Abstract
Proteins that exhibit different expression levels in normal and malignant lung cells are good candidate biomarkers to improve early diagnosis and intervention. We used a quantitative approach and compared the proteome of microdissected cells from normal human bronchial epithelium and squamous cell carcinoma tumors of histopathological grades G2 and G3. DIGE analysis and subsequent MS-based protein identification revealed that 32 non-redundant proteins were differentially regulated between the respective tissue types. These proteins are mainly involved in energy pathways, cell growth or maintenance mechanisms, protein metabolism, and the regulation of DNA and RNA metabolism. The expression of some of these proteins was analyzed by immunohistochemistry using tissue microarrays containing tissue specimen of 55 patients, including normal bronchial epithelium, squamous cell carcinomas, adenocarcinomas, and large cell carcinomas. The results of the immunohistochemical studies correlated with the proteome study data and revealed that particularly HSP47 and a group of cytokeratins (i.e. cytokeratins 6a, 16, and 17) are significantly co-regulated in squamous cell carcinoma. Furthermore cytokeratin 17 showed significantly higher abundance in G2 grade compared with G3 grade squamous cell carcinomas in both the gel-based and the immunohistochemical analysis. Therefore this protein might be used as a marker for stratification between different tumor grades.
Lung cancer is one of the most frequent cancer types and both in Europe and the United States the main cause of cancer-related mortality. In 2004 lung cancer caused 20% of all cancer-related deaths in Europe and 29% in the United States (1, 2). Lung tumors are classified into two major subtypes: small cell lung carcinoma and non-small cell lung carcinoma (NSCLC).1 NSCLC comprises squamous cell carcinoma (SCC), lung adenocarcinoma, and large cell carcinoma (LCC). The two main groups differ in growth and treatment characteristics. Small cell lung carcinoma tumors exhibit an aggressive phenotype susceptible to chemo- and radiotherapy, whereas NSCLCs are not chemosensitive and are commonly treated by surgery. SCC accounts for about 30–50% of all lung cancers cases and mainly originates from bronchial epithelium cells. Its carcinogenesis is thought to be a multistep process involving intermediate states such as metaplasia and dysplasia. Metaplastic changes are often induced by external stimuli like e.g. tobacco smoke (3). The differentiation of squamous cells starts terminally and is followed by the expression of cornified envelope precursor proteins and finally cornification (4). So far, tumorigenesis is still not well understood, but different tumor types apparently originate from different cell types. The progress in curative treatment over the last 20 years has been modest. Early detection and typing of lung cancers could cause a reduction of cancer mortality (5). Therefore specific biomarkers for efficient histological and in vitro diagnosis are highly needed.
The discovery of new tumor-associated or tumor-specific proteins could serve as a source for new diagnostic markers in serum or sputum as well as for targeted therapies. In the past, a number of efforts have been made to define specific tumor-associated proteins as diagnostic biomarkers in serum and sputum of lung cancer patients (for a review, see Ref. 6). Some groups screened lung cancer cell lines (7), whereas others used tumor tissue samples or body liquids such as sputum, blood, or pleural effusions (8–11). Besides different kinds of analyzed samples, a wide variety of methods was used to find new tumor biomarkers. Some groups used DNA- or mRNA-based technologies, but there are several reasons to apply a protein-based approach for the identification of potential tumor markers. Proteins are the functional mediators in biology and therefore are representative for the biological phenotype. Proteins are more diverse than DNA or RNA and are responsible for the huge complexity in a biological system. Alternative splicing and more than 100 post-translational modifications result in approximately 100 different proteins derived from a single gene. Furthermore many cellular events are mediated post-transcriptionally and cannot be predicted from the nucleic acid level (12).
A problem concerning the identification of lung cancer-specific proteins is the heterogeneity of cell types in lung cancer tissue. Therefore lung cancer SCC was sampled by microdissection by isolating cells from histologically defined areas to obtain material as homogeneous as possible. Unfortunately microdissection provides only a limited sample amount for subsequent proteomics analysis. We recently established a method combining two-dimensional electrophoresis (2DE) and highly sensitive DIGE saturation labeling for the analysis of limited amounts of sample obtained by microdissection (13). This technique is based on labeling proteins with fluorescence cyanine dyes (Cy3 and Cy5) via reduced thiol groups of cysteines. Here we report on the proteome analysis of microdissected material to monitor protein changes in SCC tumor progression. Confirmation of the aberrant abundance level of some of these proteins by immunohistochemistry on tissue arrays resulted in a selection of biomarker candidates to be used as SCC markers for tumor grades and diagnosis.
EXPERIMENTAL PROCEDURES
Microdissection—
The tissue samples were obtained by surgery from patients (17 male and four female) with ages between 34 and 82 years at the Department of Pathology at the University of Kiel, Germany (see supplemental Table 1) according to the permission of the local ethics committee (Number 110/99). Neoplastic and non-neoplastic peritumoral samples were immediately placed on ice, snap frozen, and stored at −80 °C. For the classification of tumors, 5-μm frozen sections prepared from tissue blocks were briefly placed in ethanol (Merck), stained with hematoxylin and eosin, and subsequently histologically analyzed by a pathologist (B. Sipos). Bronchial epithelium originated from peritumoral lung tissues from patients bearing various lung tumors (supplemental Table 1). Those samples were not matched to the G2 and G3 tumor cases. Tissue blocks containing the required tumor grades (G2 or G3) or tissue containing non-neoplastic bronchial epithelium were serially sectioned (10-μm sections). Tumor grading was performed according to Colby et al. (14). For the two-dimensional (2D) DIGE, tissue slices were stained with hematoxylin only and immediately stored at −20 °C. Tumor material and bronchial epithelium from 15–25 serial sections (10 μm) were manually microdissected under a microscope (BH2, Olympus, Wetzlar, Germany) to collect 5000 cells using a sterile injection needle (size, 0.65 × 25 mm; B. Braun, Melsungen, Germany). The number of cells to be collected by microdissection was estimated as follows. Tumor cells and bronchial epithelium cells were counted on a series of slices, and the areas comprising 5,000 cells were measured. These areas were used as a basis for the estimation of cell numbers for collecting cells during routine microdissection. Microdissected cells were collected in 50 μl of lysis buffer (30 mm Tris-Cl, 2 m thiourea, 7 m urea, 4% CHAPS, pH 8.0) and sonicated on ice (6 × 10-s pulses; ultrasonic cleaner, VWR, Darmstadt, Germany) immediately after collection.
Sample Preparation of the Reference Proteome—
To generate a reference proteome frozen tumor tissue slices were used. 25-μm-thick slices were produced that were not stained but homogenized as a whole in lysis buffer (2.4 μl of buffer/mg of tissue; 30 mm Tris-Cl, 2 m thiourea, 7 m urea, 4% CHAPS, pH 8) using a hand homogenizer. After sonication (6 × 10-s pulses on ice) and centrifugation (12,000 × g for 5 min) to remove DNA and cell debris, the protein concentration of the lysate was determined using the Bio-Rad Protein Assay. Because of limited sample access at the starting point of our study we generated our reference proteome from only four G2 and four G3 tumor samples in contrast to the classical procedure to prepare an internal standard (pool of all samples).
Protein Labeling—
For the reduction of protein cysteines 50 μl of cell lysate (from 5000 microdissected cells; ∼4 μg of protein) was incubated with 2 nmol of tris(2-carboxyethyl)phosphine hydrochloride (TCEP; Sigma) at 37 °C in the dark for 1 h. Saturation CyDyes (GE Healthcare) were diluted with anhydrous N,N-dimethylformamide pro analysi (2 nmol/μl; Sigma), and 4 nmol of CyDye was added to the TCEP-reduced sample. The samples were vortexed, centrifuged briefly, and left at 37 °C in the dark for 30 min. The labeling reaction was stopped by adding 5 μl of DTT (1.08 g/ml; Bio-Rad), and finally, before isoelectric focusing 5 μl of SERVALYT 2–4 (SERVA, Heidelberg, Germany) was added.
For the internal standardization 5 μg of the reference proteome sample was labeled with 4 nmol of Cy3 (2 nmol of TCEP). After stopping the labeling reaction with 10 μl of DDT the internal standard was mixed with the labeled microdissected sample and subjected to 2DE. For preparative gels, 400 μg of tissue lysate was labeled with 320 nmol of Cy3 (160 nmol of TCEP).
For optimizing the DIGE saturation labeling conditions we performed a series of same/same experiments as described elsewhere (13). Briefly we optimized the labeling conditions to circumvent over- or underlabeling by applying 1–4 nmol of TCEP and 2–8 nmol of CyDye (Cy3 and Cy5) and tested for identical electrophoretic mobility (overlay).
2D Gel Electrophoresis—
Carrier ampholyte-based IEF was performed in a self-made IEF chamber using tube gels (20 cm × 1.5 mm) as described elsewhere (15). Briefly after running a 21.25-h voltage gradient, the ejected tube gels were incubated in equilibration buffer (125 mm Tris, 40% (w/v) glycerol, 3% (w/v) SDS, 65 mm DTT, pH 6.8) for 10 min. The second dimension was performed in a Desaphor VA 300 system using polyacrylamide gels (15.2% total acrylamide, 1.3% bisacrylamide) as described elsewhere (15). Therefore, the IEF tube gels were placed onto the polyacrylamide gels (20 cm × 30 cm × 1.5 mm) and fixed using 1.0% (w/v) agarose containing 0.01% (w/v) bromphenol blue dye (Riedel deHaen, Seelze, Germany). For protein identification the preparative sized gel system (IEF, 20 cm × 1.5 mm; SDS-PAGE, 20 cm × 30 cm × 1.5 mm) was applied under identical conditions. Silver poststaining was performed after gel scanning using an MS-compatible protocol as described elsewhere (13).
Scanning and Image Analysis—
After 2DE, the gels were left between the glass plates, and images were acquired using the Typhoon 9400 scanner (GE Healthcare). Excitation wavelengths and emission filters were chosen specifically for each of the CyDyes according to the Typhoon user guide. Before image analysis with the DeCyder 2D 6.0 software (GE Healthcare) the images were cropped with ImageQuant™ software (GE Healthcare). The intragel spot detection was performed using the differential in-gel analysis mode of the DeCyder software, applying a spot detection algorithm co-detecting spots in the Cy3 channel and Cy5 channel spot maps. The estimated number of spots was set to 3000. An exclusion filter was applied to remove spots with a slope greater than 1.6. Then the DeCyder biological variation analysis module was used for matching spots between gels: one gel was selected as “master”, and the software mapped the other spot maps to this “master gel” facilitated by the internal standard (Cy3 channel spot map). The spot numbers of the master gel are used in the text, tables, and figures. To quantify spot volumes and perform the statistical analysis of spot volume data, the software algorithm first applies a normalization procedure resulting in normalized spot volumes for each spot map. Based on these normal volumes, standardization was performed by building ratios between the Cy5 channel and Cy3 channel (internal standard) of each spot pair. The resulting normalized and standardized spot volumes were used for further calculations: the mean volumes of matched spots were calculated for each group (G2, G3, and bronchus) and provided the basis for building spot volume ratios (G2/bronchus, G3/bronchus, G2/G3, and G2 and G3/bronchus). If this number was less than 1 the negative reciprocal is listed (e.g. 0.25 is reported as −4-fold change). Student's t tests (unpaired) were calculated on the basis of logarithmized, normalized, and standardized spot volumes by the DeCyder biological variation analysis software module. As for the -fold change, the t tests were calculated independently for the comparison of two groups (G2/bronchus, G3/bronchus, G2/G3, and G2 and G3/bronchus). Protein spots were considered as differentially regulated if the -fold change value (quotient of corresponding group means) for either the G2/bronchus or G3/bronchus or both comparisons was >3.4 or <−3.4. As an additional criterion the corresponding t test had to result in a p value <0.05.
In-gel Tryptic Digestion and Protein Identification Using MALDI-TOF-MS and Nano-HPLC/ESI-MS/MS—
Directly after gel scanning the spots of interest were manually isolated from the preparative gel and in-gel digested with trypsin (Promega, Mannheim, Germany) in 10 mm ammonium bicarbonate buffer (pH 7.8) at 37 °C overnight (16, 17).
For MALDI MS analyses, tryptic peptides were extracted from the gel matrix (17) and prepared on the MALDI target using the AnchorChip™ technique (Bruker Daltonics, Bremen, Germany) according to the manufacturer's instructions with α-cyano-4-hydroxycinnamic acid as MALDI matrix. MALDI-TOF-MS analyses were performed on an UltraFlex™ II (Bruker Daltonics) instrument according to the manufacturer's instructions. The instrument was equipped with a scout MTP™ MALDI target. The spectra were acquired in the positive ion mode according to the settings given by the manufacturer. For external calibration, a peptide standard (m/z 757.399, 1296.684, 1619.822, 2093.086, and 3147.471) was used. The MALDI-peptide mass fingerprint spectra were processed using the FlexAnalysis™ 2.4 software (Bruker Daltonics) and converted in the .xml format. For peak detection, the spectra were subjected to an internal recalibration using 13 different monoisotopic masses from autolysis products of trypsin and fragments of keratins ranging from m/z 842.509 to 2825.406 (supplemental Table 2a). The following parameters were applied: snap peak detection algorithm; signal to noise threshold, 6; maximal number of peaks, 100; quality factor threshold, 50; and base-line subtraction, TopHat. The generated mass lists were subsequently sent to the ProteinScape™ 1.3 proteome database (Bruker Daltonics), triggering database searches using ProFound (Knexus 2001.09.15) (18) and MASCOT (release version 2.2.0) (19).
The following search parameters were selected: cysteine modification with propionamide, variable modification due to methionine oxidation, one maximal missed cleavage site in the case of incomplete trypsin hydrolysis, and no details about 2DE-derived protein mass and pI. Using the Score booster function of ProteinScape the mass lists were recalibrated, and background masses were removed using a list containing 42 masses occurring in 10% of generated peak lists (supplemental Table 2b). The database searches were run with a mass tolerance of 50 ppm using a human International Protein Index (Human IPI v.3.30) decoy database containing 135,844 protein entries. This database is a composite database consisting of the human IPI and a duplicate of the same database in which the amino acid sequence of each protein entry was randomly shuffled (20). Species restriction in database searches to human was justified by the fact that human tissue specimens were analyzed. Proteins reaching a Profound score >1.5 and MASCOT score >65 were considered as identified. Using these criteria no decoy database entry was found by the search engines indicating high confidence of protein identifications. If several database entries of homologous proteins matched these criteria only the entry with the highest score is reported. Protein spots that were not successfully identified by MALDI-MS were additionally analyzed by nano-HPLC-ESI-MS/MS to readily obtain sequence-specific data of tryptic peptides that improves the probability of protein identification.
For nano-HPLC-ESI-MS/MS analyses, the generated tryptic peptides were extracted twice with 10 μl of ACN, 5% formic acid (50:50, v/v). The resulting extracts were combined before the sample volume was reduced in vacuo, and samples were acidified by addition of 5% formic acid to a final volume of 20 μl. Online reversed-phase capillary HPLC separations were performed using HPLC systems (Dionex LC Packings, Idstein, Germany) as described previously (16). ESI-MS/MS analyses were performed on a Bruker Daltonics HCT plus ion trap instrument equipped with a nanoelectrospray ion source (Bruker Daltonics) and distal coated SilicaTips (FS360-20-10-D; New Objective, Woburn, MA). The instrument was externally calibrated with standard compounds. The general mass spectrometric parameters were as follows: capillary voltage, 1400 V; plate offset, 500 V; dry gas, 10.0 liters/min; dry temperature, 160 °C; aimed ion charge control, 150,000; maximal fill time, 500 ms. For MS/MS analyses, data-dependent software (HCT plus, Esquire Control, Bruker Daltonics) was used. To generate fragment ions, low energy CID was performed on isolated multiply charged peptide ions with a fragmentation amplitude of 0.6 V. Exclusion limits were automatically placed on previously selected mass-to-charge ratios for 1.2 min. MS spectra were a sum of seven individual scans ranging from m/z 300 to 1500 with a scanning speed of 8100 (m/z)/s, whereas MS/MS spectra were a sum of four scans ranging from m/z 100 to 2200 at a scan rate of 26,000 (m/z)/s.
Peak lists of MS/MS spectra acquired on the HCT ion trap (Bruker Daltonics) were generated using the software tool DataAnalysis 3.3 with default parameters. For peptide and protein identification, peak lists were correlated with the human International Protein Index (human IPI v.3.30 decoy database) using the ProteinScape (version 1.3, Bruker Daltonics) database platform and both SEQUEST™ (21, 22) (from Bioworks 3.2 with the algorithm TurboSEQUEST-PVM Slave v.27) and MASCOT (19) algorithms.
All searches were performed with tryptic specificity allowing two missed cleavages. Cysteine modification with propionamide was considered as a fixed modification, and oxidation of methionine was considered as a variable modification. Ion trap MS/MS spectra were accepted with cutoff scores of 55 (MASCOT) and 5.5 (SEQUEST) as well as mass tolerances of 1.2 and 0.4 Da for MS and MS/MS experiments, respectively.
Proteins were assembled on the basis of peptide identifications using the ProteinExtractor Tool (version 1.0) in ProteinScape (version 1.3, Bruker Daltonics) and sorted according to their identification scores. Only proteins were considered as identified for which a minimum of two peptides had been found with both search engines and that were not obvious contaminants (keratin 1, 2, 9, or 10). The ProteinScape software automatically removes redundancies in protein entries, i.e. only one protein entry is reported if several redundant sequences are found. If several isoforms (e.g. differing in protein length) occur in the database the software might report several/different accessions of one protein. In this case all accessions given by ProteinScape are reported (supplemental Table 3b).
In the case that more than one protein was reported by ProteinExtractor, the protein entry with the highest score was considered as identified, and the number of “non-redundant proteins” was calculated on the basis of the corresponding gene symbols. Applying the search strategy with two search algorithms no decoy entry was reported by ProteinExtractor.
Hierarchical Clustering of Tumor Samples/Control Tissue—
The normalized and standardized spot volumes of all spots showing differential regulation between tumor/control tissue were used as a data basis for hierarchical clustering. Using the gene cluster software (23) a complete linkage clustering algorithm (similarity metric: uncentered correlation) was applied on the log-transformed data.
Manufacturing of Tissue Arrays—
Single (for bronchial epithelium control tissue) or duplicate (for squamous cell carcinomas, adenocarcinomas, and large cell carcinomas) 1.5-mm cores were taken from representative areas and inserted into a recipient paraffin block to create three tissue microarrays containing a total of 108 cores and additional human tonsil orientation/control cores. The construction was performed using MTA1 tissue arrayer equipment (Beecher Instruments, Sun Prairie, WI). To this end, tissue samples from 55 different patients were used (supplemental Table 1). Their lung tumors were surgically removed in the University Hospitals of Kiel. For bronchial epithelium specimens, tumor-adjacent tissue was used.
Immunohistochemistry—
Tumor tissues were fixed in formalin and routinely processed for paraffin sectioning. 5-μm-thick paraffin sections were deparaffinized and rehydrated, and immunohistochemical staining was performed according to routine methods. Before application of the primary antibody, blocking with non-immune serum was performed for 20 min. Mouse monoclonal anti-keratin 17 (MUbio Products B.V., MUB0325, clone E3, 2 μg/ml), mouse monoclonal anti-keratin 6a (Acris BM5043, clone KA12, 1:5), mouse monoclonal HSP47 (Abcam ab13510, clone M16.10A1, 1:500), and mouse monoclonal keratin 16 (Vector Laboratories VP-C412, clone LL025, 1:20) were applied as primary antibodies. All antibodies were known to recognize the corresponding human antigen. The detection was performed using a Vectastain Elite ABC kit (Vector Laboratories). For negative controls, the primary antibodies were omitted.
Evaluation of the Immunohistochemical Stains—
Sections were scored as positive if epithelial cells (bronchus) or tumor cells showed a staining reaction in the cytoplasm, plasma membrane, and/or the nucleus, according to the localization of the target antigens. First, a quantitative score was performed by estimating the percentage of immunopositive stained cells: no positive staining of cells (0% cells) obtained score 0; <10% cells, 1; 10–50% cells, 2; 50–80% cells, 3; 80–100% cells, 4. Second, the intensity of the reactions was scored as mild, moderate, or strong (score 1, 2, or 3, respectively). Finally a total score (minimum, 0; maximum, 12) was determined as a product of the quantitative and intensity scores of the stainings (24). If tissue samples from the same patient appeared in two different cores of the tissue array the mean of the two corresponding scores was determined and used for the statistical analysis. The given n numbers refer to the number of individual patients from whom the tissues were obtained.
Statistics—
The differences between the immunohistochemical scores of the different tumor specimens and bronchial epithelium were analyzed using Fisher's exact test implemented in the R statistical computing environment (The R Project for Statistical Computing) (25). Fisher's exact test was calculated for all groups (bronchus epithelium, SCC, adenocarcinoma, and large cell carcinoma) and for pairwise comparisons of two groups. For performing receiver operating characteristic analysis, the OriginPro 8G SR1 (OriginLab, Northampton, MA) software suite was used.
RESULTS
Protein Expression Profiling Using 2D DIGE Saturation labeling—
For the identification of proteins highly expressed in squamous cell carcinoma of the lung, we applied 2D DIGE saturation labeling on microdissected cells from normal bronchial epithelium and G2 and G3 grade SCC tumor specimens. Because of biological variance we analyzed tumor samples from seven patients per grade. We used needle microdissection to acquire homogeneous samples of bronchial epithelium and lung squamous carcinoma cells (Fig. 1). After microdissection protein lysates of normal bronchial epithelium and tumor tissue at grades G2 and G3 were labeled with Cy5 and co-separated with Cy3-labeled internal standard using 2DE.
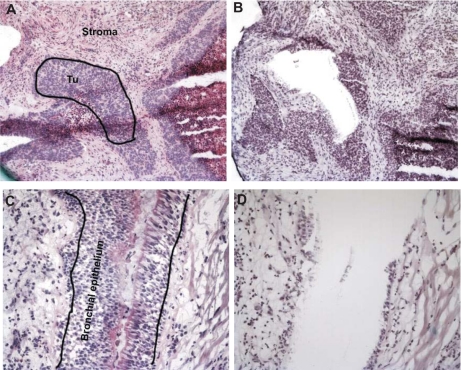
Fig. 1.
Microdissection of squamous cell carcinoma tumors and normal bronchial epithelium. Tissue sections were stained with hematoxylin and eosin. Neighboring sections stained only with hematoxylin were used for needle microdissection. Tumor (Tu) tissue (A, before and B, after needle micro-dissection) was specifically enriched and separated from surrounding stroma (C, before and D, after micro-dissection) just like bronchial epithelium was specifically enriched and separated from surrounding connective tissue and smooth muscle cells.
For protein expression profiling 21 different patient samples were analyzed: seven G2 grade tumors, seven G3 grade tumors, and seven normal bronchial epithelium samples. Thus, we reproducibly could detect a total number of about 2–500 protein spots (Fig. 2 and supplemental Fig. 3). For determination of proteins differentially regulated in low and high grade squamous cell carcinoma of the lung we individually compared normalized and standardized spot volumes of tumors at grades G2 and G2 with the normalized and standardized spot volumes of bronchial epithelium samples, respectively. Altogether we found a significant differential expression (t test p < 0.05, -fold change >3.4 or <−3.4) of 85 different protein spots. These spots appeared to be differential at least in one of the comparisons of normal bronchial epithelium and G2 or G3 grade tumors. Of these proteins spots 73 showed a differential regulation between bronchial epithelium and G2 grade tumors, and 49 showed a differential regulation between bronchial epithelium and G3 grade tumors. A comparison of a group comprising both analyzed tumor grades and control tissue resulted in 72 differentially regulated protein spots. Between the analyzed tumor grades G2 and G3 only five spots were detected as differentially regulated. Most of the protein spots (88%) were in higher abundance in tumor tissues compared with normal bronchial epithelium.
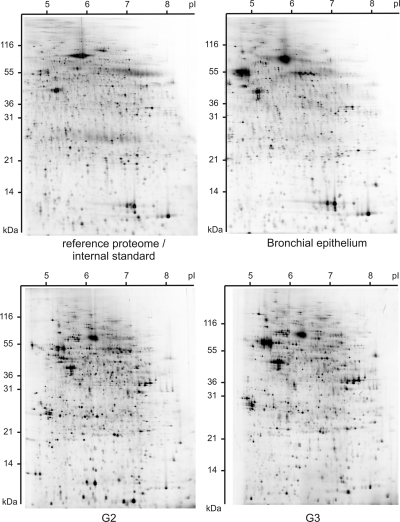
Fig. 2.
Representative 2D DIGE images showing expression profiles from 5000 microdissected cells. Protein samples of bronchial epithelium and G2 and G3 grade squamous cell carcinoma were labeled with Cy5 in a saturation labeling procedure. In each gel an internal Cy3-labeled protein standard was co-separated to improve spot matching and quantification of spot volumes. For each sample 2500 spots were revealed.
To estimate the technical variance of the DIGE method six gels were analyzed in which equal amounts of the internal standard (5 μg) were labeled using Cy3 and Cy5, respectively, and co-separated. For 200 matched spots the means of the normalized and standardized spot volumes were calculated resulting in a mean standard deviation of the spot volume means of 11%. This technical variance is much lower compared with the biological variance of proteins from different patients (supplemental Fig. 1), suggesting that 2D DIGE is an appropriate method for detecting proteomic differences between the analyzed sample groups.
Identification of Differentially Regulated Proteins—
For the identification of differentially regulated protein using different MS techniques tumor material from SCC surgery (reference proteome) was considered. The high matching rate between the reference proteome and microdissected cells (2,250 protein spots; 90%) allowed us to consider 400 μg of protein for protein identification and to save tissue material when additionally applied as internal standard. Using MALDI-MS (for information on peptide mass fingerprint spectra and peak lists see Supplement 1) and nano-HPLC/ESI-MS/MS analyses of peptides generated from 85 differentially regulated protein spots by tryptic in-gel digestion, 46 protein spots were identified (supplemental Fig. 2, Table I, and supplemental Table 3). According to the criteria defined under “Experimental Procedures” they correspond to 32 non-redundant proteins. Therefore some of the identified proteins occur on several positions in the gel suggesting the occurrence of protein isoforms/variants. The isoforms/variants of one protein might not in each case be regulated in the same manner as reflected by varying -fold change values. Of the identified proteins the cytokeratins 6a, 17, and 8 exhibited a differential regulation between the two analyzed tumor grades thereby showing a higher abundance in G2 compared with G3 samples.
Table I.
Examples of differential regulation of protein spots
-Fold change values were calculated from the mean values of the normalized and standardized spot volumes of each group (G2, G2, and bronchial epithelium). Proteins listed were shown to be differentially regulated in lung SCC (G2 and G3 grade) as compared with normal bronchial epithelium. The -fold change values and t test results for individual comparisons (G2/bronchus, G3/bronchus, G2 and G3/bronchus, and G2/G3) are provided as well as the human International Protein Index accession numbers and the GenBank™ names of the proteins. A complete list of identified proteins and potentially co-migrating proteins can be found in supplemental Table 3.
| Spot no. | Protein | Accession no. | G2/bronchus
|
G3/bronchus
|
G2 and G3/bronchus
|
G2/G3
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| -Fold change | t test < 0.05 | -Fold change | t test < 0.05 | -Fold change | t test < 0.05 | -Fold change | t test < 0.05 | |||
| 865 | KRT6A; keratin, type II cytoskeletal 6a | IPI00300725.7 | 12.5 | Yes | 2.9 | Yes | 7.7 | Yes | −4.3 | Yes |
| 954 | KRT6A; keratin, type II cytoskeletal 6a | IPI00300725.7 | 5.6 | Yes | 2.1 | No | 3.9 | Yes | −2.6 | No |
| 1006 | KRT17; KRT17 protein | IPI00747707.1 | 7.9 | Yes | 2.1 | No | 5.0 | No | −3.8 | No |
| 1025 | KRT17; keratin, type I cytoskeletal 17 | IPI00450768.7 | 19.5 | Yes | 7.5 | Yes | 13.5 | Yes | −2.6 | No |
| 1039 | KRT17; KRT17 protein | IPI00747707.1 | 33.5 | Yes | 1.7 | No | 17.6 | No | −19.2 | Yes |
| 1201 | SERPINH1; serpin H1 precursor | IPI00032140.4 | 9.0 | Yes | 7.7 | Yes | 8.4 | Yes | −1.2 | No |
| 1621 | KRT16; keratin, type I cytoskeletal 16 | IPI00217963.3 | 16.9 | Yes | 13.2 | Yes | 15.0 | Yes | −1.3 | No |
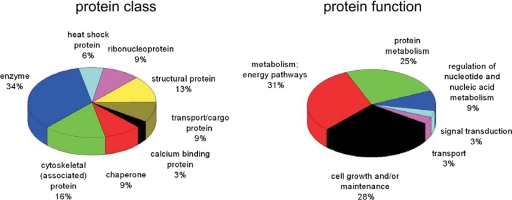
Because of the experimental design it can be expected that some of the co-regulated proteins are functionally involved in tumor progression processes. Most of the identified proteins are involved in protein metabolism (25%), metabolism and energy pathways (31%), and cell growth and maintenance (28%) (Fig. 3). All these pathways are known to be potentially altered in cancer cells. These data reflect the increased turnover of metabolites and energy requirements in tumor cells (26). Also the occurring alterations in RNA and DNA metabolism in tumor cells could be correlated with the identified proteins (9%) involved in these processes.
Fig. 3.
Classes and functions of identified proteins. The human proteome reference database was screened for classes and function of the proteins identified as differentially regulated in SCC compared with normal bronchial epithelium.
Hierarchical Clustering—
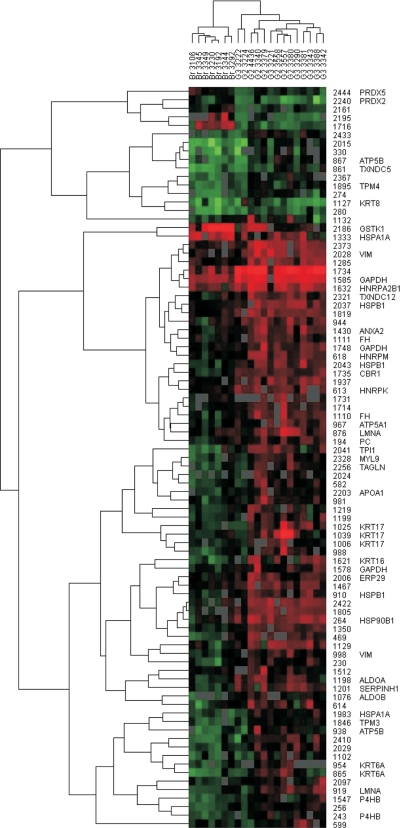
Histology-based tumor grading is often impeded because of less defined morphology and transition states. Therefore, we were interested in whether the proteome pattern reflects the different tumor stages. Hierarchical clustering was done including all 85 protein spots exhibiting a differential regulation. A hierarchical complete linkage clustering algorithm originally developed for gene array data (23) was applied to the normalized spot abundances of the proteins identified to be differentially regulated. A clear clustering of the bronchial epithelium samples in one separated branch of the clustered data suggests differences in morphological properties and protein expression of normal tissues compared with tumor tissues (Fig. 4). In the neighbor tree of the clustered data of the bronchial samples moderately differentiated (G2 grade) tumor samples are found. The clustered data of the majority of poorly differentiated (G3 grade) tumor samples are found further from the bronchial epithelium cluster as the G2 grade tumor clustered data.
Fig. 4.
Hierarchical clustering of 2D DIGE spot data. Normalized and standardized spot volumes were subjected to a hierarchical complete linkage clustering algorithm. 85 protein spots are included showing a differential regulation (t test p < 0.05, -fold change >3.4 or <−3.4) in at least one of the comparisons of normal bronchial epithelium and G2 or G3 grade tumors. The identities of differential regulated spots are given on the right side of the heat map as well as the gene symbols of proteins identified using mass spectrometry (supplemental Table 3). Red squares in the heat map represent high spot intensities, whereas green squares represent low spot intensities. The normal bronchial epithelium samples are clearly separated from the SCC (G2 and G3) samples. VIM, vimentin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HNRP, heterogeneous nuclear ribonucleoprotein; LMNA, lamin A; ALDOA, aldolase A; TAGLN, transgelin; PC, pyruvate carboxylase; FH, fumarate hydratase.
The hierarchical clustering revealed that some protein isoforms (vimentin; glyceraldehyde-3-phosphate dehydrogenase; and mitochondrial ATP synthetase, β subunit) show variable expression changes in different tumor subtypes. For instance the vimentin isoform of spot 2028 was highly up-regulated in G2 and G3 tumors, whereas the spot 998 vimentin isoform was moderately up-regulated. MS analysis of the alternative isoforms suggested different protein processing (splicing, degradation, etc.) in the different tissues analyzed: e.g. vimentin spot 2028 (showing a lower molecular weight compared with vimentin spot 998) might lack the N terminus because in this region of the N-terminal 144 amino acids no peptides were identified using MS analysis. Other proteins such as cytokeratin 17, cytokeratin 6a, and proline 4-hydroxylase are clustered close to each other suggesting a comparable regulation.
Analysis of Protein Abundance by Immunohistochemistry—
For the validation of proteins exhibiting interesting properties in respect to tumor biology and tissue diagnostics we selected HSP47, cytokeratin 6, cytokeratin 16, and cytokeratin 17. We chose immunohistochemistry to validate our proteome data of SCC in an extended series of patients, and expression of these proteins was localized on a cellular level using tissue arrays (Fig. 5 and supplemental Fig. 4). Furthermore although 2DE exhibited the highest resolution for protein mixtures, MS analysis revealed co-migration of candidate proteins, and therefore further validation by independent methods was necessary for unambiguous quantification of candidate proteins.
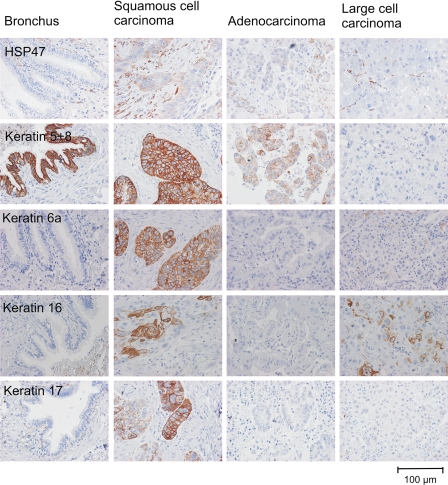
Fig. 5.
Immunohistochemistry on tissue arrays revealing the expression of proteins found to be overexpressed in squamous carcinoma. A representative picture of bronchial epithelium, squamous cell carcinoma, adenocarcinoma, and large cell carcinoma staining is shown for each antibody.
We used 55 tissue specimens including SCC (n = 15) and normal bronchial epithelium (n = 21–23). Additionally we stained tissue cores comprising adenocarcinoma (n = 9) and large cell carcinoma (n = 4–5) tissue specimens to get information about the expression of the investigated protein in these kinds of tumors.
The immunohistochemical analyses showed a high expression of HSP47 (SERPINH1), cytokeratin 6a, cytokeratin 16, and cytokeratin 17 in SCC tissue compared with non-tumor normal epithelium thereby confirming the 2D DIGE data. Fisher's exact tests revealed a significant (p < 0.001) difference between the tissue groups for all analyzed proteins. HSP47 was significantly overexpressed in SCC (n = 15) (p < 0.001) but also in adenocarcinomas (n = 9) (p < 0.001) and LCC (n = 4) (p = 0.05) as compared with normal bronchial epithelium (n = 23).
Cytokeratin 17 was found to be highly abundant in SCC cells in the 2D gel approach. The high abundance of cytokeratin 17 in these tumor cells types was confirmed by immunohistochemistry staining of SCC cells (n = 15), but also a clear expression of cytokeratin 17 was apparent in the basal cells of bronchial epithelium (n = 22). The expression difference of cytokeratin 17 (higher protein abundance in SCC) was revealed to be statistically significant between those two tissue types: p < 0.001. In contrast adenocarcinoma (n = 9) and LCC (n = 5) were not positive for cytokeratin 17 and showed a significant lower abundance of this protein as compared with bronchial epithelium (adenocarcinoma, p = 0.001; LCC, p = 0.05).
In contrast to bronchial epithelium showing no cytokeratin 6a expression (n = 22), cytokeratin 6a was highly abundant in SCC (n = 15) (p < 0.001). Also a significant higher expression of cytokeratin 6a in adenocarcinoma (n = 9) was observed as compared with bronchial epithelium (n = 22) (p = 0.004). LCC (n = 5) was not positive for cytokeratin 6a.
The higher expression of cytokeratin 16 in SCC (n = 15) as compared with bronchial epithelium (n = 21) (p < 0.001) is also in line with the 2D DIGE data. In the analyzed adenocarcinoma tissues (n = 9) we found no expression of cytokeratin 16.
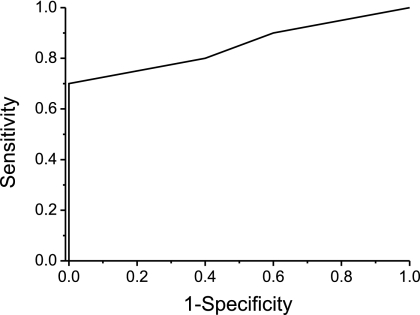
In our 2D DIGE analysis we noticed a higher expression of cytokeratin 17 (spot 1039) in G2 as compared with G3 grade SCCs. So we decided to analyze the differences in the anti-cytokeratin 17 immunoreactions in SCCs in more detail by comparing 10 G2 and five G3 grade tumor stainings. In line with our 2D DIGE results, moderately differentiated G2 grade tumors showed a higher expression cytokeratin 17 as compared with poorly differentiated G3 grade tumors. A receiver operating characteristic analysis (Fig. 6) revealed a good diagnostic accuracy with an area under the curve of 0.85 (asymptotic probability, 0.03), suggesting that cytokeratin 17 can be used as a marker protein helping to stratify G2 and G3 grade SCCs. Using our sample collective we assessed a sensitivity of 70% and a specificity of 100% if an immunohistochemical scoring cutoff of 2 (10–50% of cells mildly stained or <10% of cells moderately stained) was used.
Fig. 6.
Receiver operation characteristic curve describing the discrimination of G2 and G3 grade SCC tumors using anti-cytokeratin 17 immunostaining. 15 SCC tissues (10 G2 and five G3 grade tumors) were stained, and the staining was scored considering the staining intensity and number of stained tumor cells.
DISCUSSION
We performed a proteome analysis of human lung SCC-diseased and control tissue to quantify and identify proteins differentially regulated in the diseased areas. By selecting cell populations via microdissection we gained the advantage that proteomic differences between diseased and non-diseased tissue areas were not “diluted” by irrelevant cell types. The challenge to analyze the resulting low amounts of material was overcome by applying protein saturation labeling and 2D DIGE. By applying an internal standard, spot matching and quantification were improved in comparison with traditional 2D gel-based techniques. In addition it was feasible to identify differentially regulated proteins using MS by using a reference proteome.
In contrast to other profiling approaches used for the identification of novel biomarkers in blood or urine directly, in our approach proteins from diseased and control tissues were analyzed. It is very likely that these tumor co-regulated proteins are functionally involved in tumor progression. Combination of biological function, the abundance of these proteins as revealed by 2D DIGE, and the immunohistochemical analysis suggest a certain involvement and potential role of these proteins in tumorigenesis.
Most of the co-regulated proteins are known in the context of tumor biology and are partially described in SCC progression. They belong to several protein classes and exhibit different functions. Despite a probable methodological bias and not revealing the complete proteome this finding supports our thesis that the candidate proteins are functionally involved in tumor biology.
Heat shock proteins and chaperones are a class of proteins known to be changed in tumor cells. The proteome analysis showed HSP47 (SERPINH1) to be overexpressed in SCC tissue as compared with normal bronchial epithelium. These data were confirmed by immunohistochemistry on tissue arrays also for adenocarcinoma and LCC. To our knowledge HSP47 overexpression in this set of human NSCLC tissues has not been described before. In several organisms HSP47 is a well conserved protein associated with the binding and processing of collagens in the endoplasmic reticulum (27). It is interestingly to note that another protein known to participate in these processes was found in our study to be highly expressed in SCC tumor cells. The protein HSP90B1 (GPR94) exhibits similar expression characteristics in SCC cells as revealed by cluster analysis (Fig. 4) (28–30). These findings demonstrate the power of the 2D DIGE method used, revealing groups of proteins contributing to defined cellular processes. A functional role of HSP47 has been postulated for several different diseases. The protein is highly expressed in fibrotic tissues of kidney, lung, and liver (31–33). HSP47 autoantibodies have been found in sera of patients with rheumatic autoimmune diseases (34). Several studies have been done elucidating the role of HSP47 in cancer. The protein has been found to be overexpressed in pancreatic cancer (35), osteosarcomas (36), and squamous cell carcinoma of the head and neck. For the latter the HSP47 locus (11q13.5) is known to be frequently overexpressed (37). For squamous carcinoma of the head and neck HSP47 has been suggested as a target protein for the targeted delivery of anticancer agents (38). Even if HSP47 expression is known to be altered in a number of human cancers and overexpressed in fibrotic tissues (27), it might be an interesting task to elucidate the therapeutic and diagnostic potential of HSP47 also for lung squamous cell carcinoma in further studies.
Another group of proteins frequently altered in tumor cells are constituents of the cytoskeleton: e.g. the cytokeratin 19 marker (Cyfra 21-1) has been evaluated as a serum biomarker (39). We found a high abundance of several cytokeratins in SCC cells, whereas some of them were not found in normal bronchial epithelium.
We found e.g. overexpression of cytokeratin 6, cytokeratin 16, and cytokeratin 17. These proteins are known to play a relevant role during wound repair (40). It might be that in the development of SCC similar pathways are activated as in wound repair because cancer cells live in an environment that is more stressed and “wounded” than in an intact environment.
Cytokeratin 5/6 is a well known marker highly expressed in carcinomas of stratified epithelial origin (41). In our study keratin 6 appeared to be highly expressed in SCC tissue and not in bronchial epithelium and LCC, which is in line with the already published results. Because keratin 6 came up in our analysis in several spots, a detailed analysis of keratin 6 isoforms would be an interesting starting point for further experiments.
The intermediate filament protein cytokeratin 16 is another protein found to be highly abundant in SCC cells in our study. Overexpression of cytokeratin is found in squamous cell carcinomas and adenocarcinomas of the cervix (42) as well as in invasive squamous cell carcinomas of the skin (43).
Cytokeratin 17 has been shown to be expressed in hyperproliferative squamous epithelia and very typically in basal cells of complex epithelia (44) as confirmed in our results. Hofmann et al. (45) also found overexpression of cytokeratin 17 in an mRNA-based approach. Our finding that cytokeratin 17 was not strongly expressed in adenocarcinomas and large cell carcinomas is not in line with the data of Chu and Weiss (41) who found that 38 of 50 adenocarcinomas and four of six large cell carcinomas were positive for cytokeratin 17. These discrepancies might be due to alternating binding profiles or epitope specificity of the antibodies used. We found a staining of basal cells of normal bronchial epithelium using our cytokeratin 17 antibody; this is a good indicator for the specificity of our immunohistochemical staining using this monoclonal antibody. Interestingly induced cytokeratin 17 expression was shown to regulate cell growth in wounded stratified epithelia by binding the adaptor protein 14-3-3σ. This process, involving the Akt/mTOR (mammalian target of rapamycin) pathway, might be a common principle and could be comparable to processes in cancer cells (46).
Because of our experiment design we were in the position to analyze proteomic differences between the tumor grades G2 and G3. Besides the clear separation of the bronchial epithelium from the tumor samples using hierarchical clustering of the spot data we also could show that proteomic profiles allows G2 and G3 grade tumors to be distinguished. The G2 samples clustered near the bronchial epithelial samples, whereas most of the G3 samples clustered distant from the bronchial epithelium group. This reflects the dedifferentiation process from moderately to dedifferentiated tumor tissue. Nevertheless two G2 samples can be found in the G3 cluster. A possible reason might be that the morphological criteria for tumor grading in lung SCCs (zonation of tumor nest, grade of polymorphy, dyscohesive growth, and single cell infiltration) are not very stringent, and SCCs show variation in pattern and differentiation from one tumor area to the next. Although the grading of differentiation was performed on the first section that was applied for microdissection, it is possible that the proteome profile does not mirror the differentiation of this particular area but rather the heterogeneity of the tumor. We suggest that additional criteria like protein expression patterns might help to discriminate between tumor grades in a more significant and objective manner. In our analysis we found some proteins that might help to stratify between different tumor grades.
Analyzing single protein spots and tumors of both grades via immunohistochemistry we found cytokeratins 8, 6a, and 17 as proteins exhibiting the highest abundance in G2 grade tumors as compared with both bronchial epithelium and G3 grade tumors. We speculate that during the cell transition from bronchial epithelium to moderately differentiated tumors (G2 grade) the expression of these proteins is initially induced and declines with further dedifferentiation of the tumor cells. Interestingly Wang et al. (47) found cytokeratin 17 to be highly expressed in certain types of bronchial dysplasia. As e.g. squamous dysplasia is a well accepted preinvasive lesion for squamous lung carcinoma, the expression of cytokeratin 17 in cells with squamous dysplastic phenotype is in agreement with our hypothesis. Also bronchial epithelial dysplasia with transitional differentiation is discussed to be preneoplastic; for this lesion cytokeratin 17 expression seems to be quite heterogeneous. These and our findings make cytokeratin 17 an interesting candidate biomarker for SCC tumor formation and stratifying SCC tumor grades. As immunohistochemistry is used routinely for differential diagnosis of tumors a cytokeratin 17 staining could help to stratify SCC tumor grades on a molecular level. Further experiments in an extended number of tumor samples have to be carried out to confirm these findings.
In summary we identified 32 proteins differentially regulated between bronchial epithelium and different NSCLC subtypes and confirmed four of these using immunohistochemistry. These proteins are candidate biomarkers for the improvement of SCC diagnosis and elucidation of the biology of SCCs. One or a panel of these proteins might be useful to stratify tumor subtypes or grades of differentiation using immunohistochemistry (as shown for cytokeratin 17 and G2/G3 grade SCC tumors) or for detection of lung cancer in body fluids if the proteins or parts of them can be found to be elevated in serum, bronchial fluid, or urine. Furthermore information of the isoforms and post-translational modifications has to be acquired to evaluate their potential as a diagnostic tool.
Supplementary Material
Acknowledgments
We thank Sabine Roggenbrodt for excellent technical assistance and Klaus Jung, Katharina Podwojski, and Martin Eisenacher for supporting the statistical analysis of the data.
Footnotes
Published, MCP Papers in Press, January 27, 2009, DOI 10.1074/mcp.M800422-MCP200
The abbreviations used are: NSCLC, non-small cell lung carcinoma; 2D, two-dimensional; SCC, squamous cell carcinoma; LCC, large cell carcinoma; 2DE, two-dimensional electrophoresis; TCEP, tris(2-carboxyethyl)phosphine hydrochloride; IPI, International Protein Index; HCT, High Capacity Trap.
This work was supported by a grant from the European Commission (LCVAC, COOP-CT-2004-512855) and the Ministry of Innovation, Science, Research and Technology of Nordrhein-Westfalen.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Boyle, P., and Ferlay, J. ( 2005) Cancer incidence and mortality in Europe, 2004. Ann. Oncol. 16, 481–488 [DOI] [PubMed] [Google Scholar]
- 2.Jemal, A., Murray, T., Ward, E., Samuels, A., Tiwari, R. C., Ghafoor, A., Feuer, E. J., and Thun, M. J. ( 2005) Cancer Statistics, 2005. CA Cancer J. Clin. 55, 10–30 [DOI] [PubMed] [Google Scholar]
- 3.Basbaum, C., and Jany, B. ( 1990) Plasticity in the airway epithelium. Am. J. Physiol. 259, L38–L46 [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer, A. M., Lechner, J. F., Masui, T., Reddel, R. R., Mark, G. E., and Harris, C. C. ( 1989) Control of growth and squamous differentiation in normal human bronchial epithelial cells by chemical and biological modifiers and transferred genes. Environ. Health Perspect. 80, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patz, E. F., Jr., Rossi, S., Harpole, D. H., Jr., Herndon, J. E., and Goodman, P. C. ( 2000) Correlation of tumor size and survival in patients with stage IA non-small cell lung cancer. Chest 117, 1568–1571 [DOI] [PubMed] [Google Scholar]
- 6.Schneider, J. ( 2006) Tumor markers in detection of lung cancer. Adv. Clin. Chem. 42, 1–41 [DOI] [PubMed] [Google Scholar]
- 7.Seike, M., Kondo, T., Fujii, K., Yamada, T., Gemma, A., Kudoh, S., and Hirohashi, S. ( 2004) Proteomic signature of human cancer cells. Proteomics 4, 2776–2788 [DOI] [PubMed] [Google Scholar]
- 8.Tockman, M. S., Mulshine, J. L., Piantadosi, S., Erozan, Y. S., Gupta, P. K., Ruckdeschel, J. C., Taylor, P. R., Zhukov, T., Zhou, W. H., Qiao, Y. L., and Yao, S. X. ( 1997) Prospective detection of preclinical lung cancer: results from two studies of heterogeneous nuclear ribonucleoprotein A2/B1 overexpression. Clin. Cancer Res. 3, 2237–2246 [PubMed] [Google Scholar]
- 9.Shitrit, D., Zingerman, B., Shitrit, A. B., Shlomi, D., and Kramer, M. R. ( 2005) Diagnostic value of CYFRA 21-1, CEA, CA 19-9, CA 15-3, and CA 125 assays in pleural effusions: analysis of 116 cases and review of the literature. Oncologist 10, 501–507 [DOI] [PubMed] [Google Scholar]
- 10.Xiao, T., Ying, W., Li, L., Hu, Z., Ma, Y., Jiao, L., Ma, J., Cai, Y., Lin, D., Guo, S., Han, N., Di, X., Li, M., Zhang, D., Su, K., Yuan, J., Zheng, H., Gao, M., He, J., Shi, S., Li, W., Xu, N., Zhang, H., Liu, Y., Zhang, K., Gao, Y., Qian, X., and Cheng, S. ( 2005) An approach to studying lung cancer-related proteins in human blood. Mol. Cell. Proteomics 4, 1480–1486 [DOI] [PubMed] [Google Scholar]
- 11.Li, C., Xiao, Z., Chen, Z., Zhang, X., Li, J., Wu, X., Li, X., Yi, H., Li, M., Zhu, G., and Liang, S. ( 2006) Proteome analysis of human lung squamous carcinoma. Proteomics 6, 547–558 [DOI] [PubMed] [Google Scholar]
- 12.Anderson, L., and Seilhamer, J. ( 1997) A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18, 533–537 [DOI] [PubMed] [Google Scholar]
- 13.Sitek, B., Luttges, J., Marcus, K., Kloppel, G., Schmiegel, W., Meyer, H. E., Hahn, S. A., and Stuhler, K. ( 2005) Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics 5, 2665–2679 [DOI] [PubMed] [Google Scholar]
- 14.Colby, T. V., Koss, M. N., and Travis, W. D. ( 1995) Tumors of the lower respiratory tract, Colby, Koss, Travis (eds) Atlas of Tumor Pathology, pp. 56–67, Armed Forces Institute of Pathology, Washington
- 15.Klose, J., and Kobalz, U. ( 1995) Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis 16, 1034–1059 [DOI] [PubMed] [Google Scholar]
- 16.Schaefer, H., Chervet, J. P., Bunse, C., Joppich, C., Meyer, H. E., and Marcus, K. ( 2004) A peptide preconcentration approach for nano-high-performance liquid chromatography to diminish memory effects. Proteomics 4, 2541–2544 [DOI] [PubMed] [Google Scholar]
- 17.Schaefer, H., Chamrad, D. C., Marcus, K., Reidegeld, K. A., Bluggel, M., and Meyer, H. E. ( 2005) Tryptic transpeptidation products observed in proteome analysis by liquid chromatography-tandem mass spectrometry. Proteomics 5, 846–852 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, W., and Chait, B. T. ( 2000) ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72, 2482–2489 [DOI] [PubMed] [Google Scholar]
- 19.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. ( 1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 20.Stephan, C., Reidegeld, K. A., Hamacher, M., van Hall, A., Marcus, K., Taylor, C., Jones, P., Muller, M., Apweiler, R., Martens, L., Korting, G., Chamrad, D. C., Thiele, H., Bluggel, M., Parkinson, D., Binz, P. A., Lyall, A., and Meyer, H. E. ( 2006) Automated reprocessing pipeline for searching heterogeneous mass spectrometric data of the HUPO Brain Proteome Project pilot phase. Proteomics 6, 5015–5029 [DOI] [PubMed] [Google Scholar]
- 21.Yates, J. R., 3rd, Eng, J. K., McCormack, A. L., and Schieltz, D. ( 1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 22.Yates, J. R., iii, Eng, J. K., and McCormack, A. L. ( 1995) Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 67, 3202–3210 [DOI] [PubMed] [Google Scholar]
- 23.Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. ( 1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remmele, W., Hildebrand, U., Hienz, H. A., Klein, P. J., Vierbuchen, M., Behnken, L. J., Heicke, B., and Scheidt, E. ( 1986) Comparative histological, histochemical, immunohistochemical and biochemical studies on oestrogen receptors, lectin receptors, and Barr bodies in human breast cancer. Virchows Arch. A Pathol. Anat. Histopathol. 409, 127–147 [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team ( 2006) R: a Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria
- 26.DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G., and Thompson, C. B. ( 2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 27.Sauk, J. J., Nikitakis, N., and Siavash, H. ( 2005) Hsp47 a novel collagen binding serpin chaperone, autoantigen and therapeutic target. Front. Biosci. 10, 107–118 [DOI] [PubMed] [Google Scholar]
- 28.Ferreira, L. R., Norris, K., Smith, T., Hebert, C., and Sauk, J. J. ( 1996) Hsp47 and other ER-resident molecular chaperones form heterocomplexes with each other and with collagen type IV chains. Connect. Tissue Res. 33, 265–273 [DOI] [PubMed] [Google Scholar]
- 29.Nakai, A., Satoh, M., Hirayoshi, K., and Nagata, K. ( 1992) Involvement of the stress protein HSP47 in procollagen processing in the endoplasmic reticulum. J. Cell Biol. 117, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razzaque, M. S., and Taguchi, T. ( 1999) Role of glomerular epithelial cell-derived heat shock protein 47 in experimental lipid nephropathy. Kidney Int. Suppl. 71, S256–S259 [DOI] [PubMed] [Google Scholar]
- 31.Brown, K. E., Broadhurst, K. A., Mathahs, M. M., Brunt, E. M., and Schmidt, W. N. ( 2005) Expression of HSP47, a collagen-specific chaperone, in normal and diseased human liver. Lab. Investig. 85, 789–797 [DOI] [PubMed] [Google Scholar]
- 32.Razzaque, M. S., Kumatori, A., Harada, T., and Taguchi, T. ( 1998) Coexpression of collagens and collagen-binding heat shock protein 47 in human diabetic nephropathy and IgA nephropathy. Nephron 80, 434–443 [DOI] [PubMed] [Google Scholar]
- 33.Razzaque, M. S., Hossain, M. A., Kohno, S., and Taguchi, T. ( 1998) Bleomycin-induced pulmonary fibrosis in rat is associated with increased expression of collagen-binding heat shock protein (HSP) 47. Virchows Arch. 432, 455–460 [DOI] [PubMed] [Google Scholar]
- 34.Yokota, S., Kubota, H., Matsuoka, Y., Naitoh, M., Hirata, D., Minota, S., Takahashi, H., Fujii, N., and Nagata, K. ( 2003) Prevalence of HSP47 antigen and autoantibodies to HSP47 in the sera of patients with mixed connective tissue disease. Biochem. Biophys. Res. Commun. 303, 413–418 [DOI] [PubMed] [Google Scholar]
- 35.Maitra, A., Iacobuzio-Donahue, C., Rahman, A., Sohn, T. A., Argani, P., Meyer, R., Yeo, C. J., Cameron, J. L., Goggins, M., Kern, S. E., Ashfaq, R., Hruban, R. H., and Wilentz, R. E. ( 2002) Immunohistochemical validation of a novel epithelial and a novel stromal marker of pancreatic ductal adenocarcinoma identified by global expression microarrays: sea urchin fascin homolog and heat shock protein 47. Am. J. Clin. Pathol. 118, 52–59 [DOI] [PubMed] [Google Scholar]
- 36.Uozaki, H., Ishida, T., Kakiuchi, C., Horiuchi, H., Gotoh, T., Iijima, T., Imamura, T., and Machinami, R. ( 2000) Expression of heat shock proteins in osteosarcoma and its relationship to prognosis. Pathol. Res. Pract. 196, 665–673 [DOI] [PubMed] [Google Scholar]
- 37.Welkoborsky, H. J., Bernauer, H. S., Riazimand, H. S., Jacob, R., Mann, W. J., and Hinni, M. L. ( 2000) Patterns of chromosomal aberrations in metastasizing and nonmetastasizing squamous cell carcinomas of the oropharynx and hypopharynx. Ann. Otol. Rhinol. Laryngol. 109, 401–410 [DOI] [PubMed] [Google Scholar]
- 38.Hebert, C., Norris, K., and Sauk, J. J. ( 2003) Targeting of human squamous carcinomas by SPA470-doxorubicin immunoconjugates. J. Drug Target 11, 101–107 [DOI] [PubMed] [Google Scholar]
- 39.Pujol, J. L., Quantin, X., Jacot, W., Boher, J. M., Grenier, J., and Lamy, P. J. ( 2003) Neuroendocrine and cytokeratin serum markers as prognostic determinants of small cell lung cancer. Lung Cancer 39, 131–138 [DOI] [PubMed] [Google Scholar]
- 40.Coulombe, P. A., Tong, X., Mazzalupo, S., Wang, Z., and Wong, P. ( 2004) Great promises yet to be fulfilled: defining keratin intermediate filament function in vivo. Eur. J. Cell Biol. 83, 735–746 [DOI] [PubMed] [Google Scholar]
- 41.Chu, P. G., and Weiss, L. M. ( 2002) Keratin expression in human tissues and neoplasms. Histopathology 40, 403–439 [DOI] [PubMed] [Google Scholar]
- 42.Smedts, F., Ramaekers, F., Leube, R. E., Keijser, K., Link, M., and Vooijs, P. ( 1993) Expression of keratins 1, 6, 15, 16, and 20 in normal cervical epithelium, squamous metaplasia, cervical intraepithelial neoplasia, and cervical carcinoma. Am. J. Pathol. 142, 403–412 [PMC free article] [PubMed] [Google Scholar]
- 43.Haider, A. S., Peters, S. B., Kaporis, H., Cardinale, I., Fei, J., Ott, J., Blumenberg, M., Bowcock, A. M., Krueger, J. G., and Carucci, J. A. ( 2006) Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J. Investig. Dermatol. 126, 869–881 [DOI] [PubMed] [Google Scholar]
- 44.Moll, R. ( 1998) Cytokeratins as markers of differentiation in the diagnosis of epithelial tumors. Subcell. Biochem. 31, 205–262 [PubMed] [Google Scholar]
- 45.Hofmann, H. S., Bartling, B., Simm, A., Murray, R., Aziz, N., Hansen, G., Silber, R. E., and Burdach, S. ( 2006) Identification and classification of differentially expressed genes in non-small cell lung cancer by expression profiling on a global human 59.620-element oligonucleotide array. Oncol. Rep. 16, 587–595 [PubMed] [Google Scholar]
- 46.Kim, S., Wong, P., and Coulombe, P. A. ( 2006) A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441, 362–365 [DOI] [PubMed] [Google Scholar]
- 47.Wang, G. F., Lai, M. D., Yang, R. R., Chen, P. H., Su, Y. Y., Lv, B. J., Sun, L. P., Huang, Q., and Chen, S. Z. H. ( 2006) Histological types and significance of bronchial epithelial dysplasia. Mod. Pathol. 19, 429–437 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.