Abstract
A SISCAPA (stable isotope standards and capture by anti-peptide antibodies) method for specific antibody-based capture of individual tryptic peptides from a digest of whole human plasma was developed using a simplified magnetic bead protocol and a novel rotary magnetic bead trap device. Following off-line equilibrium binding of peptides by antibodies and subsequent capture of the antibodies on magnetic beads, the bead trap permitted washing of the beads and elution of bound peptides inside a 150-μm-inner diameter capillary that forms part of a nanoflow LC-MS/MS system. The bead trap sweeps beads against the direction of liquid flow using a continuous succession of moving high magnetic field-gradient trap regions while mixing the beads with the flowing liquid. This approach prevents loss of low abundance captured peptides and allows automated processing of a series of SISCAPA reactions. Selected tryptic peptides of α1-antichymotrypsin and lipopolysaccharide-binding protein were enriched relative to a high abundance serum albumin peptide by 1,800 and 18,000-fold, respectively, as measured by multiple reaction monitoring. A large majority of the peptides that are bound nonspecifically in SISCAPA reactions were shown to bind to components other than the antibody (e.g. the magnetic beads), suggesting that substantial improvement in enrichment could be achieved by development of improved inert bead surfaces.
MS is the method of choice for identification of peptides in digests of biological samples based on the power of MS to detect the chemically well defined masses of both peptides and their fragments produced by processes such as CID. This high level of structural specificity is also critical in improving peptide (and protein) quantitation because it overcomes the well known problems inherent in classical immunoassays related to limited antibody specificity, dynamic range, and multiplexability. In principle, a quantitative peptide assay using MRM1 detection in a triple quadrupole mass spectrometer should have nearly absolute structural specificity, a dynamic range of ∼1e+4, and the ability to multiplex measurements of hundreds of peptides per sample (1). These properties suggest that MS-based methods could ultimately replace classical immunoassay technologies in many research and clinical applications.
An important limitation of present peptide MRM measurements is sensitivity. The most sensitive widely used quantitative MS platforms use nanoflow chromatography and ESI to deliver trace amounts of peptides to the mass spectrometer. However, these processes are limited in the total amount of peptide that can be applied while retaining maximum sensitivity (typically limited to ∼1 μg of total peptide sample, i.e. the product obtained from digesting ∼14 nl of plasma). The lower cutoff for detecting proteins in a digest of unfractionated plasma by this approach appears to be in the neighborhood of 1–20 μg/ml plasma concentration, which would restrict analysis to the top 100 or so proteins in plasma (1).
The sensitivity of MS assays can be substantially increased by fractionating the sample at the level of intact proteins, the tryptic peptides derived from them, or both. For example, immunodepletion of the six most abundant plasma proteins, removes ∼85% of the protein mass (2) and results in an increase of ∼7-fold in the signal-to-noise of MRM measurements of peptides from the remaining proteins after digestion (1). Similarly chromatographic fractionation by strong cation exchange provides another major improvement in sensitivity (3). However, increased sample fractionation brings with it the disadvantages of increased cost and time, the risk of losing specific components, and the continued requirement for very high resolution (lengthy, low throughput) reversed phase nanoflow chromatography en route to the ESI source.
An alternative fractionation approach, used in the SISCAPA method, enriches specific target peptides through capture by anti-peptide antibodies, thus circumventing these disadvantages for preselected targets (4). In its initial implementation, SISCAPA used very small (∼10-nl) columns of POROS chromatography support carrying covalently bound rabbit antibodies and provided ∼100-fold enrichment of target peptides with respect to others (4). These columns were, like immunoaffinity depletion columns (2), recyclable many times. However, the potential for sample-to-sample carryover, limitations in the amount of sample digest that could be pumped over nanoaffinity columns at flow rates slow enough to permit peptide binding, and limited flexibility in changing and multiplexing antibodies were problematic. This led us to explore an alternative approach using magnetic beads as the antibody support (5). In this case, the binding reaction can be carried out off line, allowing equilibrium binding; the magnetic beads can be removed from the digest sample and washed; and the bound peptides can be eluted in 96-well plates either manually or using automated equipment such as a KingFisher Magnetic Particle Processor (ThermoFisher). One potential pitfall remains in the handling of eluted peptides. If the anti-peptide antibodies have very high selectivity, as desired in the SISCAPA approach, then in the case of low abundance peptides, only a very small amount of peptide will be eluted from the antibody. Such small amounts of peptide are easily lost through irreversible binding to the walls of vessels such as 96-well plate wells, and the smaller the amount of peptide (i.e. the more specific the capture), the worse the problem may be.
To address this issue, we report here a hybrid approach in which peptide binding occurs off line (to equilibrium), whereas the subsequent washing and elution steps are carried out within a capillary that forms part of the nanoflow LC system, thus ensuring that peptide eluted from the antibodies on the beads will not be “lost” between elution and the ESI source. Although there is extensive literature on macroscopic and microfluidic devices for manipulating magnetic beads (6–8) we were unable to find components adaptable to the small scales and high pressures required for integration into nanoflow HPLC. We therefore developed a novel “bead trap” device that satisfies the following requirements: 1) the need to retain beads in a “trap” region against the flow of liquid (loading, wash, and elution buffers for example) in a vessel of capillary dimensions, 2) the need to ensure that beads do not escape from the trap region to contaminate downstream apparatus or columns, 3) the need to ensure that beads are effectively mixed with the flowing fluids (required for efficient washing and elution), and 4) the need to ensure that all beads can be efficiently ejected from the trap region in preparation for a subsequent cycle. The device provides multiple sequential magnetic trapping regions capable of sweeping commonly used 2.8- and 1-μm magnetic beads against liquid flow to prevent escape of beads through the trapping device (i.e. the second downstream trapping zone captures beads swept by the liquid stream past the first trap and so on). In addition, the bead trap device allows the movement of these trapping regions to agitate the trapped bead mass and mix it with fluids flowing past. Finally the device allows reversal of the sweeping action to effectively eject beads from the trap into the fluid stream. The bead trap capillary can be plumbed at various points in conventional nanoflow LC systems (e.g. in place of a sample loop or connecting tube), and the device can be controlled directly by the LC-MS/MS instrument software through contact closures. We show that the bead trap provides an effective method of implementing SISCAPA experiments.
EXPERIMENTAL PROCEDURES
Reagents—
Human plasma digest was prepared from blood collected in 10-ml heparinized vials and centrifuged to liberate the plasma component. This plasma was diluted to 5 mg/ml using 25 mm ammonium bicarbonate, denatured with SDS (0.05% final concentration), and reduced with tris(2-carboxyethyl)phosphine (50 mm final concentration) for 30 min at 60 °C. The sample was cooled to room temperature before alkylating with iodoacetamide (10 mm final concentration) for 30 min at 37 °C. The reduced and alkylated plasma sample was digested with Promega sequencing grade modified porcine trypsin at an enzyme:protein ratio of 1:50 for 12 h at 37 °C. Digestion was quenched using Nα-p-tosyl-l-lysine chloromethyl ketone (100 μm final concentration), and the digest was stored at −80 °C. Rabbit polyclonal antisera to a tryptic peptide of human α1-antichymotrypsin (peptide AAC-1; Table I) were made by immunizing rabbits with keyhole limpet hemocyanin conjugates of AAC-1 peptide with added C-terminal GSGC linker as described previously (4). The resulting polyclonal antibodies were affinity-purified on an agarose column to which the respective peptide (plus linker) was conjugated. Approximate binding constants (immobilized antibody binding target tryptic peptide from solution) were measured using a Biacore 3000 system as described previously (9) and yielded values of ka = 1.91e+5 m−1 s−1, kd = 7.25e−4 s−1, and KD = 3.80e−9 m. In addition, rabbit polyclonal antisera were made to a pool of five tryptic peptides of human LPS-binding protein by immunizing rabbits with pooled keyhole limpet hemocyanin-peptide conjugates with C-terminal Cys extensions; the best responding peptide (LBP-1b; Table I) was selected, and the corresponding specific polyclonal antibody was purified on a peptide antigen affinity column as above. Approximate binding constants for this antibody were measured as above, yielding values of ka = 4.13e+5 m−1 s−1, kd = 1.41e−4 s−1, and KD = 3.42e−10 m. Protein G-coated magnetic beads (Dynabeads G, Invitrogen Dynal, catalog number 100.100.04D; 30 mg/ml reported concentration) were used to capture rabbit antibodies from solution.
Table I.
MRM transitions measured and labeled in the figures
Italics indicate a non-peptide transition for CHAPS detergent.
| Peak label | Abbreviation in text | MS1 | MS2 | Protein/detergent | Sequence |
|---|---|---|---|---|---|
| 1 | 467.3 | 660.4 | Serum albumin (HSA) | LCTVATLR | |
| 2 | 490.8 | 562.3 | Haptoglobin β chain | VGYVSGWGR | |
| 3 | AAC-1 | 531.3 | 819.5 | α1-Antichymotrypsin | EIGELYLPK |
| 4 | 559.9 | 697.4 | IgG1 heavy chain | FNWYVDGVEVHNAK | |
| 5 | HSA-1 | 575.4 | 937.4 | Serum albumin (HSA) | LVNEVTEFAK |
| 6 | 598.7 | 732.4 | IgG2 heavy chain | VVSVLTVVHQDWLNGK | |
| 7 | 599.8 | 849.5 | Plasma retinol-binding protein | YWGVASFLQK | |
| 8 | 603.4 | 997.5 | IgG1/G3/G4 heavy chain | VVSVLTVLHQDWLNGK | |
| 9 | 610.8 | 775.4 | Hemopexin | NFPSPVDAAFR | |
| 10 | 615.9 | 819.5 | CHAPS detergent | ||
| 11 | LBP-1b | 624.3 | 920.5 | LPS-binding protein | ITLPDFTGDLR |
| 12 | 751.9 | 836.5 | Ig κ light chain | DSTYSLSSTLTLSK | |
| 13 | 856.9 | 882.5 | Hemopexin | GECQAEGVLFFQGDR | |
| 14 | 923.0 | 1059.5 | α2-Macroglobulin | LLIYAVLPTGDVIGDSAK |
Liquid Chromatography—
The LC system was loaded with 0.1 m ammonium acetate, pH 7.5 (high flow solvent A); 2% acetonitrile, 0.1% formic acid (nanoflow solvent A); and 98% acetonitrile, 0.1% formic acid (nanoflow solvent B). Autosampler reagent vials were provided loaded with 0.1 m ammonium acetate, 0.5 m NaCl, 0.1% CHAPS, pH 7.5 (“wash”); 70% acetonitrile, 0.1% formic acid (“bead eluent”); and 1% CHAPS (“bead ejection solution”).
SISCAPA Magnetic Bead Protocol—
A simplified SISCAPA protocol was used in which anti-peptide antibody, free in solution, was added to a sample digest and allowed to incubate followed by addition of Dynal protein G beads and a further incubation to allow capture of the antibody on the beads. In general these additions were carried out in round bottom 96-well polypropylene plates, which were shaken on a microplate shaker (Sarstedt Monomixer MM-1) to keep the beads in suspension. Washing of beads and elution of bound molecules were carried out either manually by transfer of solutions between wells while beads were gathered on a side wall by a Novagen Magnetight HT-96 magnet array or in an automated process (using the bead trap described below) in line with the nanoflow LC system. In both approaches, the natural tendency of the beads to stick to plastic and fused silica surfaces was overcome by addition of low concentrations (0.1%) of CHAPS detergent to most solutions aside from the final wash and elution. CHAPS was selected because it elutes late in the reversed phase separation as a single major peak after most peptides and hence is more “mass spectrometer-friendly” than other detergents, most of which are polymeric, yielding many peaks when monitored by suitable MRM profiles. The presence of CHAPS was detected by a characteristic MRM (615.9/819.5). Parameters for bead capture and elution were optimized by using Alexa Fluor 488-labeled rabbit antibody (Invitrogen A11059), crude Alexa Fluor 488-labeled AAC-1 peptide (made by reacting AAC-1 immunogen peptide, including the GSGC extension, with Alexa Fluor 488 amine-reactive tetrafluorophenyl ester (Invitrogen A30005)), and AAC-1 peptide synthesized with an N-terminal fluorescein (Elim Biopharmaceuticals, Inc., Hayward, CA). Fluorescence signals were quantitated in a fluorescence plate reader (SpectraMax Gemini XS; Molecular Devices, Sunnyvale, CA). In initial optimization studies using reference capture reactions containing 1 μg of antibody and 5 μl of Dynal G beads in a 100-μl total volume, ∼70% of the antibody was captured in 4 h.
Bead Trap Device—
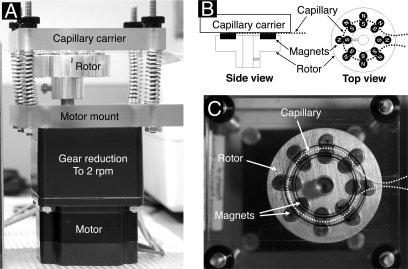
The bead trap prototype used here (Fig. 1) consisted of an aluminum rotor with eight pairs of ¼-inch diameter × 3/16-inch-thick cylindrical rare earth NdFeB permanent magnets (Amazing Magnets, Irvine, CA) that was rotated about its cylinder axis by a reversible, low speed (2 rpm) synchronous motor. The magnet faces were arranged to be approximately co-planar on their upper surfaces, each side-by-side pair (one with north side up and the other with south side up) generating a local region of high field gradient at the point of contact. The direction of motor rotation was controlled by a contact closure signal from a Spark Holland (Plainsboro, NJ) autosampler auxiliary output under software control. A length of 150-μm-inner diameter (360-μm-outer diameter) Teflon capillary tubing (Upchurch Scientific, Oak Harbor, WA) was configured to follow ∼300° of a circle of diameter the same as (and co-axial with) the circle of magnetic trapping regions (i.e. the circle defined by the contact points of the pairs of magnets as they rotate; Fig. 1). This tube was affixed in the appropriate partial circular path by a piece of thin clear adhesive tape on the underside of a tubing carrier plate of clear acrylic plastic. This plate was finally brought parallel to the upper face of the magnets on the rotor and close to them (almost touching the upper surface of the magnets), aligned so that the tube followed the path of the trap regions as the carrier rotated underneath. Alignment of the tubing mount plate parallel to and close to (but not touching) the upper face of the magnet rotor was facilitated by the use of four threaded rods with finger nuts used to force the tubing mount plate toward the upper surface of the magnet carrier against the resistance of four springs. The finger nuts were tightened until a thin sheet of paper could barely be inserted between the magnets and the capillary tubing at any point. Captured magnetic beads congregate where the tube diverges from the circle, i.e. where the effect of the magnets falls off (see supplemental video Beadtrap CIMG1277.avi).
Fig. 1.
Prototype bead trap device. A, side view; B, schematic diagram; C, top view. N, north; S, south.
LC-MS/MS System and Protocols—
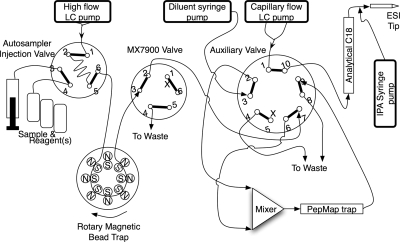
The nano-LC system (Fig. 2) consisted of an Eksigent 2-D NanoLC with Spark Holland autosampler and 10-port auxiliary valve supplemented with an additional MX-7900 six-port valve (Rheodyne), two Harvard syringe pumps (one for pretrap diluent addition and one to deliver 50 nl/min degassed 80% isopropanol added to the postcolumn flow just prior to the ESI tip (ensuring stable spray), the prototype bead trap, and a nanomixer (Upchurch N-200). The mass spectrometer was an Applied Biosystems/Sciex 4000 QTRAP controlled by Analyst software v1.4. The bead trap SISCAPA method consisted of four phases: 1) assembly of the SISCAPA binding reaction mixture and incubation, 2) loading and washing beads in the bead trap, 3) elution of peptides from the beads and transfer to the PepMap C18 trap cartridge, and 4) gradient nanoflow chromatography of the peptides and detection by MRM in the mass spectrometer. Phase 1 was carried out off line in round bottom 96-well polypropylene cell culture cluster plates (Costar 3790; Corning Life Sciences, Lowell, MA) by mixing peptide sample (usually the tryptic digest of 5 μl of human plasma); anti-peptide antibody (typically 1 μg) in PBS, pH 7.5, detergent (typically a volume of 1% CHAPS needed to yield a 0.1% final CHAPS concentration); and washed Dynal G beads (typically 5 μl (or 150 μg) of beads/μg of antibody). Prior to use, the Dynal beads were washed three times by dilution into 5 ml of 0.1 m ammonium acetate, 0.5 m NaCl, 0.1% CHAPS, pH 7.5, and vortexing for 30 s followed by magnetic recovery after which the final bead mass was resuspended in a volume of PBS equal to the volume taken from the original product vial. Plasma digest, antibody, and detergent were typically mixed and incubated for 1–8 h followed by addition of beads and further incubation for 1 h on the microplate shaker. The pH of the final mixture (7.5–8.0) was confirmed by spotting 1 μl onto pH paper. Beads suspended in sample were transferred to snap cap vials for autosampler injection and resuspended manually just prior to injection. In phase 2, a suspension of beads in sample digest (typically 5 μl) was aspirated (sandwiched between two 5-μl volumes of wash solution) into a 10- or 15-μl sample loop mounted across the autosampler injection valve and delivered into the bead trap at 10 μl/min followed by a second 10-μl injection of wash solution. The MX7900 valve directed outflow from the bead trap to waste. Although it is unconventional to aspirate particle suspensions through an autosampler sample probe and injection valve, we observed no evidence of probe clogging and minimal particle retention in the valve. In phase 3, 10 μl of bead eluent (70% acetonitrile, 0.1% FA) was injected through the bead trap at 1 μl/min, and after a 6-min delay, the MX7900 valve was switched to deliver bead trap output to the auxiliary valve and then to the mixer where the eluted peptides in 70% acetonitrile, 0.1% FA were combined with a 10 μl/min flow of 0.1% formic acid from a diluent syringe pump, resulting in eluted peptides in a final concentration of 6% acetonitrile flowing over the PepMap C18 trap cartridge for 7 min. Following transfer of peptides to the PepMap C18 trap, the MX7900 valve was switched back to waste, and a 10-μl volume of bead ejection solution was injected over the bead trap while reversing the direction of bead trap rotor rotation (to sweep in the same direction as flow) to send the beads to waste. Phase 4 was triggered at the beginning of phase 2, and after a 13-min delay, the auxiliary valve was switched to place the PepMap C18 trap (now loaded with enriched peptides) in line with the nanoflow gradient system, which delivered a linear gradient of 5–60% B at 300 nl/min over 20 min followed by a 2-min ramp to 80% B and then re-equilibration. A large series of MRMs covering the peptides of interest as well as a series of strong peptides from high abundance plasma proteins were measured with 10- or 15-ms dwell times. For comparison with unfractionated plasma, a similar protocol was developed in which a plasma digest diluted in 70% acetonitrile, 0.1% formic acid (no antibodies or magnetic beads) was injected directly through the bead trap and diluted through the mixer onto the PepMap trap cartridge. An off-line elution process was used for comparison with the bead trap. Phase 1 of the off-line process was as described above, but elution was accomplished in round bottom polypropylene 96-well plates using the reagent addition steps interspersed with bead collection with a Novagen magnet array followed by resuspension on a Sarstedt plate shaker on “microplate” setting. The protocol proceeded as follows: remove digest; wash four times in 100 μl of 0.1 m ammonium acetate, 0.5 m NaCl, 0.1% CHAPS, pH 7.5; wash one time in water; elute in 20 μl of 5% acetic acid. The 20-μl peptide eluate was placed in empty wells and dried down in a SpeedVac vacuum concentrator after which the peptides were redissolved in 20 μl of 2% acetonitrile, 0.1% formic acid and transferred to clean vials in preparation for 15-μl LC-MS/MS injection.
MRM Quantitation—
Peak areas of measured MRMs were computed using either Analyst or MultiQuant software (Applied Biosystems). MRMs for peaks labeled in the figures are shown in Table I. A “target relative abundance” was computed as the ratio within a run of the areas of the target peptide peak and either a single high abundance peptide (e.g. HSA-1) or else the total ion current reconstructed as the sum of all MRMs measured. Because we used different sets of MRMs in various experiments, we used the ratio of target to HSA-1 (measured in all experiments) unless otherwise noted. The SISCAPA enrichment factor was computed as the ratio of target relative abundances in a SISCAPA experiment versus an LC-MS/MS run of the same unfractionated plasma digest used in the SISCAPA capture.
RESULTS
Elution Method—
The ideal eluent to recover the bound peptides would 1) release peptide from antibody, 2) retain the antibody on the bead and prevent its entry into the C18 trap or analytical column, and 3) prevent binding of the peptide to tubing or valve surfaces between the bead trap and the C18 trap. A series of eluents (0.1% HCl, 0.15 m NaCl; 6 m guanidine HCl; and 0.1% TFA plus 0–70% ACN in 10% increments with water alone as a negative control) was tested in comparison with the conventional 5% acetic acid eluent in experiments carried out with Alexa Fluor 488-labeled rabbit IgG (Invitrogen) bound to Dynal protein G beads in 96-well black polypropylene plates processed using the KingFisher automated magnetic bead platform (see supplemental bar graphs). We observed that either 0.1% HCl in 0.15 m NaCl or 0.1% TFA eluted slightly more labeled IgG signal than 5% acetic acid, whereas increasing concentrations of ACN in 0.1% TFA eluted progressively less IgG. The highest ACN concentration tested (0.1% FA, 70% ACN) appeared to prevent release of ∼90% of the antibody from the protein G beads (probably by “crashing” or precipitating the Ab on the bead surface). The acidic pH of all these eluents is expected to release bound peptides from the antibodies based on wide experience with elution of many antigens (e.g. release of major plasma proteins (2) and tryptic peptides (4) bound to immobilized antibody columns) and is confirmed in the experiments reported here: 70% ACN, 0.1% TFA efficiently released Alexa Fluor-labeled AAC peptide from the beads (where it was bound by unlabeled anti-AAC peptide antibody), whereas the Alexa Fluor-labeled Ab is not released in the same eluent. Based on this crash effect and the likelihood that most peptides would not bind to tubing walls in this solvent, we elected to use 70% ACN in 0.1% FA as the standard eluent. To allow the peptides to bind to the C18 trap of the LC system, a 1 + 10 dilution of the eluted peptides with 0.1% formic acid was carried out in a nanomixer immediately before flow over the C18 trap (Fig. 2). Water alone fortunately does not elute either peptide or antibody from protein G beads and thus can serve as a final wash before elution.
Fig. 2.
LC-MS/MS plumbing diagram showing integration of bead trap into the fluidic system used. N, north; S, south; IPA, isopropyl alcohol.
Fluidic Performance of the Bead Trap—
The rotating magnetic bead trap was tested at a variety of flow rates and bead loads in the presence of 0.1% CHAPS detergent. The action of sweeping successive magnetic trapping regions against the direction of fluid flow (i.e. upstream) was found to trap and retain the nominal bead load (5 μl of the standard Dynal G package concentration of ∼30 mg/ml) at flow rates up to 50 μl/min. A video clip of 5 μl of Dynal G beads maintained in a capillary against a 10 μl/min flow (from left to right) is included as supplemental video Beadtrap CIMG1277.avi. Larger amounts of beads (up to 15 μl) could be retained although with less efficient mixing as the capillary in the primary magnetic trap region was packed with a “solid” but undulating mass of beads. In the absence of detergent, a portion of the bead load formed a more flocculent mass (instead of the smoothly flowing mass seen with detergent) and occasionally adhered to the walls of the bead trap capillary. However, this effect did not appear to impede final washing or elution and was instantly reversed when a bolus of detergent was injected to release the beads. Reversal of rotation of the bead trap rotor, so that the magnetic trap regions moved in the same sense as the liquid flow, resulted in complete ejection of all the beads.
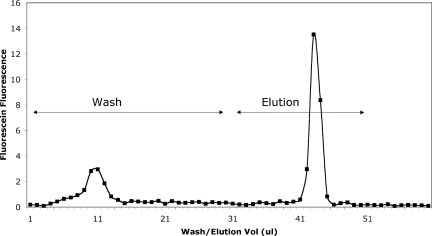
We examined the efficiency of washing and elution using protein G beads loaded with rabbit anti-AAC-1 affinity-purified antibody that in turn bound fluorescein-labeled AAC-1 peptide (Fig. 3). Outflow from the loaded bead trap was collected in 1-μl aliquots during a wash step and subsequent elution with 50% acetonitrile, 0.1% TFA; neutralized; and read in a fluorescence plate reader. The width of the peak of eluted peptide at half-maximum is ∼2 μl, or ∼1/10 of the minimum volume in which the beads can be processed off line (either manually or with a robot such as the KingFisher).
Fig. 3.
Elution of fluorescein-labeled peptide from magnetic beads during washing and elution in bead trap.
Bead Trap SISCAPA—
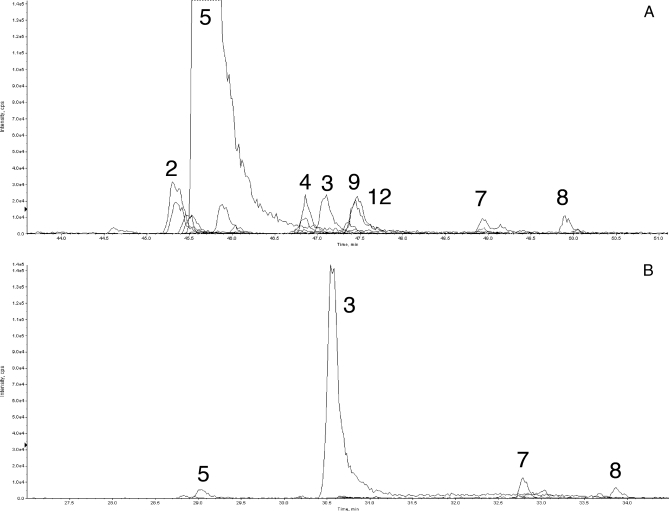
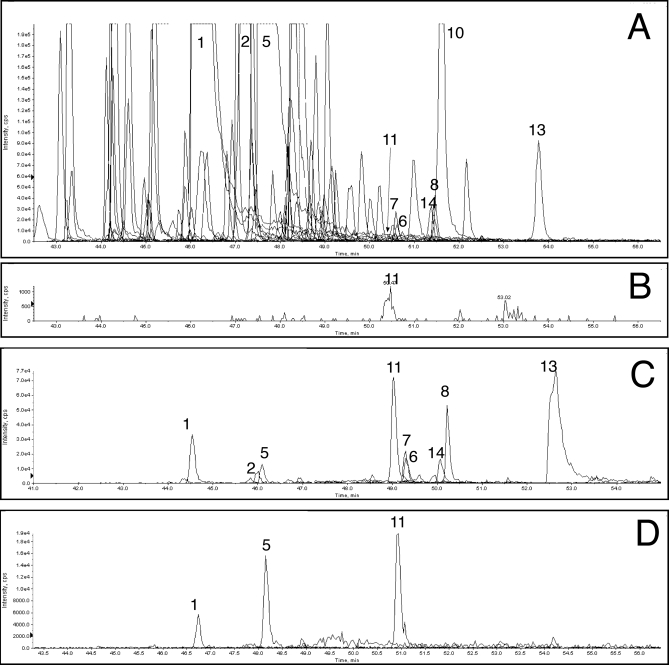
The bead trap device was evaluated in an operating nanoflow LC-MS/MS system by introducing it in place of a transfer line between autosampler injection and LC valves (Fig. 2). A SISCAPA enrichment was carried out by loading the bead trap with a 5-μl mixture containing 1) human plasma digest equivalent to 160 nl of plasma, 2) 0.6 μg of affinity-purified rabbit polyclonal anti-AAC peptide antibody, and 3) 2.8 μl of washed Dynal protein G beads all in 0.1 m ammonium acetate and 0.1% CHAPS. A total of 57 MRMs were monitored by the 4000 QTRAP in triple quadrupole mode, including the AAC-1 peptide and a series of peptides representing other high abundance plasma proteins such as albumin. As an unfractionated control, a sample containing the unfractionated digest of 14 nl of human plasma diluted to 10 μl in 70% acetonitrile, 0.1% formic acid (∼1 μg of total peptide; no antibodies or beads) was injected through the same plumbing system and diluted prior to capture on the C18 PepMap trap to mimic the procedure used for the SISCAPA run. Fig. 4 presents the results of all 57 MRMs for both samples. In Fig. 4A (the unfractionated digest experiment), AAC-1 peptide represents a small peak (labeled 3), and the HSA-1 peptide (peak 5) is off scale, whereas in Fig. 4B (the SISCAPA experiment), the AAC-1 peptide is almost 7-fold higher in intensity, and the HSA-1 peptide is reduced to near base line. The ratio of AAC-1 to HSA-1 (the target relative abundance) is 0.015 in the unfractionated digest and 27.5 in the SISCAPA experiment, yielding a change in ratios (the overall enrichment of AAC-1 relative to HSA-1 peptide) of 1,800. In the SISCAPA run, the peak area for AAC-1 was increased 6.4-fold, indicating an efficiency of antibody capture of about 55% from an 11.5-fold greater amount of plasma digest.
Fig. 4.
Enrichment of medium abundance AAC-1 peptide. Traces of MRM signals during LC-MS/MS analysis of unfractionated plasma digest (A) and peptides eluted in bead trap from beads carrying anti-AAC-1 antibody (B) are shown. Peak 5 is a major albumin peptide (HSA-1), and peak 3 is AAC-1 peptide; others are identified in Table I. cps, counts/s; XIC, extracted ion chromatogram.
Enrichment of a Lower Abundance Peptide, LBP-1b—
A tryptic peptide of LPS-binding protein (LBP-1b, whose plasma abundance is ∼50-fold lower than AAC; Table II) was captured by an affinity-purified rabbit polyclonal antibody (having an affinity about 10 times better than the AAC-1 Ab) from the digest of 5 μl of human plasma. Because SISCAPA enrichment in previous experiments did not appear to be perfect (i.e. eliminating all off-target MRM peaks) when capturing a lower abundance peptide from larger amounts of plasma digest (the typical case for SISCAPA assays) we screened a larger series of MRMs to monitor a wider range of high abundance peptides from the plasma digest. Using the extended MRM set, more peaks were observed (Fig. 5A). We also introduced an MRM for CHAPS detergent and showed that after bead washing its effect was minimal. LPB-1b was barely detected in the unfractionated plasma digest (its MRM is shown in Fig. 5B), whereas it was a dominant peak after antibody capture and bead trap elution (Fig. 5C). The ratio of LBP-1b (peak 11) to HSA-1 (peak 5) areas was 3.2e−4 and 5.7 in the unfractionated plasma and SISCAPA capture, respectively, yielding an enrichment of 18,000. The LBP-1b peak area was 54-fold greater after capture from 357-fold more digest, giving a capture efficiency of ∼15%.
Table II.
Normal plasma abundances of proteins compared
Fig. 5.
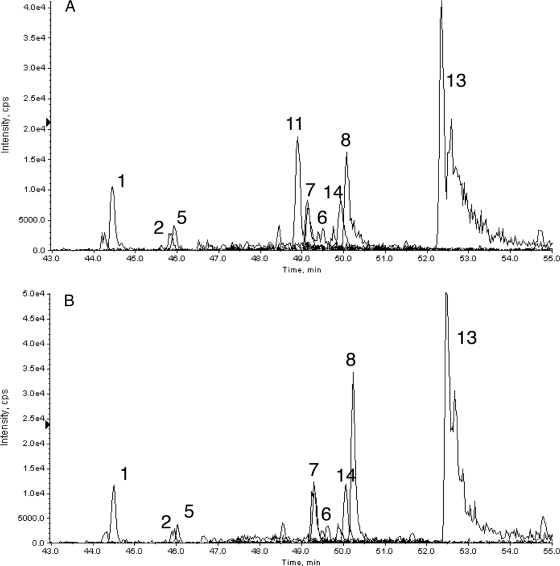
Enrichment of lower abundance LBP-1b peptide. A, unfractionated digest using a more comprehensive MRM set; B, isolated LBP-1b MRM trace from A; C, peptides eluted in bead trap from beads carrying anti-LBP-1b antibody; D, peptides eluted off line from beads carrying anti-LBP-1b antibody. Peptide peaks are identified in Table I. cps, counts/s; XIC, extracted ion chromatogram.
Bead Trap Versus Off-line Elution—
The off-target peptides recovered were different if, after capture, the peptides were eluted from the beads off line with 5% acetic acid and the peptide supernatant was dried and reconstituted in 2% acetonitrile, 0.1% formic acid for LC-MS/MS (off-line processing; Fig. 5D). The peak area observed for the LBP-1b peptide after off-line elution (1.7e+5) was ∼25% of that observed after bead trap elution (6e+5) using identical capture aliquots processed to yield equal volumes of sample loaded. The enrichment factor for LPB-1b eluted off line was 4,500 compared with HSA-1. The direct comparison of bead trap versus off-line elution was done in two different experiments, each of which included two such comparisons (i.e. two bead trap elutions versus two off-line elutions of the same SISCAPA capture samples). The appearance of Fig. 5D is typical of all four off-line elution runs, and Fig. 5C is typical of the bead trap elutions (e.g. compare with Fig. 6A). Bead trap elution using 5% acetic acid gave a pattern of eluted peptides very similar to that observed with 70% acetonitrile, 0.1% formic acid (data not shown), indicating that the eluents yield similar sets of eluted peptides but that in-line (bead trap) and off-line elution methods differ significantly in peptide recovery. Most peptides, and especially the later eluting more hydrophobic peptides, were substantially reduced in off-line processing as opposed to bead trap elution.
Fig. 6.
Bead trap elution of a SISCAPA capture reaction (A) and a mock capture omitting antibody (B). cps, counts/s; XIC, extracted ion chromatogram.
Using the bead trap we observed the same nonspecifically bound tryptic peptides from high abundance plasma proteins in experiments with a variety of other anti-peptide antibodies, indicating that these peptides must be bound at sites distinct from the peptide binding sites of the antibodies. To test whether off-target peptides were bound by the antibodies (at some other site than the antigen binding site) or by other components such as the bead itself, we compared SISCAPA capture reactions using the LBP-1b antibody (Fig. 6A) with an identical reaction from which antibody was omitted (Fig. 6B). The only peptide peak showing a significant difference was the LBP-1b target peptide (peak 11) as expected if the nonspecific binding sites are associated with the surfaces of the magnetic beads or capillary tubing rather than the capture antibody itself.
DISCUSSION
Extending the capability of mass spectrometry to accurately measure concentrations of clinically interesting proteins is a major goal of biomarker proteomics. Quantitative MRM-MS offers potentially absolute specificity and large scale multiplexing with excellent analytical reproducibility. MS sensitivity remains an issue, however, with the best immunoassays offering detection limits several orders of magnitude lower. For a typical 50-kDa protein target, the 100-amol current best practice limit of MRM quantitation via nano-LC-ESI-MS/MS is equivalent to 5 pg of the protein or 0.1 pg of a typical tryptic peptide derived from the protein. If the digest of 1 ml of plasma could be loaded onto such an LC-MS/MS system, this would give an analytical sensitivity of 5 pg/ml protein, equal to the best immunoassays and satisfying most known biomarker requirements. Unfortunately nanoflow LC typically accommodates only ∼1 μg of total peptide load, equivalent to the tryptic digest of ∼10 nl of plasma: 1/100,000 ml. Using such an analytical system to analyze digests of unfractionated plasma, we are thus 5 orders of magnitude shy of the sensitivity goal.
Immunoaffinity depletion (2) and strong cation exchange fractionation (3) can each improve sensitivity by about 10× per fractionation “dimension” as can other fractionation approaches (10–14), together providing MRM measurements approaching ng/ml protein sensitivities. These methods have the advantage of being able to process large volumes of plasma or serum, and predigestion depletion reduces the scale of tryptic digestion substantially. The attendant disadvantages are the multistep nature of such methods (with increased cost and possibility of protein/peptide loss) and the multiplicative increase in the number of LC-MS/MS analyses that may be required to cover an arbitrary number of protein/peptide targets. Combination of more than two such stages to achieve subnanogram sensitivity is probably impractical for analysis of large clinical sample sets and thus does not solve the general problem of verifying or qualifying biomarker candidates.
The SISCAPA (4) method takes a different approach, using peptide-specific affinity reagents (anti-peptide antibodies) to enrich identified target peptides from a complex digest (e.g. unfractionated, digested plasma). Recent work has focused on magnetic bead-based implementations (5) of this method because such single use capture reagents 1) eliminate the possibility of carryover inherent in the fixed immunoadsorbent column format, 2) allow capture from larger volumes of digest than can conveniently be flowed over a fixed nanoaffinity column, 3) facilitate reuse of a sample digest for later capture of additional peptides, and 4) allow off-line reagent addition to many samples in parallel, thereby providing enough time for the antibody-peptide reaction to achieve equilibrium binding. In this scheme, capturing target peptides (with good recovery) from a digest of 10 or 100 μl of plasma should provide 1e+3 or 1e+4 improvement in MRM sensitivity compared with unfractionated digest essentially by scaling up the equivalent plasma volume of the LC-MS/MS sample from 10 nl to 10–100 μl. Although 1-ml digests are certainly possible, cost (largely trypsin) and volume available from sample banks become limiting issues. Sample sizes of 10 and 100 μl of plasma would give calculated sensitivities (at 100-amol limit of quantitation) of 50–500 pg/ml protein, approaching the sensitivity of high performance clinical immunoassays. Anticipated advances in triple quadrupole MS sensitivity should drive this limit down a further order of magnitude to 5–50 pg/ml protein, opening the door for MS to replace almost all existing clinical immunoassays while providing better specificity (i.e. better quality) with facile multiplexing (lower cost).
Our magnetic bead implementation of SISCAPA aimed to achieve maximum flexibility and simplicity by avoiding covalent coupling of antibody to the beads. This approach allows easy mixing of antibody mixtures and is particularly well suited to screening antibodies (including monoclonal antibody-containing supernatants). Commercially available magnetic beads coated with protein G are added, either with the antibody or later, to bind the antibodies from solution and transport them and their peptide cargo out of the digest for subsequent washing and elution. In a reference capture reaction, we found that 1 μg of antibody (sufficient to capture most peptide targets from a 100-μl volume) can be retrieved with high (>70%) efficiency by this two-step capture process. A potential disadvantage with this “free antibody” approach is the release of the antibodies from the beads with the peptides on elution and attendant possibility of clogging of the C18 trap or analytical column. Several approaches are available to circumvent such a problem. First, the antibodies could be biotinylated at the cost of additional effort in preparing each antibody, allowing them to be bound essentially irreversibly by streptavidin-coated beads. Second, a modified C18 trap could be used that is capable of binding peptides but not proteins, analogous to the size-selective media used by Rule and Henion (15) to bind small molecules but not antibodies. Third, elution conditions could be tailored to release peptides from the antibodies but not the antibodies from the beads, which is the strategy pursued here. We found that acidic solutions (<pH 3.5) effectively release the peptides, and that addition of high organic content (e.g. 70% acetonitrile) effectively precipitates (“crashes”) the antibodies on the beads. In theory this approach offers the optimal transport of peptides from the magnetic bead surface to the C18 trap, sweeping all the intervening tubing with a high-organic solvent that prevents peptide adsorption. Because highly organic solvent would prevent the released peptides from binding to C18 chromatography supports downstream, we introduced a 1 + 10 dilution step immediately prior to the C18 trap.
By capturing specific peptides, SISCAPA drastically simplifies the peptide sample presented for MS analysis, in principal reducing a mixture of hundreds of thousands of peptides to a handful. This simplification results in two additional advantages: reduction of ion suppression and potential for shortened reversed phase chromatography (and hence increased sample throughput). Our experience (data not shown) suggests that ion suppression for peptides of interest in an unfractionated plasma digest is significant (an average 2–4-fold reduction in response) but not an overwhelming factor in total sensitivity for nano-LC-MS/MS. The potential for shortening the final reversed phase chromatography step (e.g. from the current 20–40 to 2–4 min) is an obvious throughput advantage, the potential of which has yet to be fully exploited.
The advantages of isolating target peptides from a complex digest come at the risk of losing material during processing; 100 amol (60 million molecules or ∼0.1 pg) of a possibly “sticky” target peptide is very easy to lose without a carrier to saturate surface binding sites. The well known difficulty of recovering peptides from in-gel digests of small two-dimensional gel spots is largely attributable to such losses on the walls of 96-well plates. Similarly the probability of efficiently recovering 1 fmol of a pure synthetic peptide from a storage vial is generally thought to be low.
Here we have described improvements in the SISCAPA method aimed at preventing peptide losses after capture. We used a rotating magnetic bead trap device to minimize loss of peptides captured by anti-peptide antibodies on magnetic particles, carrying out all the washing and elution steps inside a capillary flow system that can be purged completely by highly organic solvents. By continuously sweeping magnetic trapping regions against the direction of liquid flow (i.e. upstream) and by ensuring that a continuous succession of trap regions are available to capture any beads escaping from the primary trap zone, the device can hold a mass of magnetic beads in a 150-μm-inner diameter capillary indefinitely while liquid flows by at rates up to 10–50 μl/min. Movement of the trap regions also stirs the beads continuously, facilitating interaction with passing solvents. Using this system, we could elute bound peptides in a 2-μl volume (at 1 μl/min flow), which is less than 1/10 the minimum volume practical in plate-based magnetic bead systems. The device has only two controls (on-off and direction of rotation) and hence is easily interfaced with LC-MS/MS equipment through software-controlled contact closures. The remaining uncaptured digest peptides can be replaced in the original sample vial, available for subsequent capture of other peptides.
In experiments with two peptide/polyclonal antibody pairs (AAC-1 and LBP-1b) captured from an unfractionated human plasma digest, we obtained enrichment factors of 1,800 and 18,000, respectively, versus a high abundance albumin peptide. These factors represent an underestimate of the true enrichment by the antibody because the HSA-1 peptide, like the other non-target peptides, was carried through the capture by nonspecific interaction with the beads, not the antibody. In fact if this nonspecific binding could be eliminated, it appears (Fig. 6, A and B) that only the target peptide among the MRMs measured would show a peak, approaching the ideal for purification of the target peptide.
The approach described can be further improved by several additional developments. First it is clear that magnetic beads with lowered nonspecific peptide binding are needed because more peptide is currently bound nonspecifically than specifically via the antibody. Current beads have been developed to minimize protein binding, as required in conventional immunoassays, but not specifically to minimize peptide binding. It is likely that the beads we used (based on polystyrene) retain a hydrophobic surface character and thus are likely to bind a variety of sticky peptides. Smaller beads (e.g. 1 μm or less) would also be advantageous because they would remain in suspension and hence not require resuspension during capture or immediately before aspiration into the bead trap. A second area for development is the fluid flow system. Although the bead trap has been successfully integrated into existing nanoflow LC systems, a purpose-built fluid system for delivering wash and elution solvents would allow faster cycle times and simpler method development. In the future, it should be possible to overcome the need for long reversed phase separations to spread peptides out and focus instead on very short separations aimed primarily at stacking a few essentially pure peptides into very sharp peaks or ultimately to eliminate the reversed phase separation entirely by spraying the SISCAPA capture eluent directly into the mass spectrometer. The third important area for development concerns antibodies. Equilibrium binding calculations suggest that efficient capture (i.e. >50%) of low abundance peptides from large digest volumes requires antibodies with dissociation constants of <1e−9. The AAC and LBP-1b rabbit polyclonal Abs used here have measured KD values of 3.8 and 0.34 nm, respectively, with the better (LBP-1b) Ab giving ∼10× better enrichment ratio. We have now demonstrated comparable or better affinities with a variety of monoclonal rabbit antibodies specific for several peptide targets (9), indicating that the required affinity is likely to be generally achievable. The bead trap approach provides a convenient interface between magnetic particle fractionation methods generating small quantities of purified analytes and the preferred nanoflow LC-MS/MS systems providing precise quantitation and absolute structural specificity.
Supplementary Material
Acknowledgments
We thank Matt Pope and Martin Soste in T. W. Pearson's laboratory for Biacore measurements of antibody affinities. We acknowledge Applied Biosystems for loan of MS equipment. The bead trap device was developed and constructed by Anderson Forschung Group LLC.
Footnotes
Published, MCP Papers in Press, February 4, 2009, DOI 10.1074/mcp.M800446-MCP200
The abbreviations used are: MRM, multiple reaction monitoring; SISCAPA, stable isotope standards and capture by anti-peptide antibodies; FA, formic acid; HSA, human serum albumin; LPS, lipopolysaccharide; AAC, α1-antichymotrypsin; Ab, antibody; LBP, LPS-binding protein; ESI, electrospray ionization.
This work was supported, in whole or in part, by National Institutes of Health Grant U24-CA126476-01 from the NCI Clinical Proteomic Technology Assessment for Cancer program. This work was also supported by a Canary Foundation seed grant.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Anderson, L., and Hunter, C. L. ( 2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 2.Pieper, R., Su, Q., Gatlin, C. L., Huang, S. T., Anderson, N. L., and Steiner, S. ( 2003) Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 3, 422–432 [DOI] [PubMed] [Google Scholar]
- 3.Keshishian, H., Addona, T., Burgess, M., Kuhn, E., and Carr, S. A. ( 2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, N. L., Anderson, N. G., Haines, L. R., Hardie, D. B., Olafson, R. W., and Pearson, T. W. ( 2004) Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 5.Whiteaker, J. R., Zhao, L., Zhang, H. Y., Feng, L. C., Piening, B. D., Anderson, L., and Paulovich, A. G. ( 2007) Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal. Biochem. 362, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safarik, I., and Safarikova, M. ( 2004) Magnetic techniques for the isolation and purification of proteins and peptides. Biomagn. Res. Technol. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pamme, N. ( 2006) Magnetism and microfluidics. Lab Chip 6, 24–38 [DOI] [PubMed] [Google Scholar]
- 8.Rida, A., and Gijs, M. A. ( 2004) Manipulation of self-assembled structures of magnetic beads for microfluidic mixing and assaying. Anal. Chem. 76, 6239–6246 [DOI] [PubMed] [Google Scholar]
- 9.Pope, M. E., Soste, M. V., Eyford, B. A., Anderson, N. L., and Pearson, T. W. ( 2009) Anti-peptide antibody screening: selection of high affinity monoclonal reagents by a refined surface plasmon resonance technique. J. Immunol. Methods 341, 86–96 [DOI] [PubMed] [Google Scholar]
- 10.Pieper, R., Gatlin, C. L., Makusky, A. J., Russo, P. S., Schatz, C. R., Miller, S. S., Su, Q., McGrath, A. M., Estock, M. A., Parmar, P. P., Zhao, M., Huang, S. T., Zhou, J., Wang, F., Esquer-Blasco, R., Anderson, N. L., Taylor, J., and Steiner, S. ( 2003) The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 3, 1345–1364 [DOI] [PubMed] [Google Scholar]
- 11.Dayarathna, M. K., Hancock, W. S., and Hincapie, M. ( 2008) A two step fractionation approach for plasma proteomics using immunodepletion of abundant proteins and multi-lectin affinity chromatography: application to the analysis of obesity, diabetes, and hypertension diseases. J. Sep. Sci. 31, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 12.Regnier, F. E., Riggs, L., Zhang, R., Xiong, L., Liu, P., Chakraborty, A., Seeley, E., Sioma, C., and Thompson, R. A. ( 2002) Comparative proteomics based on stable isotope labeling and affinity selection. J. Mass Spectrom. 37, 133–145 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, Y., Aebersold, R., and Zhang, H. ( 2007) Isolation of N-linked glycopeptides from plasma. Anal. Chem. 79, 5826–5837 [DOI] [PubMed] [Google Scholar]
- 14.Faca, V., Pitteri, S. J., Newcomb, L., Glukhova, V., Phanstiel, D., Krasnoselsky, A., Zhang, Q., Struthers, J., Wang, H., Eng, J., Fitzgibbon, M., McIntosh, M., and Hanash, S. ( 2007) Contribution of protein fractionation to depth of analysis of the serum and plasma proteomes. J. Proteome Res. 6, 3558–3565 [DOI] [PubMed] [Google Scholar]
- 15.Rule, G. S., and Henion, J. D. ( 1992) Determination of drugs from urine by on-line immunoaffinity chromatography-high-performance liquid chromatography-mass spectrometry. J. Chromatogr. 582, 103–112 [DOI] [PubMed] [Google Scholar]
- 16.Cunningham, S. C., Malone, D. L., Bochicchio, G. V., Genuit, T., Keledjian, K., Tracy, J. K., and Napolitano, L. M. ( 2006) Serum lipopolysaccharide-binding protein concentrations in trauma victims. Surg. Infect. (Larchmt.) 7, 251–261 [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki, S., Iwamura, K., Itakura, M., Kamiguchi, H., and Katsunuma, T. ( 1981) A clinical evaluation of serum α-1-antichymotrypsin levels in liver disease and cancers. Gastroenterol. Jpn. 16, 582–591 [DOI] [PubMed] [Google Scholar]
- 18.Hollander, C., Westin, U., Wallmark, A., Piitulainen, E., Sveger, T., and Janciauskiene, S. M. ( 2007) Plasma levels of α1-antichymotrypsin and secretory leukocyte proteinase inhibitor in healthy and chronic obstructive pulmonary disease (COPD) subjects with and without severe α1-antitrypsin deficiency. BMC Pulm. Med. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.