Abstract
The overarching goal of the NIF (Neuroscience Information Framework) project is to be a one-stop-shop for Neuroscience. This paper provides a technical overview of how the system is designed. The technical goal of the first version of the NIF system was to develop an information system that a neuroscientist can use to locate relevant information from a wide variety of information sources by simple keyword queries. Although the user would provide only keywords to retrieve information, the NIF system is designed to treat them as concepts whose meanings are interpreted by the system. Thus, a search for term should find a record containing synonyms of the term. The system is targeted to find information from web pages, publications, databases, web sites built upon databases, XML documents and any other modality in which such information may be published. We have designed a system to achieve this functionality. A central element in the system is an ontology called NIFSTD (for NIF Standard) constructed by amalgamating a number of known and newly developed ontologies. NIFSTD is used by our ontology management module, called OntoQuest to perform ontology-based search over data sources. The NIF architecture currently provides three different mechanisms for searching heterogeneous data sources including relational databases, web sites, XML documents and full text of publications. Version 1.0 of the NIF system is currently in beta test and may be accessed through http://nif.nih.gov.
Keywords: ontology, data federation, neuroscience resource
Introduction
Today, there are thousands of neuroscience information resources created by a wide range of information providers including research groups, funding agencies, vendor groups and public data initiatives that publish information in one form or another. A neuroscience information resource is any electronically accessible site that provides information of interest to neuroscience. A neuroscience information resource can be a digital library of publications like PubMed; it can be the web site of a neuroscience research group that publishes its research detail as web pages; it can be a tissue bank that allows a potential neuroscientist customer to navigate through its samples; it can be a database that houses experimental research results, and allows users to query it; it can even be a software tool that enables a user to perform a computation online. Unfortunately, despite the growing body of information resources, the problem of finding just the right information from one or more of these has not become easier, and it may be very hard for a general neuroscientist to locate a relevant information resource if she does not know about its existence. Let us consider the following example to illustrate the problem. Assume that a neuroscientist is looking for resources that might provide cDNA for mouse models. Typically, she would use the Google search engine with the keyword combination like “mouse, model, cDNA”. Figure 1(a) shows the first result page returned by Google. Although the results are indeed about mouse models and cDNA, they are mostly URLs of Google-indexed publications and general web sites.
Figure 1.
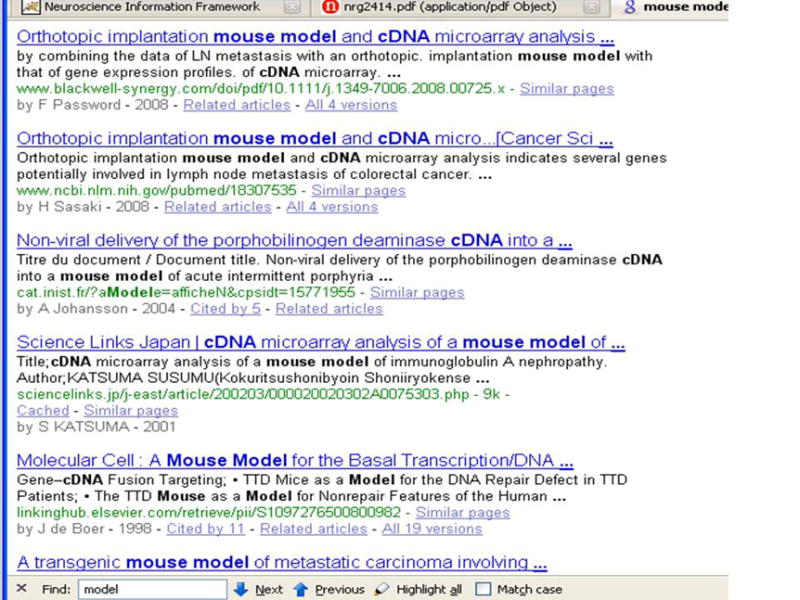
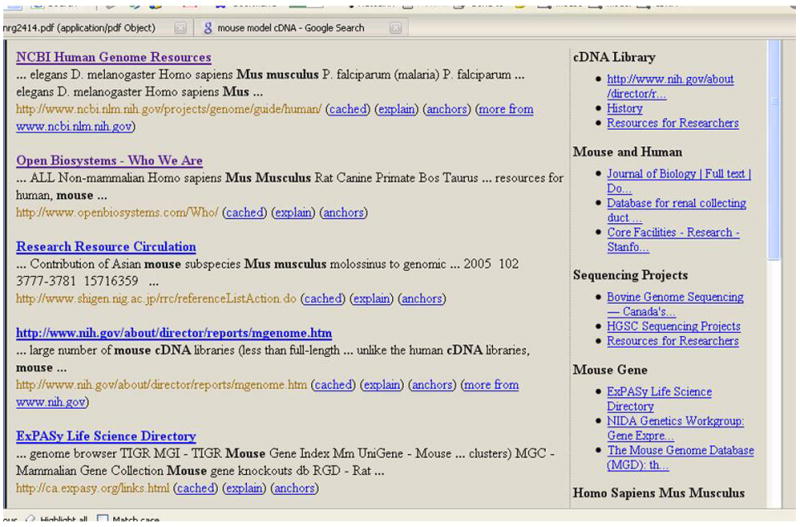
Figure 1(a). The result of the query (mouse model cDNA) against Google. The top results are very general, and mostly from papers that are indexed by Google.
Figure 1(b). The same query as in Figure 1(a) now executed against NIF. In contrast with Google, the selective web crawling coverage of NIF enables it to return results that are more closely related to Neuroscience.
A more focused search that might actually help the neuroscientist better is shown in Figure 1(b). This result is mostly about resources like Open Biosystems that can be used as a resource for cDNA libraries for mouse models of human diseases. The resource finding problem gets compounded if the neuroscientist wants to search for the information not only from the web, but over any kind of information resource mentioned in the previous paragraph, because there are no search tools that provide adequate functionality to satisfy the information needs of our neuroscientist.
The Problem
There are a number of underlying factors behind the resource finding problem. These factors are not specific to the domain of neuroscience, but come into play whenever a discipline-specific information seeker tries to locate information resources that have been created for very different goals, have heterogeneous content, provide heterogeneous access mechanisms, and have not been put into a common information framework. In the following, we list the contributing factors that need to be overcome to allow an information seeker find meaningful results quickly over these heterogeneous data sources.
Although a domain user searches for resources using keywords, the intent of the search is conceptual. Thus, although the search term astrocytoma, there is an implicit expectation that a resource about astrocytic glioma or glial malignancy will be part of the result. Most search engines do not provide a semantic search facility, which includes not only search by synonyms but by terms that are notionally related to the search terms. As another example, the user searching for hippocampal formation would possibly also be interested in the cell types found there because they are semantically related.
-
The web is an important class of information resource, but discipline-specific web search suffers from three significant limitations:
Most web search engines are not discipline specific and are based on a general pagerank like mechanism. Therefore, the result of a query is more likely to use the general popularity of a page instead of finding its discipline-specific relevance when returning a search result. Thus a query on knockout is likely to rank web pages on “knockout matches” in sports higher than “knockout animals”, which are more relevant to biological sciences.
The problem of the “deep web”, whereby a resource does not expose the content of the database but allows a user to access it only through forms or some other functional interface, is not yet solved. It is an active research area among information management and information retrieval researchers.
The web is not a coherently designed information system. So it does not resolve or correlate an information entity found in one source to another information entity found in another source, even though might refer to the same real world entity. Thus two web sites referring to the same publication are considered to be different pieces of information, and will typically produce duplicated results for a query.
Keyword-based search is the most natural form of information search when the user knows very little about the structure and content of an information resource. However, not all information resources provide a facility for keyword search. For example, database systems or data files may support very limited form of keyword search if any (although academic researchers are working in this area e.g., Hristidis et al (2003)). On the other hand, as the user gets to know an information source better, the user prefers some other mode of information access including data browsing, queries, or special purpose techniques like atlas exploration. There are no such search/query tools that provide the user with the ability to uniformly search over all types of heterogeneous information resources and then refine the search procedure as the user gains the ability to perform deeper search.
To address this problem, we have developed an information federation framework called NIF (Neuroscience Information Framework) where heterogeneous information resources can be accessed through a shared ontology. This framework is designed to admit resources that provide different degrees of access to their data content. An extensible OWL (Web Ontology Language, see http://www.w3.org/TR/owl-ref/) ontology called NIFSTD (NIF Standard) has been constructed based on sound ontological principles. We have constructed OntoQuest, an ontology management system that permits a user to store, search and navigate any number of OWL-structured ontologies. A fully functional web-accessible system, NIF version 1.0, currently in beta release, (available through http://nif.nih.gov) has been developed.
In the following sections we describe the overall architecture and different components of the NIF system. More details on the background of the NIF project is covered in the NIF white paper in this issue.
The NIF System Architecture
Recently, the term dataspace has been introduced in Franklin et al (2005) to refer to an information management scenario where the data resides not just within the custody of a managed storage-and-retrieval software like a DBMS (DataBase Management System like MySQL or Oracle), but in text files, emails, software-produced documents, and yet provides a set of common services for search and information organization. The NIF architecture is an example of a dataspace system that provides search and data exploration services over heterogeneous information systems whose capabilities and ontological descriptions are registered to a few central catalogs.
The NIF system currently consists of a set of tools and services to search collectively across different types of neuroscience resources through a simple interface (Fig. 2). The system also includes a set of registration tools to make resources known to the NIF data integration system. A few of the components are described in further detail in the following paragraphs. Let us first define a few terms used in the rest of the paper.
Figure 2.
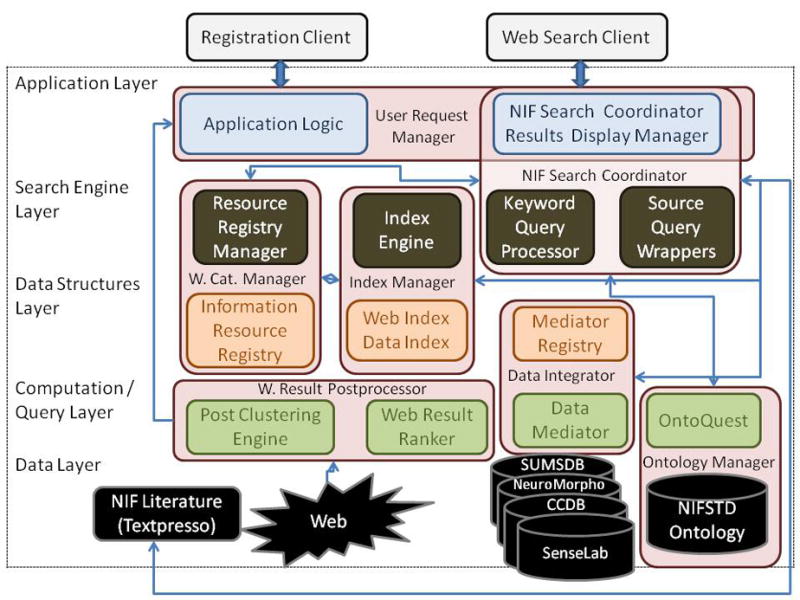
The architecture of the NIF system is organized by layers; the clients at the top of the diagram, the data and ontology sources are at the bottom. The middle layers contain the modules for supporting search, data and index structures, and the different query handlers. The word Web has been abbreviated to W. The combination of the databases, Textpresso, the web resources are collectively called “NIF Data Resources”.
A NIF Web Resource is a web site that has information relevant to Neuroscientists. Such a resource can be an informational web site that only allows browsing, a web site that allows browsing and queries through web forms, software sites, sites for chemicals like reagents, and so on.
A NIF Data Resource is a database that enables an external application to send a query using a query API or a query language.
A Data Mediator is a data integration engine, developed in the context of the BIRN (Biomedical Informatics Research Network) project that allows one to query a set of distributed relational databases, and computation engines, by creating a single virtual database on top of them.
A Mediated NIF Data Resource is a database that can be queried by NIF only through the Data Mediator.
The NIF Literature Resource is a text processing system that parses publications, extracts its metadata, marks its content from a known vocabulary and allows the NIF system to search for publications through keyword and metadata queries. Currently, the NIF literature resource is assembled through the Textpresso text indexing system (see Müller et al. in this issue).
The NIF Ontology is a human curated, semi-automatically assimilated OWL-structured ontology called NIFSTD (NIF Standard, see Bug et al (2008) for details) that contains terms and inter-term relationships relevant to neuroscience researchers.
The overall architecture of the NIF system is shown in Figure 2. The different building blocks are discussed below.
Registration Client
The client software that allows an authorized user to add a new resource name to the NIF Web Catalog.
Web Search Client
This is the web client that is used by the simple and the advanced query interfaces.
The Application Layer facilitates the user’s interaction with the system and contains the following:
User Request Manager: The Request Manager is the entry point of the system where the users can either add a new entry for the NIF Web Catalog, and more importantly performs a conceptual search operation. The application logic handles the request, for example, by passing on the query to the Search Coordinator, described next. It also controls the display of the results.
NIF Search Coordinator: An integral part of the application logic, the NIF Search Coordinator takes the user’s keyword query and in the most common case, performs an ontological search to retrieve conceptual terms that closely match the terms in the ontology, and if desired, the neighborhood of these ontological terms. This process of exploring the ontology to find related terms is performed interactively. When the user settles on the final query terms, the keyword module uses the index to locate sources that have the data or web documents satisfying the keywords. Once the data sources are located, the source query wrapper module transforms the query into queries against all sources and broadcasts these transformed queries. The process of transformation converts the query keywords into SQL (or HTTP calls and so on) for structured data sources, XML requests, search against the web index and so forth. If the user’s search terms are not found in the ontology, the search coordinator allows the query to be posted directly against the sources as a string search.
The Search Engine Layer performs the tasks needed to transform the user’s query to actual search instructions within the NIF system. It contains the following components.
Keyword Query Processor: This module manipulates the user’s keyword queries to an internal form
Index Manager: We use the term Index Manager to refer to the indexing engine and the controlling program surrounding it. The NIF system uses the Lucene indexing engine from Apache to create an inverted index of the results of the web crawl. The Lucene index is also used to index all readable data sources, both relational and XML. The index manager contains the methods to create, update and access the index, and is primarily used by the NIF Search Coordinator.
-
NIF Web Catalog Manager: The NIF Web Catalog (also called the “NIF Registry”) is a repository of NIF Web Resources. For each resource, NIF maintains a number of attributes that characterize the resource. Of these, some like the URL of the resource, or the rough classification of the source are mandatory, while others, like the detailed description of the Web Resource are optional. In the current version of the NIF system, the category assigned to a Web Resource comes from a simple hierarchical vocabulary (e.g., a neural modeling resource comes under the category software resource) assembled by the NIF team. In the current implementation, both the catalog and the vocabulary are structured as XML documents. The Catalog Handler is a set of methods and index structures that enable searching of the catalog information. Currently, both keyword queries and XML queries are supported by the handler.
To include more web pages, we developed “NIF Web”, which uses a web crawler to traverse the web sites contained in the NIF catalog. This expands the scope of NIF search beyond the web sites of the NIF catalog, but still keeps the scope within the realms of Neuroscience. Using these seed sites, the Nutch web crawler from Apache (http://lucene.apache.org/nutch/) is used to crawl the links to a depth of 15. Even as the number of seed sites grows, we have found that the 15-deep crawl provides a sufficiently broad coverage and yet retrieves web pages that largely contain information relevant to Neuroscience. The results from the web crawl are harvested and sent to the Index manager.
The Data Structure Layer contains different index structures to make queries faster. A technical description of this layer is beyond the scope of this paper.
The Computation and Query Layer refers to the modules that actually performs queries against the various sources, and manages the results that are returned. The main modules in this layer are:
NIF Ontology Manager: The NIF Ontology (NIFSTD) is a large and growing OWL entity that is itself a combination of several ontologies (see Bug et al (2008)). These ontologies are stored in OntoQuest (Chen et al (2006)). Partly inspired by the IODT framework from IBM (Mei et al 2006) OntoQuest stores all distinguished relationships permitted by OWL (e.g., subclass-of, allValuesFrom, disjoint etc.) in separate tables, while all user-defined relation names are stored in a quad-store. Logically, OntoQuest views the ontology as a graph and performs graph-like operations (e.g., finding the k-neighborhood) on it. It contains specialized indexes (see Chen et al (2005)) to quickly find ancestor-descendant like relationships for transitive relationships like subclass-of and part-of. OntoQuest contains its own query processing engine to support ontological queries.
Structured Data Integrator: We use the term “structured data” to refer to relational databases that can be accessed in any of the following ways – 1) directly by querying an SQL database (e.g., Cell Centered Database, Martone et al (2003)), 2) through an HTTP GET or POST operation executed against a database exposed through a web form (e.g., the CRISP grants database from NIH), 3) invoking a function or a web service, 4) by querying the BIRN mediator (Gupta et al (2003)), which in turn integrates multiple databases (e.g., the Senselab database from Yale). The structured data integrator module uses the mediator’s data integration registry to find the schemas of the databases, and performs a federated query by sending SQL queries created in the manner described below. The result of the federated query is sent back to the Search Coordinator Module.
Web Result Post-processor: For the NIF Web, the results of the keyword search are passed through two additional steps. The first step ranks the results, placing higher importance on the title and the relative frequency (the tf-idf score) of the query keywords in the content of the document than, for instance, on its recency. The results are also sent to post-clustering module, currently implemented with the fuzzyAnts algorithm in Weiss (2006) of the Carrot Clustering engine (see http://demo.carrot2.org) to organize the results into groups of related web sites whose pages significantly share common terms.
The Data Layer contains the actual data that are queried including the ontology, the web resources, and literature. We briefly describe the NIF Literature source:
The Textpresso Subsystem: The NIF Literature search system provides the ability to search text from publications. This is performed through the Textpresso subsystem, which indexes full-text publications and categorizes all non-trivial terms against predefined term categories. The user’s keyword query is posed against the Textpresso system to retrieve publication with the search terms and synonyms highlighted. In the NIF infrastructure, Textpresso is accessed as a set of web services. The web services are implemented as a two-step process. The first step is to run a search on the server; the second is to retrieve results from the server. Such a process is necessary because the search results (in XML format) may be on the order of several megabytes. Forming the XML file may take more time than the time out limit for the client. Also the client may not need all the documents that the search resulted in. In most cases, users are interested only in the documents (and the sentences therein) that have the maximum scores, similar to how users look only through the first few pages of a Google or Yahoo search. The current set-up allows the client to retrieve only a maximum of 500 documents in one call. For retrieving more than 500 documents, the client needs to send more queries with appropriate document range. This system, currently indexing about 67000 papers, is described in more detail in Müller et al. (2008).
How the NIF System Works
The user of the NIF system can use either a simple interface or an advanced interface. With the simple interface, the user issues a query with one or more keywords (a multi-term keyword like “tissue bank” is quoted). While the NIF system allows a user to put in a negated keyword (“not dopamine” or “– dopamine”), not all data sources (e.g., CRISP) allows queries with negative terms. Currently, only the NIF Web and the database entries allow negations. For the simple interface, the system performs term expansion, by including its synonyms from the ontology. Thus the query with the two keywords “Parkinson’s mouse” can expand to ((“Parkinson’s disease” OR “Parkinson disease” OR “Paralysis Agitans” OR PD OR “Parkinson’s syndrome”) AND (Mouse OR “Mus musculus” OR “house mouse”)). Term expansion can be seen in Figure 3. After the terms are expanded, the search manager broadcasts the query to the wrappers for all resources (the NIF Web, the databases, and so on). These wrappers transform the keyword query to the respective query languages of the individual sources, and bring back the results in different result panels. If some of the query terms do not exist in the ontology, or if a term exists but has no synonyms, the terms are sent directly as part of the query without expansion.
Figure 3.
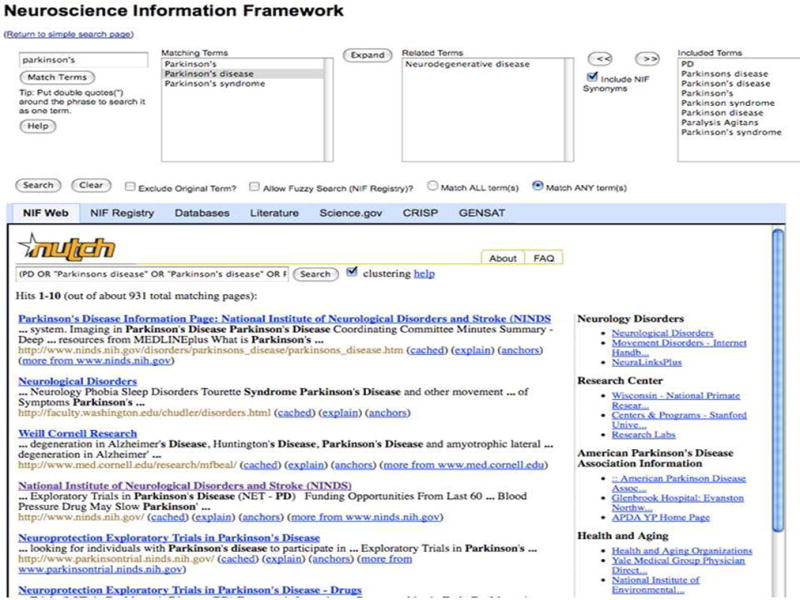
The advanced search query interface allows ontological expansion and synonym selection for query terms. The results of the NIF Web are ranked by a number of criteria including both content and recency of documents.
The advanced interface exposes the ontology search to the user. It also provides the user with the choice of using (or not using) term expansion. In this interface, the query terms are matched with the syntactically close terms in the ontology and displayed to the user. Thus the partial word “neuro” will map to “neuron”, “neuropil”, “neuroma” and so forth. In the next step, the user may choose the appropriate terms from the display. Once the desired terms are chosen, the user can choose to search around the ontology for related terms. This results in a neighborhood search in the ontology. At this time, the search is confined to only the closest terms. Thus an ontological expansion on “neuron” will produce both its superclass “nerve cell” and its direct subclasses like “pyramidal cell” and “cerebellar granule cell”, but will not produce a term like “axon” because the concept axon is under the concept “neuron compartment” which is related to “neuron” through a part-of relationship, and is hence more than one step removed from the term “neuron”.. The ontological expansion will be made more comprehensive in future versions of the system. As the user selects one or more terms from this expanded list, she can opt to use synonyms by checking the “use NIF synonyms” box. These terms are then broadcast to the NIF resources as described before. The user may include the original search terms (for example, to include terms that did not match the ontology) by checking the appropriate check box. After this step, the query processing occurs as in the case of the simple interface. One possible sequence of invocation of the different software modules in query processing is shown in Figure 4.
Figure 4.
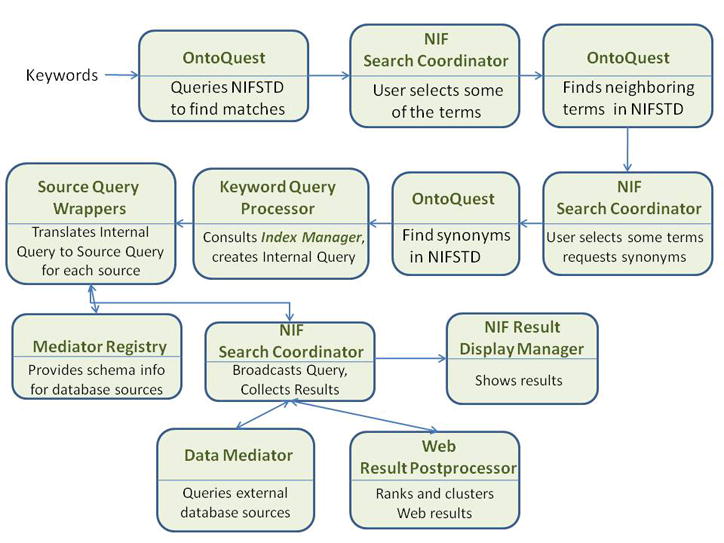
A “data flow” trace that can occur while a keyword query is processed. To avoid clutter, we did not show the invocation of the index manager in a separate module. The mediator registry is connected to the Source Query Wrapper with a bidirectional connection because the registry is queried by the (database) wrapper and gets an answer back from it. For the same reason there is a bidirectional connection between the NIF Search Coordinator and the Web Result Postprocessor. Other variants of this trace are possible depending on the choices made by the user.
At present, the NIF system is only partially capable of performing more complex matching strategies where, for example, a search on “neuron” will also match “neural”. We have implemented such a “fuzzy search” on the NIF Web Catalog content on an experimental basis – the user may optionally use this feature. Based on community feedback, and the response time needed to perform such a search on large volumes of data, we might add this feature to other resources in future versions of the system.
The results of the NIF search are organized in tabbed result panes. There is a tab for each type of source (NIF Web, NIF Registry, Databases …) and an additional level of tabbed panes for databases (for CCDB, Neuromorpho etc.). Resources with positive query results are highlighted in black; resources that lack query results are grayed out. Clicking on each of the tabs returns a simple table of the search results for the corresponding source. Note that since NIF is a federated system, we do not maintain the neuroscience resources at the NIF; rather, we provide some description of the content in the search result page, and then provide a link to the host resource. In the example shown in Figure 5, the search string was “drosophila”, and the results from the NIF Web are shown in the main window. Clicking on any of the links below will take the user to the individual resource within the NIF search results window. Notice the results of the post-clustering on the right panel – the results get grouped different data and research resources for drosophila.
Figure 5.
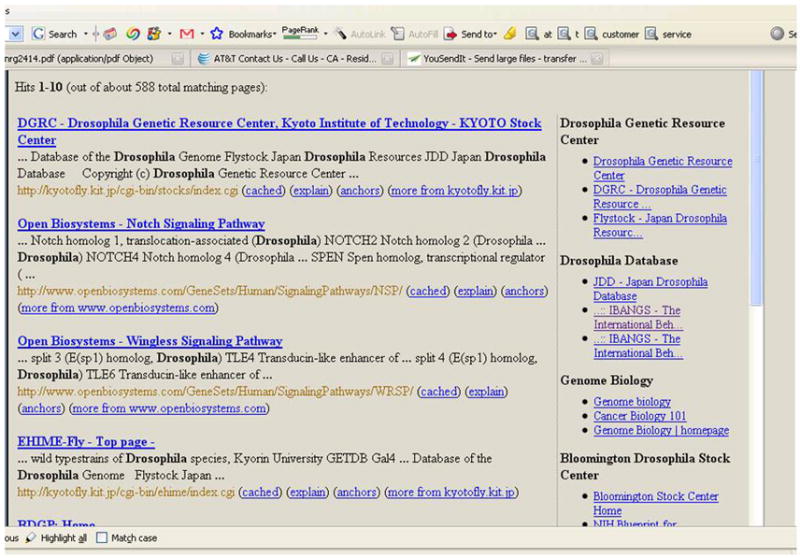
The right panel of a NIF Web search shows a meaningful clustering of the total result set. The Bloomington Drosophila Stock Center was not in the NIF Registry but is an example of an important resource that was picked up by our focused crawling strategy.
An important feature of the NIF Web is the ability to control factors used to rank results. Because this is a web index built specifically for neuroscientists, we can develop appropriate criteria for determining the rank order of returned results. We envision that such a system could be tuned by different groups hosting a NIF site depending upon their constituents. For example, the NIF Web may be tuned to rank NIH Blueprint-sponsored resources higher than non-Blueprint resources so that they appear higher in the returned list in the NIF Web. Many of these resources are small and do not have the web traffic to rank highly in the commercial search engines. However, through the NIF, these resources can be given more weight.
For the NIF Registry (NIF Web Catalog), we provide a link to the host resource and also to the description in the NIF registry for that resource (Figure 6). The query here is “antibody”. Notice that with the “Fuzzy search” option checked, the number of results returned is 7 instead of 4 that would have resulted with the option turned off. For example, the first result would not have come up because it contains the term “antibodies” rather than antibody.
Figure 6.
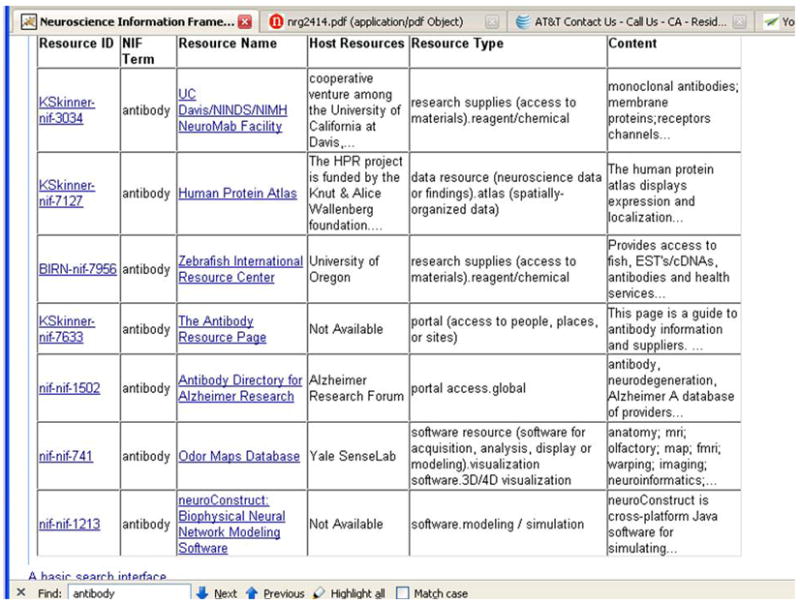
The NIF Registry is human curated and hence prone to variations in spelling, classifications, and general characterization of a resource. The use of fuzzy search is an effective way to find approximately matching terms, and thus improves result recall despite the variation in data.
Figure 7 shows the results of the query “hippocampus” on the federated database. The query received responses from three of the five databases included. For any database, the results are designed to include information that would allow a user to go to the actual website of any source and open the corresponding record. If the source provides any web accessible applications, they can be launched as well. Figure 7 shows how the user can open the WebCaret brain surface visualization tool (http://brainmap.wustl.edu/caret/) provided by SUMSDB site showing a hippocampal surface. One issue with the current data federation is that the databases themselves are not thematically characterized under groups like “image containing database”, or “genetic information containing database”. In future, if the number of databases grows significantly, the user will potentially like to select the kind of database resources over which their search should be performed, thereby reducing the extent of search performed by the system.
Figure 7.
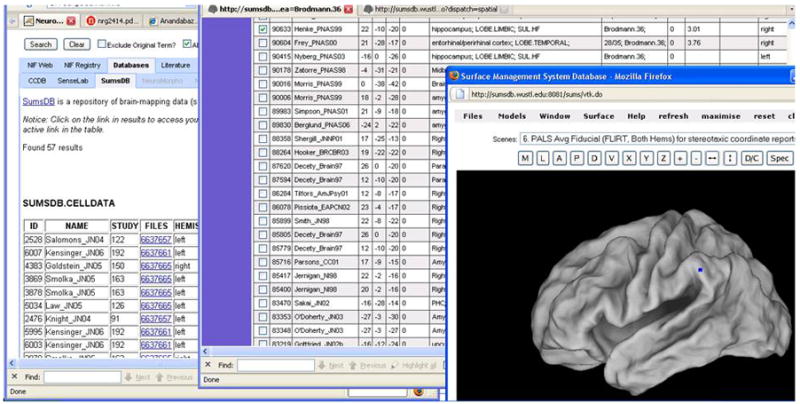
Federated search of relational data sources allows the NIF system to take advantage of the schema registration process. Since the schema is registered, it is easier to design the result page to show meaningful tables and columns. It also allows the result designer to choose output data in such a way that the database results can be hyperlinked to the original data records and to any web-accessible tools exposed by the data sources. For SUMSDB, the NIF search on “hippocampus” leads to the display of the brain surfaces in the WebCaret tool.
For NIF literature, the Textpresso system returns an XML result. The NIF system not only displays the data, but automatically constructs links to PubMed and Google Scholar from which the articles can be downloaded if the appropriate permissions are in place. It also links the results to the Textpresso-annotated records at the Textpresso site where the full capabilities of the Textpresso web site can be utilized.
Adding a New Data Resource to NIF
An important aspect of NIF is that new information resources can be added to it without having to change the infrastructure. NIF allows one to add two categories of information sources – those that are accessed as web sites (e.g., CRISP), and those that are accessed as databases.
To add a new web resource, the NIF system needs to determine how to convert a keyword query posed by a user to an equivalent HTTP query to the web resource. At this time, this is accomplished semi-automatically. The new website entry points are analyzed to determine how an HTTP GET or and HTTP POST can be constructed for the specific web site with the keywords. Sometimes, as in the case of CRISP, additional parameters need to be supplied (e.g., number of results desired); a set of default values are used for this purpose. In future versions, these parameters can be made user-selectable. This information is stored in a site wrapper specifically created for that source. In our experience, in most cases, this step takes at most a couple of hours for each new source.
Adding a new database source to NIF is a little more involved and requires an IT personnel like a database administrator who goes through a process called database registration, and then optionally, a step called concept mapping. The database registration step is based on the information integration mechanism developed for the NIH/NCRR funded BIRN (Biomedical Informatics Research Network) project. The registration maker uses a tool called Fuente that connects to the database being registered. Fuente operates by first connecting to the database to be registered and reading the full schema into a visual tool. From this schema the registration maker determines which tables and columns should be accessed by the integration engine, and how to map the data types of the database to the data types known to the integration engine.
Once, this mapping is specified, Fuente exports it to the integration system, which in turn stores the schema in a registry. When a new schema is deposited in the registry, the NIF system makes an update in its configuration so that the next time a query is made, it would also be broadcast to the new schema, and the results would be reported in a new panel on the interface. The configuration can be modified by the NIF operators to decide which tables and columns should be visible to the NIF user. When a new database is registered, the NIF indexing mechanism updates the NIF indexes so that the keyword queries can operate efficiently against the new data source. In our experience, the whole process of adding a new database takes between 2 to 4 hours, depending on the size of the database, and the efficiency of the manual part of the process. Currently, Fuente can connect to MySQL, PostGreSQL, Oracle, SQL Server, and a couple of smaller DBMS systems.
The concept mapping step can occur after a database has been registered. The goal of this step is to create a mapping from the field names and terms used within the database to the terms known to the NIFSTD ontology. For example, if the database has the term “electron tomography” and the ontology does not have this term, then a knowledgeable and authorized user of the database can map it to a nearby term in the ontology like “electron microscopic imaging technique”. If such a mapping is created, a query on an ontological term like “electron microscopic imaging technique” will also retrieve the data record on “electron tomography” which would have been otherwise impossible to retrieve. In NIF we have created the first version of a concept mapping tool that can map one term of a database to one term of the ontology. The mappings are stored in one part of the NIF infrastructure called the Term Index Source. We estimate that the concept mapping process currently requires between a few hours to several days effort, depending on the complexity of the information to be shared. In future versions, this tool will be upgraded to add further automation, and the ability to specify more complex mappings.
Information Content of the NIF System
The version 1.0 of the NIF system has been developed to capture a relatively small, but representative portion of the total amount of Neuroscience information available through the internet. In the following, we describe the content accessible from different parts of the system. We leave the ontology and the Textpresso contents out of this discussion; they are described in companion papers. Although accessible through the NIF system, we also do not further discuss the contents of the three web resources, viz. science.gov (http://www.science.gov), GenSAT (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gensat), the NIH database for gene expression images and CRISP (http://crisp.cit.nih.gov), the NIH grants database, because they are well known to the readers.
The NIF Web Catalog: The content of the NIF Web catalog is created by expert contributors, by selecting web sites that represent different forms of Neuroscience resources. Each entry of the NIF Catalog (NIF Registry) is annotated with high level descriptors from a controlled vocabulary that describes the resource type, its general content and other information about the resource. As of this writing, there are a total of 388 resources registered to the NIF. A breakdown of these resources according to the high level categories established by NIF is given in Figure 3. Many of these resources were imported directly from the Internet Analysis Tool Registry (IATR) (http://www.cma.mgh.harvard.edu/iatr/), an existing resource that maintained a list of software tools for neuroscience, leading to a heavy representation of software tools. In addition, the NIF was selective in the types of resources that it catalogued: no commercial sites or products were included; the resources had to provide information or tools directly relevant to performing neuroscientific research.
The NIF Web: The limited web crawling process outlined in the previous section has turned out to be quite effective. The crawling depth of 15 almost always captures the content of the entire web site of the seed sites. In 90% of the time, the links connecting outside of the seed sites turn out to be neuroscience relevant sites, pointing to NIH sites for instance. The crawler also indexes Word and PDF documents accessible from the web site. This gives us the extra benefit that even if these pages were not initially marked up through the ontology, the system can still perform ontological search on them after the indexed text content has been brought into the NIF system. At the present time about 10 million relevant web pages are indexed and are searchable. In 10% of the cases, however, our current crawling strategy produces extraneous content not connected to Neuroscience. For example, a pointer to a newspaper article about a neuroscientific discovery, may further link to other unrelated content from the same newspaper article. One hindrance encountered in operating the NIF Web crawler is that some very informative web sites like The Antibody Resource Page (http://www.antibodyresource.com/) have explicit directives for crawlers not to crawl the site. Since we have to respect such provider directives, we cannot complete cover all content through the NIF Web. Further note that the current version of the NIF system does not address the “hidden web problem”.
External Databases: Currently, relatively few neuroscience resources use a well-designed robust relational database system. Even those that do, usually do not allow external systems to query their databases directly. However, we believe that data sharing, including database sharing, will be much more common for Neuroscience in the future. To illustrate how such a community sharing might occur, we chose five databases, each with unique but overlapping content. The Cell-centered database at UCSD provides access to multi-resolution cellular data captured by different imaging and volume reconstruction techniques. The Senselab system at Yale provides access to physiological models of neuronal circuits. The SUMSDB database at Washington University provides access to cortical maps of human, macaque and rodent brains. The Neuromorph database at George Mason University provides synthetically constructed neuron models (see Ascoli et al in this issue). The NeuroMAB database at University of California Davis is an antibody supply catalog for mouse models that was included because it is a “facilities” type of resource that can be accessed based on molecular targets like potassium channels, transporters and scaffold proteins.
Beta Testing the NIF version 1.0
The beta version of the NIF 1.0 was released on January 15, 2008, with the goal of getting some feedback on the user experience with the system. To help testers understand the system, a series of tutorials and user materials were created to explain the NIF project and provide instruction on how to use the user interface to search the NIF. A link was also provided to the NIF public wiki, where more information on the NIF architecture and technical details could be found. Recommended by members of the NIF technical team, NIH Program Team and the NIF Advisory Committee, beta testers were recruited from multiple groups in order to gain a diverse set of opinions on the NIF system: including undergraduates, graduate students, post-docs, junior and senior scientists and science librarians, with expertise in multiple areas of neuroscience. Two on-line questionnaires were provided for feedback; email responses were also accepted. Many of the responses involved simple changes of the user interfaces that could be readily addressed, and were added to the system during the testing period itself. The primarily findings from the beta testing process that could not be addressed immediately were the following:
The simple user interface was used by more people initially, but there was a steady increase in the use of the advanced interface with time. This illustrated that with some degree of experience, the users, particularly the more knowledgeable users, found the use of ontology to be more useful.
A number of users pointed out that it will be beneficial if they could (a) have the option to select the sources they wanted to search over, and (b) specify the type of results they wanted (e.g., results with image content only).
A number of users showed cases where the NIF Web retrieves some data pages that are not in the domain of neuroscience.
The users almost unanimously stated that they wanted the results of the queries to be organized by ontological terms instead of (or in addition to) by resource type. The primary argument was that an ontology-based result presentation will scale much better as more data sources are added.
A number of users wanted a greater variety of content to be covered, ranging from genetic data to the latest imaging techniques to drugs used for neurological disorders. For these cases, the users found that Google had better coverage than NIF. We verified that this observation comes from the facts that (a) the NIF Registry sites we have used to seed the NIF Web search did not have a well-rounded coverage, while Google’s coverage, albeit not focused, is much more universal and (b) sometimes Google’s ranking of the results were preferred by users compared to the ranking produced by NIF web.
While users liked the fact that NIF Web results were clustered, the quality of clustering produced mixed reactions because for some searches the grouping produced by the clustering algorithm were considered “not useful”.
The response of some of the system components like Textpresso and science.gov became slower as the number of query terms was increased. This could be partly rectified in the beta testing period.
These findings, albeit coming from a non-rigorous beta testing process, do indeed highlight the mismatches between the users’ expectations and the current capabilities of the NIF version 1.0 system.
Conclusion and Future Work
In this paper we have described the technical design and functionality of the version 1.0 of NIF system. We expect this system to evolve in the future with a number of enhancements besides the ones listed under the beta test feedback. We plan to include genetic and proteomic data and computational resources related to Neuroscience. For instance, web-accessible genetic data from NCBI, mouse model data from the Jackson Laboratory (http://www.jax.org), QTL data from University of Tennessee (http://www.genenetwork.org/), are likely to be added to NIF. The NIF will also be able to query and access RDF–formatted (RDF stands for Resource Description Framework, which is an emerging standard for representing semantic information for the web) from the Neurocommons project (http://neurocommons.org). We also plan to experiment with different variations of user interface for different categories of users to determine the differences in the intended behavior of system for different audiences.
Acknowledgments
The NIF was supported by the NIH Blueprint through contract HHSN271200577531C administered by NIDA.
References
- Astakhov V, Gupta A, Grethe JS, Ross E, Little D, Yilmaz A, Qian X, Santini S, Martone ME, Ellisman M. Semantically based data integration environment for biomedical research. Proc. 19th IEEE Symposium on Computer-Based Medical Systems; IEEE Computer Society, Washington, DC, USA. 2006. pp. 171–176. [Google Scholar]
- Bug W, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird A, Larson S, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: Building comprehensive ontologies for neuroscience. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9032-z. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gupta A, Kurul ME. Stack-based algorithms for pattern matching on dags. Proc. 31st Int. Conf. on Very Large Databases (VLDB); Stockholm. 2005. pp. 493–504. [Google Scholar]
- Chen L, Martone ME, Gupta A, Fong L, Wong-Barnum M. Ontoquest: Exploring ontological data made easy. Proc. 31st Int. Conf. on Very Large Databases (VLDB); 2006. pp. 1183–1186. [Google Scholar]
- Franklin MJ, Halevy AY, Maier D. From databases to dataspaces: a new abstraction for information management. SIGMOD Record. 2005;34(4):27–33. [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, Goldberg DH, Grafstein B, Grethe JS, Gupta A, Halavi M, Kennedy DN, Marenco L, Martone ME, Miller PL, Müller H-M, Robert A, Shepherd GM, Sternberg PW, van Essen DC, Williams RW. The Neuroscience Information Framework: A Data and Knowledge Environment for Neuroscience. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9024-z. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Ludaescher B, Martone ME. BIRN-M: a semantic mediator for solving real-world neuroscience problems. Proc. SIGMOD Int. Conf. on Management of Data; ACM Press, New York, NY, USA. 2003. pp. 678–678. [Google Scholar]
- Halavi M, Polavaram S, Donohue DE, Hamilton G, Hoyt J, Smith KP, Ascoli GA. NeuroMorpho. Org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9030-1. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nature Neuroscience. 2004 May;7(5):483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Hristidis V, Gravano L, Papakonstantinou Y. Efficient IR-style keyword search over relational databases. Proc. 29th International Conference on Very large data bases; 2003. pp. 850–861. [Google Scholar]
- Lam HY, Marenco L, Clark T, Gao Y, Kinoshita J, Shepherd G, Miller P, Wu E, Wong GT, Liu N, Crasto C, Morse T, Stephens S, Cheung KH. Alzpharm: integration of neurodegeneration data using RDF. BMC Bioinformatics. 2007;8:S4. doi: 10.1186/1471-2105-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone ME, Zhang S, Gupta A, Qian X, He H, Price D, Wong M, Santini S, Ellisman M. The Cell-Centered Database: A Database for Multiscale Structural and Protein Localization Data from Light and Electron Microscopy. Neuroinformatics. 2003;1(4):379–396. doi: 10.1385/NI:1:4:379. [DOI] [PubMed] [Google Scholar]
- Mei J, Ma L, Pan Y. Ontology query answering on databases. International Semantic Web Conference; 2006. pp. 445–458. [Google Scholar]
- Müller H-M, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9031-0. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor MJ, Shankar RD, Tu SW, Das AK, Parrish DB, Nyulas CI, Musen MA. Efficiently querying relational databases using OWL and SWRL. 1st Int. Conf. on Web Reasoning and Rule Systems; LNCS 4524. Springer. 2007a. pp. 361–363. [Google Scholar]
- O’Connor MJ, Shankar RD, Tu SW, Parrish DB, Das AK, Musen MA. Using semantic web technologies for knowledge-driven querying of biomedical data. Proc. 11th Conf. on Artificial Intelligence in Medicine (AIME2007), LNAI 4594; Springer. 2007b. pp. 267–276. [Google Scholar]
- Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, Doherty D, Forsberg K, Gao Y, Kashyap V, Kinoshita J, Luciano J, Marshall M, Ogbuji C, Rees J, Stephens S, Wong G, Wu E, Zaccagnini D, Hongsermeier T, Neumann E, Herman I, Cheung K-H. Advancing translational research with the semantic web. BMC Bioinformatics. 2007:8. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. PhD thesis. Poznan University of Technology; Poznan, Poland: 2006. Descriptive clustering as a method for exploring text collections. [Google Scholar]


