the N terminus or C terminus of APP or residues 1–16 of amyloid-β peptide (Fig. 6). When medium conditioned by macrophages for 6 or 24 h was probed with an antibody that binds to the N-terminal extracellular domain of APP, a band of ∼100 kDa was readily detected in M9A samples but was much less prominent in the WT samples (Fig. 6A). In addition, the apparent molecular mass of the immunoreactive protein in medium from M9A cells was lower than that of the immunoreactive protein detected in lysates of WT and M9A cells (Fig. 6C, left panel). In contrast, the antibody specific for the C terminus failed to detect protein in medium conditioned by either M9A or WT macrophages, but it did react with full-length APP in both cell populations and a low molecular weight band in M9A lysates (Fig. 6C, right panel). These data show that cleavage of APP leaves a fragment containing the cytoplasmic domain associated with cells. When M9A cells were cultured in the presence of the metalloprotease inhibitor GM6001, APP shedding was significantly inhibited (Fig. 6B).
Fig. 6.
Activated MMP-9 promotes APP shedding and Aβ1–40 degradation by macrophages. Equal amounts of protein from medium conditioned by M9A or WT cells were fractionated by SDS-PAGE and then immunoblotted with antibodies specific for the N-terminal or C-terminal domains of APP or the secreted form of APP containing Aβ1–40 (sAPPα). A, immunoblot analysis of conditioned medium from M9A and WT cells with antibody specific for the N terminus. B, effect of the metalloproteinase inhibitor GM6001 on levels of APP detected with an N terminus-specific antibody in medium conditioned by macrophages for 6 h. C, immunoblot analysis of macrophage-conditioned medium (24 h) with antibodies specific for the ectodomain (N terminus; left) or cytoplasmic domain (C terminus; right). Note that APP was detected in the medium only with the ectodomain antibody and that the band was shifted to a lower apparent molecular weight. In contrast, both antibodies detected equivalent amounts of full-length APP in macrophage lysates. In addition, a low molecular weight protein detected by an antibody to the cytoplasmic domain was significantly elevated in M9A lysates. D, immunoblot analysis of medium conditioned for 24 h by M9A or WT cells with an antibody for the 16-amino acid region contained in sAPPα but absent in sAPPβ. Note that the α-secretase product of APP (sAPPα) is enriched in medium from M9A cells. E, quantification of Aβ1–40 (Ab40) peptide in medium from M9A and WT macrophages. Similar results were observed in three independent analyses. Veh, vehicle.
The two secreted forms of APP, sAPPα and sAPPβ, are generated when α- and β-secretase, respectively, cleave the full-length protein (62). The product of α-secretase is 16 amino acids longer than that of β-secretase. SDS-PAGE and immunoblot analysis with an antibody specific for the region containing those 16 amino acids revealed a single band in medium from M9A cells (Fig. 6D). In contrast, no immunoreactive band was detected in WT medium, suggesting that shedding requires α-secretase activity. Furthermore Aβ1–40 levels were lower in medium from macrophages expressing autoactivating MMP-9 (Fig. 6E). Together the proteomics analysis and biochemical studies provide strong evidence that MMP-9 sheds the ectodomain of APP into the medium, perhaps with α-secretase specificity.
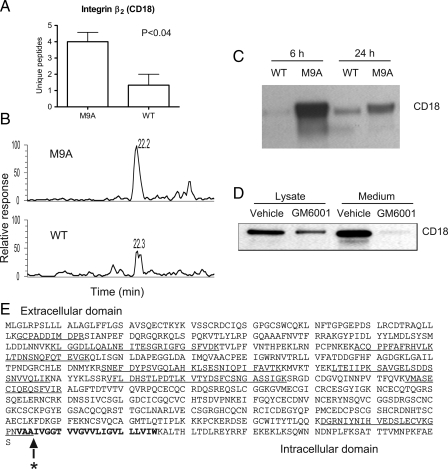
β2 integrin subunit (CD18) forms a heterodimer with various α integrin subunits on leukocytes (65). Like APP, the β2 subunit is a type I transmembrane protein with a large ectodomain and a comparatively small cytosolic domain. All of the β2 peptides detected by 2D LC-MS/MS in M9A and WT medium were derived from the extracellular domain of the subunit (Fig. 7E). In three independent experiments, we observed 4 times as many unique peptides in medium exposed for 24 h to M9A macrophages as in medium from WT macrophages (Fig. 7A). Importantly one of the peptides that was detectable only in the medium of M9A macrophages contained a non-tryptic cleavage site, raising the possibility that it was a proteolytic target for autoactivating MMP-9. Similar results were observed after the macrophages were incubated for 6 h when a mean of 11 unique peptides derived from β2 integrin were detected in M9A medium compared with none in WT medium (three independent analyses).
Fig. 7.
The extracellular domain of β2 integrin subunit (CD18) is shed by macrophages expressing autoactivating MMP-9. A, the number of unique peptides derived from β2 integrin and identified by 2D LC-MS/MS in medium conditioned by M9A macrophages for 24 h. Results represent means and S.D. of three independent experiments. B, XIC for peptide LTDNSNQFQTEVGK (m/z 791.2, [M + 2H]2+) of β2 integrin. The peptide was detected in a single salt gradient fraction. Similar results were observed in three independent experiments. C, immunoblot analysis of β2 integrin in medium conditioned by M9A macrophages for 6 or 24 h. Similar results were observed in three independent experiments. D, effect of metalloproteinase inhibitor GM6001 on β2 integrin shedding. E, identified peptides derived from β2 integrin (underlined) identified in medium from M9A macrophages. Note that all peptides come exclusively from the ectodomain of the protein. The predicted transmembrane domain of the integrin is indicated by boldface type. The star indicates the MMP-9 cleavage site identified by peptide substrate mapping.
β2 integrin can form heterodimers with four different α subunits (66), and more unique peptides derived from one of those α subunits (αM/CD11b) were detected in medium from M9A macrophages compared with WT macrophages. However, the difference did not reach statistical significance. When medium incubated with M9A macrophages for 6 h was analyzed, we detected a mean of 5.4 unique peptides derived from integrin αM in three separate experiments. In contrast, a mean of only 0.2 peptides was detected in medium of WT cells (p = 0.009, Student's t test). These observations suggest that MMP-9 might target the CD11b/CD18 complex for proteolysis.
XICs confirmed that peptides derived from the β2 integrin subunit were more abundant in medium from M9A macrophages than in medium from WT macrophages after either a 6- or 24-h incubation. For example, LTDNSNQFQTEVGK (m/z 791.2, [M + 2H]2+) produced a readily detectable peak in the XIC of medium harvested from M9A macrophages at both time points (Fig. 7B). In contrast, a much smaller peak was observed in medium from WT macrophages, and it was detected only in medium collected at 24 h. We used a β2 integrin-specific antibody to confirm these observations. β2 subunit was markedly more abundant at both 6 and 24 h in M9A-conditioned medium than in medium conditioned by WT cells (Fig. 7C), but protein levels in the cell lysates did not differ (data not shown). Addition of 50 μm GM6001, a hydroxamate compound that inhibits MMPs by binding to the zinc at the active site of the enzyme, significantly decreased the shed levels of the β2 subunit (Fig. 7D). These observations indicate that activated MMP-9 directly or indirectly mediates shedding of β2 integrin subunit from the surface of macrophages.
MMP-9 Directly Cleaves Only One β2 Integrin Juxtamembrane Peptide—
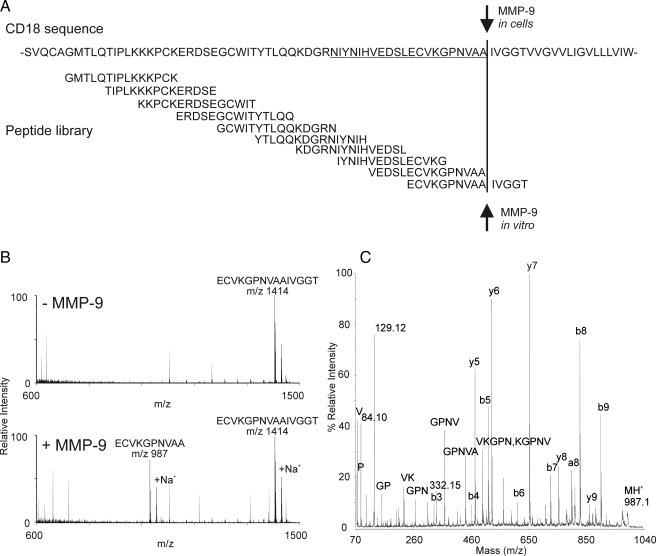
To determine whether β2 integrin subunit is a direct substrate for MMP-9, we synthesized an overlapping library of 10 15-amino acid peptides whose sequences covered the 60 residues adjacent to the extracellular side of the putative transmembrane domain of β2 integrin (Gly651–Thr710; Fig. 8A). Each peptide was incubated with active MMP-9, and the reaction mixture was analyzed by MALDI-TOF-MS and MALDI-TOF-MS/MS.
Fig. 8.
A library of synthetic peptides identifies a candidate MMP-9 cleavage site in β2 integrin. A, a library of 10 overlapping peptides based on the extracellular domain of β2 integrin adjacent to its putative transmembrane region was synthesized. B, each synthetic peptide was incubated in PBS with or without active MMP-9. Reaction mixtures were subjected to MALDI-TOF-MS. This approach demonstrated that a single peptide, ECVKGPNVAAIVGGT, was cleaved proteolytically between Ala and Ile to yield a product peptide of m/z 987. C, MALDI-TOF/TOF analysis of the cleavage product.
Of the 10 peptides in the library, only ECVKGPNVAAIVGGT (where the bold letters indicate the cleavage site) was detectably proteolyzed by active MMP-9 with cleavage between its Ala and Ile residues (Fig. 8B). Those residues correspond to Ala705 and Ile706 in β2 integrin subunit. The identity of the cleavage site was confirmed by MS/MS analysis (Fig. 8C).
We next searched for β2 integrin peptides containing this cleavage site in the medium of M9A and WT cells. Our shotgun proteomics analysis of tryptic digests of M9A medium collected at 24 h had identified a semitryptic peptide, DGRNIYNIHVEDSLECVKGPNVAA (where the underline indicates the sequence shared with the synthetic peptide cleaved by MMP-9), with high confidence as assessed by two different algorithms (PeptideProphet (probability = 1.0, Xcorr = 5.7, ΔCn = 0.424) and MASCOT (MOWSE (molecular weight search) score 40, p < 0.05)). The peptide contained the same C-terminal sequence and the Ala705 non-tryptic cleavage site as the synthetic peptide (ECVKGPNVAAIVGGT) that was cleaved by active MMP-9. Importantly we detected this peptide only in medium from cells expressing autoactivating MMP-9.
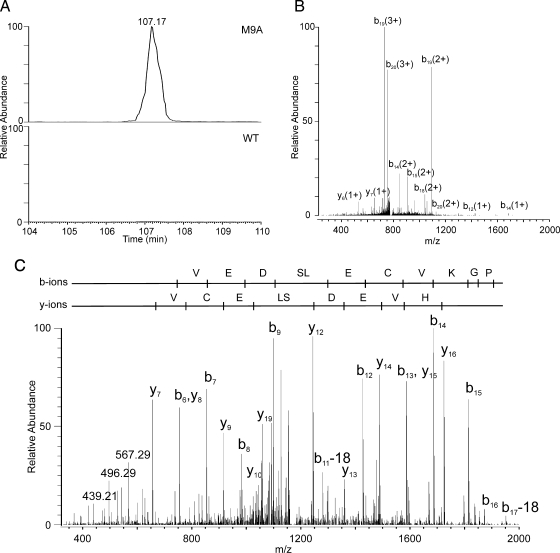
We used targeted LC-MS/MS analysis to search for other proteolytic products derived when MMP-9 cleaved β2 integrin between Ala705 and Ile706. We focused our studies on macrophages incubated for 6 h because medium harvested at this time point contained the highest level of immunoreactive β2 integrin (Fig. 7C). This approach identified two semitryptic peptides ([DGRNIYNIHVEDSLECVKGPNVAA + 3H]3+, [NIYNIHVEDSLECVKGPNVAA + 3H]3+, and [NIYNIHVEDSLECVKGPNVAA + 2H]2+). Both peptides contained the C-terminal Ala residue represented in the synthetic peptide ECVKGPNVAAIVGGT that active MMP-9 cleaved in vitro. Again the peptides were detected in the medium of M9A cells but not in the medium of WT cells. The XICs for NIYNIHVEDSLECVKGPNVAA in conditioned medium harvested from M9A and WT macrophages are shown in Fig. 9A. In two independent experiments, the peptide was detectable only in medium harvested from M9A macrophages. Using MS/MS analysis, we confirmed the identity of the semitryptic peptide in both the triply (Fig. 9B) and doubly (Fig. 9C) charged states. Collectively these observations provide strong evidence that autoactivated MMP-9 can directly shed β2 integrin subunit from the macrophage surface.
Fig. 9.
The β2 integrin subunit of macrophages is a direct proteolytic target for autoactivating MMP-9. Medium conditioned by M9A or WT macrophages for 6 h was monitored for β2 integrin peptides containing the non-tryptic MMP-9 cleavage site identified in the peptide substrate mapping studies (Fig. 8). A, XICs for the semitryptic peptide NIYNIHVEDSLECVKGPNVAA ([M + 3H]3+) in medium from M9A or WT macrophages. Similar results were observed in two independent experiments. B and C, tandem mass spectra of the [M + 3H]3+ ions and [M + 2H]2+ ions of NIYNIHVEDSLECVKGPNVAA in the medium of M9A macrophages. Note the almost complete coverage of b and y ions, confirming the identity of the peptide with high confidence.
DISCUSSION
In a search for cellular substrates of MMP-9, we used subtractive proteomics to identify plasma membrane-associated proteins that had been shed into culture medium of M9A cells, a macrophage cell line expressing an autoactivating form of pro-MMP-9. We focused on proteins with known or predicted transmembrane domains because ectodomain shedding has been implicated in inflammation, wound healing, and host-pathogen interactions (67, 68). Furthermore MMP-9 localizes to the cell surface of activated leukocytes (14), suggesting that it might proteolyze membrane-associated proteins.
Using stringent criteria for protein identification and requiring the detection of ≥3 times as many unique peptides in medium conditioned by M9A cells as in medium from control cells, we identified 18 proteins that appeared to be enriched in the medium of macrophages expressing autoactivating MMP-9 (Table II). The identification of such a limited number of proteins in M9A expressing macrophages suggests that overexpression of active MMP-9 in our system does not result in nonspecific proteolysis. Four of the identified proteins (APP, E-selectin ligand 1, immunoglobulin superfamily member 8, and β2 integrin subunit) have transmembrane domains, and another (retinoic acid early inducible protein 1α) associates with the cell surface via a glycosylphosphatidylinositol anchor. The medium of M9A macrophages was also enriched in proteins with hydrolase, amylase, hexosaminidase, or sulfuric ester hydrolase activity (Fig. 4). The mechanism for enriching lysosomal proteins in the medium of M9A cells is unclear, but this finding raises the possibility that MMP-9 promotes the secretion of lysosomal proteins.
The β2 integrin subunit, E-selectin ligand 1, and immunoglobulin superfamily member 8 (CD316; EWI-2) have all been implicated in cellular adhesion (69–71), suggesting that their shedding might affect macrophage recruitment during inflammation. Moreover natural killer cell activation is regulated by β2 integrin engagement and retinoic acid early inducible protein 1α (72–74). Thus, proteolytic shedding of these two cell surface proteins may modulate the activity of natural killer cells and the interaction of those cells with macrophages. Importantly the relative abundance of these proteins in macrophage medium has not been shown previously to be regulated by MMP-9.
We selected the β2 integrin subunit and APP for detailed biochemical characterization because both are transmembrane proteins that have been implicated in human pathology (64, 75). β2 integrin subunit, also termed CD18, is a glycosylated type I membrane protein that is part of a leukocyte-specific membrane receptor that is critical for host defense (76).
Human leukocytes shed integrin αLβ2 integrin in a model of acute tissue injury (77), but the protease responsible for that cleavage remains unknown. It is therefore of interest that we detected 4 times as many unique β2 integrin peptides in medium from M9A macrophages as in medium from WT macrophages. Moreover all of the detected peptides originated exclusively from the extracellular domain of the integrin. SDS-PAGE and immunoblot analysis with a β2 integrin-specific antibody confirmed that immunoreactive protein was much more abundant in M9A medium than in WT medium and that its abundance in M9A medium was reduced by a metalloprotease inhibitor. These observations suggest that expressing autoactivating pro-MMP-9 in macrophages promotes the shedding of β2 integrin subunit from the cell surface.
We used a two-tiered strategy to demonstrate that β2 integrin is a direct proteolytic substrate for autoactivating MMP-9. First we synthesized a library of 10 peptides with overlapping sequences that spanned 60 residues (Gly651–Thr710) of the transmembrane and extracellular membrane-associated domains of β2 integrin. We incubated each peptide with active MMP-9 and used MALDI-TOF and -TOF/TOF to identify proteolytic cleavage products. This approach revealed a single peptide, ECVKGPNVAAIVGGT, that was cleaved between its Ala and Ile residues. Significantly this site corresponded to a site in a semitryptic peptide that we identified with high confidence in our initial proteomics analysis of M9A medium. Moreover the peptide was detected only in medium from macrophages expressing the autoactivating form of MMP-9, suggesting that the protease was shedding β2 integrin from the cell surface.
To confirm that β2 integrin subunit is a direct substrate for proteolysis by autoactivating MMP-9, we used targeted MS/MS analysis to determine whether macrophage-conditioned medium contained peptides that contained this same cleavage site. Peptides NIYNIHVEDSLECVKGPNVAA and DGRNIYNIHVEDSLECVKGPNVAA were readily identified in the medium of M9A macrophages but were undetectable in the medium of WT macrophages. It is noteworthy that the proteolytic cleavage site for MMP-9 resides within or near the putative transmembrane domain of β2 integrin, indicating that the protease acts at a site very close to the plasma membrane. Collectively our observations provide strong evidence that β2 integrin subunit is a direct target for proteolysis by autoactivating pro-MMP-9.
Other lines of evidence support the proposal that MMPs might promote integrin shedding. For example, both partners of the Mac-1 complex, integrin αMβ2 integrin, bind pro-MMP-9 in vitro (78). Interestingly the active form of MMP-9 lacks this ability (70). It is noteworthy that unique peptides derived from integrin αM were significantly enriched in the medium of M9A following a 6-h incubation, although the difference in peptide abundance was only borderline significant in medium exposed to M9A and WT cells for 24 h. This observation raises the possibility that MMP-9 sheds the αMβ2 integrin complex.
The other candidate MMP-9 substrate we characterized, APP, is the precursor of amyloid-β peptide, the major component of the extracellular plaques found in the brain in Alzheimer disease (79). Recent studies demonstrate that overexpression of activated MMP-9 by HEK/APP695 cells promotes the release of APP (80). Many types of cells, including macrophages, express this glycosylated type I integral membrane protein (64). Our MS/MS analysis detected only peptides derived from its ectodomain in medium from M9A macrophages; this is consistent with proteolytic shedding of APP. Immunoblot analysis demonstrated that an apparently truncated form of APP was much more abundant in M9A medium than in WT medium; this is also consistent with shedding by a protease. Moreover a low molecular weight protein reactive with an antibody to the cytoplasmic domain of APP was detected only in lysates of M9A cells. These observations suggest that cleavage of APP releases the extracellular domain of the protein into the medium, leaving a small fragment containing the cytoplasmic domain associated with the macrophage. Together our proteomics analysis and biochemical studies provide strong evidence that MMP-9 is involved in shedding the APP ectodomain.
Two competing proteolytic pathways lead to shedding of APP from the cell surface. The amyloidogenic pathway, which releases the neurotoxic amyloid-β peptide from APP, involves a β-secretase (an aspartyl protease) and a γ-secretase (62). The non-amyloidogenic α-secretase pathway involves a metalloproteinase, which cleaves APP in the amyloid-β domain and precludes amyloid-β peptide formation. Immunohistochemical studies with antibodies specific for the various regions of APP suggest that MMP-9 may also have α-secretase-like activity. Another metalloproteinase we identified in macrophage-conditioned medium, ADAM-10, has α-secretase activity (81). However, we did not observe a significant difference between the number of unique peptides of ADAM-10 in the medium of control and M9A macrophages. Differential expression or release of ADAM-10 is therefore unlikely to be responsible for increased shedding of APP by M9A cells.
An α-secretase-like cleavage of APP by MMP-9 would reduce amyloid-β levels. In agreement with this proposal, we observed lower amyloid-β levels in M9A medium than in WT medium. Additionally higher levels of amyloid-β peptide levels are seen in the brains of mice deficient in MMP-9 (82). Moreover MMP-9 can proteolytically degrade amyloid-β peptides (63, 82). This raises the intriguing possibility that MMP-9 might reduce cerebral amyloid burden by two complementary mechanisms: by impairing production and by increasing the degradation of amyloid-β peptide (63, 80, 82, 83).
Previous studies have identified a variety of cell surface proteins as substrates of MMP-9, including CD166 (ALCAM) (28), c-kit ligand (84), galectin-3 (85), IL-2 receptor α (86), syndecan-1 and -4 (52), and interleukin-1β (87). Our proteomics studies identified two of those proteins, CD166 and syndecan-4, but their concentrations were similar in M9A and WT media. This discrepancy may reflect a number of factors, including variations in protein expression by different cell types. It is also important to note that data-dependent scanning typically selects the most abundant peptide ions for MS/MS analysis: therefore, protein sampling in complex mixtures is incomplete, and the ability to detect low abundance proteins, such as interleukin-1β, is limited (38).
In summary, our observations indicate that subtractive proteomics, which requires no a priori knowledge of enzyme specificity, in concert with peptide substrate mapping of candidate proteolytic cleavage sites is a powerful strategy for identifying proteolytic substrates in cells. In future studies, it will be of great interest to use both macrophages deficient in MMP-9 and mouse models of inflammation to determine whether shedding of the candidate MMP-9 substrates we have identified is physiologically relevant.
Supplementary Material
Acknowledgments
Mass spectrometry experiments were performed by the Mass Spectrometry Core (Diabetes Endocrinology Research Center) and the Mass Spectrometry Resource (Department of Medicine, University of Washington).
Footnotes
Published, MCP Papers in Press, December 30, 2008, DOI 10.1074/mcp.M800449-MCP200
The abbreviations used are: MMP, matrix metalloproteinase; 2D, two-dimensional; APP, amyloid protein precursor; CD18, β2 integrin subunit; CD11b, integrin αM; XIC, extracted ion chromatogram; ADAM, a disintegrin and a metalloproteinase; WT, wild type; DAVID, Database for Annotation, Visualization, and Integrated Discovery; BiNGO, Biological Networks Gene Ontology tool; ID, identification number; ALCAM, activated leukocyte cell adhesion molecule; PF4, platelet factor 4; sAPP, secreted APP; Aβ, amyloid-β.
This work was supported, in whole or in part, by National Institutes of Health Grants DK002456, P01 HL030086, P01 HL018645, P50 HL073996, P30 ES07083, P30 DK017047, R01 HL081785, R01 HL078527, and N01 HV28179 (to the Seattle Proteome Center).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Woessner, J. F., and Nagase, H. ( 2000) Matrix metalloproteinases and TIMPs, Oxford University Press, Oxford
- 2.Mott, J. D., and Werb, Z. ( 2004) Regulation of matrix biology by matrix metalloproteinases. Curr. Opin. Cell Biol. 16, 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parks, W. C., Wilson, C. L., and Lopez-Boado, Y. S. ( 2004) Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 4, 617–629 [DOI] [PubMed] [Google Scholar]
- 4.Shapiro, S. D. ( 2003) Proteolysis in the lung. Eur. Respir. J. Suppl. 44, 30s–32s [DOI] [PubMed] [Google Scholar]
- 5.Thompson, R. W. ( 1996) Basic science of abdominal aortic aneurysms: emerging therapeutic strategies for an unresolved clinical problem. Curr. Opin. Cardiol. 11, 504–518 [DOI] [PubMed] [Google Scholar]
- 6.Galis, Z. S., and Khatri, J. J. ( 2002) Matrix metalloproteinases in vascular remodeling and atherogenesis: the good, the bad, and the ugly. Circ. Res. 90, 251–262 [PubMed] [Google Scholar]
- 7.Birkedal-Hansen, H., Moore, W. G., Bodden, M. K., Windsor, L. J., Birkedal-Hansen, B., DeCarlo, A., and Engler, J. A. ( 1993) Matrix metalloproteinases: a review. Crit. Rev. Oral Biol. Med. 4, 197–250 [DOI] [PubMed] [Google Scholar]
- 8.Libby, P., and Aikawa, M. ( 1998) New insights into plaque stabilisation by lipid lowering. Drugs 56, Suppl. 1, 9–13, 33 [DOI] [PubMed] [Google Scholar]
- 9.Pepper, M. S. ( 2001) Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler. Thromb. Vasc. Biol. 21, 1104–1117 [DOI] [PubMed] [Google Scholar]
- 10.La Fleur, M., Underwood, J. L., Rappolee, D. A., and Werb, Z. ( 1996) Basement membrane and repair of injury to peripheral nerve: defining a potential role for macrophages, matrix metalloproteinases, and tissue inhibitor of metalloproteinases-1. J. Exp. Med. 184, 2311–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, M. J. ( 1996) Stability and instability: two faces of coronary atherosclerosis. The Paul Dudley White Lecture 1995. Circulation 94, 2013–2020 [DOI] [PubMed] [Google Scholar]
- 12.Thompson, R. W., and Parks, W. C. ( 1996) Role of matrix metalloproteinases in abdominal aortic aneurysms. Ann. N. Y. Acad. Sci. 800, 157–174 [DOI] [PubMed] [Google Scholar]
- 13.Overall, C. M., and Dean, R. A. ( 2006) Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 25, 69–75 [DOI] [PubMed] [Google Scholar]
- 14.Owen, C. A., Hu, Z., Barrick, B., and Shapiro, S. D. ( 2003) Inducible expression of tissue inhibitor of metalloproteinases-resistant matrix metalloproteinase-9 on the cell surface of neutrophils. Am. J. Respir. Cell Mol. Biol. 29, 283–294 [DOI] [PubMed] [Google Scholar]
- 15.Parks, W. C., Lopez-Boado, Y. S., and Wilson, C. L. ( 2001) Matrilysin in epithelial repair and defense. Chest 120, (suppl.) 36S–41S [DOI] [PubMed] [Google Scholar]
- 16.Guo, L., Eisenman, J. R., Mahimkar, R. M., Peschon, J. J., Paxton, R. J., Black, R. A., and Johnson, R. S. ( 2002) A proteomic approach for the identification of cell-surface proteins shed by metalloproteases. Mol. Cell. Proteomics 1, 30–36 [DOI] [PubMed] [Google Scholar]
- 17.Overall, C. M., and Blobel, C. P. ( 2007) In search of partners: linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 8, 245–257 [DOI] [PubMed] [Google Scholar]
- 18.Van den Steen, P. E., Proost, P., Wuyts, A., Van Damme, J., and Opdenakker, G. ( 2000) Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-α and leaves RANTES and MCP-2 intact. Blood 96, 2673–2681 [PubMed] [Google Scholar]
- 19.McQuibban, G. A., Gong, J. H., Wong, J. P., Wallace, J. L., Clark-Lewis, I., and Overall, C. M. ( 2002) Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood 100, 1160–1167 [PubMed] [Google Scholar]
- 20.Park, H. I., Ni, J., Gerkema, F. E., Liu, D., Belozerov, V. E., and Sang, Q. X. ( 2000) Identification and characterization of human endometase (matrix metalloproteinase-26) from endometrial tumor. J. Biol. Chem. 275, 20540–20544 [DOI] [PubMed] [Google Scholar]
- 21.Knauper, V., Cowell, S., Smith, B., Lopez-Otin, C., O'shea, M., Morris, H., Zardi, L., and Murphy, G. ( 1997) The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J. Biol. Chem. 272, 7608–7616 [DOI] [PubMed] [Google Scholar]
- 22.Overall, C. M. ( 2002) Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 22, 51–86 [DOI] [PubMed] [Google Scholar]
- 23.Overall, C. M., McQuibban, G. A., and Clark-Lewis, I. ( 2002) Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biol. Chem. 383, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 24.Hwang, I. K., Park, S. M., Kim, S. Y., and Lee, S. T. ( 2004) A proteomic approach to identify substrates of matrix metalloproteinase-14 in human plasma. Biochim. Biophys. Acta 1702, 79–87 [DOI] [PubMed] [Google Scholar]
- 25.Tam, E. M., Morrison, C. J., Wu, Y. I., Stack, M. S., and Overall, C. M. ( 2004) Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U. S. A. 101, 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenlee, K. J., Corry, D. B., Engler, D. A., Matsunami, R. K., Tessier, P., Cook, R. G., Werb, Z., and Kheradmand, F. ( 2006) Proteomic identification of in vivo substrates for matrix metalloproteinases 2 and 9 reveals a mechanism for resolution of inflammation. J. Immunol. 177, 7312–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, D., Suenaga, N., Edelmann, M. J., Fridman, R., Muschel, R. J., and Kessler, B. M. ( 2008) Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol. Cell. Proteomics 7, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bech-Serra, J. J., Santiago-Josefat, B., Esselens, C., Saftig, P., Baselga, J., Arribas, J., and Canals, F. ( 2006) Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol. Cell. Biol. 26, 5086–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean, R. A., and Overall, C. M. ( 2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 30.Dieguez-Acuna, F. J., Gerber, S. A., Kodama, S., Elias, J. E., Beausoleil, S. A., Faustman, D., and Gygi, S. P. ( 2005) Characterization of mouse spleen cells by subtractive proteomics. Mol. Cell. Proteomics 4, 1459–1470 [DOI] [PubMed] [Google Scholar]
- 31.Schirmer, E. C., Florens, L., Guan, T., Yates, J. R., III, and Gerace, L. ( 2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 32.Garton, K. J., Ferri, N., and Raines, E. W. ( 2002) Efficient expression of exogenous genes in primary vascular cells using IRES-based retroviral vectors. BioTechniques 32, 830, 832, 834 passim [DOI] [PubMed] [Google Scholar]
- 33.Gough, P. J., Gomez, I. G., Wille, P. T., and Raines, E. W. ( 2006) Macrophage expression of active MMP-9 induces acute plaque disruption in apoE-deficient mice. J. Clin. Investig. 116, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shipley, J. M., Wesselschmidt, R. L., Kobayashi, D. K., Ley, T. J., and Shapiro, S. D. ( 1996) Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc. Natl. Acad. Sci. U. S. A. 93, 3942–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaisar, T., Pennathur, S., Green, P. S., Gharib, S. A., Hoofnagle, A. N., Cheung, M. C., Byun, J., Vuletic, S., Kassim, S., Singh, P., Chea, H., Knopp, R. H., Brunzell, J., Geary, R., Chait, A., Zhao, X. Q., Elkon, K., Marcovina, S., Ridker, P., Oram, J. F., and Heinecke, J. W. ( 2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 37.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 38.Liu, H., Sadygov, R. G., and Yates, J. R., III ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 39.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 40.Lu, P., Vogel, C., Wang, R., Yao, X., and Marcotte, E. M. ( 2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 41.Fu, X., Gharib, S. A., Green, P. S., Aitken, M. L., Frazer, D. A., Park, D. R., Vaisar, T., and Heinecke, J. W. ( 2008) Spectral index for assessment of differential protein expression in shotgun proteomics. J. Proteome Res. 7, 845–854 [DOI] [PubMed] [Google Scholar]
- 42.Green, P. S., Vaisar, T., Pennathur, S., Kulstad, J. J., Moore, A. B., Marcovina, S., Brunzell, J., Knopp, R. H., Zhao, X.-Q., and Heinecke, J. W. ( 2008) Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation 118, 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chelius, D., and Bondarenko, P. V. ( 2002) Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Proteome Res. 1, 317–323 [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., Zhou, H., Lin, H., Roy, S., Shaler, T. A., Hill, L. R., Norton, S., Kumar, P., Anderle, M., and Becker, C. H. ( 2003) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818–4826 [DOI] [PubMed] [Google Scholar]
- 45.Resing, K. A., Meyer-Arendt, K., Mendoza, A. M., Aveline-Wolf, L. D., Jonscher, K. R., Pierce, K. G., Old, W. M., Cheung, H. T., Russell, S., Wattawa, J. L., Goehle, G. R., Knight, R. D., and Ahn, N. G. ( 2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem. 76, 3556–3568 [DOI] [PubMed] [Google Scholar]
- 46.Feutren, G., Lacour, B., and Bach, J. F. ( 1984) Immune lysis of hepatocytes in culture: accurate detection by aspartate aminotransferase release measurement. J. Immunol. Methods 75, 85–94 [DOI] [PubMed] [Google Scholar]
- 47.Kulstad, J. J., Green, P. S., Cook, D. G., Watson, G. S., Reger, M. A., Baker, L. D., Plymate, S. R., Asthana, S., Rhoads, K., Mehta, P. D., and Craft, S. ( 2006) Differential modulation of plasma β-amyloid by insulin in patients with Alzheimer disease. Neurology 66, 1506–1510 [DOI] [PubMed] [Google Scholar]
- 48.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. ( 2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 49.Maere, S., Heymans, K., and Kuiper, M. ( 2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 50.Bolshev, L. N. ( 1962) On the comparison of parameters of Poisson distributions. Theory Probab. Appl. 7, 113–114 [Google Scholar]
- 51.Fisher, K. E., Fei, Q., Laird, E. R., Stock, J. L., Allen, M. R., Sahagan, B. G., and Strick, C. A. ( 2002) Engineering autoactivating forms of matrix metalloproteinase-9 and expression of the active enzyme in cultured cells and transgenic mouse brain. Biochemistry 41, 8289–8297 [DOI] [PubMed] [Google Scholar]
- 52.Brule, S., Charnaux, N., Sutton, A., Ledoux, D., Chaigneau, T., Saffar, L., and Gattegno, L. ( 2006) The shedding of syndecan-4 and syndecan-1 from HeLa cells and human primary macrophages is accelerated by SDF-1/CXCL12 and mediated by the matrix metalloproteinase-9. Glycobiology 16, 488–501 [DOI] [PubMed] [Google Scholar]
- 53.Washburn, M. P., Ulaszek, R. R., and Yates, J. R., III ( 2003) Reproducibility of quantitative proteomic analyses of complex biological mixtures by multidimensional protein identification technology. Anal. Chem. 75, 5054–5061 [DOI] [PubMed] [Google Scholar]
- 54.Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 55.Fernandes, J. M., and Smith, V. J. ( 2002) A novel antimicrobial function for a ribosomal peptide from rainbow trout skin. Biochem. Biophys. Res. Commun. 296, 167–171 [DOI] [PubMed] [Google Scholar]
- 56.Hiemstra, P. S., van den Barselaar, M. T., Roest, M., Nibbering, P. H., and van Furth, R. ( 1999) Ubiquicidin, a novel murine microbicidal protein present in the cytosolic fraction of macrophages. J. Leukoc. Biol. 66, 423–428 [DOI] [PubMed] [Google Scholar]
- 57.Stinchcombe, J. C., and Griffiths, G. M. ( 1999) Regulated secretion from hemopoietic cells. J. Cell Biol. 147, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stinchcombe, J. C., and Griffiths, G. M. ( 2001) Normal and abnormal secretion by haemopoietic cells. Immunology 103, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sondag, C. M., and Combs, C. K. ( 2004) Amyloid precursor protein mediates proinflammatory activation of monocytic lineage cells. J. Biol. Chem. 279, 14456–14463 [DOI] [PubMed] [Google Scholar]
- 60.Stefanidakis, M., and Koivunen, E. ( 2006) Cell-surface association between matrix metalloproteinases: role of the complexes in leukocyte migration and cancer progression. Blood 108, 1441–1450 [DOI] [PubMed] [Google Scholar]
- 61.Postina, R. ( 2008) A closer look at α-secretase. Curr. Alzheimer Res. 5, 179–186 [DOI] [PubMed] [Google Scholar]
- 62.De Strooper, B., and Annaert, W. ( 2000) Proteolytic processing and cell biological functions of the amyloid precursor protein. J. Cell Sci. 113, 1857–1870 [DOI] [PubMed] [Google Scholar]
- 63.Backstrom, J. R., Lim, G. P., Cullen, M. J., and Tokes, Z. A. ( 1996) Matrix metalloproteinase-9 (MMP-9) is synthesized in neurons of the human hippocampus and is capable of degrading the amyloid-β peptide (1–40). J. Neurosci. 16, 7910–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bauer, J., Konig, G., Strauss, S., Jonas, U., Ganter, U., Weidemann, A., Monning, U., Masters, C. L., Volk, B., Berger, M., and Beyreuther, K. ( 1991) In-vitro matured human macrophages express Alzheimer's β A4-amyloid precursor protein indicating synthesis in microglial cells. FEBS Lett. 282, 335–340 [DOI] [PubMed] [Google Scholar]
- 65.Ley, K., Laudanna, C., Cybulsky, M. I., and Nourshargh, S. ( 2007) Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 66.Mazzone, A., and Ricevuti, G. ( 1995) Leukocyte CD11/CD18 integrins: biological and clinical relevance. Haematologica 80, 161–175 [PubMed] [Google Scholar]
- 67.Garton, K. J., Gough, P. J., and Raines, E. W. ( 2006) Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J. Leukoc. Biol. 79, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 68.Huovila, A. P., Turner, A. J., Pelto-Huikko, M., Karkkainen, I., and Ortiz, R. M. ( 2005) Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 69.Mayadas, T. N., and Cullere, X. ( 2005) Neutrophil β2 integrins: moderators of life or death decisions. Trends Immunol. 26, 388–395 [DOI] [PubMed] [Google Scholar]
- 70.Wild, M. K., Huang, M. C., Schulze-Horsel, U., van der Merwe, P. A., and Vestweber, D. ( 2001) Affinity, kinetics, and thermodynamics of E-selectin binding to E-selectin ligand-1. J. Biol. Chem. 276, 31602–31612 [DOI] [PubMed] [Google Scholar]
- 71.Stipp, C. S., Kolesnikova, T. V., and Hemler, M. E. ( 2003) EWI-2 regulates α3β1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerwenka, A., Bakker, A. B., McClanahan, T., Wagner, J., Wu, J., Phillips, J. H., and Lanier, L. L. ( 2000) Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 12, 721–727 [DOI] [PubMed] [Google Scholar]
- 73.Bryceson, Y. T., March, M. E., Ljunggren, H. G., and Long, E. O. ( 2006) Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamerman, J. A., Ogasawara, K., and Lanier, L. L. ( 2004) Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J. Immunol. 172, 2001–2005 [DOI] [PubMed] [Google Scholar]
- 75.Weber, C. ( 2003) Novel mechanistic concepts for the control of leukocyte transmigration: specialization of integrins, chemokines, and junctional molecules. J. Mol. Med. 81, 4–19 [DOI] [PubMed] [Google Scholar]
- 76.Lee, S. H., and Corry, D. B. ( 2004) Homing alone? CD18 in infectious and allergic disease. Trends Mol. Med. 10, 258–262 [DOI] [PubMed] [Google Scholar]
- 77.Evans, B. J., McDowall, A., Taylor, P. C., Hogg, N., Haskard, D. O., and Landis, R. C. ( 2006) Shedding of lymphocyte function-associated antigen-1 (LFA-1) in a human inflammatory response. Blood 107, 3593–3599 [DOI] [PubMed] [Google Scholar]
- 78.Stefanidakis, M., Ruohtula, T., Borregaard, N., Gahmberg, C. G., and Koivunen, E. ( 2004) Intracellular and cell surface localization of a complex between αMβ2 integrin and promatrix metalloproteinase-9 progelatinase in neutrophils. J. Immunol. 172, 7060–7068 [DOI] [PubMed] [Google Scholar]
- 79.Hardy, J., and Selkoe, D. J. ( 2002) The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 80.Talamagas, A. A., Efthimiopoulos, S., Tsilibary, E. C., Figueiredo-Pereira, M. E., and Tzinia, A. K. ( 2007) Aβ(1–40)-induced secretion of matrix metalloproteinase-9 results in sAPPα release by association with cell surface APP. Neurobiol. Dis. 28, 304–315 [DOI] [PubMed] [Google Scholar]
- 81.Allinson, T. M. J., Parkin, E. T., Turner, A. J., and Hooper, N. M. ( 2003) ADAMs family members as amyloid precursor protein α-secretases. J. Neurosci. Res. 74, 342–352 [DOI] [PubMed] [Google Scholar]
- 82.Yin, K. J., Cirrito, J. R., Yan, P., Hu, X., Xiao, Q., Pan, X., Bateman, R., Song, H., Hsu, F. F., Turk, J., Xu, J., Hsu, C. Y., Mills, J. C., Holtzman, D. M., and Lee, J. M. ( 2006) Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-β peptide catabolism. J. Neurosci. 26, 10939–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan, P., Hu, X., Song, H., Yin, K., Bateman, R. J., Cirrito, J. R., Xiao, Q., Hsu, F. F., Turk, J. W., Xu, J., Hsu, C. Y., Holtzman, D. M., and Lee, J. M. ( 2006) Matrix metalloproteinase-9 degrades amyloid-β fibrils in vitro and compact plaques in situ. J. Biol. Chem. 281, 24566–24574 [DOI] [PubMed] [Google Scholar]
- 84.Heissig, B., Hattori, K., Dias, S., Friedrich, M., Ferris, B., Hackett, N. R., Crystal, R. G., Besmer, P., Lyden, D., Moore, M. A., Werb, Z., and Rafii, S. ( 2002) Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell 109, 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ortega, N., Behonick, D. J., Colnot, C., Cooper, D. N., and Werb, Z. ( 2005) Galectin-3 is a downstream regulator of matrix metalloproteinase-9 function during endochondral bone formation. Mol. Biol. Cell 16, 3028–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheu, B. C., Hsu, S. M., Ho, H. N., Lien, H. C., Huang, S. C., and Lin, R. H. ( 2001) A novel role of metalloproteinase in cancer-mediated immunosuppression. Cancer Res. 61, 237–242 [PubMed] [Google Scholar]
- 87.Ito, A., Mukaiyama, A., Itoh, Y., Nagase, H., Thogersen, I. B., Enghild, J. J., Sasaguri, Y., and Mori, Y. ( 1996) Degradation of interleukin 1β by matrix metalloproteinases. J. Biol. Chem. 271, 14657–14660 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.