Abstract
The actin-binding protein filamin A (FLNa) affects the intracellular trafficking of various classes of receptors and has a potential role in oncogenesis. However, it is unclear whether FLNa regulates the signaling capacity and/or down-regulation of the activated epidermal growth factor receptor (EGFR). Here it is shown that partial knockdown of FLNa gene expression blocked ligand-induced EGFR responses in metastatic human melanomas. To gain greater insights into the role of FLNa in EGFR activation and intracellular sorting, we used M2 melanoma cells that lack endogenous FLNa and a subclone in which human FLNa cDNA has been stably reintroduced (M2A7 cells). Both tyrosine phosphorylation and ubiquitination of EGFR were significantly lower in epidermal growth factor (EGF)-stimulated M2 cells when compared with M2A7 cells. Moreover, the lack of FLNa interfered with EGFR interaction with the ubiquitin ligase c-Cbl. M2 cells exhibited marked resistance to EGF-induced receptor degradation, which was very active in M2A7 cells. Despite comparable rates of EGF-mediated receptor endocytosis, internalized EGFR colocalized with the lysosomal marker lysosome-associated membrane protein-1 in M2A7 cells but not M2 cells, in which EGFR was found to be sequestered in large vesicles and subsequently accumulated in punctated perinuclear structures after EGF stimulation. These results suggest the requirement of FLNa for efficient EGFR kinase activation and the sorting of endocytosed receptors into the degradation pathway.
Filamin A expression plays an important role in the activation and degradation of EGF receptors.
Filamin A (FLNa; ABP280) is a member of the family of ubiquitously expressed actin-binding proteins that has been implicated in many processes including proliferation, cell migration, the formation of blood vessels, and signaling pathways that mediate organogenesis in multiple tissues (reviewed in Refs. 1 and 2). The binding of FLNa to actin helps to form the orthogonal branching of actin filaments that make up the cytoskeleton. FLNa also links actin to a number of receptors at the plasma membrane to regulate their functions within the cell (3,4,5,6). Emerging evidence suggests that filamin has an important role in recruiting costimulatory molecules to cell surface receptors present in specialized lipid microdomains of the plasma membrane, thus affecting signaling events and cellular responses induced by external stimuli (7,8). A significant role for FLNa has been proposed in carcinogenesis: for example, the metallopeptidase activity of prostate-specific membrane antigen is inhibited on binding to FLNa within prostate cancer cells (9), and the anticancer activity of 1α,25-dihydroxyvitamin D (3) is associated with up-regulation of FLNa in human SW480-ADH colon cancer cells (10). FLNa has also been implicated in human melanoma cell migration (11,12). In head and neck squamous cell carcinoma, activation of CD44 by hyaluronan increases migration via changes in filamin and activation of the epidermal growth factor receptor (EGFR) (13). However, the mechanistic link between filamin and early signaling events associated with malignancy remains elusive.
The EGFR family of receptor tyrosine kinases encompasses four members (also known as erbB-1 or EGFR, erbB-2 or HER2/neu, erbB-3 and erbB-4) that control important aspects of cell proliferation, differentiation, motility, and survival, and their deregulation is implicated in oncogenesis (reviewed in Ref. 14). The regulation of the pleiotropic responses of EGFR occurs at multiple levels, including receptor compartmentalization in lipid microdomains (15,16,17), ligand-induced receptor dimerization, and endocytosis of activated receptors, which can result in lysosomal degradation of the receptor and termination of the signal or its recycling back to the cell surface (18,19). Ligand-mediated down-regulation of EGFR requires recruitment of the endocytic machinery for efficient endocytosis. It has become increasingly evident that the distribution of EGFR between various microdomains of the plasma membrane is playing a role in the control of the rate of internalization and degradation of this receptor. In addition to the classical pathways (e.g. clathrin coated pits and uncoated vesicles containing caveolin-1), ligand-induced internalization of EGFR has been also shown to occur by a nonclassical pathway through circular dorsal ruffles (20).
In this study, we examined the possible relationship between EGFR and FLNa expression in established human melanoma cell lines with varying metastatic potential and in primary cultures of human melanoma biopsies. We then investigated the role of FLNa as putative regulator of ligand-mediated activation and down-regulation of EGFR in human melanoma cells. Our results indicate that knockdown of FLNa expression resulted in the internalization and vesicular sequestration of ligand-bound EGFRs, and their accumulation to a perinuclear location away from the degradation machinery.
Materials and Methods
Materials
Mammalian expression vector pXER-EGFR encoding the EGFR-green fluorescent protein (GFP) construct was obtained from Dr. Alexander Sorkin (University of Colorado Health Sciences Center, Aurora, CO). The mouse monoclonal antihuman lysosome-associated membrane protein (LAMP)-1 antibody developed by Drs. J. Thomas August and James E. K. Hildreth was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa (Iowa City, IA).
Analysis of the Mannheim data set
The Mannheim data are publicly available from National Center for Biotechnology Information’s gene expression omnibus (www.ncbi.nlm. nih.gov/geo) under GEO series accession no. GSE4845. This data set was generated from microarray analysis of 45 primary cultures of melanoma biopsies, using HGU133 microarray chips (Affymetrix, Santa Clara, CA). Analysis of the data set for expression of FLNa and EGFR was performed using a Student’s two-tailed t test (assuming equal variance) to determine statistical significance between cohorts.
Maintenance of cell lines
The human M2 melanoma cell line and the stable M2 subclone, M2A7 cells, were previously described (6). The M2A7 cells were cultured with the addition of 500 μg/ml of Geneticin (Life Technologies, Carlsbad, CA) to maintain FLNa expression. Geneticin was removed from the medium when M2A7 cells were seeded for all experiments. Maintenance of the G-361, UACC903, M93-047, and UACC647 cell lines were described previously (21). Franklin Square cell lines were cultured in RPMI 1640 (Life Technologies) supplemented with 4 mm glutamine, penicillin/streptomycin, and 10% fetal bovine serum (HyClone, Logan, UT).
RNA extraction and cDNA synthesis
RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) and an RNeasy minikit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. Using 1 μg RNA, cDNA was synthesized using SuperScript III first strand synthesis SuperMix for quantitative RT-PCR (Invitrogen).
Quantitative real-time PCR
Gene expression was quantified using the SYBR green (Applied Biosystems, Foster City, CA) method of real-time PCR and mRNA levels were compared with standard curves and normalized to 18S mRNA. PCRs were performed in triplicate with universal 18S primers (Ambion, Austin, TX) or 100 nm of each gene-specific primer. The primers for human EGFR [forward (5′-AGGACCAAGCAACATGGTCA-3′) and reverse (5′-CCTTGCAGCTGTTTTCACCT-3′)], FLNa [forward (5′-GTCGCTCTCAGGAACAGCAG-3′) and reverse (5′-AGGGGACGGCCCTTTAAT-3′)], and HER-2 [forward (5′-GTCTCTGCCTTCTACTCTCTACC-3′) and reverse (5′-GACAGGTCAACAGCCACATGA-3′)] (Integrated DNA Technologies, Coralville, IA) were designed to cross intron-exon junctions. Real-time PCR was performed on an ABI Prism 7300 sequence detection system.
Scratch assays
These assays were carried out as described previously (21). Once confluent, cells were left untreated or treated with 20 nm epidermal growth factor (EGF; Upstate Biotechnologies, Temecula, CA) or 100 nm AG1478 (Calbiochem, San Diego, CA). Images of the same field were taken every 12 h (G-361 cells) or 3 h (M93-047 cells) until closure of the scratch.
Small interfering RNA (siRNA) transfection
Cells were grown to approximately 80% confluency, washed with PBS, serum starved for 1 h, and then transfected with 150 nm of control or FLNa siRNA (QIAGEN) using LipofectAMINE reagent (Invitrogen). After 5 h, the transfection media was removed and replaced with complete α-MEM containing 10% fetal bovine serum. The cells were routinely used 72 h after transfection. The sequences used for the FLNa siRNA were sense reverse (GGAAGAAGAUCCAGCAGAA)dTdT and antisense reverse (UUCUGCUGGAUCUUCUUCC)dAdC and the nonsilencing control sense reverse (UUCUCCGAACGUGUCACGU)dTdT and antisense reverse (ACGUGACACGUUCGGAGAA)dTdT. These HP-validated siRNAs have been demonstrated by QIAGEN to perform efficient knockdown with minimal off-target effects.
EGF binding and internalization assays
EGF binding and internalization assays were carried out with cells grown to confluency on 12-well tissue culture plates. Details are provided in supplemental methods (published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo. endojournals.org).
Immunoprecipitation and Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer as described previously (22). Insoluble material was removed by centrifugation, and the precleared lysates were immunoprecipitated with 2 μg of either anti-EGFR (Upstate Biotechnologies) or anti-c-Cbl (Santa Cruz Biotechnology, Santa Cruz, CA) antibody overnight at 4 C. Immune complexes and total cell lysates were subjected to immunoblotting as previously described (22). Primary affinity-purified antibodies against ubiquitin, c-Cbl, c-Src, EGFR (sc-03), CD82/KAI-1, HSP70, ERK1/2, and inhibitory-κB kinase-α/β were from Santa Cruz Biotechnology. Antibody against β-tubulin was from Cell Signaling Technology (Danvers, MA), whereas antibodies against FLNa and phosphotyrosine (clone 4G10) were from Research Diagnostics (Concord, MA) and Upstate Biotechnologies, respectively.
Flow cytometry to analyze cell surface EGFR levels
Cells were serum starved for 3–4 h and then incubated with vehicle or 20 nm EGF for 1 h at 37 C. Cells were washed with ice-cold PBS and then treated with an acid-stripping solution to remove prebound EGF. After washing, cells were processed for flow cytometry as outlined in supplemental methods.
Immunofluorescence microscopy
Cells were seeded at 7 × 105 cells/well in complete medium into chamber slides (Lab-Tek; Nunc, Rochester, NY). Transient transfections were performed with pXER-EGFR using LipofectAMINE and Plus reagents (Invitrogen). Forty-eight hours later, cells were maintained in serum-free medium for 3–4 h and then incubated with EGF (10 nm) for the indicated times, after which cells were washed, fixed with ice-cold 95% methanol, and blocked as previously described (21). EGFR-GFP was labeled with Alexa Fluor 488-conjugated rabbit anti-GFP antibody (Molecular Probes, Carlsbad, CA), and LAMP-1 with a mouse monoclonal antibody (Developmental Studies Hybridoma Bank). Anti-LAMP-1 antibody was detected with Alexa Fluor 555-conjugated goat antimouse IgG (Molecular Probes). Cells were mounted in Prolong Gold antifade reagent containing 4′,6′-diamino-2-phenylindole (Invitrogen) and then analyzed by confocal microscopy using an inverted LSM 510 microscope (Zeiss, Thornwood, NY). Images were processed using the Zeiss LSM Image Browser.
Results
Positive correlation between FLNa and EGFR expression in metastatic melanomas from patients and cell lines
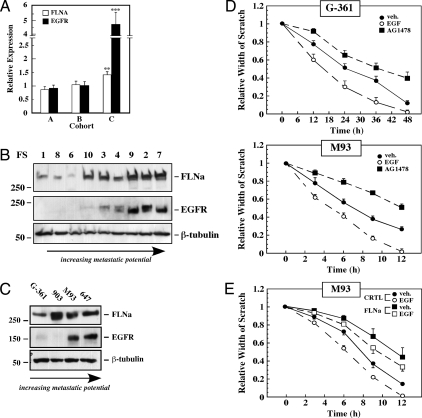
In a recent study, Hoek et al. (23) performed DNA microarray analyses on 45 cultures of melanoma samples (known as the Mannheim data set), and identified three cohorts (A, B, and C), which represent melanoma groups of differing metastatic potential. Cohorts A and B are susceptible to TGF-β mediated inhibition of proliferation and have low motility (low metastatic potential), whereas melanomas from cohort C are resistant to TGF-β and are highly metastatic. Analysis of the Mannheim data set demonstrated that both FLNa and EGFR were significantly up-regulated in cohort C compared with cohorts A and B (Fig. 1A). Western blot analysis was used to validate these observations in another set of melanoma cell lines derived from nine patient biopsies (Fig. 1B). The results demonstrated a positive correlation between FLNa and EGFR expression (R2 = 0.65, P < 0.01) (supplemental Fig. 1). Finally, expression of FLNa and EGFR was also examined in a panel of low (G-361), moderate (UACC903), and highly (M93-047 and UACC647) metastatic melanoma cell lines. There was a 5.2 ± 1.5- and 4.3 ± 1.0-fold induction of FLNa in M93-047 and UACC647 cells vs. G-361 cells (P < 0.05), respectively (Fig. 1C). Expression of the EGFR protein was markedly higher in the more metastatic cell lines (Fig. 1C).
Figure 1.
FLNa and EGFR expression in metastatic human melanomas from patient biopsies and cell lines. A, FLNA and EGFR mRNA expression in the three cohorts of the Mannheim data set (23). Cohorts A (n = 19) and B (n = 10) are described as low metastatic cell lines, whereas cells from cohort C (n = 16) were found to be highly metastatic. The values are expressed as the mean ± sem. Statistical analysis was performed on cohort A vs. C with values of **, P = 0.003 for FLNa and ***, P = 3.38E-05 for EGFR. B, Western blot analysis was carried out on nine melanoma patient cell lines [Franklin Square lines (FS)] to detect FLNa and EGFR. β-Tubulin was used as a loading control. The position of the molecular mass markers (in kilodaltons) is shown on the right. C, Western blot analysis using antibodies against FLNa or EGFR in human melanoma cell lines of increasing metastatic potential. The cells are G-361, UACC903 (903), M93-047 (M93), and UACC647 (647). β-Tubulin was used as a loading control. Data shown are representative of four independent experiments, each performed in duplicate dishes. D, Scratch assays were performed with low metastatic G-361 cells and high metastatic M93-047 cells in the absence or presence of EGF (20 nm) or AG1478 (100 nm) for the indicated times. The relative scratch width was measured over time and plotted. E, M93-047 cells were transfected with control or FLNa siRNA for 72 h and then subjected to scratch assays in the absence (filled symbols) or presence of EGF (open symbols). All results from scratch assays are expressed as mean ± sd (n = 4).
To examine whether EGFR status correlates with melanoma motility, we performed a wound-healing assay with the low metastatic G-361 cells and the highly metastatic M93-047 cells. As anticipated, M93-047 cells were markedly more motile than G-361 cells, with 50% closure of the wound at 7 and 26 h, respectively (Fig. 1D, filled circles). In both cell lines, the scratch closed faster in the presence of EGF, with 50% closure of the wound at 5 and 17 h, respectively (Fig. 1D, open circles). Furthermore, the EGFR antagonist, AG1478, sharply reduced the basal wound-healing rate in both G-361 and M93-047 melanoma cell lines (Fig. 1D, filled squares), indicating that locally produced EGF or other members of the EGF family also contributed to the constitutive motility of human melanoma cells.
The importance of FLNa in efficient wound-healing capacities of the metastatic M93-047 cells was then investigated. The rate of basal and EGF-induced wound closure was markedly reduced when knockdown of FLNa by RNA interference was performed, compared with cells treated with control siRNA (Fig. 1E).
Role of FLNa in ligand-induced EGFR degradation in metastatic melanoma cells
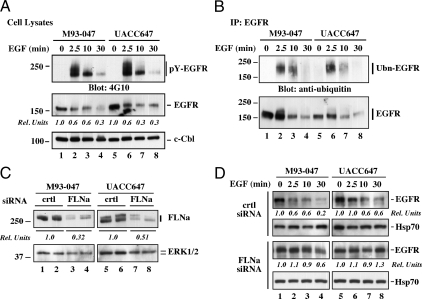
The binding of EGF induces dimerization, autophosphorylation, and activation of EGFR intrinsic tyrosine kinase, which then undergoes ubiquitination and proteolytic degradation (24). Before assessing the importance of FLNa in effective down-regulation of activated EGFR, we determined the kinetics of ligand-induced receptor activation in M93-047 and UACC647 cells, which express high levels of endogenous EGFR. Addition of EGF elicited rapid increase in the levels of tyrosine phosphorylated EGFR (pY-EGFR), with a maximum at 2.5 min and declining thereafter (Fig. 2A, top panel). The levels of total EGFR were decreased over the time course, with more than 50% loss 10 min after EGF stimulation (Fig. 2A, middle panel). Reprobing the membrane with anti-c-Cbl confirmed equal protein load in each lane (Fig. 2A, bottom panel). To examine the kinetics of EGFR ubiquitination, M93-047 and UACC647 cells were stimulated with EGF, lysed, and EGFR immunoprecipitates were analyzed by Western blotting. The amount of ubiquitin incorporation in EGFR was maximal at 2.5 min after EGF treatment and declined thereafter (Fig. 2B, top panel), consistent with the phosphorylation data. The total amount of immunoprecipitated EGFR was also rapidly decreased on EGF addition (Fig. 2B, bottom panel). These data illustrate efficient ligand-induced activation and degradation of EGFR in these cells.
Figure 2.
Effect of EGF on ligand-mediated EGFR activation and down-regulation in highly metastatic human melanoma cell lines. A, M93-047 and UAC647 cells were fed serum-free medium for 3 h and then left alone or incubated with EGF (20 nm) for 2.5, 10, and 30 min. Cells were lysed and analyzed by Western blot with antibodies against phosphotyrosine (pY, clone 4G10), EGFR, or c-Cbl as a loading control. Densitometry analysis was carried out and normalized to c-Cbl. The EGFR signal in unstimulated cells (second panel, lanes 1 and 5) was arbitrarily given the value of 1.0. B, M93-047 and UACC647 cells were treated as in A. EGFR immunoprecipitates (IP) prepared from cell lysates were resolved by SDS-PAGE and then analyzed by Western blot with antibodies against ubiquitin, or EGFR. Ubn-EGFR, Ubiquitinated EGFR. C, M93-047 and UACC647 cells were transfected with either control (crtl) or FLNa siRNA for 72 h, lysed and then analyzed by Western blot with antibodies against FLNa or ERK1/2 as a loading control. Densitometry analysis was carried out and normalized to ERK. The FLNa signal in crtl siRNA-transfected cells was arbitrarily given the value of 1.0. D, Time course of EGF stimulation was carried out in cells transfected with control (crtl) or FLNa siRNA. Cell lysates were prepared and immunoblotted for total EGFR or Hsp70 as a loading control. Densitometry analysis was carried out and normalized to Hsp70. The EGFR signal in unstimulated cells was arbitrarily given the value of 1.0. Data shown in C and D are representative of two independent experiments, each performed in duplicate dishes.
We next studied whether suppression of FLNa levels was sufficient to alter ligand-induced EGFR down-regulation. M93-047 and UACC647 cells were incubated with control or FLNa siRNA for 48 h, after which EGF was added for periods up to 30 min. As anticipated, there was more than 50% knockdown in FLNa expression after cell incubation with FLNa siRNA, compared with the nonsilencing control siRNA (Fig. 2C). Reprobing the membrane with anti-ERK1/2 has been performed to exclude the possibility of off-target effects of the FLNa siRNA (Fig. 2C). EGF had only a minimal effect in promoting EGFR degradation in FLNa-depleted cells (Fig. 2D), indicating that the lack of FLNa impairs either EGFR kinase activation, its intracellular sorting to lysosomes, or both.
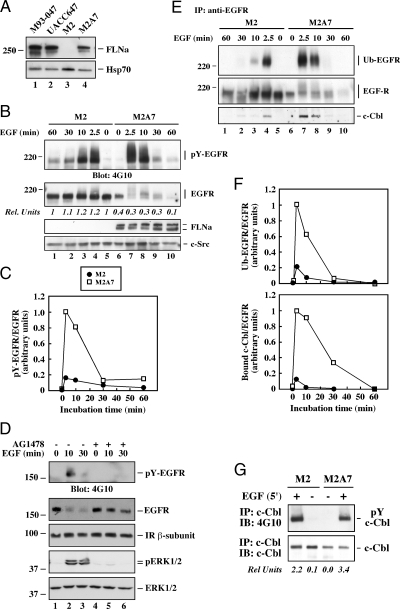
Based on our findings, the human M2 melanoma cell model was then used to independently assess the importance of FLNa in ligand-induced activation and sorting of EGFR. M2 melanoma cells express undetectable levels of endogenous FLNa, whereas the stable M2 subclone, M2A7, expresses human FLNa cDNA (25). A comparison of the expression profile of FLNa, EGFR, and HER2 mRNAs was carried out with the six human melanoma cell lines used in this study (Table 1) and enabled us to determine that the FLNa mRNA levels in M2A7 cells were similar to those of the highly metastatic melanoma M93-047 and UACC647 cells and were nearly undetectable in M2 cells. The level of FLNa protein expression in M2A7 cells approximated that observed in M93-047 and UACC647 cells (Fig. 3A). Nonetheless, the EGFR expression levels in M2 cells resembled those found in M2A7, M93-047 and UACC647 cells, with more than 10- to 25-fold higher expression than in the low metastatic G-361 cells. Of interest, no difference in the levels of the protooncogene HER2/neu was observed among the cell lines studied (Table 1).
Table 1.
Quantitative real-time PCR data of FLNa, EGFR, and HER-2 mRNA expression in six human melanoma cell lines
| Gene name | Cell line
|
|||||
|---|---|---|---|---|---|---|
| G-361 | UACC903 | M93-047 | UACC647 | M2 | M2A7 | |
| FLNA | 1.00 ± 0.17 | 1.42 ± 0.62 | 44.58 ± 4.72 | 41.15 ± 3.08 | 0.13 ± 0.08 | 31.54 ± 2.78 |
| EGFR | 1.00 ± 0.14 | 0.48 ± 0.13 | 17.47 ± 2.44 | 25.73 ± 1.12 | 24.94 ± 2.88 | 10.28 ± 1.24 |
| HER2 | 1.00 ± 0.21 | 1.04 ± 0.17 | 1.08 ± 0.15 | 1.04 ± 0.21 | 0.96 ± 0.23 | 0.71 ± 0.16 |
The relative expression of each gene was normalized to 18S, and the signal associated with G-361 cells was arbitrarily given the value of 1.00. The normalized values for FLNa, EGFR, and HER-2 mRNA levels in G-361 cells were 0.036, 0.140, and 0.529, respectively. Results are the mean± sem of three independent analyses.
Figure 3.
Role of FLNa in ligand-induced EGFR degradation in human melanoma cell lines. A, Lysates from M2, M2A7, M93-047, and UACC647 cells were analyzed by Western blot with antibodies against FLNa or Hsp70 as a loading control. B, Human melanoma M2 and M2A7 cells were serum starved for 3 h and then incubated with EGF (20 nm) for the indicated periods of time at 37 C. Western blot analysis was performed with a portion of cell lysates using antibodies against pY (4G10 clone), EGFR, FLNa, or c-Src, shown here as a loading control. The OD of EGFR in unstimulated M2 cells (second panel, lane 5) was arbitrarily given the value of 1.0. C, Quantitated ratio of pY-EGFR to total EGFR in EGF-stimulated M2A7 cells (2.5 min) was arbitrarily given the value of 1.0. D, Serum-starved M2A7 cells were pretreated with dimethylsulfoxide (0.1%) or AG1478 (200 nm) for 30 min followed by the addition of EGF for 0, 10, or 30 min. Cell lysates were analyzed by Western blotting with antibodies against pY (4G10 clone), EGFR, insulin receptor (IR) ß-subunit, or phosphorylated and total ERK1/2. E, EGFR immunoprecipitates (IP) were separated by SDS-PAGE and analyzed by Western blot with antibodies against ubiquitin, EGFR, or c-Cbl. F, Quantitated ratios of ubiquitinated EGFR (Ub-EGFR; upper panel) and bound c-Cbl (lower panel) to total EGFR in EGF-stimulated M2A7 cells (2.5 min) were arbitrarily given the value of 1.0. G, Lysates from control and EGF-stimulated cells were immunoprecipitated (IP) with anti-c-Cbl antibody and immunoblotted (IB) with 4G10 monoclonal antibody or c-Cbl antibody. Quantifications of the ratio of phosphorylated to total c-Cbl proteins are shown. All data shown are representative of three independent experiments, each performed in duplicate dishes.
Based of these results, M2 and M2A7 melanoma cells were used to determine the contribution of FLNa to EGFR activation and degradation. Scatchard analysis indicated similar binding constants but 4.5-fold higher cell surface EGF receptor numbers for M2 cells compared with M2A7 cells (supplemental Fig. 2). Nonetheless, cells were treated with EGF for various times, and extracts were analyzed by Western blot for changes in the levels of phosphorylated EGFR (pY-EGFR) and total EGFR levels. In both cell lines, addition of EGF elicited rapid tyrosine phosphorylation of EGFR with a maximum at 2.5 min and declining thereafter (Fig. 3B, top panel). The amount of intact EGFR was sharply reduced in a time-dependent fashion in M2A7 cells after EGF treatment but not in M2 cells (Fig. 3B, second panel, lanes 6–10 vs. 1–5). It is noteworthy that the basal levels of EGFR expression in M2 cells were 2.5-fold higher compared with M2A7 cells (Fig. 3B, second panel, lane 5 vs. 6). In several instances, FLNa migrates as a double band, which results from proteolytic cleavage at hinge 2 of the molecule (2). Quantifications of the ratio of phosphorylated to total EGFR proteins showed that the ligand-induced increase in pY-EGFR levels in M2 cells was about 20% compared with M2A7 cells (Fig. 3C). Taken together, these results are consistent with the previous data showing that the knockdown of FLNa levels by siRNA can reduce the extent of EGF-mediated receptor proteolysis (Fig. 2D), and illustrate a mechanism that limits ligand-induced degradation of EGFR. No parallel change in insulin receptor content was detectable (Fig. 3D). We inferred specificity to EGFR because no changes of insulin receptor levels were observed in M2 an M2A7 cells stimulated with insulin (6). Pretreatment of M2A7 cells with the selective EGFR kinase inhibitor, AG1478, blocked EGF-induced EGFR degradation (Fig. 3D), consistent with a requirement for EGFR kinase activity in this process.
Next, the kinetics of EGFR ubiquitination was studied. Control and EGF-stimulated cells were lysed, and EGFR immunoprecipitates were analyzed for the presence of ubiquitinated EGFR by Western blotting. The amount of ubiquitin incorporation was maximal at 2.5 min after EGF treatment and declined thereafter (Fig. 3E). Ligand-induced EGFR ubiquitination was significantly lower (∼4-fold) in M2 cells compared with M2A7 cells both before (Fig. 3E) and after quantifications of the ratio of ubiquitinated to total EGFR protein levels (Fig. 3F, top panel), in agreement with the phosphorylation data. The ubiquitination of EGFR requires physical association with the ubiquitin ligase c-Cbl (26,27). The ligand-induced interaction between EGFR and c-Cbl was markedly lower in M2 cells compared with M2A7 cells, both before (Fig. 3E, lower panel) and after quantification of the ratio of bound c-Cbl to total EGFR proteins (Fig. 3F, lower panel). The kinetics of dissociation of c-Cbl from EGFR was linked with receptor deubiquitination. It is noteworthy that the extent of c-Cbl phosphorylation in response to EGF was comparable in both cell lines (Fig. 3G). Taken together, these studies indicate that a significant portion of cell surface EGFR appears refractory to ligand-induced activation and degradation in FLNa-deficient M2 cells.
FLNa-dependent sequestration and sorting of endocytosed EGFR to lysosomes
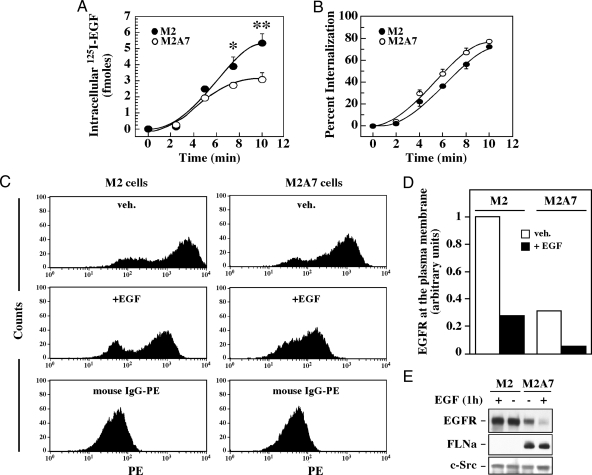
Impaired degradation of ligand-induced EGFR in M2 cells could result from inefficient endocytosis and/or poor transfer of endocytosed receptors to lysosomes. To address whether the expression of FLNa has any effect on the rate of receptor-mediated EGF uptake, a bolus of exogenous 125I-labeled EGF was given at 4 C to bind cell surface EGFR. Cells were returned for different time points at 37 C, which allows for occupancy-dependent EGFR trafficking. After glycine stripping treatment to remove surface-bound 125I-labeled EGF, the residual radioactivity was counted and used as an index of EGFR internalization. After 10 min, the relative amount of intracellular 125I-EGF was nearly doubled in M2 cells compared with M2A7 cells (Fig. 4A), probably as a consequence of increased cell surface EGFR expression in M2 cells (see above). When normalized to their respective maximum, the initial rate of uptake was slightly faster in M2A7 cells compared with M2 cells (Fig. 4B). The presence of sucrose, a known endocytosis inhibitor, counteracted EGFR-induced 125I-EGF uptake in both cell lines, indicating the importance of the endocytic process in the internalization of EGFR (data not shown).
Figure 4.
Effect of FLNa expression on ligand-induced EGFR internalization. A, M2 (filled circles) and M2A7 (open circles) cells were incubated for 3 h at 4 C in the presence of 125I-EGF (3 ng/ml) and washed rapidly with cold PBS to remove nonassociated label. Cells were transferred for different times at 37 C to promote EGF internalization and then treated with low pH buffer to remove plasma membrane-associated 125I-EGF. The remaining acid-inaccessible internalized label was measured in a γ-counter. Specific binding was determined as total binding minus the radioactivity associated with cells incubated in the presence of an excess of unlabeled EGF. B, Intracellular 125I-EGF was expressed as a percentage of total cell-associated radioactivity. Results are means ± sd (n = 3). *, P < 0.05, **, P < 0.01 compared with M2A7 cells. C, Cells were incubated with vehicle (veh.) or 20 nm EGF for 1 h at 37 C, followed by the addition of the low pH buffer to remove prebound surface-associated EGF. Cells were fixed, and the amount of EGFR present at the plasma membrane was measured by flow cytometry with a minimum of 10,000 cells counted. A group of cells were stained with phycoerythrin-conjugated mouse IgG as controls. D, Quantification of the mean peak values of cell surface EGFR before and after EGF treatment (from the fluorescence-activated cell sorter data shown in C). The level of cell surface EGFR in unstimulated M2 cells was arbitrarily given the value of 1.0. Data shown are representative of duplicate experiments. E, Lysates from untreated and EGF-stimulated M2 and M2A7 cells were immunoblotted with antibodies against EGFR, FLNa, or c-Src as a loading control.
To independently examine EGFR uptake, cells were fixed, stained with an antibody that recognizes the extracellular domain of EGFR, and then analyzed by flow cytometry (Fig. 4C). The fluorescence-activated cell sorter data revealed the presence of two fluorescent peaks, with the lower one corresponding to the phycoerythrin conjugated IgG control. The mean peak values of cell surface EGFR are represented in the graph shown in Fig. 4D. Unstimulated M2 cells exhibited about 3.5-fold higher levels of cell surface EGFR expression as compared with M2A7 cells (Fig. 4D, open bars), consistent with the Scatchard analysis (see above). Cells were also treated with EGF after which an acid wash step was performed to remove prebound cell surface EGF before fixing. The amount of EGFR present at the plasma membrane was markedly reduced in both cell types (≥70%) on EGF stimulation (Fig. 4, C and D, filled bars). Control experiments indicated that the acid wash procedure effectively stripped prebound 125I-EGF (data not shown).
To further confirm the differential ligand-induced EGFR stability between M2 and M2A7 cells, a second set of cells was incubated for 1 h in the presence or absence of EGF, lysed, and analyzed by Western blot. There was no ligand-induced receptor degradation in M2 cells, in sharp contrast to the near complete depletion of EGFR after EGF treatment of M2A7 cells (Fig. 4E). Combined with the results from Fig. 4, A–D, these data illustrate that EGFR is internalized normally in both cell types in the presence of EGF but that endocytosed EGFR is sorted into the degradation pathway only in FLNa-expressing M2A7 cells.
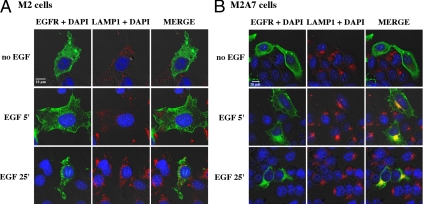
To study intracellular EGFR itinerary, M2 and M2A7 cells were transiently transfected with EGFR-GFP, and its subcellular distribution was analyzed by immunofluorescence and confocal microscopy. After fixation, the cells were immunostained for GFP and LAMP-1, a membrane protein marker of late endosomes and lysosomes. Experiments conducted without ligand revealed the presence of EGFR on the cell surface (Fig. 5). In M2 cells, EGF promoted a time-dependent endocytosis of EGFR in vesicles that were LAMP-1 negative (Fig. 5A, right panels), whereas in M2A7 cells, a good portion of the EGFRs was confined to LAMP-1 positive vesicles 5 min after EGF treatment (Fig. 5B, right panels). By 25 min of stimulation, EGFR-GFP-containing vesicles moved away from the cell surface and segregated as punctuated structures at the perinuclear region of M2 cells (Fig. 5A, bottom panels). The absence of detectable EGFR proteolytic degradation in M2 cells was associated with a distinctive pattern of LAMP-1 staining compared with FLNa-expressing M2A7 cells. Similar perinuclear LAMP staining was observed in M2A7, M93-047, and UACC647 cells (supplemental Fig. 3). These data indicate that FLNa could have an important role in EGFR delivery into the degradation pathway.
Figure 5.
Ligand-induced cellular redistribution of EGFR. M2 cells (A) and M2A7 cells (B) were transiently transfected with EGFR-GFP and then stimulated with EGF (10 nm) for the indicated time intervals at 37 C. Cells were then fixed and immunolabeled with polyclonal anti-GFP (green, left panels) and a monoclonal anti-LAMP-1 (red, middle panels) antibodies. Nuclei were counterstained with 4′,6′-diamino-2-phenylindole (DAPI; blue). Merge images are shown in the right panels. Areas of colocalization between EGFR and LAMP-1 appear in yellow in M2A7 cells, which were largely absent in M2 cells. Bar, 10 μm. Similar results were obtained in three independent experiments.
Biological activity of EGFR in M2 and M2A7 cells
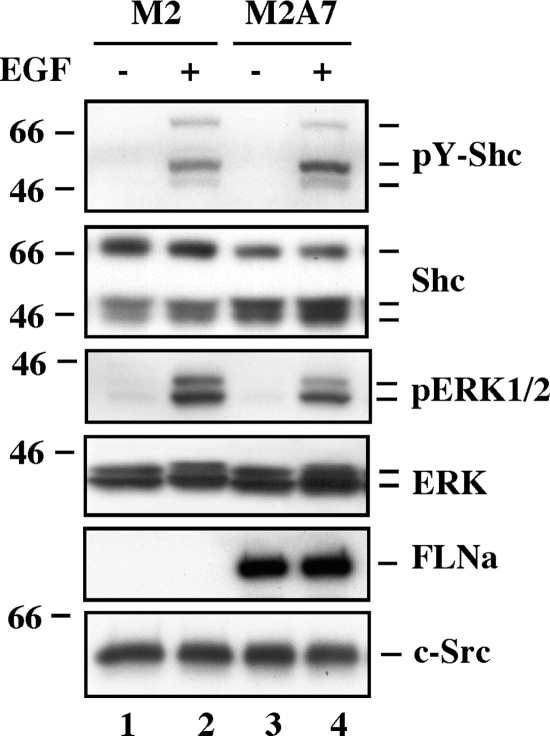
To determine whether absolute number or fraction of activated cell surface receptors dictates the intensity of EGF signal, M2 and M2A7 cells were stimulated with EGF for 10 min, lysed, and the lysates analyzed by Western blotting. The amount of EGF-induced Shc and ERK1/2 phosphorylation was comparable in both cell lines, despite higher EGFR expression in FLNa-deficient M2 cells (Fig. 6).
Figure 6.
Role of FLNa in EGF-induced activation of downstream signaling. Serum-starved M2 and M2A7 cells were left untreated or treated with EGF (20 nm) for 10 min, lysed, and analyzed by Western blot using antibodies against tyrosine phosphorylated Shc (pY-Shc), total Shc, phosphoactive ERK1/2 (pERK), ERK, FLNa, or c-Src as a loading control. Data shown are representative of three independent experiments.
Discussion
In this work, EGFR expression was found to be up-regulated in highly metastatic melanomas from patient biopsies and cell lines, with a modest increase in FLNa levels. Both the basal and EGF-stimulated rate of cell motility, as assessed by wound-healing assays, correlated with the expression of endogenous EGFR, consistent with earlier reports showing that EGFR activation plays an important role in melanoma cell motility (28,29). Accordingly, pharmacological inhibition of EGFR significantly reduced the basal rate of gap closure, which supports the notion that endogenously produced members of the EGF family contribute to this EGFR response. However, the difference in the rate of cell motility between G-361 (low metastatic) and M93-047 (highly invasive) cells persisted in the absence of EGFR kinase activity (Fig. 1D), indicating that different levels of EGFR expression contribute only partly in melanoma cell migration. Of importance, because FLNa emerges from this study as a player for both basal and EGF-induced cell motility (Fig. 1E), its regulatory role in the activation and intracellular trafficking of EGFR may be quite extensive.
The results we present identify FLNa as a major component of the down-regulation process of EGFR. Incidentally, siRNA-mediated knockdown of FLNa expression interferes with the mechanism of ligand-dependent degradation of the activated receptors in the two highly metastatic melanoma lines, M93-047 and UACC647 (Fig. 2D). Because of high levels of FLNa expression in these cells, the attenuation in FLNa expression was only partial. To circumvent effects reflecting partial silencing of FLNa, we used human M2 melanoma cell model in all following experiments. This model has been extensively used to study the role of FLNa in various aspects of cell signaling and biological processes (4,5,6,11,30,31,32,33,34,35,36,37).
Ectopic expression of FLNa in M2A7 cells led to an approximately 2.5-fold reduction in EGFR mRNA (Table 1) and protein (Fig. 3) levels when compared with the isogenic M2 cells. It is possible that FLNa has several roles in EGFR gene expression, such that in the absence of FLNa (e.g. M2 cells), increased mRNA stability, mRNA localization, and/or translational efficiency of EGFR may occur. Further investigation of such mechanism falls outside the scope of this paper, as EGFRs were found to be inserted in the plasma membrane and bind EGF with the same affinity, irrespective of the presence or absence of FLNa. Nevertheless, the potential impact that varying levels of EGFR expression has in some of the observed differences in EGF signaling between M2 and M2A7 cells (irrespective of the expression level of FLNa) cannot be ignored.
Our data demonstrate the presence of a large pool of cell surface EGFR in M2 cells (FLNa deficient) that is refractory to ligand-induced phosphorylation and ubiquitination when compared with its M2A7 subclone stably expressing FLNa cDNA. Such inefficient activation of the receptor autophosphorylation may prevent conformational changes required for EGFR to associate with molecules viewed as critical for endocytosis through clathrin-coated pits (38,39,40,41,42). However, the rate of ligand-induced EGFR internalization was found to be unaffected by the level of FLNa expression. The simplest interpretation is that nonclathrin pathways may be implicated in the rapid sequestration and cellular entry of large numbers of EGFR in response to EGF, independent of the actin-binding properties of FLNa. This observation is consistent with the study of Orth et al. (20), who reported that various cell types form a transient structure, known as circular dorsal ruffles, that promotes ligand-induced internalization of EGFR.
Our present study shows that the levels of ligand-induced EGFR ubiquitination and association with the ubiquitin ligase, c-Cbl, are also significantly reduced in the absence of FLNa. It is likely that this actin-binding protein elicits a key role in EGFR function by catalyzing the formation of EGF-inducible complexes of c-Cbl with interacting molecules that is necessary for both the transmission and ultimate termination of EGFR signaling. It should be noted that c-Cbl-mediated ubiquitination is not needed for EGFR internalization but is required for the exit of the receptor from early endosomes and its subsequent trafficking to the lysosomes (43,44). Besides c-Cbl, many accessory proteins interact with a subset of tyrosine phosphorylated residues within the cytoplasmic domain of activated EGFR; these phosphotyrosine moieties serve as docking sites for SH2 domain-containing signaling molecules, including the protein tyrosine phosphatase SHP-1, phospholipase C-γ1, c-Cbl, and protein kinase Cε (45,46,47,48). The latter site (Tyr845) does not belong to the autophosphorylation sites but appears to be phosphorylated by c-Src post-EGF stimulation (49). Possibly, the absence of FLNa prevents ligand-induced phosphorylation at specific receptor sites, thereby limiting EGFR association with signaling intermediates involved in protein-protein interactions responsible for intracellular EGFR processing.
Ligand-activated endocytosed EGFR was not efficiently transferred to lysosomes in M2 cells and was specifically targeted for lysosomal degradation in FLNa-expressing M2A7 cells. This conclusion was reached based on independent lines of evidence. First, EGF-mediated EGFR uptake was accompanied by minimal degradation of the receptor in the absence of FLNa. Second, confocal microscopy analysis in M2 cells revealed that endocytosed EGFR distributed with intracellular vesicles that were negative for LAMP-1, a lysosomal marker (Fig. 5). Like FLNa, EGFR is an actin-binding protein, as revealed by in vitro and in vivo interactions with F-actin (50,51). Using cell fractionation assays, others have demonstrated sorting from the degradation pathway of an EGFR mutant that lacks its actin-binding domain (52). We speculate that the lack of FLNa could interfere with EGFR binding to actin and therefore hamper proper EGFR sorting. Interestingly, Liu et al. (53) showed that the sorting of furin from the cell surface to early endosomes is independent of FLNa but that the trafficking of molecules into late endocytic and lysosomal compartments requires FLNa. The association of filamin with the lipid phosphatase SHIP-2 allows its translocation from the cytosol to membrane ruffles after growth factor stimulation (8), whereas the endocytic sorting and recycling of the internalized G protein-coupled calcitonin receptor are deficient in the absence of FLNa (54). Finally, μ-opioid receptor down-regulation and intracellular trafficking depends on the presence of FLNa (32). Therefore, these data support the concept that FLNa plays an important role in generic steps in vesicular transport and protein trafficking. It follows therefore that FLNa deficiency may elicit a pleiotropic defect in late endosome/lysosomal organelle distribution rather than a more specific defect in EGFR lysosomal targeting.
As shown in the study herein, the fraction of activated EGFR was markedly lower in M2 vs. M2A7 cells, whereas the absolute number of activated receptors (Fig. 3B, 3E) and EGF responsiveness (Fig. 6) appeared to be comparable. This points to the presence of a significant pool of cell surface EGFRs in M2 cells that binds EGF with high affinity but is unable to undergo activation. These refractory receptors could provide a reserve for subsequent signaling and support the concept of spare receptors (55,56).
In conclusion, the molecular mechanism behind the function of FLNa in ligand-induced activation and intracellular sorting of EGFR remains unclear. Perhaps the association of FLNa with other structural proteins is required to facilitate ligand-induced EGFR phosphorylation and its recruitment by clathrin-coated vesicles for proper trafficking into the lysosomal pathway. It is possible that abrogation of normal actin polymerization and impaired formation of protein scaffolds affects EGFR localization to various microdomains at the plasma membrane and their trafficking. In this regard, the coat protein caveolin-1 has been shown to constitutively bind EGFR and attenuate ligand-induced EGFR activity (57,58). Moreover, the segregation of caveolin-1 controls the formation of caveolae and the regulation of raft-dependent endocytosis (59). Through its scaffolding properties, FLNa likely provides a platform for protein-protein interactions with membrane coat proteins and the cytoskeleton network, which in turn ensures efficient EGFR activation, intracellular trafficking, and degradation.
Acknowledgments
We thank Dr. A. Sorkin for cDNA encoding EGFR-GFP.
Footnotes
This work was supported entirely by the Intramural Research Program of the National Institutes of Health, National Institute on Aging. A portion of that support was through a research and development contract with MedStar Research Institute.
Current address for T.-N.Z.: Department of Endocrinology, Fourth Hospital, Hebei Medical University, 12 Jiankang Road, Shijiazhuang, Hebei 050011, People’s Republic of China.
Disclosure Summary: The authors have nothing to disclose.
First Published Online February 12, 2009
Abbreviations: EGF, Epidermal growth factor; EGFR, EGF receptor; FLNa, filamin A; GFP, green fluorescent protein; LAMP, lysosome-associated membrane protein; pY-EGFR, tyrosine phosphorylated EGFR; siRNA, small interfering RNA.
References
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA 2006 Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA 103:19836–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowicz GM, Schleicher M, Noegel AA, Holak TA 2006 Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci 31:411–419 [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH 2001 Increased filamin binding to β-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol 3:1060–1068 [DOI] [PubMed] [Google Scholar]
- Awata H, Huang C, Handlogten ME, Miller RT 2001 Interaction of the calcium-sensing receptor and filamin, a potential scaffolding protein. J Biol Chem 276:34871–34879 [DOI] [PubMed] [Google Scholar]
- Lin R, Karpa K, Kabbani N, Goldman-Rakic P, Levenson P 2001 Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc Natl Acad Sci USA 98:5258–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HJ, Kole S, Kwon YK, Crow MT, Bernier M 2003 Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem 278:27096–27104 [DOI] [PubMed] [Google Scholar]
- Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U 2000 Physical and functional interaction of filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J Biol Chem 275:271–278 [DOI] [PubMed] [Google Scholar]
- Dyson JM, O'Malley CJ, Becanovic J, Munday AD, Berndt MC, Coghill ID, Nandurkar HH, Ooms LM, Mitchell CA 2001 The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembranous actin. J Cell Biol 155:1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar G, Rajasekaran SA, Wang S, Hankinson O, Bander NH, Rajasekaran AK 2003 Prostate-specific membrane antigen association with filamin A modulates its internalization and NAALADase activity. Cancer Res 63:2645–2648 [PubMed] [Google Scholar]
- Pálmer HG, Sánchez-Carbayo M, Ordóñez-Morán P, Larriba MJ, Cordón-Cardó C, Muñoz A 2003 Genetic signatures of differentiation induced by 1α,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res 63:7799–7806 [PubMed] [Google Scholar]
- Flanagan LA, Chou J, Falet H, Neujahr R, Hartwig JH, Stossel TP 2001 Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J Cell Biol 155:511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaile E, Müller MM, Kannicht C, Singer BB, Lucka L 2005 CEACAM1 functionally interacts with filamin A and exerts a dual role in the regulation of cell migration. J Cell Sci 118:5513–5524 [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P 2006 Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase Cε-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem 281:14026–14040 [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS 2006 Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366:2–16 [DOI] [PubMed] [Google Scholar]
- Waugh MG, Lawson D, Hsuan JJ 1999 Epidermal growth factor receptor activation is localized within low-buoyant density, non-caveolar membrane domains. Biochem J 337:591–597 [PMC free article] [PubMed] [Google Scholar]
- Ringerike T, Blystad FD, Levy FO, Madshus IH, Stang E 2002 Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J Cell Sci 115:1331–1340 [DOI] [PubMed] [Google Scholar]
- Roepstorff K, Thomsen P, Sandvig K, van Deurs B 2002 Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibit ligand binding. J Biol Chem 277:18954–18960 [DOI] [PubMed] [Google Scholar]
- Leahy DJ 2004 Structure and function of the epidermal growth factor (EGF/ErbB) family of receptors. Adv Protein Chem 68:1–27 [DOI] [PubMed] [Google Scholar]
- Carpenter G 2000 The EGF receptor: a nexus for trafficking and signaling. Bioessays 22:697–707 [DOI] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Weller SG, McNiven MA 2006 A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res 66:3603–3610 [DOI] [PubMed] [Google Scholar]
- Dissanayake SK, Wade M, Johnson CE, O'Connell MP, Leotlela PD, French AD, Shah KV, Hewitt KJ, Rosenthal DT, Indig FE, Jiang Y, Nickoloff BJ, Taub DD, Trent JM, Moon RT, Bittner M, Weeraratna AT 2007 The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem 282:17259–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HJ, Zhu TN, Xie Y, Fan J, Kole S, Saxena S, Bernier M 2006 Pyrrolidine dithiocarbamate inhibits interleukin-6 signaling through impaired STAT3 activation and association with transcriptional coactivators in hepatocytes. J Biol Chem 281:31369–31379 [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Uqurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, Schadendorf D, Dummer R 2006 Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res 19:290–302 [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW 2003 Epidermal growth factor receptor: mechanisms of activation and signaling. Exp Cell Res 284:31–53 [DOI] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP 1992 Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255:325–327 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y 1998 c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev 12:3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill NL, Douillard P, Awward RA, Ota S, Lupher Jr MI, Miyake S, Meissner-Lula N, Hsu VW, Band H 2000 The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem 275:367–377 [DOI] [PubMed] [Google Scholar]
- Xie H, Pallero MA, Gupta K, Chang P, Ware MF, Witke W, Kwiatkowski DJ, Lauffenburger DA, Murphy-Ullrich JE, Wells A 1998 EGF receptor regulation of cell motility: EGF induces disassembly of focal adhesions independently of the motility-associated PLCγ signaling pathway. J Cell Sci 111:615–624 [DOI] [PubMed] [Google Scholar]
- Gordon-Thomson C, Jones J, Mason RS, Moore GP 2005 ErbB receptors mediate both migratory and proliferative activities in human melanocytes and melanoma cells. Melanoma Res 15:21–28 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K 2001 Filamin associates with Smads and regulates transforming growth factor-β signaling. J Biol Chem 276:17871–17877 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shen Z 2001 Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem 276:48318–48324 [DOI] [PubMed] [Google Scholar]
- Onoprishvili I, Andria ML, Kramer HK, Ancevska-Taneva N, Hiller JM, Simon EJ 2003 Interaction between the μ-opioid receptor and filamin A is involved in receptor regulation and trafficking. Mol Pharmacol 64:1092–1100 [DOI] [PubMed] [Google Scholar]
- Meng X, Yuan Y, Maestas A, Shen Z 2004 Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280. J Biol Chem 279:6098–6105 [DOI] [PubMed] [Google Scholar]
- Zhang M, Breitwieser GE 2005 High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J Biol Chem 280:11140–11146 [DOI] [PubMed] [Google Scholar]
- Zhu TN, He HJ, Kole S, D'Souza T, Agarwal R, Morin PJ, Bernier M 2007 Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J Biol Chem 282:14816–14826 [DOI] [PubMed] [Google Scholar]
- Kim EY, Ridgway LD, Dryer SE 2007 Interactions with filamin A stimulate surface expression of large-conductance Ca2+-activated K+ channels in the absence of direct actin binding. Mol Pharmacol 72:622–630 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Park JS, Um SJ 2007 Filamin A negatively regulates the transcriptional activity of p73α in the cytoplasm. Biochem Biophys Res Commun 362:1101–1106 [DOI] [PubMed] [Google Scholar]
- Jiang X, Huang F, Marusyk A, Sorkin A 2003 Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol Biol Cell 14:858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Sorkin A 2005 Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol Biol Cell 16:1268–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ 2003 A cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem 278:1805–1813 [DOI] [PubMed] [Google Scholar]
- Kurten RC, Cadena DL, Gill GN 1996 Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science 272:1008–1010 [DOI] [PubMed] [Google Scholar]
- Gullapalli A, Garrett TA, Paing MM, Griffin CT, Yang Y, Trejo J 2004 A role for sorting nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting nexin 1 and 2 in protein trafficking. Mol Biol Cell 15:2143–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, Rao N, Donoghue S, Rogers RA, Bowtell D, Naramura M, Gu H, Band V, Band H 2003 Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem 278:28950–28960 [DOI] [PubMed] [Google Scholar]
- Ravid T, Heidinger JM, Gee P, Khan EM, Goldkorn T 2004 c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J Biol Chem 279:37153–37162 [DOI] [PubMed] [Google Scholar]
- Keilhack H, Tenev T, Nyakatura E, Godovac-Zimmermann J, Nielsen L, Seedorf K, Böhmer FD 1998 Phosphotyrosine 1173 mediates binding of the protein-tyrosine phosphatase SHP-1 to the epidermal growth factor receptor and attenuation of receptor signaling. J Biol Chem 273:24839–24846 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, Vecchi M, Ji Q, Mernaugh R, Carpenter G 1999 The role of individual SH2 domains in mediating association of phospholipase C-γ1 with the activated EGF receptor. J Biol Chem 274:26091–26097 [DOI] [PubMed] [Google Scholar]
- Grøvdal LM, Stang E, Sorkin A, Madshus IH 2004 Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp Cell Res 300:388–395 [DOI] [PubMed] [Google Scholar]
- Valkova C, Maerz S, Imhof D, Liebmann C 2007 Protein kinase Cε may act as EGF-inducible scaffold protein for phospholipase Cγ1. Cell Signal 19:1830–1843 [DOI] [PubMed] [Google Scholar]
- Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ 1999 c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem 274:8335–8343 [DOI] [PubMed] [Google Scholar]
- den Hartigh JC, van Bergen en Henegouwen PM, Verkleij AJ, Boonstra J 1992 The EGF receptor is an actin-binding protein. J Cell Biol 119:349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Heyden MA, Nievers M, Verkleij AJ, Boonstra J, Van Bergen en Henegouwen PM 1997 Identification of an intracellular domain of the EGF receptor required for high-affinity binding of EGF. FEBS Lett 410:265–268 [DOI] [PubMed] [Google Scholar]
- Stoorvogel W, Kerstens S, Fritzsche I, den Hartigh JC, Oud R, van der Heyden MA, Voortman J, van Bergen en Henegouwen PM 2004 Sorting of ligand-activated epidermal growth factor receptor to lysosomes requires its actin-binding domain. J Biol Chem 279:11562–11569 [DOI] [PubMed] [Google Scholar]
- Liu G, Thomas L, Warren RA, Enns CA, Cunningham CC, Hartwig JH, Thomas G 1997 Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J Cell Biol 139:1719–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck T, Baron R, Horne WC 2003 Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J Biol Chem 278:10408–10416 [DOI] [PubMed] [Google Scholar]
- Sjödin L, Dahlén HG, Viitanen E 1992 Binding of epidermal growth factor to receptors in preparations of enriched porcine parietal cells and inhibition of aminopyrine uptake. Scand J Gastroenterol 27:495–500 [DOI] [PubMed] [Google Scholar]
- Nandagopal K, Popp DM, Niyogi SK 2001 Utilization of a receptor reserve for effective amplification of mitogenic signaling by an epidermal growth factor mutant deficient in receptor activation. J Cell Biochem 83:326–341 [DOI] [PubMed] [Google Scholar]
- Couet J, Sargiacomo M, Lisanti MP 1997 Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem 272:30429–30438 [DOI] [PubMed] [Google Scholar]
- Park WY, Park JS, Cho KA, Kim DI, Ko YG, Seo JS, Park SC 2000 Up-regulation of caveolin attenuates epidermal growth factor signaling in senescent cells. J Biol Chem 275:20847–20852 [DOI] [PubMed] [Google Scholar]
- Lajoie P, Nabi IR 2007 Regulation of raft-dependent endocytosis. J Cell Mol Med 11:644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]