Abstract
In poplar (Populus spp.), the major defense phenolics produced in leaves are the flavonoid-derived proanthocyanidins (PAs) and the salicin-based phenolic glycosides. Transcriptional activation of PA biosynthetic genes leading to PA accumulation in leaves occurs following herbivore damage and mechanical wounding as well as infection by the fungal biotroph Melampsora medusae. In this study, we have identified a poplar R2R3 MYB transcription factor gene, MYB134, that exhibits close sequence similarity to the Arabidopsis (Arabidopsis thaliana) PA regulator TRANSPARENT TESTA2 and that is coinduced with PA biosynthetic genes following mechanical wounding, M. medusae infection, and exposure to elevated ultraviolet B light. Overexpression of MYB134 in poplar resulted in transcriptional activation of the full PA biosynthetic pathway and a significant plant-wide increase in PA levels, and electrophoretic mobility shift assays showed that recombinant MYB134 protein is able to bind to promoter regions of PA pathway genes. MYB134-overexpressing plants exhibited a concomitant reduction in phenolic glycoside concentrations and other minor alterations to levels of small phenylpropanoid metabolites. Our data provide insight into the regulatory mechanisms controlling stress-induced PA metabolism in poplar, and the identification of a regulator of stress-responsive PA biosynthesis constitutes a valuable tool for manipulating PA metabolism in poplar and investigating the biological functions of PAs in resistance to biotic and abiotic stresses.
Plant secondary metabolites play important ecological roles and in many plants constitute a critical component of defenses against biotic and abiotic stress. Many secondary metabolic pathways are responsive to environmental conditions and can be rapidly activated by stresses such as pathogen infection, elevated light, and herbivory. The phenylpropanoid pathway in particular leads to the synthesis of a large and diverse class of plant secondary metabolites, many of which are stress induced (Dixon and Paiva, 1995). Synthesis of phenylpropanoids and other secondary metabolites following stress is typically mediated by the transcriptional activation of suites of biosynthetic genes coordinately regulated by transcription factor proteins (Weisshaar and Jenkins, 1998; Davies and Schwinn, 2003). The possibility of identifying transcription factors that control entire pathways is motivating many studies in plant stress biology, since such regulators would be valuable for the metabolic engineering of plants for both plant and human health (Dixon, 2005; Sharma and Dixon, 2005; Yu and McGonigle, 2005).
Populus species (cottonwoods, poplars, and aspens, hereafter referred to collectively as poplar) are often ecological foundation species and include the most widely distributed trees in the Northern Hemisphere. The phenolic metabolites produced by poplar are thought to be important determinants of community structure and ecosystem dynamics (Lindroth and Hwang, 1996; Schweitzer et al., 2004; Bailey et al., 2005; LeRoy et al., 2006; Whitham et al., 2006). Poplar leaves typically accumulate several classes of phenolic metabolites, including the salicylate-derived phenolic glycosides (PGs), flavonoids such as flavonol glycosides, anthocyanins, and proanthocyanidins (PAs; or condensed tannins), and numerous small phenolic acids and their esters (Pearl and Darling, 1971; Klimczak et al., 1972; Palo, 1984; Lindroth and Hwang, 1996; Fig. 1). PGs and PAs are generally the most abundant foliar phenolic metabolites in poplar and together can constitute more than 30% of leaf dry weight (Lindroth and Hwang, 1996). PGs are constitutively produced in poplar leaves and function as potent anti-insect herbivore compounds (Hwang and Lindroth, 1997; Osier and Lindroth, 2001). PG levels can exhibit considerable genotypic variability and are influenced by light and nutrient availability, but they do not typically increase rapidly following herbivore damage (Osier and Lindroth, 2001, 2006; Stevens and Lindroth, 2005). PAs are also constitutively produced in poplar leaves, but their biosynthesis is often up-regulated by stresses such as insect herbivory, mechanical wounding, and pathogen infection (Peters and Constabel, 2002; Stevens and Lindroth, 2005; Miranda et al., 2007). PA accumulation following wounding and herbivory occurs both locally at the site of damage and systemically in distal leaves (Peters and Constabel, 2002). The strong systemic activation of the PA biosynthetic pathway in poplar following insect herbivory suggests that these compounds function in herbivore defense. However, experimental evidence indicates that poplar leaf PAs may not be strong, broad-spectrum antiherbivore compounds (Hemming and Lindroth, 1995; Ayres et al., 1997). In addition to biotic stresses, nutrient limitation and high light levels have also been found to result in greater PA concentrations in poplar (Hemming and Lindroth, 1999; Osier and Lindroth, 2001), hinting at broader biological roles.
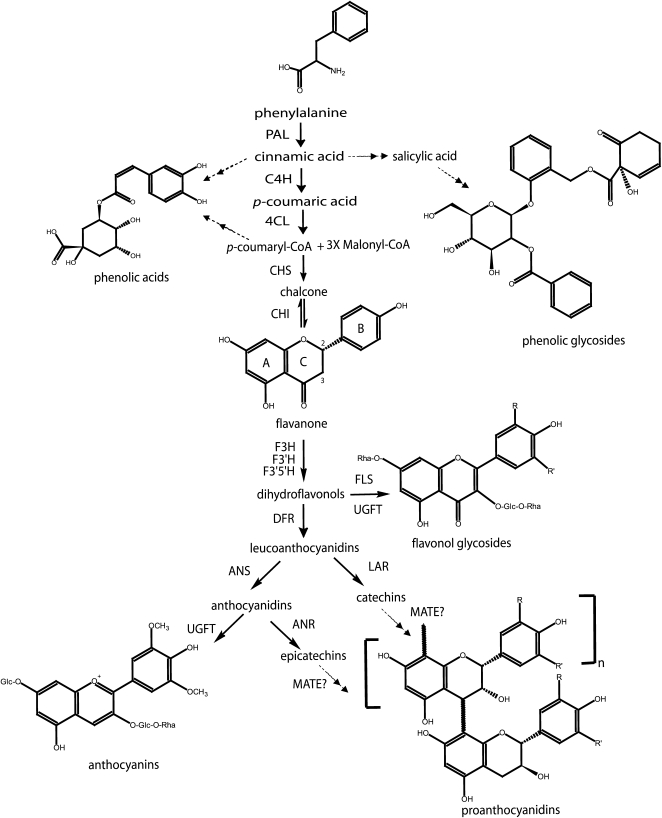
Figure 1.
Biosynthetic pathway for the production of flavonoids, proanthocyanidins, and other classes of small phenolic compounds produced in poplar leaves (phenolic acids are represented by chlorogenic acid and phenolic glycosides are represented by tremulacin). The flavonone illustrates the basic 15-carbon flavonoid structure with three rings (A–C). The C-2 and C-3 carbons at which PA starter units differ in stereochemistry are labeled. PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA-ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; DFR, dihydroflavonol reductase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; MATE, multidrug and toxic compound extrusion transporter; UFGT, UDP-Glc flavonoid glucosyltransferase.
PAs are polymers of flavan-3-ols or flavan-3,4-diols condensed with flavan-3-ol starter units and usually deposited in the vacuole (Stafford, 1990; Xie and Dixon, 2005; Fig. 1). With the exception of late biosynthetic steps, including the transport and condensation of PA monomers, the enzymatic steps in PA biosynthesis have been well characterized, as most are also involved in the production of other well-studied flavonoid classes such as anthocyanins (Xie and Dixon, 2005; Fig. 1). The Arabidopsis (Arabidopsis thaliana) transparent testa (tt) and tannin-deficient seed mutants have been invaluable in identifying genes involved in PA biosynthesis and its regulation (Lepiniec et al., 2006). Recent major advances in PA biochemistry include the identification of the PA-specific enzymes anthocyanidin reductase (ANR; encoded by the BANYULS [BAN] gene in Arabidopsis) and leucoanthocyanidin reductase (LAR), which are involved in the production of the flavan-3-ol PA starter units with 2,3-cis- or 2,3-trans stereochemistry of the heterocyclic C ring, respectively (Tanner et al., 2003; Xie et al., 2003; Fig. 1). Additional genes implicated in PA biosynthesis in Arabidopsis include TT12, encoding a multidrug and toxic compound extrusion (MATE) family transporter (Debeaujon et al., 2001), TT19, encoding a glutathione S-transferase (Kitamura et al., 2004), AUTO-INHIBITED H+-ATPase ISOFORM10 (AHA10), encoding a proton pump involved in vacuolar biogenesis (Baxter et al., 2005), and TT10, a predicted laccase-like polyphenol oxidase functioning in PA oxidation (Pourcel et al., 2005). Recently, UGT72L1, a glycosyltransferase specifically active toward the PA precursor (−)-epicatechin, was identified in hairy roots of Medicago truncatula in which the Arabidopsis PA regulator TT2 was expressed (Pang et al., 2008). This study illustrates the utility of using PA regulatory factors to discover new genes involved in the PA biosynthetic pathway.
Transcriptional regulation of flavonoid and PA biosynthetic genes involves combinatorial interactions between several classes of transcription factor proteins (Mol et al., 1998; Nesi et al., 2001; Winkel-Shirley, 2001). These include members of the R2R3 MYB domain, basic helix-loop-helix (bHLH) domain, and WD-repeat (WDR) families (Lepiniec et al., 2006). In Arabidopsis seed testa, PA biosynthesis is regulated by a MYB-bHLH-WDR ternary complex composed of the TT2, TT8, and TTG1 proteins (Nesi et al., 2000, 2001; Debeaujon et al., 2003; Baudry et al., 2004). The MYB factor (TT2) confers target gene specificity to the complex, activating the late PA biosynthetic genes, including DFR, BAN, TT12, and AHA10 (Nesi et al., 2001; Baudry et al., 2004; Sharma and Dixon, 2005). The DNA sequences bound by TT2 have not been elucidated, although the closely related maize (Zea mays) COLORLESS1 (C1) protein, a regulator of anthocyanin metabolism, has been shown to bind to AC-rich motifs known as AC elements present in the regulatory regions of numerous phenylpropanoid genes (Sainz et al., 1997; Hernandez et al., 2004). Canonical AC elements were defined in the Phaseolus vulgaris promoter (Hatton et al., 1995), and they have been identified in the promoters of a variety of phenylpropanoid genes, including those involved in lignin metabolism and different branches of flavonoid metabolism, such as flavonols and anthocyanins (Sainz et al., 1997; Hartmann et al., 2005; Rogers and Campbell, 2004). The minimal Arabidopsis BAN promoter characterized by Debeaujon et al. (2003) contains motifs that match the animal c-myb consensus (CNGTTR) and bHLH domain protein-binding consensus (CANNTG) as well as two AC element-like motifs.
The R2R3 MYBs constitute large gene families in plants, with 126 members in Arabidopsis (Stracke et al., 2001) and 192 in poplar (Wilkins et al., 2009). Although many remain functionally uncharacterized, numerous R2R3 MYB proteins are implicated in the regulation of plant-specific developmental and physiological processes, including the regulation of phenylpropanoid metabolism (Stracke et al., 2001). R2R3 MYB proteins are characterized by two imperfectly repeated N-terminal MYB domains each forming DNA-binding helix-turn-helix structures. Outside of the R2R3 MYB domain, the proteins are highly divergent except for short conserved amino acid sequence motifs. These motifs, together with sequence homology within the MYB domains, form the basis for their classification into different subgroups (Stracke et al., 2001; Jiang et al., 2004).
Constitutive expression of TT2 in Arabidopsis does not induce plant-wide PA accumulation, indicating that PA synthesis is controlled by the tissue- and cell type-specific expression patterns of additional regulatory genes (Nesi et al., 2001). A related PA regulatory R2R3 MYB protein, MYBPA1, has been characterized in grapevine (Vitis vinifera; Bogs et al., 2007). Unlike Arabidopsis TT2, MYBPA1 also regulates the early flavonoid biosynthetic genes. Constitutive expression of grapevine MYBPA1 in the Arabidopsis tt2 mutant not only complemented the PA-deficient seed phenotype but also induced ectopic PA production in a cell-specific manner (Bogs et al., 2007). TT2-like R2R3 MYB factors appear to be involved in the regulation of PA metabolism in other species, as homologous genes have recently been described in Brassica napus (Wei et al., 2007), lotus (Lotus japonicus; Yoshida et al., 2008), and grapevine (Terrier et al., 2009).
We previously showed that the stress induction of PAs in poplar leaves follows the transcriptional activation of PA biosynthetic genes (Peters and Constabel, 2002; Miranda et al., 2007) and therefore hypothesized that a TT2-like R2R3 MYB protein regulates this process. Here, we have identified MYB134 as a candidate PA regulator that is consistently coregulated with PA biosynthetic genes. Constitutive expression of MYB134 in transgenic poplar resulted in a specific activation of PA pathway genes, leading to a dramatic increase in PA concentrations, suggesting that this gene is indeed a poplar PA regulator. MYB134 was found to activate both early and late PA biosynthetic genes but not non-PA phenylpropanoid structural genes. Recombinant MYB134 protein was shown to bind to promoter regions of both early and late PA pathway genes containing predicted MYB binding sites. These findings provide insight into the regulatory mechanisms mediating stress-induced PA biosynthesis, and the PA-modified poplar trees produced here represent a valuable tool for investigating the functions of carbon-based allelochemicals in poplar.
RESULTS
Identification of Putative PA Regulatory R2R3 MYB Genes in Poplar
Based on our knowledge that PA synthesis is activated by wounding and of the central role of R2R3 MYB factors in regulating flavonoid metabolism, we began a search for a MYB regulator of wound-induced PA metabolism. We first identified an EST sequence predicted to encode an R2R3 MYB domain transcription factor in an EST library made from systemically wounded poplar leaves (GenBank accession no. CN192773; Christopher et al., 2004). The predicted protein encoded by this transcript exhibits high sequence similarity to Arabidopsis TT2 as well as maize C1, both of which belong to the N08 MYB subgroup (Jiang et al., 2004), also referred to as subgroup 5 (Stracke et al., 2001) or subgroup C32 (Wilkins et al., 2009). In order to identify additional candidate regulators of stress-induced PA metabolism in poplar, we searched public poplar EST databases for additional homologs of TT2. This search resulted in the identification of four genes predicted to encode R2R3 MYB transcription factors of subgroup N08. cDNAs corresponding to these four candidate PA regulatory MYB genes were cloned from Populus tremuloides, a species with particularly high PA induction. With the completion of the Populus trichocarpa genome sequence (Tuskan et al., 2006), the corresponding genomic sequences were identified and found to correspond to P. trichocarpa MYB097, MYB086, MYB134, and MYB183 (Wilkins et al., 2009). The genome contains an additional copy of the MYB086 gene with more than 98% nucleotide identity within the coding sequences, MYB087.
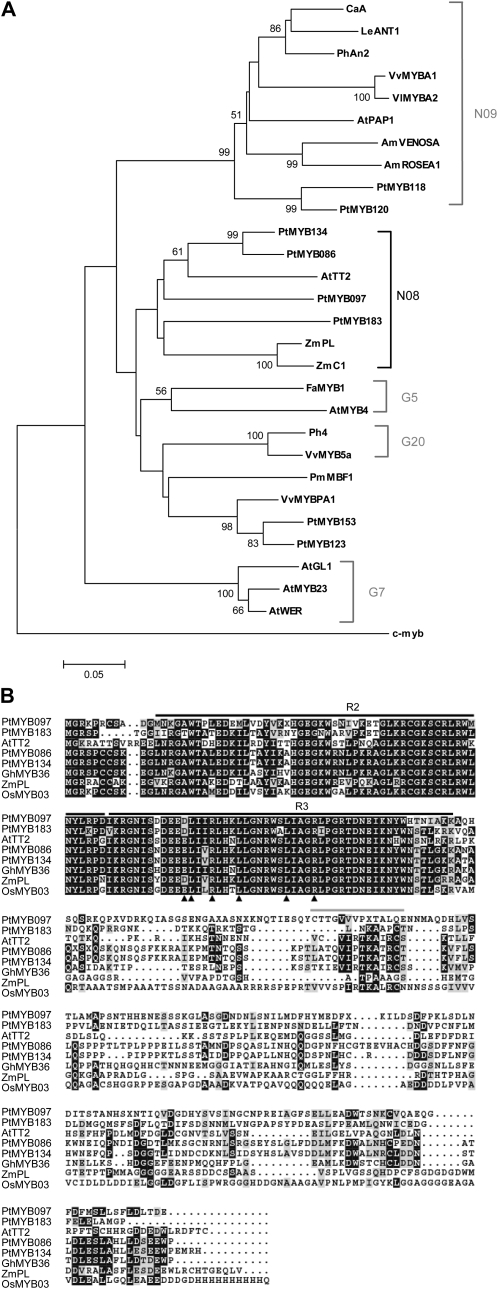
A phylogenetic tree was constructed using the predicted amino acid sequences of the R2R3 MYB domains of putative flavonoid regulatory MYBs from poplar as well as a number of R2R3 MYB proteins from other species (Fig. 2A). The four candidate MYBs (MYB097, MYB086, MYB134, and MYB183) are most closely related to members of the N08 group rather than to other flavonoid regulatory MYB subgroups such as N09, G20, or G5 (Fig. 2A). These findings are congruent with a recently published phylogenetic analysis of the entire P. trichocarpa R2R3 MYB family (Wilkins et al., 2009). The N09 subgroup is composed of MYB proteins that function as anthocyanin biosynthetic gene activators (Quattrocchio et al., 1999; Borevitz et al., 2000; Schwinn et al., 2006). Members of subgroup G5 include negative regulators of phenylpropanoid and flavonoid metabolism (Jin et al., 2000; Aharoni et al., 2001), while the G20 subgroup includes proteins involved in controlling vacuolar pH and flavonoid biosynthetic gene activation (Deluc et al., 2006; Quattrocchio et al., 2006). Examination of the P. trichocarpa genome led to the identification of additional MYB genes sharing high sequence similarity to other activators of flavonoid metabolism, including MYB123, MYB153, MYB118, and MYB120 (Fig. 2A). MYB118 and MYB120 cluster with anthocyanin regulators of subgroup N09, while the MYB123 and MYB153 genes are closely related to the grapevine PA regulator MYBPA1 and thus may be involved in PA regulation in poplar (Fig. 2A). However, we focused our analyses on the homologs of Arabidopsis TT2 in subgroup N08, as this was the only known PA regulator when our study was initiated.
Figure 2.
A, Phylogenetic analysis of putative flavonoid regulatory poplar R2R3 MYB domain proteins with selected R2R3 MYB domain proteins from other species. R2R3 MYB domains were aligned using ClustalW, and the phylogenetic tree was constructed using MEGA 3.1 (Kumar et al., 2004) using the minimum evolution test and p-distance model with 1,000 bootstrap replicates. Bootstrap values higher than 50% are shown. Human c-myb and Arabidopsis subgroup G7 MYBs are included as outgroups. The N08, N09, G5, and G20 subgroups of Jiang et al. (2004) are indicated. GenBank accession numbers are as follows: CaA (Capsicum annuum A, AJ608992), LeANT1 (Lycopersicon esculentum [now Solanum lycopersicum] ANT1, AAQ55181), PhAN2 (Petunia × hybrida AN2, AAF66727), VvMYBA1 (Vitis vinifera MYBA1, BAD18977), VvMYBA2 (V. vinifera MYBA2, BAD18978), AtPAP1 (Arabidopsis thaliana PAP1/MYB75, AAG42001), AmVENOSA (Antirrhinum majus VENOSA, ABB83828), AmROSEA1 (A. majus ROSEA1, ABB83826), AtTT2 (Arabidopsis TT2/MYB123, Q9FJA2), ZmC1 (Zea mays C1, AAK09327), ZmPL (Z. mays PL, AAB67721), FaMYB1 (Fragaria × ananassa MYB1, AAK84064), AtMYB4 (Arabidopsis MYB4, NP_850879), Ph4 (Petunia × hybrida PH4, AAY51377), VvMYB5a (V. vinifera MYB5a, AAS68190), PmMBF1 (Picea mariana MBF1, AAA82943), VvMYBPA1 (V. vinifera MYBPA1, AM259485), AtGL1 (Arabidopsis GLABROUS1, P27900), AtMYB23 (Arabidopsis MYB23, NP_198849), AtWER (Arabidopsis WEREWOLF1, NP_196979), and c-myb (Homo sapiens c-myb, AAB49039). B, Alignment of predicted poplar PA regulatory MYB proteins with the Arabidopsis PA regulator TT2, maize PL, and uncharacterized MYB proteins from Oryza sativa (OsMYB3, BAA23339) and Gossypium hirsutum (GhMYB36, AAK19617) containing conserved motifs C terminal to the MYB DNA-binding domain. The residues involved in the interaction with bHLH cofactors are indicated by arrowheads. The gray bar indicates the Vx2IRTKA[IL]RC[SN] motif found in AtTT2 and OsMYB03, and the black bars indicate the R2 and R3 repeats of the MYB domain.
MYB134 and MYB086 encode proteins containing a sequence motif similar to the Vx2IRTKA[IL]RC[SN] motif located C terminal to the R2R3 MYB domain in Arabidopsis TT2 (Nesi et al., 2001; Fig. 2B). A similar conserved sequence motif is found in other N08 subgroup proteins, including the recently identified lotus TT2 proteins (Yoshida et al., 2008), the grapevine MYBPA2 (Terrier et al., 2009), and several uncharacterized predicted MYB proteins including cotton (Gossypium hirsutum) MYB36 (GenBank accession no. AAK19617), Fagus crenata MYB251 (BAG75107), and Malus × domestica MYB11 (AAZ20431) and MYB9 (ABB84757). The [DE]Lx2[RK]x3Lx6Lx3R motif involved in the interaction of MYB proteins with bHLH partners (Grotewold et al., 2000; Zimmermann et al., 2004) is present in all four putative poplar PA regulatory MYBs, which indicates that, like TT2, these MYBs likely require the presence of bHLH cofactors to function (Fig. 2B).
Candidate PA Regulatory MYBs Exhibit Distinct Expression Profiles following Flavonoid Pathway-Activating Stresses
Mechanical wounding of poplar leaves has been shown to induce the up-regulation of flavonoid biosynthetic genes leading to PA accumulation (Peters and Constabel, 2002; Tsai et al., 2006b). In order to identify candidate regulators of stress-induced PA biosynthesis, we profiled MYB and flavonoid structural gene expression in mechanically wounded leaves using northern-blot analysis (Fig. 3A, left and center). The flavonoid structural gene family members analyzed in these experiments had been identified as herbivore and pathogen induced in previous work (Peters and Constabel, 2002; Miranda et al., 2007) and are named according to the annotations of Tsai et al. (2006b).
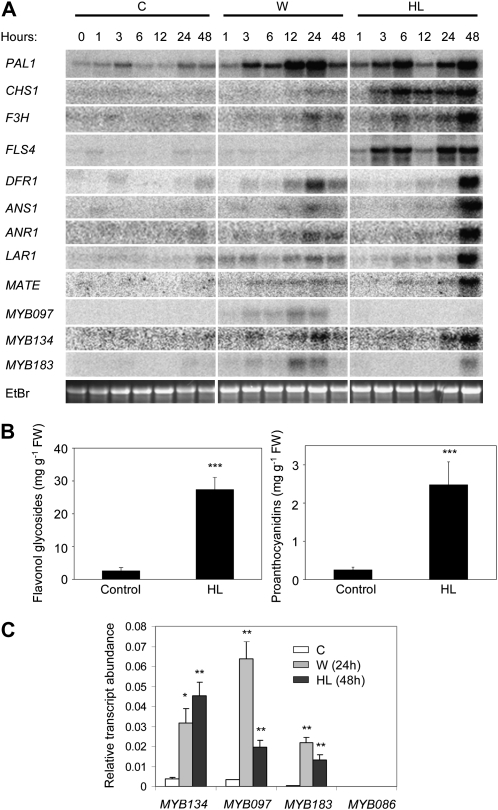
Figure 3.
Analysis of flavonoid pathway activation and MYB gene expression in poplar leaves after wounding and exposure to elevated light. A, Northern-blot analysis of phenylpropanoid and flavonoid genes as well as putative PA regulatory MYB genes in control (C), mechanically wounded (W), and HL-exposed (HL) leaves. EtBr, Ethidium bromide. B, Analysis of PAs and flavonol glycosides in leaves after 7 d of HL treatment. PAs were quantified using the acid-butanol method, and total flavonol glycosides were quantified as rutin equivalents using HPLC. Bars indicate means of four (flavonol glycoside analysis) or seven (PA analysis) trees per treatment, with error bars indicating se. FW, Fresh weight. C, Real-time PCR analysis of relative transcript abundance (arbitrary units) for putative PA regulatory MYB genes in leaves of untreated control, mechanically wounded (after 24 h), and HL-exposed (after 48 h) plants. Asterisks indicate significant differences using Student's t test (** P < 0.01, *** P < 0.001).
As expected, mechanical wounding of leaf margins resulted in an up-regulation of phenylpropanoid and flavonoid genes (Fig. 3A). Gene expression was monitored at 1, 3, 6, 12, 24, and 48 h after wounding. PAL1, the wound-inducible flavonoid-associated PAL gene characterized by Kao et al. (2002), was rapidly up-regulated (Fig. 3A). General flavonoid biosynthetic genes, including CHS, F3H, ANS1, and the DFR1 gene previously implicated in herbivore-induced PA accumulation (Peters and Constabel, 2002), were up-regulated within 24 h of wounding, as were the PA-specific ANR1 and LAR1 genes. FLS4, a flavonol synthase gene, was not responsive to wounding. A putative poplar homolog of the Arabidopsis MATE transporter gene TT12, previously found to be coinduced with PA biosynthetic genes following infection of poplar leaves with the fungal biotroph Melampsora medusae (Miranda et al., 2007), was also up-regulated following wounding (Fig. 3A). The putative PA regulatory genes MYB097, MYB134, and MYB183 all exhibited wound-induced transcript accumulation that correlated with the activation of flavonoid structural genes (Fig. 3A).
In order to analyze MYB gene expression under a flavonoid-activating stress different from mechanical wounding, we moved plants from the greenhouse into full natural sunlight, where they were exposed to elevated levels of both visible and UV-B (290–320 nm) radiation (together termed “high light” [HL]). Intense visible and UV-B radiation are known to stimulate multiple branches of phenylpropanoid and flavonoid metabolism (Jordan, 1996; Grace and Logan, 2000; Winkel-Shirley, 2002; Kimura et al., 2003; Treutter, 2005). This experiment was conducted at the same time and with the same cohort of plants as the wounding experiment described above. Analysis of gene expression at the same time points as for the wounded plants revealed two distinct patterns of flavonoid structural gene activation in HL-exposed poplar leaves (Fig. 3A, left and right). A rapid activation of PAL1, CHS, F3H, and the flavonol synthase gene FLS4 suggests that flavonol glycoside biosynthesis was up-regulated very early. The activation of flavonol biosynthetic genes exhibited a reduction at 12 h, which may have been caused by the absence of the stimulus during the night or by circadian regulation. Flavonol glycosides are known to be important UV-B protective compounds (Li et al., 1993; Landry et al., 1995), and increased flavonol accumulation in leaves has been observed in poplar and other trees under elevated UV-B light (Warren et al., 2002, 2003; Keski-Saari et al., 2005; Turtola et al., 2005). HPLC analysis of extracts from leaves after 7 d of HL exposure revealed a large increase in total flavonol glycosides (Fig. 3B), corresponding to the strong activation of the flavonol biosynthetic genes. Unexpectedly, the early activation of flavonol biosynthetic genes was followed by a strong activation of PA biosynthetic genes by 48 h, including the PA-specific genes ANR1 and LAR1 (Fig. 3A). The subsequent accumulation of PAs following HL-induced PA pathway activation was confirmed using acid-butanol PA assays (Porter et al., 1986; Fig. 3B).
None of the candidate PA regulatory MYB genes was coactivated with the rapidly induced flavonol biosynthetic genes. However, MYB134 and MYB183 both exhibited coactivation with the late flavonoid and PA biosynthetic genes after HL exposure (Fig. 3A). MYB086 and MYB134 are similar at the nucleotide sequence level, such that expression may not be reliably distinguishable using northern-blot analysis. Therefore, we analyzed their expression and stress responsiveness using real-time PCR with primers specific to MYB134 or MYB086. Expression of MYB097 and MYB183 was also monitored. We determined relative transcript abundance in untreated control leaf tissue as well as leaves 24 h after wounding and 48 h after movement into HL conditions, with triplicate biological and technical replication (Fig. 3C). MYB086 exhibited a very low, constitutive level of expression, while MYB134 was found to be wound and HL induced (Fig. 3C; MYB086 expression was too low relative to the other genes to be visible with the scale of this graph, but it was visible with gel electrophoresis). The wound- and HL-induced expression of MYB097 and MYB183 was also confirmed (Fig. 3C).
To pursue the distinct HL-induced patterns of early and late flavonoid structural gene expression in more detail, we examined flavonoid gene expression in poplar leaves exposed to increased UV-B irradiance in a growth chamber. Northern-blot analysis of gene expression revealed an activation of flavonol biosynthetic genes by 3 h, followed by up-regulation of PA-specific biosynthetic genes by 48 h (Fig. 4A, left and center). PA and total flavonol glycoside concentrations were determined after 7 d of stress treatment and were again found to have increased significantly (Fig. 4B). None of the poplar MYB genes tested exhibited a coinduction with FLS4 and the early flavonoid biosynthetic genes. Of the wound- and light-induced MYB genes, only MYB134 was coinduced with the late flavonoid biosynthetic genes under elevated UV-B light.
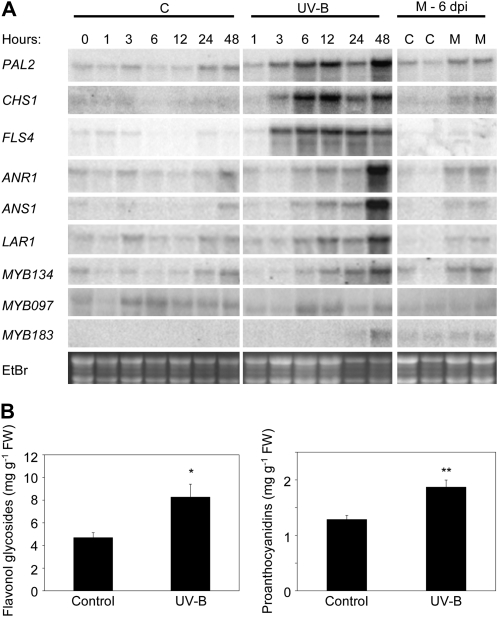
Figure 4.
UV-B light- and M. medusae-induced activation of the flavonoid biosynthetic pathway and putative PA regulatory MYB genes. A, Northern-blot analysis of phenylpropanoid and flavonoid structural genes as well as putative PA regulatory MYB genes in control (C) and UV-B-treated (UV-B) poplar leaves as well as control and M. medusae-infected leaves at 6 d after inoculation (M - 6 dpi). EtBr, Ethidium bromide. B, Phytochemical changes in UV-B-treated leaves after 7 d. PAs were quantified using the acid-butanol method, and total flavonol glycosides were quantified as rutin equivalents using HPLC. Bars indicate means of four trees per treatment, with error bars indicating se. Asterisks indicate significant differences using Student's t test (* P < 0.05, ** P < 0.01). FW, Fresh weight.
Since we observed the coactivation of PA biosynthetic genes and putative MYB regulators following both wounding and exposure to elevated light levels, we sought to determine whether this coactivation would occur following an additional PA-activating stress. We had previously found that infection of poplar leaves by the fungal biotroph M. medusae results in a significant up-regulation of PA biosynthetic genes and a corresponding increase in foliar PAs (Miranda et al., 2007). Profiling of flavonoid and putative PA regulatory MYB gene expression in leaf tissue 6 d after inoculation revealed that MYB134 was again most strongly coinduced with PA structural genes (Fig. 4A, right). As with mechanical wounding, FLS4 was not strongly up-regulated by M. medusae infection, confirming that expression of MYB134 is not correlated with the activation of flavonol metabolism (Fig. 4A).
Overall, our data show that several members of the poplar N08 MYB subgroup exhibit some stress-induced coactivation with PA biosynthetic genes. However, MYB134 expression was the most correlated with PA activation under all conditions analyzed. Of the inducible MYB genes, the predicted protein encoded by MYB134 also exhibited the highest homology to TT2 within the R2R3 MYB domain and shared a C-terminal sequence motif. Based on these observations, we hypothesized that MYB134 may be a regulator of stress-induced PA metabolism and sought to further characterize its functions using transgenic plants.
Effects of MYB134 Overexpression in Poplar on Phenolic Metabolism and Gene Expression
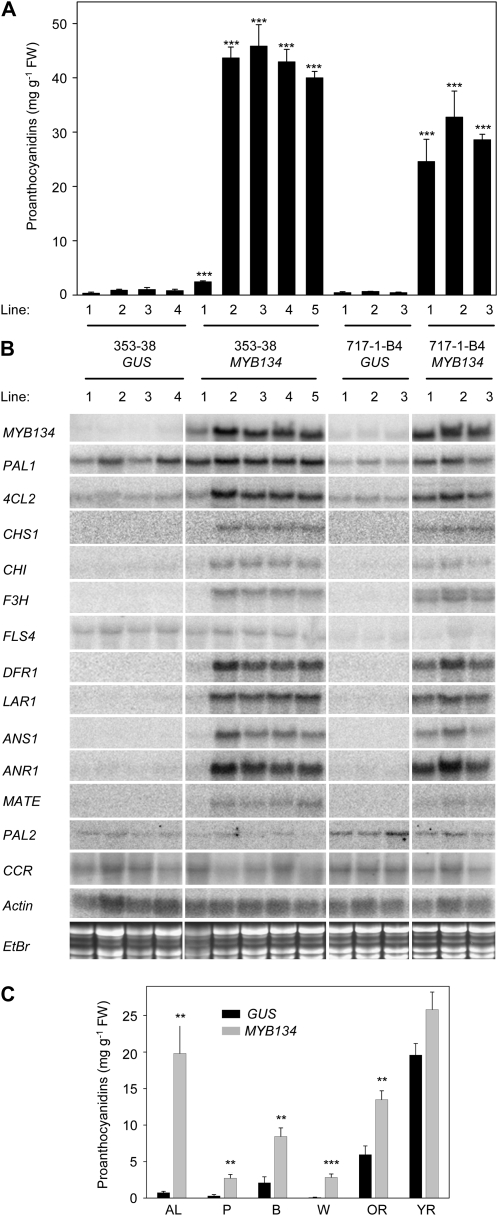
In order to investigate the role of MYB134 in the regulation of stress-induced PA metabolism, we overexpressed this gene in poplar under the control of a double cauliflower mosaic virus 35S promoter. GUS-overexpressing lines were also produced as controls. Two genotypes, Populus tremula × P. tremuloides clone 353-38 and P. tremula × Populus alba clone 717-1-B4, were chosen for Agrobacterium tumefaciens-mediated transformation because of differences they exhibit in wound-induced PA metabolism. Under our growth conditions, the stress-induced transcriptional activation of the PA biosynthetic pathway is stronger in the 353-38 clone than in the 717-1-B4 clone (R.D. Mellway and C.P. Constabel, unpublished data). Both are characterized by relatively low leaf PA levels when grown under standard greenhouse conditions. Multiple independently transformed lines were generated for both genotypes, and confirmed transgenics were moved to a greenhouse for analysis. For plants of both genotypes, grown and maintained under our standard greenhouse conditions for up to 6 months (see “Materials and Methods”), MYB134 overexpression did not lead to any discernible gross phenotypic abnormalities (Supplemental Fig. S1). However, analysis of PA levels in leaves revealed that MYB134 overexpression resulted in a dramatic increase in PA concentrations (Fig. 5A). The PA levels found in these MYB134 overexpressor plants were similar to the levels found in the leaves of outdoor P. tremuloides trees and much higher than we have observed for any Populus species under our greenhouse conditions (R.D. Mellway and C.P. Constabel, unpublished data). One of the 353-38 MYB134 overexpressor lines exhibited a much lower but still significant (P = 0.003) increase in PA levels (353-38 MYB134 line 1; Fig. 5A). Southern-blot analysis using a probe complementary to the neomycin phosphotransferase II gene present on the T-DNA revealed that this line contained five or six T-DNA insertions, compared with one or two copies for the other lines (data not shown). This suggested that the lower increase in PA levels in this line may be the result of silencing of the transgene due to the high number of T-DNA insertions (Tang et al., 2007). Consistent with this, northern-blot analysis revealed a correspondingly lower level of transgene expression in this line (Fig. 5B). The correlation between the level of MYB134 expression and PA accumulation further supports our conclusion that MYB134 is a regulator of the PA pathway.
Figure 5.
Effects of MYB134 overexpression on PA concentration and phenylpropanoid gene expression in transgenic poplar. A, PA accumulation in leaves of independently transformed GUS and MYB134 overexpressor 353-38 and 717-1-B4 plants. Bars indicate means of at least three trees per line, with error bars indicating se. FW, Fresh weight. B, Northern-blot analysis showing expression of phenylpropanoid and flavonoid structural genes in leaves of the same lines analyzed in A. EtBr, Ethidium bromide. C, Comparison of PA levels in tissues of control and MYB134 overexpressor poplar determined using the acid-butanol assay. AL, Apical leaves; P, petioles of leaves (LPI 9–11); B, bark; W, woody stem with bark removed; OR, old root (within 4 cm of shoot base); YR, young root (within 4 cm of tip). Bars indicate means of four independently transformed lines (353-38 GUS lines 1–4 and MYB134 overexpressor lines 2–5), with error bars indicating se. Asterisks indicate significant differences using Student's t test (** P < 0.01, *** P < 0.001).
In order to confirm that MYB134 activates genes of the PA biosynthetic pathway, we examined phenylpropanoid and flavonoid structural gene expression in leaves of control and MYB134-overexpressing plants. In both the 353-38 and 717-1-B4 clones, MYB134 overexpression was found to activate the entire phenylpropanoid pathway leading to PA production, including the flavonoid-specific general phenylpropanoid genes PAL1 and 4CL2 (Kao et al., 2002; Fig. 5B). Both early (CHS1, CHI1, and F3H) and late (DFR1, ANR1, ANS1, and LAR1) flavonoid biosynthetic genes were more highly expressed in the MYB134-overexpressing plants, but the level of late biosynthetic gene up-regulation was much greater (Fig. 5B). The stress-inducible MATE gene was also activated in the MYB134 overexpressors, suggesting that it is a poplar homolog of Arabidopsis TT12 and that it is important for PA biosynthesis. Consistent with our hypothesis that MYB134 functions specifically to regulate PA metabolism and does not directly regulate other flavonoid pathways, the light stress- and UV-B-induced flavonol synthase gene FLS4 was not up-regulated by MYB134 overexpression. Rather, FLS4 was expressed in leaves of both control and MYB134 overexpressor plants at similarly low levels (Fig. 5B). The expression of PAL2 and CCR (for cinnamoyl CoA-reductase), genes involved in lignin production (Kao et al., 2002; Li et al., 2005), was likewise not altered in leaves of MYB134 overexpressors (Fig. 5B). MYB097 and MYB183 were also not up-regulated in MYB134-overexpressing plants (Supplemental Fig. S2). To determine if tissues other than mature leaves also respond to MYB134 overexpression with activation of the PA biosynthetic pathway, we conducted a tissue survey of PA levels in four high-MYB134 transgene-expressing lines (353-38 MYB134 overexpressor lines 2–5). Acid-butanol PA assays revealed a significant increase in PA concentrations in all tissues analyzed (P < 0.01), with the exception of young roots (P = 0.055; Fig. 5C).
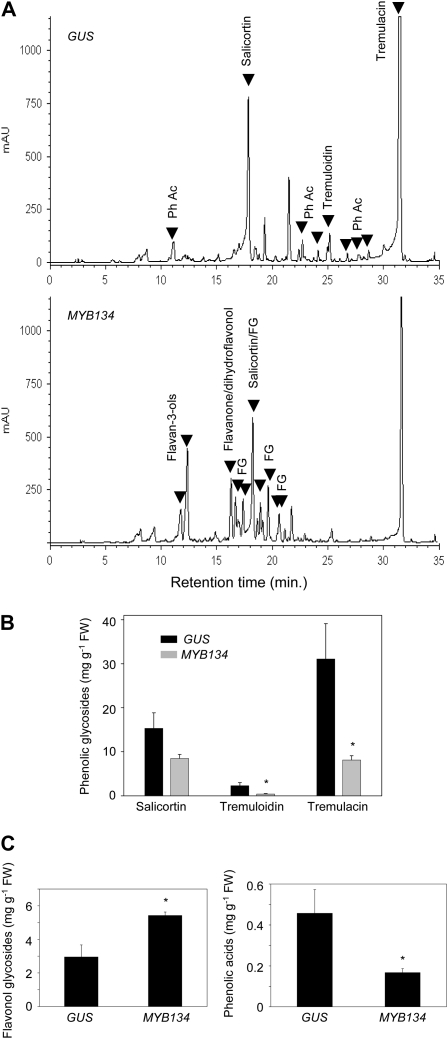
In addition to the large increase in PA concentrations, HPLC analysis revealed that MYB134 overexpression also caused unexpected secondary alterations to phenolic metabolism (Fig. 6). The most pronounced effect was a reduction in PG concentrations. Total PG levels were reduced from a mean of 46.7 ± 12.2 mg g−1 fresh weight to 17.0 ± 1.9 mg g−1 fresh weight, and levels of all individual PGs (salicortin, tremuloidin, and tremulacin) reflect this change (Fig. 6B). We also observed an increase in concentrations of total flavonol glycosides and a decrease in levels of nonflavonoid phenolic acids (Fig. 6C). These latter changes were statistically significant (P < 0.05), but they were minor in comparison with the increase in PAs. Anthocyanins were detected in neither control nor MYB134 overexpressor leaves. A number of peaks with absorption spectra and retention times corresponding to PA biosynthetic intermediates, including flavan-3-ols (catechin and epicatechin) and a flavanone/dihydroflavonol, were observed in MYB134 overexpressor leaf extracts but were undetectable in controls (Fig. 6A). The same pattern of moderately reduced nonflavonoid phenylpropanoids (PGs and phenolic acids) and a small increase in non-PA flavonoids (flavonol glycosides and PA biosynthetic intermediates) was also found in the 717-1-B4 MYB134 overexpressor lines (data not shown). Overall, the combined decreases in nonflavonoid phenylpropanoids were less than the total increase in PA and flavonoid levels; thus, MYB134 overexpression led to a net increase in total soluble phenolics in all tissues (Supplemental Table S1).
Figure 6.
HPLC analysis of soluble phenolics in control 353-38 GUS (top) and MYB134 overexpressor (bottom) leaf extracts. A, Representative maxplot chromatograms showing each peak at its λmax (note that some peaks present in both chromatograms are labeled in one or the other). Ph Ac, Phenolic acids; FG, flavonol glycosides. B, Phenolic glycoside (salicortin, tremuloidin, and tremulacin) concentrations in leaves of 353-38 GUS controls and MYB134 overexpressors. FW, Fresh weight. C, Total phenolic acid and flavonol glycoside concentrations in leaves of 353-38 GUS controls and MYB134 overexpressors. Bars indicate means of four independently transformed lines (GUS lines 1–4 and MYB134 overexpressor lines 2–5), with error bars indicating se. Asterisks indicate significant differences using Student's t test (* P < 0.05).
To determine PA localization in MYB134-overexpressing and control poplar plants, leaf, petiole, and stem sections were stained with dimethylaminocinnamaldehyde (DMACA), which reacts specifically with PAs and flavan-3-ols to form a blue chromophore (Feucht and Treutter, 1990). As expected, DMACA stained the MYB134 overexpressor tissues more intensely than control tissues (Fig. 7). In leaves of control plants, DMACA staining revealed PA accumulation primarily in the abaxial epidermis, with very light staining in the adaxial epidermis (Fig. 7A). In MYB134 overexpressor leaves, PAs were also present in both the adaxial and abaxial epidermal layers but at much greater concentrations. Most notably, DMACA staining was also very abundant in the upper layer of palisade mesophyll cells and sporadically within the spongy mesophyll (Fig. 7B). In petioles, staining was observed only in the epidermal cells in controls (Fig. 7C), while in MYB134 overexpressors, strong staining was observed in the epidermis with sporadic staining in the cortex and vascular tissues (Fig. 7D). In stem sections (fourth internode), PAs were localized only to the epidermis of control plants (Fig. 7E), while staining was observed in the epidermis, cortex, xylem, and pith in the MYB134 overexpressors (Fig. 7F). The pattern of PA distribution within the MYB134-overexpressing plants thus includes multiple cell types. Although our control plants showed relatively faint staining, based on previous work (Kao et al., 2002) it appears that in MYB134 overexpressors many of the same cell types that are competent to produce PAs are now stimulated to produce much higher levels.
Figure 7.
Localization of PAs in tissues of control and MYB134 overexpressor plants. Leaf (LPI 10), petiole (LPI 10), and stem (fourth internode) cross sections were stained with the PA-specific stain DMACA (blue). A, C, and E, Control leaf, petiole, and stem, respectively. B, D, and F, MYB134 overexpressor leaf, petiole, and stem, respectively. Bars = 200 μm (A and B) and 400 μm (C–F). c, Cortex; e, epidermis; pa, palisade mesophyll; ph, phloem; pi, pith; s, spongy mesophyll; x, xylem.
MYB134 Binds to Promoter Regions of PA Biosynthetic Genes
Consistent with the hypothesis that PA biosynthetic pathway genes function as downstream targets of MYB134, electrophoretic mobility shift assays (EMSAs) revealed that recombinant MYB134 protein bound to 180-bp DNA regions residing upstream of the transcriptional start of the three putative target genes (Fig. 8). These target genes were chosen to represent general phenylpropanoid/early PA metabolism (PAL1), late flavonoid metabolism (DFR1), and the PA-specific branch of flavonoid metabolism (ANR2). Candidate MYB134-binding sites in the regulatory regions of these genes were identified by visual examination of the upstream genomic sequence and comparison with characterized phenylpropanoid promoters as well as with a search of the PLACE plant cis-element database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) using SIGNAL SCAN (Prestridge, 1991; Higo et al., 1998). The promoter regions of the target genes were found to contain motifs similar to the adenosine- and cytosine-rich AC elements found in the regulatory regions of biosynthetic genes of different branches of phenylpropanoid metabolism, including both flavonoid and lignin biosynthesis (Hatton et al., 1995; Rogers and Campbell, 2004; Hartmann et al., 2005). AC elements are bound by the maize C1 protein, the most closely related MYB protein to MYB134 for which DNA-binding sites have been defined, as well as several MYB proteins involved in the regulation of lignin metabolism (Hatton et al., 1995; Patzlaff et al., 2003; Rogers and Campbell, 2004). The 180-bp ANR2 promoter region analyzed also contains a motif matching the CNGTTR consensus sequence bound by the vertebrate c-myb (Luscher and Eisenman, 1990; Fig. 8A). A c-myb consensus site is similarly present in the Arabidopsis BAN minimal promoter (Debeaujon et al., 2003). Inspection of these representative promoter sequences also revealed the presence of bHLH protein consensus-binding sites (CANNTG) in close proximity to the putative MYB-binding sites (Fig. 8A). The upstream region of poplar PAL1 contains two overlapping AC element sequences identical to the high-affinity P-binding site (ACCTACCAACC) identified in the maize A1 (encoding dihydroflavonol reductase) promoter sequence that is bound by maize C1 and the maize P protein, an R2R3 MYB protein that regulates the biosynthesis 3-deoxy flavonoids and phlobaphenes (Sainz et al., 1997). Within the 180-bp regions analyzed, poplar DFR1 and ANR2 both contain motifs that are quite similar to the AC elements defined by Hatton et al. (1995) in the P. vulgaris PAL2 promoter (GCCTACC and ACCTACA, respectively; Fig. 8A). The Arabidopsis BAN minimal promoter characterized by Debeaujon et al. (2003) also contains a motif differing by one nucleotide from canonical AC elements that is located within a region of high similarity to the maize A1 high-affinity P-binding site.
Figure 8.
MYB134 binds to the promoters of putative downstream target genes. A, Schematic representation of 1,000 bp of 5′ noncoding sequences for three putative MYB134 downstream target genes. + and − indicate the orientations of AC element-like motifs relative to the sense coding strand; numbers indicate the positions of these motifs relative to the putative transcriptional start. Arrows above each line indicate bHLH consensus sites (CANNTG), while arrows below each line indicate c-myb consensus sites (CNGTTR). Light gray horizontal lines under the sequences correspond to the location of the DNA sequence used as the binding target in the EMSA conducted in B. B, MYB134 binding to 5′ noncoding sequences of the three putative target genes as determined by EMSA. Recombinant MYB134 bound to all three 5′ noncoding sequences, as determined by a gel shift of the probe (arrows), which could be outcompeted with increasing quantities of unlabeled DNA corresponding to a canonical R2R3 MYB-binding site, known as an AC element motif (AC; 5′-ATTGTTCTTCCTGGGGTGACCGTCCACCTAACGCTAAAAGCCGTCGCGGGATAAGCCTGTCTG-3′). C, MYB134 binding to the AC-rich canonical R2R3 MYB-binding site motif as determined by EMSA. Binding of recombinant MYB134 to radiolabeled AC can be outcompeted by cold competitor AC (left) but not by the nonspecific competitor poly(dIdC).
EMSA experiments showed that the recombinant MYB134 protein specifically bound the 180-bp upstream regulatory sequences (Fig. 8B). Two shifted bands were observed for the PAL1 and ANR2 180-bp probes, while only one was seen with the DFR1 probe (Fig. 8B). It is possible that the MYB134 protein binds both of the overlapping AC elements in the PAL1 promoter and both the AC element-like sequence and the c-myb-binding site in the ANR2 promoter. A sequence containing a canonical AC element was an effective competitor and eliminated MYB134 binding (Fig. 8B), and recombinant MYB134 also bound to this element in a specific manner (Fig. 8C). Thus, MYB134 appears to bind to the gene regulatory regions of putative target genes in an AC motif-dependent fashion.
DISCUSSION
Identification of MYB134, a Stress-Induced Regulator of PA Metabolism in Poplar
PAs are important molecules for plant adaptation to the environment and for human health, and there is considerable interest in the control of their biosynthesis. Here, we present evidence that MYB134 plays a role in the regulation of PA synthesis in poplar leaves, the first example, to our knowledge, of such a regulator implicated in stress induction of PAs in vegetative tissues. We believe MYB134 to be a PA regulator for the following reasons. First, of all the predicted R2R3 MYB genes in the P. trichocarpa genome, MYB134 exhibits the highest sequence similarity to TT2, the Arabidopsis MYB required for PA synthesis in the seed coat. Second, transcript profiling following stresses that induce the PA biosynthetic pathway demonstrated that, unlike other TT2-like poplar MYB genes, MYB134 was consistently coinduced with the PA biosynthetic pathway genes, including the PA-specific genes LAR1 and ANR1. Third, overexpression of MYB134 in transgenic poplar behind a strong constitutive promoter led to a large plant-wide accumulation of PAs. This was the result of the activation of the full PA biosynthetic pathway, including general phenylpropanoid genes, early and late flavonoid genes, and PA-specific genes. Fourth, our EMSA experiments show that MYB134 binds to representative early and late PA pathway gene regulatory regions containing putative MYB-binding sites, indicating that these genes may be directly activated by MYB134.
Our data indicate that MYB134 functions specifically in regulating PA biosynthesis and is unlikely to be involved in activating other flavonoid branches. MYB134 transcripts were not coinduced with FLS4 and the other early flavonoid biosynthetic genes by elevated light and UV-B stress (Figs. 3 and 4). Similarly, the absence of FLS4 induction in wounded and M. medusae-infected leaves (in which MYB134 expression was induced), as well as the lack of FLS4 up-regulation in the MYB134 overexpressor plants, indicates that it is not a target of MYB134 regulation. In addition, in MYB134 overexpressors, transcript levels of the lignin-specific PAL2 (Kao et al., 2002) and CCR (Li et al., 2005) genes were unaffected. Therefore, for all marker genes tested, MYB134 targets appear to be restricted to genes required for PA synthesis. A more in-depth delineation of the suite of genes regulated by MYB134 will require analysis of the transgenic plants with whole-genome arrays. Such experiments will also be useful for the identification of novel genes related to PA biosynthesis and may shed light on unresolved aspects of PA biosynthesis, such as PA precursor transport and polymerization. The activation of the putative TT12 homologue in the MYB134 overexpressors validates this approach and suggests that TT12-like MATE transporters may play an important role in PA biosynthesis in poplar as well.
While MYB134 overexpression is sufficient for activation of PA biosynthetic genes, up-regulation of predicted target genes in stably transformed plants does not in itself demonstrate a direct regulation by MYB134, and it is thus possible that this protein activates some downstream genes indirectly via other factors. Data generated using stable transformation of plants with genes encoding transcription factors must be interpreted with caution, as the resulting phenotype may be confounded by off-target and indirect effects (Broun, 2004). We note that we used a strong constitutive promoter, which is likely to lead to expression in additional cell types where MYB134 is not normally expressed. Nevertheless, we believe that when our data are considered as a whole, including the stress-responsive expression profiling, promoter binding assays, and the specificity of phenylpropanoid gene activation in MYB134-overexpressing plants, as well as the apparent conservation of function of flavonoid regulatory MYB proteins across species, the role of MYB134 as a stress-responsive regulator of PA biosynthetic genes is strongly supported.
MYB134 overexpression results in a differential degree of activation of early and late PA biosynthetic genes (Fig. 5B). This suggests that precise regulation of the full pathway in planta could involve additional MYB or other regulatory proteins (see below). The expression of the other two stress-induced poplar N08 MYB genes (MYB183 and MYB097) was unaltered in the MYB134 overexpressors (Supplemental Fig. S2), indicating that they are not required for pathway activation by MYB134. Overexpression of MYB183 and MYB097 in transgenic poplar does not result in activation of the full PA biosynthetic pathway (R.D. Mellway and C.P. Constabel, unpublished data). Since many of the flavonoid enzymes in poplar are encoded by small gene families that exhibit differential stress inducibility (Tsai et al., 2006b), it is possible that the other stress-induced MYB proteins regulate more restricted sets of biochemically distinct flavonoid structural gene family members. Such fine regulation mediated by a group of closely related but functionally distinct transcription factors would permit poplar to tailor biologically important features of PA chemistry to specific stress conditions (i.e. hydroxylation patterns or stereochemistry). Analogously, the closely related members of the anthocyanin regulatory MYB family in Antirrhinum majus have distinct target gene specificities; in this case, differences in their expression control the anthocyanin pigmentation patterns observed in different Antirrhinum species (Schwinn et al., 2006).
The Predicted Poplar MYB134 Protein Shares Features with Other Known PA Regulators
In addition to this work, Arabidopsis and lotus TT2 and grapevine MYBPA1 and MYBPA2 are the only functionally characterized R2R3 MYBs shown to be involved specifically in the regulation of PA metabolism. While Arabidopsis TT2, grapevine MYBPA2, and poplar MYB134 are very similar and fall within subgroup N08, MYBPA1 does not fall within the same phylogenetic cluster (Fig. 2A). Rather, it is more closely related to poplar MYB factors such as MYB115, MYB201, and MYB153. Another well-characterized N08 member, maize C1 (and paralogues), is an anthocyanin regulator, although C1 was found to strongly activate the Arabidopsis BAN (ANR) promoter when coexpressed with the maize bHLH protein Sn in transient expression experiments (Baudry et al., 2004). By contrast, for the anthocyanin regulators of the N09 subgroup, phylogenetic relationship has proven to be a strong predictor of function, with all characterized members functioning as activators of the anthocyanin pathway (Quattrocchio et al., 1999; Borevitz et al., 2000; Mathews et al., 2003; Morita et al., 2006; Schwinn et al., 2006; Takos et al., 2006). Our work indicates that high sequence similarity to TT2 can be used to link MYB gene function to PA pathway regulation.
Like Arabidopsis TT2, the predicted poplar MYB134, MYB097, MYB183, and MYB086 proteins contain residues in the R3 domain involved in mediating interactions with members of the bHLH transcription factor family (Fig. 2B); thus, these MYBs likely function with bHLH partners in transcription factor complexes. The accumulation of PAs in diverse organs and cell types of MYB134-overexpressing plants suggests that the additional factors required to form a functional MYB-bHLH-WDR ternary transcription complex are already present or are themselves regulated by MYB134, at least in those cell types within each tissue in which PA accumulation was observed. DMACA staining of MYB134 overexpressors revealed that PA accumulation was not uniform within tissues (Fig. 7). For example, in leaves, PAs consistently accumulated in the upper layer of palisade mesophyll cells and not in the lower layer (Fig. 7B). It is likely that cell type-specific expression patterns of additional factors, such as interacting WDR or bHLH proteins, may be integral in the regulation of the PA pathway, as has been found in Arabidopsis (Baudry et al., 2004).
In silico analysis has shown that the promoter regions of the poplar flavonoid and PA biosynthetic genes contain cis elements matching the consensus sequences recognized by phenylpropanoid regulatory R2R3 MYB proteins (Tsai et al., 2006a). MYB134 was shown to bind to promoter fragments containing motifs similar to the AC elements found in a wide variety of phenylpropanoid biosynthetic gene promoters (Fig. 8B). MYB134 was also shown to bind to a DNA sequence containing a canonical AC element (ACCTAAC; Fig. 8C). These results suggest that such motifs are bound by MYB134 in vivo, although these results do not rule out the involvement of other putative MYB binding sites, such as the animal c-myb recognition site found in the ANR2 promoter. AC element-like motifs are present within the 2-kb 5′ noncoding sequence of most poplar flavonoid genes (Tsai et al., 2006a). Given that AC elements are widely distributed in the regulatory regions not just of PA biosynthetic genes but of genes involved in other branches of flavonoid and phenylpropanoid metabolism, interactions with cofactors such as bHLH domain proteins that require the presence of additional binding sites likely contribute to the specific activation of different branch pathways (Hartmann et al., 2005). Consistent with specific bHLH cofactor binding sites contributing to MYB134 target gene specificity, putative bHLH-binding sites are present in all poplar PA pathway genes (R.D. Mellway and C.P. Constabel, unpublished data).
In activating the full suite of early and late flavonoid as well as PA biosynthetic genes, MYB134 differs from Arabidopsis TT2, which regulates a more limited set of late PA structural genes (Nesi et al., 2001; Sharma and Dixon, 2005). A wider target gene set for MYB134, in conjunction with the natural constitutive PA production in a wider range of poplar tissues, may account for the different effects of TT2 overexpression in Arabidopsis compared with MYB134 overexpression in poplar. Unlike poplar, Arabidopsis produces PAs only in the seed testa, and ectopic expression of TT2 does not result in plant-wide PA accumulation (Nesi et al., 2001). When combined with constitutive expression of the anthocyanin regulator PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), TT2 overexpression did not lead to PA synthesis outside of cell types in which the BAN promoter is naturally expressed (Sharma and Dixon, 2005). Constitutive expression of grapevine MYBPA1 alone in Arabidopsis resulted in organ- and cell-specific PA accumulation similar to that observed following coexpression of TT2 and PAP1 in Arabidopsis (Bogs et al., 2007). Analysis of the poplar MYB gene family has revealed that in addition to the small family of TT2-related poplar MYBs, there are also a number of MYB genes closely related to the grapevine MYBPA1 (Wilkins et al., 2009). It is possible that some of the effects of MYB134 overexpression are mediated indirectly through activation of one or more of these MYBPA1 homologs, and a more detailed elucidation of how the pathway is regulated will require functional characterization of the members of both MYB gene families as well as identification and analysis of the additional interacting proteins such as the bHLH and WDR proteins.
MYB134 Overexpression Affects Other Branches of Phenolic Metabolism in Poplar
In addition to the large increase in PA levels, the up-regulation of PA biosynthesis by MYB134 overexpression resulted in other changes to the phenolic metabolite profile. Concentrations of the phenolic glycosides were consistently reduced in leaves of the MYB134 overexpressor plants, resulting in an approximately 3-fold overall decrease relative to controls. The phenolic glycosides are the most abundant soluble phenolic compounds in leaves of the control plants, so this reduction represents a significant shift in carbon flux. Although less abundant overall, levels of phenolic acids were also reduced in the MYB134 overexpressor leaves, while non-PA flavonoids were increased slightly. These differences were consistently observed in different genotypes and transformed lines.
Based on the specificity of previously characterized flavonoid regulatory R2R3 MYB factors, it seems unlikely that MYB134 controls the PG and phenolic acid pathways directly. However, it is possible that these alterations are caused by competition for common cofactors between MYB134 and MYB factors regulating other branches of phenolic metabolism. Because the biosynthetic pathway of the phenolic glycosides is as yet unknown, we do not currently have molecular probes for enzymes of this pathway that would allow us to determine if transcriptional regulation is involved. However, it seems probable that the high rate of PA synthesis is diverting metabolic resources from other phenolic pathways, so that the observed changes in non-PA phenylpropanoid levels may be the result of competition for common phenolic precursors between the up-regulated PA pathway and other branches of phenolic metabolism. Holton et al. (2003) reported decreased PG levels corresponding to increased PA accumulation in aspen grown under elevated ozone, supporting the idea of a metabolic trade-off between these two major allelochemical pools.
The increase in flavonol glycoside levels but not other non-PA flavonoid end products such as flavones and anthocyanins is also consistent with the hypothesis that the high flux into PAs has indirect consequences on other pathways. The increased flux into the flavonoid pathway resulting from MYB134 overexpression and the accumulation of PA intermediates should provide higher levels of substrates for branch pathways that are already active in leaves (i.e., flavonol biosynthesis). Our data show that FLS4 was equally expressed in controls and MYB134 overexpressors (Fig. 5B) and that flavonol glycosides were present in leaves of both controls and MYB134 overexpressors (Fig. 7). With an enhanced synthesis of shared PA and flavonol intermediates such as dihydroflavonols, substrate availability for flavonol synthase should also increase. This would result in increased flavonol production without the activation of FLS4 or other flavonol-specific genes. Flavonoid branch pathways not already active in these tissues (i.e. anthocyanins) would be unaffected. However, other levels of regulation cannot be excluded.
The observation that MYB134 overexpression can directly or indirectly affect multiple phenylpropanoid pathways and end products has implications for analyses of metabolic flux and control points in phenolic metabolism, and these plants may be useful tools for such studies. However, the modification of several biologically active metabolites also complicates the interpretation of the biological effects of this overexpression.
A Diversity of Biological Functions for PAs?
PAs have long been investigated in the context of tree defense against insect pests, but their importance is still debated in the literature. Some of the controversy is likely due to observations that the biological activities of PAs are dependent upon subtle differences in their chemical structures, which show variation among different plants (Ayres et al., 1997; Xie and Dixon, 2005). Alternative functions for foliar PAs have been proposed, including defense against microbes, protection from photodamage, and storage of excess carbon (Feucht and Treutter, 1999; Hemming and Lindroth, 1999; Close and McArthur, 2002; Dixon, 2005). PAs may well be multifunctional, so these roles could all be important in some plants or under some conditions. Research aimed at evaluating proposed stress-protective or other roles of PAs will benefit from transgenic plants with altered PA or phenolic levels as we have produced here.
The strong up-regulation of PA biosynthetic genes and accumulation of PAs following light stress and UV-B exposure was a novel result of our experiments. PAs are not usually considered to be light stress- or UV-B-protective compounds. To our knowledge, rapid induction of PA biosynthesis following UV-B stress has not been previously reported in poplar, although Lavola (1998) reported that in birch (Betula pendula) saplings grown in growth chambers, elevated UV-B light resulted in increased foliar PA levels. In contrast, flavonols have often been found to be responsive to elevated UV-B light, and increased sensitivity of flavonol-deficient mutants to UV-B light has been demonstrated (Li et al., 1993).
Whether PAs can protect leaves of poplar, birch, and other plants against excess UV-B light directly as “sun screens” is unclear. Correlations between light levels and concentrations of PAs or other high-Mr polyphenols have been observed (Mole et al., 1988), and PA concentrations in poplar leaves are strongly influenced by light levels (Hemming and Lindroth, 1999). Like many other flavonoids, PAs exhibit strong antioxidant capacity in vitro (Hagerman et al., 1998) and may contribute to stress resistance by scavenging oxygen radicals. Visible light levels in excess of photosynthetic capacity can result in oxidative stress from the production of reactive oxygen species (Hideg et al., 2000; Demmig-Adams and Adams, 2006), and UV-B light can also lead to increased oxygen radicals in addition to directly damaging nucleic acids, proteins, and lipids (Rozema et al., 1997; Jordan et al., 1998; Mackerness et al., 1999). Oxidative stress is also associated with pathogen attack and herbivory and is thus a common theme in the stresses that induce PA synthesis in poplar. Although an antioxidant function must be reconciled with the vacuolar location of PAs in planta (Edreva, 2005), a protective role against oxygen radicals could explain why this pathway is up-regulated under a variety of stress conditions.
CONCLUSION
The extensive genomics resources combined with the complexity and biological importance of phenylpropanoid metabolism in poplar make it a useful system for investigating this pathway. In this report, we describe work identifying a gene encoding an R2R3 MYB transcription factor that appears to play an important role in controlling PA biosynthesis under a variety of stress conditions. We have established that MYB factors not only regulate developmental PA production but also function to control PA synthesis in response to stress. Identifying transcriptional regulators of biosynthetic pathway genes is an important goal for metabolic engineering of secondary metabolism in plants, and the identification of a putative regulator of PA metabolism in poplar may permit new experimental approaches for evaluating the biological functions of PAs.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Populus tremuloides clone A2 was collected from the vicinity of Edmonton, Alberta, Canada (Haruta et al., 2001). Populus tremula × P. tremuloides clone INRA 353-38 was provided by Steve Strauss (Oregon State University) and Richard Meilan (Purdue University), P. tremula × Populus alba clone INRA 717-1-B4 was provided by David Ellis (CellFor), and Populus trichocarpa × Populus deltoides clone H11-11 was provided by Gary Radamaker (Washington State University). All clones were micropropagated in vitro on solid Murashige and Skoog medium (Sigma), except clone H11-11, which was macropropagated from greenwood cuttings. Plants were maintained in the University of Victoria's Bev Glover Greenhouse with supplemental fertilizer and light as described (Major and Constabel, 2006). Twelve-week-old plants were used for stress experiments and analysis of transgenics. All stress treatments were applied beginning at 10:00 am. Upon harvest of leaves, midveins and necrotic tissue were removed and tissues were frozen in liquid nitrogen and stored at −80°C until analyzed. Leaves within the range of LPI 9 to 15 (Leaf Plastochron Index; Larson and Isebrands, 1971) were used for all stress experiments.
For wounding experiments, leaf margins totaling approximately one-fifth of the area of each leaf were crushed with pliers. For HL exposure experiments, trees were moved from the greenhouse (mean maximum photosynthetically active radiation, 400–700 nm; 377 mol m−2 s−1; biologically effective UV-B irradiance [UV-Bbe], 0.26 kJ m−2 d−1) into full natural sunlight during August in Victoria, British Columbia, Canada (mean maximum photosynthetically active radiation, 1,655 mol m−2 s−1; UV-Bbe, 3.48 kJ m−2 d−1). The wounding and light stress experiment shown was conducted in August 2005 in Victoria and replicated with equivalent results in 2006. For UV-B treatments, trees were acclimated for 1 week in a growth chamber (16-h/8-h photoperiod, 19–25°C) equipped with F40T12 UV-B lamps (Phillips Lighting) with presolarized cellulose acetate filters to block UV-C light transmission. Plants were exposed to 0.21 kJ m−2 d−1 UV-Bbe before UV-B lamps were activated and to 1.45 kJ m−2 d−1 UV-Bbe after activation. The UV-B experiment shown was conducted in October 2006. Measurements of UV-Bbe were made with an IL1700 radiometer equipped with an IL782A high-gain photomultiplier (International Light) using weighting factors from the Caldwell action spectrum normalized to 300 nm (Bjorn and Teramura, 1993; L'Hirondelle and Binder, 2002). Clone 353-38 was used for all wounding, light stress, and UV-B experiments. For Melampsora medusae infections, urediospores were collected from foliage and used to inoculate leaves of clone H11-11 as described (Miranda et al., 2007). The experiment shown was conducted in October 2006 and was a replicate of an experiment conducted in March 2005.
Phylogenetic Analysis and Cloning of Putative Poplar PA Regulatory R2R3 MYB Genes
Primers for amplifying full-length sequences were designed using Vector NTI Advance version 9.0 (Invitrogen). Primer sequences are listed in Supplemental Table S2. Full-length coding sequences were amplified from P. tremuloides (clone A2) leaf cDNA and cloned into pGEM-T Easy (Promega) for sequencing. For multiple sequence alignment and phylogenetic analysis, sequences were aligned using ClustalW (Chenna et al., 2003). The phylogenetic tree was constructed using the neighbor-joining method with the minimum evolution test and p-distance model with 1,000 bootstrap replicates using the Molecular Evolutionary Genetics Analysis package version 3.1 (Kumar et al., 2004).
Cloning of Poplar MYB134 and Plant Transformation
The coding sequence of MYB134 was PCR amplified from a P. tremuloides (clone A2) cDNA library with primers (Supplemental Table S2) containing restriction linker sites for subcloning into the vector pBI-524 between the double cauliflower mosaic virus 35S promoter with α-mosaic virus RNA4 transcriptional enhancer sequence and the nopaline synthase terminator sequence (Datla et al., 1992; Wang and Constabel, 2004). This overexpression cassette was then subcloned into the pRD400 binary plasmid carrying the neomycin phosphotransferase II gene for kanamycin resistance (Datla et al., 1992). The binary vector pRD400-MYB134 was transferred to the Agrobacterium tumefaciens strain C58 (pMP90) (Koncz and Schell, 1986). The pRD410 plasmid containing the GUS gene was used as a control construct (Datla et al., 1992). The 353-38 and 717-1-B4 clones were transformed using the method of Leplé et al. (1992). Positive independently transformed lines were identified by selection of shoots from separate explants on kanamycin-containing rooting medium and confirmed by PCR and Southern-blot analyses.
Phytochemical Assays and HPLC Analysis
For HPLC analysis, 0.50 g of frozen leaf tissue was ground in liquid nitrogen and extracted for 4 h in 10 mL of 80% methanol. Extracts were centrifuged to remove solid debris, and methanol was removed using a rotary evaporator, followed by cleanup with Strata-X 33-μm solid-phase extraction columns, according to the manufacturer's instructions (Phenomenex). Compounds were eluted in 2 mL of methanol:acetonitrile (1:1, v/v), and 30 μL was injected onto an HPLC system (Beckman Coulter System Gold 126 solvent module with a System Gold 168 diode array detector) with a reverse-phase Luna C18(2) column (250 × 60 mm, 5 μm; Phenomenex). Separation was performed with a linear elution gradient from 90% solvent A (0.5% methanol in 0.01 m phosphoric acid, v/v) to 40% solvent B (100% acetonitrile) over 30 min at a flow rate of 1.5 mL min−1. Although HPLC with diode array detection does not permit the precise structural identification of compounds, the subclasses of phenolic compounds present in poplar leaves are well characterized and peaks can be confidently assigned to different subclasses based on their distinctive UV/visible absorption spectra and quantified using representative standards (Mabry et al., 1970; Markham, 1982; Maatta et al., 2001; Santos-Buelga et al., 2003). All standards were from Sigma unless specified otherwise. Total phenolic acids were quantified as chlorogenic acid equivalents. Total flavonol glycosides were quantified as rutin equivalents. Phenolic glycosides were quantified using purified tremulacin, tremuloidin, and salicortin kindly provided by Richard Lindroth and Thomas Clausen. Proanthocyanidins were assayed using the acid-butanol assay, as described (Porter et al., 1986; Peters and Constabel, 2002), using purified P. tremuloides proanthocyanidin as a standard. Relative levels of total soluble phenolics were determined using the Folin-Ciocalteau method (Singleton and Rossi, 1965).
RNA Extraction and Expression Analysis
RNA for northern-blot and PCR analyses was isolated from leaf tissue using the cetyltrimethylammonium bromide method as described (Haruta et al., 2001). Northern-blot analysis was performed using 32P-labeled DNA probes using standard protocols (Church and Gilbert, 1984; Sambrook and Russell, 2001). 32P-labeled probes were synthesized with the Rediprime II labeling kit (Amersham) using Qiaquick-purified (Qiagen) DNA template fragments. For phenylpropanoid biosynthetic gene probes, cDNA sequence fragments were amplified and cloned into pGEM-T Easy (Promega) from clone 353-38 using primers designed using sequences available from the Department of Energy Joint Genome Project database version 1.1 (http://genome.jgi-psf.org/Poptr1_1) or in GenBank (http://www.ncbi.nlm.nih.gov; for all primer sequences, see Supplemental Table S2). The DFR1 and PAL2 probes were synthesized using the cloned fragments described by Peters and Constabel (2002).
For real-time PCR analysis, 25 ng of total RNA was treated with DNase I (Invitrogen) according to the manufacturer's instructions. Five nanograms of DNase I-treated RNA was then was used for reverse transcription with SuperScript II reverse transcriptase (Invitrogen). Following validation experiments, real-time PCR analysis was performed using a Stratagene Mx4000. Triplicate reactions were run on triplicate independent experiments. Reactions (15 μL) consisted of the QuantiTect SYBRGreen mix (Qiagen) with 0.67 μm gene-specific primers and 6.25 ng of cDNA template per reaction. The amplification protocol was 95°C for 15 min followed by 40 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 60 s. Dissociation curves were obtained and reaction products visualized using agarose gel electrophoresis to confirm that single, specific products were produced in each reaction. Cycle threshold (Ct) values were determined by Mx4000 software at a manually set fluorescence threshold of 0.019, and relative transcript abundances (2−ΔCt) were determined after normalization to a constitutively expressed Actin gene (for all real-time PCR primer sequences, see Supplemental Table S2).
Histochemical Staining
Fresh plant material was placed into Tissue-Tek O.C.T. Compound Embedding Medium (Sakura Finetek) and left overnight in a −20°C freezer before slicing 20-μm-thick (leaf) or 40-μm-thick (petiole and stem) sections using a Microm HM 500 cryomicrotome. PAs and flavan-3-ols were detected by staining sections for 5 min with DMACA (1% [w/v] in ethanol:6 n HCl, 1:1 [v/v]). Images were recorded using a Spot RT KE digital camera (Diagnostic Instruments) mounted on a Zeiss Universal compound microscope for leaf blade sections or a Wild M420 macroscope for stem and petiole sections.
EMSA
Recombinant MYB134 protein was produced in Escherichia coli using the coding sequence cloned in-frame into the NdeI and BamHI sites of the pET15b vector (Novagen). Recombinant MYB134 protein was produced, extracted, and affinity purified as described previously for pine (Pinus spp.) MYB proteins (Patzlaff et al., 2003). EMSA conditions were exactly as described previously (Patzlaff et al., 2003; Gomez-Maldonado et al., 2004) except that recombinant MYB134 protein was used in place of pine MYB protein.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ573150, FJ573151, FJ573152, and FJ588548.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Photographs of transgenic and control plants described in this study.
Supplemental Figure S2. Northern-blot analysis of MYB183 and MYB097 gene expression in MYB134-overexpressing poplar.
Supplemental Table S1. Percentage increase in total soluble phenolics in tissues of high-PA-accumulating 353-38 MYB134 overexpressors relative to GUS control plants.
Supplemental Table S2. Sequences of primers used in this study.
Supplementary Material
Acknowledgments
We thank Brent Gowen (University of Victoria) for assistance with the preparation of tissue for histochemical staining, George Newcombe (University of Idaho) for the M. medusae isolate, Richard Lindroth (University of Wisconsin-Madison) and Thomas Clausen (University of Alaska) for the purified phenolic glycosides, Silvia L'Hirondelle (Ministry of Forests and Range) for the IL1700 radiometer, and Nici T. Darychuk (University of Victoria) for critical reading of the manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery Grants to C.P.C. and M.M.C., and Undergraduate, Canada Graduate, and Postgraduate Scholarships to R.D.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: C. Peter Constabel (cpc@uvic.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aharoni A, De Vos CHR, Wein M, Sun ZK, Greco R, Kroon A, Mol JNM, O'Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28 319–332 [DOI] [PubMed] [Google Scholar]
- Ayres MP, Clausen TP, MacLean SF, Redman AM, Reichardt PB (1997) Diversity of structure and antiherbivore activity in condensed tannins. Ecology 78 1696–1712 [Google Scholar]
- Bailey JK, Deckert R, Schweitzer JA, Rehill BJ, Lindroth RL, Gehring C, Whitham TG (2005) Host plant genetics affect hidden ecological players: links among Populus, condensed tannins, and fungal endophyte infection. Can J Bot 83 356–361 [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 [DOI] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorn LO, Teramura AH (1993) Simulation of daylight ultraviolet radiation and effects of ozone depletion. In AR Young, LO Bjorn, J Moan, W Nultsch, eds, Environmental UV Photobiology. Plenum Press, New York, pp 41–71
- Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7 202–209 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher ME, Miranda M, Major IT, Constabel CP (2004) Gene expression profiling of systemically wound-induced defenses in hybrid poplar. Planta 219 936–947 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close DC, McArthur C (2002) Rethinking the role of many plant phenolics: protection from photodamage not herbivores? Oikos 99 166–172 [Google Scholar]
- Datla RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W (1992) Modified binary plant transformation vectors with the wild-type gene encoding nptII. Gene 122 383–384 [DOI] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE (2003) Transcriptional regulation of secondary metabolism. Funct Plant Biol 30 913–925 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Merillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW (2006) Photoprotection in an ecological context: the remarkable complexity of thermal energy dissipation. New Phytol 172 11–21 [DOI] [PubMed] [Google Scholar]
- Dixon RA (2005) Engineering of plant natural product pathways. Curr Opin Plant Biol 8 329–336 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva A (2005) The importance of non-photosynthetic pigments and cinnamic acid derivatives in photoprotection. Agric Ecosyst Environ 106 135–146 [Google Scholar]
- Feucht W, Treutter D (1990) Flavan-3-ols in trichomes, pistils and phelloderm of some tree species. Ann Bot (Lond) 65 225–230 [Google Scholar]
- Feucht W, Treutter D (1999) The role of flavan-3-ols and proanthocyanidins in plant defense. In KMM Dakshini, CL Foy, eds, Principles and Practices in Plant Ecology: Allelochemical Interactions. CRC Press, New York, pp 307–338
- Gomez-Maldonado J, Avila C, de la Torre F, Canas R, Canovas FM, Campbell MM (2004) Functional interactions between a glutamine synthetase promoter and MYB proteins. Plant J 39 513–526 [DOI] [PubMed] [Google Scholar]
- Grace SC, Logan BA (2000) Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Philos Trans R Soc Lond B Biol Sci 355 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman AE, Riedl KM, Jones GA, Sovik KN, Ritchard NT, Hartzfeld PW, Riechel TL (1998) High molecular weight plant polyphenolics (tannins) as biological antioxidants. J Agric Food Chem 46 1887–1892 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57 155–171 [DOI] [PubMed] [Google Scholar]
- Haruta M, Major IT, Christopher ME, Patton JJ, Constabel CP (2001) A Kunitz trypsin inhibitor gene family from trembling aspen (Populus tremuloides Michx.): cloning, functional expression, and induction by wounding and herbivory. Plant Mol Biol 46 347–359 [DOI] [PubMed] [Google Scholar]
- Hatton D, Sablowski R, Yung MH, Smith C, Schuch W, Bevan M (1995) 2 classes of cis sequences contribute to tissue-specific expression of a pal2 promoter in transgenic tobacco. Plant J 7 859–876 [DOI] [PubMed] [Google Scholar]
- Hemming JDC, Lindroth RL (1995) Intraspecific variation in aspen phytochemistry: effects on performance of gypsy moths and forest tent caterpillars. Oecologia 103 79–88 [DOI] [PubMed] [Google Scholar]
- Hemming JDC, Lindroth RL (1999) Effects of light and nutrient availability on aspen: growth, phytochemistry, and insect performance. J Chem Ecol 25 1687–1714 [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E (2004) Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J Biol Chem 279 48205–48213 [DOI] [PubMed] [Google Scholar]
- Hideg E, Kalai T, Hideg K, Vass I (2000) Do oxidative stress conditions impairing photosynthesis in the light manifest as photoinhibition? Philos Trans R Soc Lond B Biol Sci 355 1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26 358–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton MK, Lindroth RL, Nordheim EV (2003) Foliar quality influences tree-herbivore-parasitoid interactions: effects of elevated CO2, O−3, and plant genotype. Oecologia 137 233–244 [DOI] [PubMed] [Google Scholar]
- Hwang SY, Lindroth RL (1997) Clonal variation in foliar chemistry of aspen: effects on gypsy moths and forest tent caterpillars. Oecologia 111 99–108 [DOI] [PubMed] [Google Scholar]
- Jiang CZ, Gu X, Peterson T (2004) Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol 5 R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BR (1996) The effects of ultraviolet-B radiation on plants: a molecular perspective. In JA Callow, ed, Advances in Botanical Research, Vol 22. Academic Press, Toronto, pp 98–149
- Jordan BR, James PE, Mackerness SAH (1998) Factors affecting UV-B-induced changes in Arabidopsis thaliana L. gene expression: the role of development, protective pigments and the chloroplast signal. Plant Cell Physiol 39 769–778 [DOI] [PubMed] [Google Scholar]
- Kao YY, Harding SA, Tsai CJ (2002) Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol 130 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keski-Saari S, Pusenius J, Julkunen-Tiitto R (2005) Phenolic compounds in seedlings of Betula pubescens and B. pendula are affected by enhanced UVB radiation and different nitrogen regimens during early ontogeny. Glob Change Biol 11 1180–1194 [Google Scholar]
- Kimura M, Yamamoto YY, Seki M, Sakurai T, Sato M, Abe T, Yoshida S, Manabe K, Shinozaki K, Matsui M (2003) Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol 77 226–233 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37 104–114 [DOI] [PubMed] [Google Scholar]
- Klimczak M, Kahl W, Grodzins Z (1972) Studies on phenolic acids, derivatives of cinnamic acid, in plants. 1. Phenolic acids in poplar (Populus). Dissert Pharm Pharma 24 181–185 [Google Scholar]
- Koncz C, Schell J (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson PR, Isebrands JG (1971) The plastochron index as applied to developmental studies of cottonwood. Can J For Res 1 1–11 [Google Scholar]
- Lavola A (1998) Accumulation of flavonoids and related compounds in birch induced by UV-B irradiance. Tree Physiol 18 53–58 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57 405–430 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Brasileiro ACM, Michel MF, Delmotte F, Jouanin L (1992) Transgenic poplars: expression of chimeric genes using 4 different constructs. Plant Cell Rep 11 137–141 [DOI] [PubMed] [Google Scholar]
- LeRoy CJ, Whitham TG, Keim P, Marks JC (2006) Plant genes link forests and streams. Ecology 87 255–261 [DOI] [PubMed] [Google Scholar]
- L'Hirondelle SJ, Binder WD (2002) Ultraviolet-B Radiation Impacts on Tree Seedlings in British Columbia. Land Management Handbook, Vol 49. Forest Science Program, British Columbia Ministry of Forests, Victoria, British Columbia
- Li JY, Oulee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LG, Cheng XF, Lu SF, Nakatsubo T, Umezawa T, Chiang VL (2005) Clarification of cinnamoyl co-enzyme A reductase catalysis in monolignol biosynthesis of aspen. Plant Cell Physiol 46 1073–1082 [DOI] [PubMed] [Google Scholar]
- Lindroth RL, Hwang SY (1996) Diversity, redundancy, and multiplicity in chemical defense systems of aspen. In JT Romeo, JA Saunders, P Barbosa, eds, Phytochemical Diversity and Redundancy in Ecological Interactions. Plenum Press, New York, pp 25–56
- Luscher B, Eisenman RN (1990) New light on Myc and Myb. II. Myb. Genes Dev 4 2235–2241 [DOI] [PubMed] [Google Scholar]
- Maatta K, Kamal-Eldin A, Torronen R (2001) Phenolic compounds in berries of black, red, green, and white currants (Ribes sp.). Antioxid Redox Signal 3 981–993 [DOI] [PubMed] [Google Scholar]
- Mabry TJ, Markham KR, Thomas MB (1970) The Systematic Identification of Flavonoids. Springer-Verlag, New York
- Mackerness SAH, Surplus SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B (1999) Ultraviolet-B-induced stress and changes in gene expression in Arabidopsis thaliana: role of signalling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 22 1413–1423 [Google Scholar]
- Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172 617–635 [DOI] [PubMed] [Google Scholar]
- Markham KR (1982) Techniques of Flavonoid Identification. Academic Press, Toronto
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Ralph SG, Mellway R, White R, Heath MC, Bohlmann J, Constabel CP (2007) The transcriptional response of hybrid poplar (Populus trichocarpa × P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol Plant Microbe Interact 20 816–831 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3 212–217 [Google Scholar]
- Mole S, Ross JAM, Waterman PG (1988) Light-induced variation in phenolic levels in foliage of rain-forest plants. 1. Chemical changes. J Chem Ecol 14 1–21 [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47 457–470 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier TL, Lindroth RL (2001) Effects of genotype, nutrient availability, and defoliation on aspen phytochemistry and insect performance. J Chem Ecol 27 1289–1313 [DOI] [PubMed] [Google Scholar]
- Osier TL, Lindroth RL (2006) Genotype and environment determine allocation to and costs of resistance in quaking aspen. Oecologia 148 293–303 [DOI] [PubMed] [Google Scholar]
- Palo RT (1984) Distribution of birch (Betula spp), willow (Salix spp), and poplar (Populus spp) secondary metabolites and their potential role as chemical defense against herbivores. J Chem Ecol 10 499–520 [DOI] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105 14210–14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36 743–754 [DOI] [PubMed] [Google Scholar]
- Pearl IA, Darling SF (1971) Studies on leaves of family Salicaceae. 16. Phenolic extractives of leaves of Populus balsamifera and of P. trichocarpa. Phytochemistry 10 2844–2847 [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32 701–712 [DOI] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG (1986) The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 25 223–230 [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS (1991) Signal Scan: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7 203–206 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Campbell MM (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164 17–30 [DOI] [PubMed] [Google Scholar]
- Rozema J, van de Staaij J, Bjorn LO, Caldwell M (1997) UV-B as an environmental factor in plant life: stress and regulation. Trends Ecol Evol 12 22–28 [DOI] [PubMed] [Google Scholar]
- Sainz MB, Grotewold E, Chandler VL (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Santos-Buelga C, Garcia-Vigera C, Tomas-Barberan FA (2003) On-line identification of flavonoids by HPLC coupled to diode array detection. In C Santos-Buelga, G Williamson, eds, Methods in Polyphenol Analysis. The Royal Society of Chemistry, Cambridge, UK, pp 92–127
- Schweitzer JA, Bailey JK, Rehill BJ, Martinsen GD, Hart SC, Lindroth RL, Keim P, Whitham TG (2004) Genetically based trait in a dominant tree affects ecosystem processes. Ecol Lett 7 127–134 [Google Scholar]
- Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SB, Dixon RA (2005) Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J 44 62–75 [DOI] [PubMed] [Google Scholar]
- Singleton VL, Rossi JA (1965) Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Enol Vitic 16 144–158 [Google Scholar]
- Stafford HA (1990) Pathway to proanthocyanidins (condensed tannins), flavan-3-ols, and unsubstituted flavans. In Flavonoid Metabolism. CRC Press, Boca Raton, FL, pp 63–100
- Stevens MT, Lindroth RL (2005) Induced resistance in the indeterminate growth of aspen (Populus tremuloides). Oecologia 145 298–306 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58 545–554 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278 31647–31656 [DOI] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verries C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7 581–591 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, El Kayal W, Harding SA (2006. a) Populus, the new model system for investigating phenylpropanoid complexity. Int J Appl Sci Eng 4 221–233 [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan YN (2006. b) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172 47–62 [DOI] [PubMed] [Google Scholar]
- Turtola S, Rousi M, Pusenius J, Yamaji K, Heiska S, Tirkkonen V, Meier B, Julkunen-Tiitto R (2005) Clone-specific responses in leaf phenolics of willows exposed to enhanced UVB radiation and drought stress. Glob Change Biol 11 1655–1663 [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wang J, Constabel CP (2004) Polyphenol oxidase in transgenic Populus enhances resistance to forest tent caterpillar (Malacosoma disstria) herbivory and is activated in the insect gut. Planta 220 87–96 [DOI] [PubMed] [Google Scholar]
- Warren JM, Bassman JH, Eigenbrode S (2002) Leaf chemical changes induced in Populus trichocarpa by enhanced UV-B radiation and concomitant effects on herbivory by Chrysomela scripta (Coleoptera:Chrysomelidae). Tree Physiol 22 1137–1146 [DOI] [PubMed] [Google Scholar]
- Warren JM, Bassman JH, Fellman JK, Mattinson DS, Eigenbrode S (2003) Ultraviolet-B radiation alters phenolic salicylate and flavonoid composition of Populus trichocarpa leaves. Tree Physiol 23 527–535 [DOI] [PubMed] [Google Scholar]
- Wei YL, Li JN, Lu J, Tang ZL, Pu DC, Chai YR (2007) Molecular cloning of Brassica napus TRANSPARENT TESTA 2 gene family encoding potential MYB regulatory proteins of proanthocyanidin biosynthesis. Mol Biol Rep 34 105–120 [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1 251–257 [DOI] [PubMed] [Google Scholar]
- Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, Leroy CJ, Lonsdorf EV, Allan GJ, DiFazio SP, Potts BM, et al (2006) A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet 7 510–523 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5 218–223 [DOI] [PubMed] [Google Scholar]
- Xie DY, Dixon RA (2005) Proanthocyanidin biosynthesis: still more questions than answers? Phytochemistry 66 2127–2144 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299 396–399 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49 157–169 [DOI] [PubMed] [Google Scholar]
- Yu O, McGonigle B (2005) Metabolic engineering of isoflavone biosynthesis. Adv Agron 86 147–190 [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.