Abstract
Galectins are proteins that regulate immune responses through the recognition of cell-surface glycans. We present evidence that 16 human galectin genes are expressed at the maternal–fetal interface and demonstrate that a cluster of 5 galectin genes on human chromosome 19 emerged during primate evolution as a result of duplication and rearrangement of genes and pseudogenes via a birth and death process primarily mediated by transposable long interspersed nuclear elements (LINEs). Genes in the cluster are found only in anthropoids, a group of primate species that differ from their strepsirrhine counterparts by having relatively large brains and long gestations. Three of the human cluster genes (LGALS13, -14, and -16) were found to be placenta-specific. Homology modeling revealed conserved three-dimensional structures of galectins in the human cluster; however, analyses of 24 newly derived and 69 publicly available sequences in 10 anthropoid species indicate functional diversification by evidence of positive selection and amino acid replacements in carbohydrate-recognition domains. Moreover, we demonstrate altered sugar-binding capacities of 6 recombinant galectins in the cluster. We show that human placenta-specific galectins are predominantly expressed by the syncytiotrophoblast, a primary site of metabolic exchange where, early during pregnancy, the fetus comes in contact with immune cells circulating in maternal blood. Because ex vivo functional assays demonstrate that placenta-specific galectins induce the apoptosis of T lymphocytes, we propose that these galectins reduce the danger of maternal immune attacks on the fetal semiallograft, presumably conferring additional immune tolerance mechanisms and in turn sustaining hemochorial placentation during the long gestation of anthropoid primates.
Keywords: adaptive evolution, glycocode, maternal–fetal immune tolerance, PP13, preeclampsia
The genetic differences between the mother and the fetal semiallograft necessitate immune tolerance at the maternal–fetal interface to reduce the danger of destructive maternal immune attacks on fetal alloantigens in eutherian pregnancies (1–4). Species with invasive hemochorial placentation have a maternal–fetal interface in which extravillous trophoblasts invade uterine decidual tissues and interact with maternal immune cells residing on mucosal surfaces, whereas villous trophoblasts residing on placental surfaces are bathed in maternal blood and are in direct contact with maternal leukocytes (2). Among species with hemochorial placentas, anthropoid primates (i.e., Old and New World monkeys and apes, including humans) generally have a long gestation and large brain relative to their body size (5). Humans have a more invasive placentation in which extravillous trophoblasts invade the inner third of the myometrium, and villous trophoblasts are in intimate and extended contact with maternal blood, challenging the maternal immune system and possibly requiring additional tolerance mechanisms (6).
Recent studies in humans and other primates show that natural killer (NK) cell–extravillous trophoblast interactions at uterine mucosal surfaces depend on adequate ligand binding between glycosylated HLA antigens on trophoblasts and NK cell receptors (e.g., KIRs) (2, 7). At the villous trophoblast–blood barrier, the syncytiotrophoblast apical membrane is densely covered with glycoproteins that may confer tolerance (e.g., Fas-ligand/CD178) (8). Indeed, this glycosylated trophoblast membrane inhibits activated maternal leukocytes (9, 10) and affects compatibility between mother and offspring (11).
Galectins are proteins that bind and cross-link glycans on leukocyte surfaces. They function through transmembrane signaling and the regulation of adaptive and innate immune responses (12–15). Galectin-1 and galectin-13 (PP13) are also present on the syncytiotrophoblast apical membrane, and their placental expression is altered in preeclampsia (16–21), a syndrome linked to immune maladaptation (2–4, 7, 22). Other galectins (-3, -9, -14) are also expressed at the maternal–fetal interface (16, 18, 23, 24); many induce the apoptosis of activated T cells and other leukocytes, thereby conferring immune tolerance (25–27). Indeed, galectin-1 plays a central role in maternal–fetal immune tolerance by promoting the generation of tolerogenic dendritic cells and regulatory T (Treg) cells in mice (28) and by inducing apoptosis of T cells in humans (27).
Based on the importance of galectins in immune tolerance and their abundant placental expression, we aimed to study the evolution, structure, and immune function of galectins predominantly expressed in the human placenta. The data we present suggests that anthropoid primates evolved a cluster of galectins as additional immunoregulatory molecules at the maternal–fetal interface in conjunction with the evolution of highly invasive placentation and long gestation, which were essential for human evolution.
Results and Discussion
Abundant and Tissue-Specific Expression of Galectins at the Maternal–Fetal Interface.
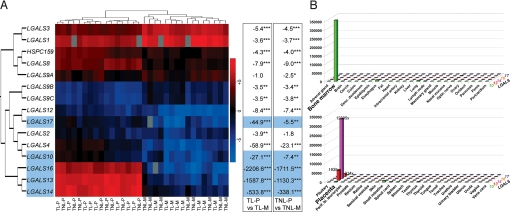
To determine which galectin genes are expressed at the maternal–fetal interface, we performed qRT-PCR expression profiling on human placenta and fetal membranes. All 16 genes, including 2 predicted loci (LOC148003 and LOC400696, which we name LGALS16 and LGALS17) contained within a cluster of 5 galectin-like genes on Chr19, are expressed at the maternal–fetal interface irrespective of labor status (Fig. 1A). LGALS1 and LGALS3 are strongly and predominantly expressed in fetal membranes. LGALS7 is not detected in the placenta, and its expression in fetal membranes is low. Three genes in the Chr19 cluster (LGALS13, -14, and -16) have 338–2,206-fold higher (P < 10−16) expression in the placenta than in fetal membranes.
Fig. 1.
Galectins have abundant expression in human placenta and fetal membranes; 3 genes in a Chr19 cluster are placenta-specific. (A Left) Heatmap represents the expression of 15 genes in placenta (P) and fetal membranes (M) in normal pregnant woman at term, in labor (TL), or not in labor (TNL). Color key is assigned for −ΔCt values; gray depicts missing values. Three genes in a Chr19 cluster have strong placental expression. (Right) Fold-change differences between mean gene expression levels in placenta and fetal membranes are shown separately for laboring (left box) and nonlaboring (right box) women. Positive numbers show higher expression in the placenta. ∗, P < 0.01; ∗∗, P < 0.001; ∗∗∗, P < 0.00001. (B) qRT-PCR on a human 48-tissue cDNA panel reveals that LGALS13, -14, and -16 are highly and solely expressed in the placenta (Lower), LGALS17 has low expression in the placenta and other tissues, and LGALS10 is predominantly expressed in the bone marrow (Upper). The y axis shows expression level, and the numbers depict fold-change difference between placental and mean gene expression levels of all other tissues.
GenBank searches revealed only placental ESTs for LGALS13, -14, and -16; thus, we hypothesized that these genes would have predominant placental expression. We explored gene expression in the Chr19 cluster by profiling 48 human tissues and found that LGALS13, -14, and -16 are highly and chiefly expressed in the placenta (Fig. 1B). LGALS17 is weakly expressed in 19 tissues including placenta. LGALS10 is predominantly expressed in bone marrow, a finding in accord with previous data demonstrating galectin-10 only in eosinophils, basophils, and Treg cells (29, 30).
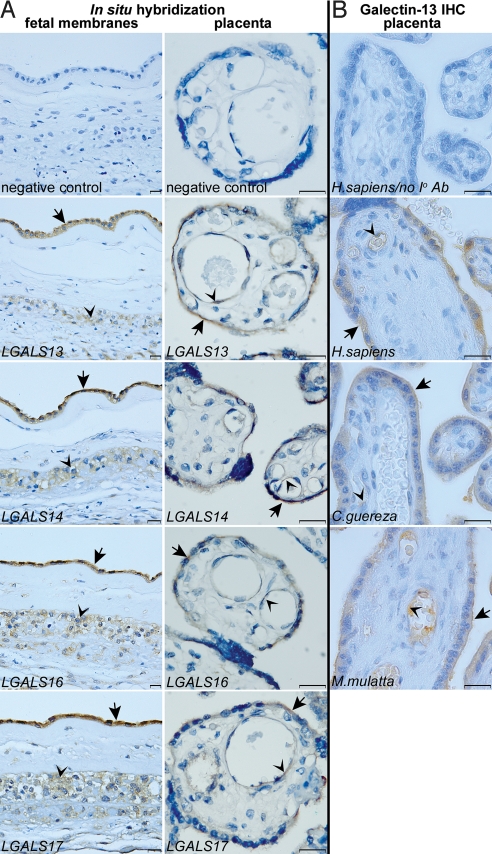
To localize LGALS13, -14, -16, and -17 in the placenta and fetal membranes, we performed mRNA in situ hybridization (Fig. 2A). In fetal membranes, these genes are expressed in the amnion and extravillous trophoblasts, where maternal–fetal immune interactions occur (2). In the placenta, they are mainly expressed in the syncytiotrophoblast and also in the endothelia of fetal vessels, in cells of epithelial and endothelial origin. We also detected galectin-13 protein in the syncytiotrophoblast and fetal endothelia in human, colobus, and macaque placentas. The syncytiotrophoblast microvillous membrane, another location of maternal–fetal interactions, is immunopositive in all tested species (Fig. 2B). The clinical relevance of these genes has yet to be fully appreciated, but the diminished expression of LGALS13 has previously been associated with preeclampsia (20, 21).
Fig. 2.
Chr19 cluster galectins have a similar expression pattern at the maternal–fetal interface. (A Left) In situ hybridization reveals LGALS13, -14, -16, and -17 expression in the amnion (arrows) and chorionic trophoblasts (arrowheads) in fetal membranes of nonlaboring women. (Right) In the placenta, these genes are predominantly expressed by the syncytiotrophoblast (arrows) and fetal endothelia (arrowheads). (B) Galectin-13 immunostaining is conserved in the syncytiotrophoblast, its apical membrane (arrows), and the endothelia (arrowheads) of human and anthropoid primate placentas. (Scale bars: 20 μm.)
Birth and Death of Genes in the Chr19 Galectin Cluster.
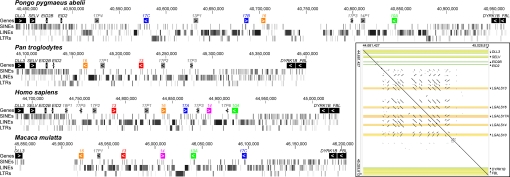
To determine the evolutionary origin of placenta-specific genes in the Chr19 cluster, we combined BLAT and BLAST searches of assembled and nonassembled genomes with the generation of new sequence data from cDNA and genomic DNA. We defined the cluster as an ≈300-kb region of human Chr19q13.2 between EID2 and DYRK1B (Fig. 3). We collected 24 primate sequences and annotated an additional 49 gene and pseudogene sequences from Whole Genome Shotgun (WGS) data [supporting information (SI) Dataset S1]. We collected cDNA sequence data from placentas of human, baboon, macaque, and Spider monkey. We were able to find evidence for the presence of genes in the cluster in apes (human, common chimpanzee, bonobo, gorilla, and orangutan), Old World monkeys (macaque, baboon, colobus), and New World monkeys (Spider monkey and marmoset), but not in the genomes of prosimian primates (tarsier, bushbaby, mouse lemur) or nonprimates. LGALS14 is absent in bonobos and common chimpanzees, and LGALS10 has been expanded to include 3 functional marmoset genes. Sheep LOC443162 encodes a protein termed “galectin-14” (31), but this gene is closely related to LGALS9 and distantly related to the genes in the cluster (15). LGALS15, a gene found in artiodactyls, shares the most sequence identity with genes in the anthropoid cluster (32) and is located on cow Chr18 between EID2 and DYRK1B, suggesting the possiblity that genes encoding galectins were already present in the region at the time of the last common ancestor of cows and primates.
Fig. 3.
Comparative genomic map of the Chr19 cluster in humans and nonhuman primates. Boxes below chromosome coordinates show genes, inside (colors) or outside (black) of the cluster, and pseudogenes (gray); arrows indicate orientations. SINEs are prominent outside the cluster, and genes in the cluster are surrounded by LINEs and LTRs. (Inset) PipMaker alignment of the human cluster with itself shows numerous duplications and inversions. Numbers indicate chromosomal locations; short arrows show coding strand orientations. Positions of genes (introns, yellow; exons in the cluster, red; exons outside of the cluster, green) are also depicted.
The alignment of the human cluster with itself shows extensive duplications and inversions (Fig. 3). Additional genomic rearrangements and deletions are apparent from the pair-wise alignments of the 4 clusters (Fig. S1). Fig. 3 shows that short interspersed transposable elements (SINEs) are the most frequent repetitive transposable elements directly outside the boundaries of the cluster; however, genes and pseudogenes in the cluster are surrounded by LINEs and long terminal repeats (LTRs). Analysis of assembled genomes revealed LINE elements at the majority of boundaries of large inversions and gene duplication units, suggesting that these elements have primarily mediated the extensive rearrangements within the cluster (Fig. 3).
Genes in the anthropoid cluster include LGALS10, -13, -14, -16, -17, -19, and -20. As with other galectin genes, they have a conserved 4-exon structure. GenBank searches and our cDNA sequences show that human LGALS17 does not transcribe the canonical fourth exon but instead transcribes 5 other exons. The cluster contains a large number of pseudogenes in the studied species, 19 of 38 are LGALS17-related sequences (Dataset S2). Pseudogenes are truncated by missing exons, mutations of the exon–intron boundaries, and the introduction of 1 or more in-frame premature stop codons. Strikingly, 18 pseudogenes contain a premature stop codon at the site encoding residue 55, which would result in a truncated protein that lacks the carbohydrate-recognition domain (CRD).
Although errors in sequencing and genome assembly can cause misannotation of genes, and duplicated regions are also prone to assembly error (33, 34), we propose that the Chr19 galectin cluster is the result of a repeat mediated birth and death process. This process, in which some genes are duplicated and some of the duplicated genes are lost, is a primary means by which species gain novel phenotypes and adapt to their environment (35); thus, anthropoids may have evolved placental phenotypes associated with the emergence of the Chr19 cluster.
Phylogenetic Analysis Shows Positive Selection Preceded Purifying Selection in the Cluster.
To study the phylogeny of the 79 coding sequences included in our analysis, we inferred the optimal Bayesian tree (ln L = −10321.79) (Fig. S2), which is mostly congruent with the ML tree (In L = −10392.67). There are 2 main clades within genes and pseudogenes in the cluster. One clade contains 5 genes with predominant placental expression, including the previously identified LGALS13 (17, 36–38) and LGALS14 (23), as well as related pseudogenes. LGALS14 is found in all 3 major anthropoid clades (apes, Old World monkeys, and New World monkeys), whereas LGALS13 is found only in catarrhines (Old World monkeys and apes). A third gene, LGALS16 (LOC148003), is also restricted to catarrhines. The final 2 genes, LGALS19 and LGALS20, appear to be restricted to New World monkeys. The other clade contains genes and related pseudogenes of LGALS10 (29, 30), LGALS17 (LOC400696), and LGALS18.
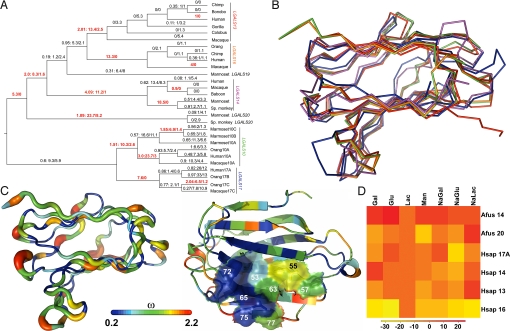
To examine the effects of natural selection on genes in the cluster, we conducted phylogenetic likelihood ratio tests of dN/dS (i.e., ω). There is significant variation in this commonly used metric of adaptive evolution; a null model in which dN/dS does not vary (ln L = −3267.62) is a worse fit of the data than a model where the ratio is allowed to vary on each branch (ln L = −3202.94; P = 3.3 × 10− 7). There is evidence for positive selection on 15 branches of the gene tree (Fig. 4A), including the branches directly after the duplicative origin of each individual gene, with the exception of LGALS19; ω values associated with the emergence of these genes are as follows: LGALS10 = 1.51, LGALS13 = 2.01, LGALS14 = 4.09, LGALS16 = ∞, LGALS17 = ∞, and LGALS20 = 1.09. Branch-site tests on these newly duplicated lineages further indicate positive selection suggestive of neofunctionalization (P = 1 × 10−16). LGALS10 appears to break this rule, but close examination shows that there are multiple LGALS10 paralogs.
Fig. 4.
Evolutionarily changes leading to structural and functional diversification in Chr19 cluster galectins. (A) Numbers above the branches of the gene tree represent ω, N*dN (nonsynonymous substitutions), and S*dS (synonymous substitutions), respectively, shown in red on branches with evidence for positive selection. (B) Superposition of human galectin structures (10, green; 13, red; 14, purple; 16, orange; 17, blue) demonstrates their conserved β-sandwich structure composed of 2 antiparallel sheets. Loop regions show more structural variability. (C Left) The width of the ribbon representing the molecular backbone of galectin-16 varies in proportion with site-specific ω values for all cluster galectins. ω, also indicated by color spectrum depicted on the bar, is the smallest along β-strands and highest in loop regions. (Right) The same color coding shows that 4 residues in the CRD of cluster galectins (residues 53, 65, 72, and 75) are under strong purifying selection, others on the opposite side (residues 55, 57, 63, and 77) show more variability. (D) Heatmap shows the percentage of competitively eluted proteins from lactose-agarose beads relative to lactose.
Proteins in the Cluster Are Structurally Related to Galectins.
We examined amino acid sequence evolution and modeled protein structures to infer the function of the proteins encoded by the cluster genes. Evolutionary constraints are still acting on these genes as follows. (i) Purifying selection (ω < 1) characterizes 75 of 142 residues in the encoded proteins. (ii) Homology modeling of human proteins encoded by LGALS14, -16, and -17 revealed their prototype galectin structure (39, 40). This conserved β-sandwich is composed of 2 antiparallel sheets that include the CRD, whereas loop regions show more structural variability (Fig. 4B). (iii) The mean ω for residues in the core is significantly lower than for those on the surface (0.67 vs. 1.05, P < 0.001), especially for those in loop regions (Fig. 4C). (iv) From the 8 conserved residues in the CRD of galectins that are involved in sugar binding, N65, W72, and E75 were subject to strong purifying selection in all proteins. These residues form a pocket in one side of the CRD and are important for the overall sugar binding by H-bond (N65, Q75) or stacking interactions (W72) (Fig. 4C) (39, 40).
Residues 55, 57, 63, and 77 on the opposing side of the CRDs were replaced including K→T77 (galectin-13), K→N77 (galectin-16), V→I63 (galectin-14), and K→R77 (galectin-17) in several lineages following gene duplications, which have contributed to differences in the CRDs (Fig. 4C). As these residues are crucial for the binding of galactose or glucose moieties (39, 40), their replacements may have resulted in the neofunctionalization of the newly evolved proteins. ω = 0.68 in H53, which is involved in H-bond interactions, and replacements also occurred at this residue in galectins-13 (R) and -14 (E). Twenty of 24 residue replacements involving cysteines have experienced positive selection (ω > 1). Cysteines can form intra- or intermolecular disulphide bridges that confer the redox regulation of the structural and functional properties of galectins (41); therefore, the redox regulatory potential of the cluster proteins might have frequently changed. Based on their evolutionary history we would expect differential sugar-binding capacities for the cluster proteins.
Diversification of Galectin Activity in Cluster Proteins.
To study how their structural divergence gave rise to functional divergence, we cloned and expressed human and Ateles proteins to examine their sugar-binding characteristics. As negative control, a truncated human galectin-13 that lacks the CRD was generated. First, we found that all full-length recombinant proteins bound to lactose-agarose beads (Fig. 4D), but the truncated galectin-13 lacked this lectin activity. We conclude that the sugar-binding capacity of these proteins has been in place for at least the last 40 million years, since the time of the last common ancestor of Ateles and Homo. Because of their affinity for lactose and their shared structural features, we can call the proteins in the cluster galectins.
To see whether these galectins have differential sugar-binding capabilities, we tested the ability of various sugars to elute bound galectins (Fig. 4D) with the following results. (i) β-galactosides eluted proteins, whereas buffer did not. (ii) Lactose, a common ligand for galectins (12, 13), was effective in eluting all proteins. Galectin-16, which has the most (5:8) conserved residues in its CRD with the consensus galectin sequence, had the strongest affinity for lactose. (iii) Differences at residues 53, 63, and 77 in the CRDs and other adjacent residues result in differences in the binding profiles. (iv) Ateles galectin-14 and -20 have different sugar-binding profiles, suggesting that functional diversity also exists in Ateles galectins. (v) Placenta-specific galectins preferentially bind N-acetyl-lactosamine, a molecule commonly present on syncytiotrophoblast apical membranes (11). We suggest that when bound to these glycans at the syncytiotrophoblast apical membrane, oligomerized galectins could act as immune surveillance agents that cross-link and interact with syncytiotrophoblast and immune cells.
Placenta Expressed Galectins Regulate Immune Responses at the Maternal–Fetal Interface.
The genes that evolved during primate descent are primarily expressed in the placenta and bone marrow, tissues known to be involved in the regulation of immune responses. We have shown that LGALS10 is most abundantly expressed in bone marrow, the birthplace of immune cells. Human galectin-10 is highly expressed in mature eosinophils and basophils (29) and is considered the most potent indicator of Treg cells' suppressive function (30). It is tempting to hypothesize that the expanded number of functional marmoset LGALS10 genes may play a role in regulating their chimeric immune system (42).
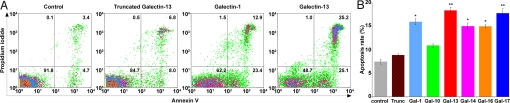
To examine their function, we asked whether cluster galectins might regulate adaptive immune response by inducing T cell apoptosis, because other galectins (-1, -2, -3, -4, -8, and -9) have also been shown to kill activated T cells predominantly through apoptosis (25–27). T cells freshly isolated from healthy donors were incubated with recombinant galectins, and galectin-1, a strong T cell apoptosis inducer (25–27), served as positive control. Because of the differential sugar-binding affinity of these proteins, we could not rely on a single sugar for inhibition assays. Instead, we used the truncated galectin-13 to examine whether the apoptotic effects of these proteins are related to the CRD. Indeed, we did not see the apoptotic effect of the truncated galectin-13. Comparable to the galectin-1 effect, placental galectins, but not galectin-10, induced a significantly higher rate of apoptosis than the truncated protein or what was detected in activated, but not treated, cells (Fig. 5).
Fig. 5.
Placental galectins induce apoptosis of human CD3+ T cells. The effect of placental galectins is comparable with that of galectin-1, whereas truncated galectin-13 does not have effect when proteins are applied in 8 μM. (A) Numbers in quadrants indicate percent of CD3+ T cells. (B) Apoptosis rate is the percentage of Annexin V and propidium iodide double-positive cells. Data are the mean ± SEM of 9 independent experiments. *, P < 0.05; **, P < 0.01. Trunc: truncated galectin-13.
Taken together, these findings suggest the following: (i) placenta and bone marrow-specific galectins in the cluster have different functional characteristics; (ii) placental galectins are strong apoptosis inducers of T cells, and this induction is conferred through their CRD because the truncated protein that lacks the CRD lacks apoptotic activity. Low placental expression and truncation of galectin-13 because of polymorphisms in patients with preeclampsia suggests that altered function of the cluster galectins may be associated with adverse pregnancy outcome (20, 21, 43).
Summary and Implications.
This study demonstrates that human galectins are expressed at the maternal–fetal interface. Three of the 5 human genes clustered on Chr19 are chiefly expressed in the placenta, and cDNA evidence shows that cluster genes are also expressed in the placentas of Old and New World monkeys. This gene cluster arose during primate evolution, was in place by the time of the last common ancestor of anthropoids 40 million years ago, and has diversified in anthropoids via a birth and death process. Genes in the cluster encode proteins that have a conserved tertiary structure as well as a definite but varying capacity to bind beta-galactosides, and thus, can be considered galectins. Placenta-specific cluster galectins cause immune cell death via apoptotic pathways as demonstrated by flow-cytometry of activated T cells.
Anthropoids and strepsirrhines evolved different reproductive strategies during their evolutionary descent from a common ancestor. Strepsirrhines evolved a less invasive epitheliochorial placenta and retained a bicornate uterus, resulting in relatively short gestations and small offspring (5). As an alternative reproductive strategy, anthropoids retained the ancestral invasive hemochorial placentation and evolved a simplex uterus (5, 44–46). The consequences of this anthropoid “evolutionary choice” were long gestations, large offspring, and an increased brain to body size ratio. In conjunction with these developmental and anatomical changes, anthropoids have expanded gene clusters chiefly expressed in the placenta, including those containing CG, growth hormones, siglecs (47–50), and the galectins described in this study. Functional divergence among paralogs can be established relatively quickly following gene duplication (35, 51, 52), and these duplicated genes may have conferred advantages during anthropoid pregnancies.
Gene clustering may facilitate the coordinated, tissue-specific, and developmental expression of genes, as well as their functional diversification (35, 52). Indeed, 3 of the human galectin cluster genes are expressed in the differentiated syncytiotrophoblast. The strong selective forces acting on these galectin genes in anthropoids results in a rapid birth and death process, in which genes are lost and gained. Of importance, galectin-13 has decreased placental expression and maternal serum concentrations in the first trimester in women who subsequently develop preeclampsia (20, 21, 43), a pregnancy-specific syndrome linked to immune maladaptation (2–4, 7, 22). We propose that the immunosuppressive properties of placental galectins in the Chr19 cluster confer additional mechanisms of maternal–fetal immune tolerance, which were necessary for sustaining hemochorial placentation during the long gestation of anthropoid primates.
Materials and Methods
Human tissues were retrieved from the bank of biological specimens of the Perinatology Research Branch. Fresh-frozen tissues were used for RNA isolation, cDNA synthesis, qRT-PCR or sequence analysis; formalin-fixed tissues were applied for immunohistochemistry and in situ hybridization. Human T cells were isolated from the peripheral blood of healthy individuals and used for apoptosis assays. Written informed consent was obtained from all women before the collection of samples, and the research was approved by the Internal Review Boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and Wayne State University.
Formalin-fixed primate placentas were used for immunostaining; RNA later-preserved placentas were used for RNA isolation, cDNA synthesis, and sequence analysis. Genomic DNA was also used as PCR template. Galectin expression profiling on human placentas, fetal membranes, and on 48 human tissues was performed by qRT-PCR. Expression of 4 genes in human placentas and fetal membranes was localized by in situ hybridization. Homo sapiens, Colobus guereza, and Macaca mulatta placentas were immunostained for galectin-13.
Sequences analyzed in this study are shown in Dataset S1. The positions of genes and pseudogenes in the Chr19 cluster and genomic rearrangements were determined by using PipMaker (http://bio.cse.psu.edu), Spidey (http://www.ncbi.nlm.nih.gov/spidey), and University of California Santa Cruz Genome Browser (http://genome.ucsc.edu). Maximum likelihood algorithms were used for phylogenetic analyses of 79 sequences. Phylogenetic trees were generated by using Bayesian inference and codon-model-based methods to examine selection pressures. Protein structures were determined by homology modeling.
One truncated and eight full-length galectins were cloned, expressed in Escherichia coli and purified with affinity chromatography. Their functional characteristics were revealed by sugar-binding assays, apoptotic effect on freshly isolated human T cells by flow-cytometry. All methods are described in detail in SI Methods, and additional data are shown in Dataset S3, Dataset S4, Dataset S5, and Dataset S6.
Supplementary Material
Acknowledgments.
We thank Dr. Prasenjit Das, Sivasakthy Sivalogan, and Hong Meng for assistance; Dr. Sue Land (Wayne State University, Applied Genomic Technology Center) for running qRT-PCR; Drs. Howard Petty, Sally Madsen-Bouterse, Maik Hüttemann, and Raghavendra Navath for helpful advice; and Sara Tipton for critically reading the manuscript. We also thank Drs. Caro-Beth Stewart (State University of New York, Albany, NY) and Kathy Neiswanger (University of Pittsburgh, Pittsburgh, PA), the New England Regional Primate Center (Southborough, MA), and the Duke University Primate Center (Durham, NC) for providing DNA and/or tissue samples. This research was supported by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services; by National Science Foundation Grants BCS-0751508 (to D.E.W.) and BCS-0550209 (to M.G.); and by the Hungarian Scientific Research Fund (OTKA) Grants NK77978 (to P.Z.) and PD73096 (to A.Sz.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ613334–FJ613357 and TPA: BK006815–BK006863).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903568106/DCSupplemental.
References
- 1.Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;44:320–338. [Google Scholar]
- 2.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 3.Trowsdale J, Betz AG. Mother's little helpers: Mechanisms of maternal–fetal tolerance. Nat Immunol. 2006;7:241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 4.Terness P, et al. Tolerance signaling molecules and pregnancy: IDO, galectins, and the renaissance of regulatory T cells. Am J Reprod Immunol. 2007;58:238–254. doi: 10.1111/j.1600-0897.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 5.Richards AF. Primates in Nature. New York: W. H. Freeman; 1985. [Google Scholar]
- 6.Goodman M. The role of immunochemical differences in the phyletic development of human behavior. Hum Biol. 1961;33:131–162. [PubMed] [Google Scholar]
- 7.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 8.Uckan D, et al. Trophoblasts express Fas ligand: A proposed mechanism for immune privilege in placenta and maternal invasion. Mol Hum Reprod. 1997;3:655–662. doi: 10.1093/molehr/3.8.655. [DOI] [PubMed] [Google Scholar]
- 9.Arkwright PD, et al. Suppression of allogeneic reactivity in vitro by the syncytiotrophoblast membrane glycocalyx of the human term placenta is carbohydrate dependent. Glycobiology. 1994;4:39–47. doi: 10.1093/glycob/4.1.39. [DOI] [PubMed] [Google Scholar]
- 10.Petty HR, Kindzelskii AL, Espinoza J, Romero R. Trophoblast contact deactivates human neutrophils. J Immunol. 2006;176:3205–3214. doi: 10.4049/jimmunol.176.5.3205. [DOI] [PubMed] [Google Scholar]
- 11.Jones CJ, Aplin JD. Glycosylation at the fetomaternal interface: Does the glycocode play a critical role in implantation? Glycoconj J. 2008;26:359–366. doi: 10.1007/s10719-008-9152-6. [DOI] [PubMed] [Google Scholar]
- 12.Barondes SH, et al. Galectins: A family of animal beta-galactoside-binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 13.Cooper DN. Galectinomics: Finding themes in complexity. Biochim Biophys Acta. 2002;1572:209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 14.Rabinovich GA, et al. Galectins and their ligands: Amplifiers, silencers, or tuners of the inflammatory response? Trends Immunol. 2002;23:313–320. doi: 10.1016/s1471-4906(02)02232-9. [DOI] [PubMed] [Google Scholar]
- 15.Houzelstein D, et al. Phylogenetic analysis of the vertebrate galectin family. Mol Biol Evol. 2004;21:1177–1187. doi: 10.1093/molbev/msh082. [DOI] [PubMed] [Google Scholar]
- 16.Vicovac L, Jankovic M, Cuperlovic M. Galectin-1 and -3 in cells of the first trimester placental bed. Hum Reprod. 1998;13:730–735. doi: 10.1093/humrep/13.3.730. [DOI] [PubMed] [Google Scholar]
- 17.Than NG, et al. Functional analyses of placental protein 13/galectin-13. Eur J Biochem. 2004;271:1065–1078. doi: 10.1111/j.1432-1033.2004.04004.x. [DOI] [PubMed] [Google Scholar]
- 18.Jeschke U, et al. Expression of galectin-1, -3 (gal-1, gal-3) and the Thomsen–Friedenreich (TF) antigen in normal, IUGR, preeclamptic, and HELLP placentas. Placenta. 2007;28:1165–1173. doi: 10.1016/j.placenta.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Than NG, et al. Severe preeclampsia is characterized by increased placental expression of galectin-1. J Matern Fetal Neonatal Med. 2008;21:429–442. doi: 10.1080/14767050802041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Than NG, et al. Placental protein 13 (galectin-13) has decreased placental expression but increased shedding and maternal serum concentrations in patients presenting with preterm preeclampsia and HELLP syndrome. Virchows Arch. 2008;453:387–400. doi: 10.1007/s00428-008-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekizawa A, et al. PP13 mRNA expression in trophoblasts from preeclamptic placentas. Reprod Sci. 2008;16:408–413. doi: 10.1177/1933719108328615. [DOI] [PubMed] [Google Scholar]
- 22.Saito S, et al. Inadequate tolerance induction may induce preeclampsia. J Reprod Immunol. 2007;76:30–39. doi: 10.1016/j.jri.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Yang QS, et al. Cloning and expression of a novel human galectin cDNA, predominantly expressed in placenta(1) Biochim Biophys Acta. 2002;1574:407–411. doi: 10.1016/s0167-4781(01)00319-0. [DOI] [PubMed] [Google Scholar]
- 24.von Wolff M, Wang X, Gabius HJ, Strowitzki T. Galectin fingerprinting in human endometrium and decidua during the menstrual cycle and in early gestation. Mol Hum Reprod. 2005;11:189–194. doi: 10.1093/molehr/gah144. [DOI] [PubMed] [Google Scholar]
- 25.Perillo NL, Pace KE, Seilhamer JJ, Baum LG. Apoptosis of T cells mediated by galectin-1. Nature. 1995;378:736–739. doi: 10.1038/378736a0. [DOI] [PubMed] [Google Scholar]
- 26.Hsu DK, Yang RY, Liu FT. Galectins in apoptosis. Methods Enzymol. 2006;417:256–273. doi: 10.1016/S0076-6879(06)17018-4. [DOI] [PubMed] [Google Scholar]
- 27.Kopcow HD, et al. T cell apoptosis at the maternal–fetal interface in early human pregnancy, involvement of galectin-1. Proc Natl Acad Sci USA. 2008;105:18472–18477. doi: 10.1073/pnas.0809233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blois SM, et al. A pivotal role for galectin-1 in fetomaternal tolerance. Nat Med. 2007;13:1450–1457. doi: 10.1038/nm1680. [DOI] [PubMed] [Google Scholar]
- 29.Ackerman SJ, et al. Molecular cloning and characterization of human eosinophil Charcot-Leyden crystal protein (lysophospholipase). Similarities to IgE binding proteins and the S-type animal lectin superfamily. J Immunol. 1993;150:456–468. [PubMed] [Google Scholar]
- 30.Kubach J, et al. Human CD4+CD25+ regulatory T cells: Proteome analysis identifies galectin-10 as a novel marker essential for their anergy and suppressive function. Blood. 2007;110:1550–1558. doi: 10.1182/blood-2007-01-069229. [DOI] [PubMed] [Google Scholar]
- 31.Dunphy JL, et al. Isolation and characterization of a novel eosinophil-specific galectin released into the lungs in response to allergen challenge. J Biol Chem. 2002;277:14916–14924. doi: 10.1074/jbc.M200214200. [DOI] [PubMed] [Google Scholar]
- 32.Lewis SK, et al. Galectin 15 (LGALS15): A gene uniquely expressed in the uteri of sheep and goats that functions in trophoblast attachment. Biol Reprod. 2007;77:1027–1036. doi: 10.1095/biolreprod.107.063594. [DOI] [PubMed] [Google Scholar]
- 33.Bailey JA, Eichler EE. Primate segmental duplications: Crucibles of evolution, diversity, and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 34.Green P. 2x genomes–does depth matter? Genome Res. 2007;17:1547–1549. doi: 10.1101/gr.7050807. [DOI] [PubMed] [Google Scholar]
- 35.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohn H, Kraus W, Winckler W. Purification and characterization of two new soluble placental tissue proteins (PP13 and PP17) Oncodev Biol Med. 1983;4:343–350. [PubMed] [Google Scholar]
- 37.Than NG, et al. Isolation and sequence analysis of a cDNA encoding human placental tissue protein 13 (PP13), a new lysophospholipase, homologue of human eosinophil Charcot-Leyden Crystal protein. Placenta. 1999;20:703–710. doi: 10.1053/plac.1999.0436. [DOI] [PubMed] [Google Scholar]
- 38.Burger O, et al. Placental protein 13 (PP-13): Effects on cultured trophoblasts, and its detection in human body fluids in normal and pathological pregnancies. Placenta. 2004;25:608–622. doi: 10.1016/j.placenta.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Leonidas DD, et al. Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins. Structure. 1995;3:1379–1393. doi: 10.1016/s0969-2126(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 40.Visegrady B, et al. Homology modelling and molecular dynamics studies of human placental tissue protein 13 (galectin-13) Protein Eng. 2001;14:875–880. doi: 10.1093/protein/14.11.875. [DOI] [PubMed] [Google Scholar]
- 41.Than NG, et al. Emergence of hormonal and redox regulation of galectin-1 in placental mammals: Implication in maternal–fetal immune tolerance. Proc Natl Acad Sci USA. 2008;105:15819–15824. doi: 10.1073/pnas.0807606105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benirschke K, Anderson JM, Brownhill LE. Marrow chimerism in marmosets. Science. 1962;138:513–515. doi: 10.1126/science.138.3539.513. [DOI] [PubMed] [Google Scholar]
- 43.Than NG, et al. Prediction of preeclampsia - a workshop report. Placenta. 2008;29(Suppl A):S83–S85. doi: 10.1016/j.placenta.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter AM, Mess A. Evolution of the placenta in eutherian mammals. Placenta. 2007;28:259–262. doi: 10.1016/j.placenta.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Elliot MG, Crespi BJ. Placental invasiveness mediates the evolution of hybrid inviability in mammals. Am Nat. 2006;168:114–120. doi: 10.1086/505162. [DOI] [PubMed] [Google Scholar]
- 46.Wildman DE, et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci USA. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallis OC, Zhang YP, Wallis M. Molecular evolution of GH in primates: Characterisation of the GH genes from slow loris and marmoset defines an episode of rapid evolutionary change. J Mol Endocrinol. 2001;26:249–258. doi: 10.1677/jme.0.0260249. [DOI] [PubMed] [Google Scholar]
- 48.Maston GA, Ruvolo M. Chorionic gonadotropin has a recent origin within primates and an evolutionary history of selection. Mol Biol Evol. 2002;19:320–335. doi: 10.1093/oxfordjournals.molbev.a004085. [DOI] [PubMed] [Google Scholar]
- 49.Brinkman-Van der Linden EC, et al. Human-specific expression of Siglec-6 in the placenta. Glycobiology. 2007;17:922–931. doi: 10.1093/glycob/cwm065. [DOI] [PubMed] [Google Scholar]
- 50.Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–181. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- 51.Grus WE, Zhang J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol Biol Evol. 2008;25:1593–1601. doi: 10.1093/molbev/msn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trowsdale J. The gentle art of gene arrangement: The meaning of gene clusters. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-comment2002. comment2002.1–comment2002.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.