Abstract
Two types of branches, rachis branches (i.e. nonfloral) and spikelets (i.e. floral), are produced during rice (Oryza sativa) inflorescence development. We previously reported that the ABERRANT PANICLE ORGANIZATION1 (APO1) gene, encoding an F-box-containing protein orthologous to Arabidopsis (Arabidopsis thaliana) UNUSUAL FLORAL ORGANS, suppresses precocious conversion of rachis branch meristems to spikelets to ensure generation of certain number of spikelets. Here, we identified four dominant mutants producing an increased number of spikelets and found that they are gain-of-function alleles of APO1. The APO1 expression levels are elevated in all four mutants, suggesting that an increase of APO1 activity caused the delay in the program shift to spikelet formation. In agreement with this result, ectopic overexpression of APO1 accentuated the APO1 gain-of-function phenotypes. In the apo1-D dominant alleles, the inflorescence meristem starts to increase in size more vigorously than the wild type when switching to the reproductive development phase. This alteration in growth rate is opposite to what is observed with the apo1 mutants that have a smaller inflorescence meristem. The difference in meristem size is caused by different rates of cell proliferation. Collectively, these results suggest that the level of APO1 activity regulates the inflorescence form through control of cell proliferation in the meristem.
The inflorescence is one of the major determinants of appearance in flowering plants. The basic architecture of the grass inflorescence is defined by spatial arrangement of spikelets, small branches containing flowers, and rachis branches; both are born as lateral meristems during inflorescence development. Lateral meristems receive a specific identity at their initiation according to the position and timing of their occurrence. They then progress through a genetically defined developmental program by changing identity in response to internal and external cues, and finally acquire the determinate floral state (Irish, 1997; McSteen et al., 2000).
The timing of meristem phase change is crucial in the control of inflorescence architecture in grass species (McSteen et al., 2000; Bortiri and Hake, 2007; Wang and Li, 2008). In rice (Oryza sativa), transition to the reproductive phase evokes transformation of the shoot apical meristem (SAM) to the inflorescence meristem (IM), which initiates several lateral meristems that grow as primary rachis branches (PBs; Ito et al., 2005). Then, the IM loses its activity, leaving a vestige at the base of the uppermost primary branch. Thus, in rice, the timing of IM abortion determines the number of primary branches in the inflorescence. The meristem of the PB produces next order branches as lateral meristems. The few initially formed meristems grow as secondary rachis branches, while later borne meristems directly become spikelet meristems. A spikelet is also a small branch that contains a variable number of flowers and is a basic unit defining inflorescence architecture in grasses. The spikelet in rice consists of a single flower (Hoshikawa, 1989; Ito et al., 2005).
All rachis branches in the rice inflorescence are determinate and are programmed to finally change their state from an indeterminate rachis branch meristem to the determinate spikelet meristem after producing a certain number of lateral meristems. Therefore, spikelets in the wild-type rice inflorescence are classified into two types, lateral spikelets and terminal spikelets, according to their origins. Lateral spikelets differentiate directly from newly borne lateral meristems while terminal spikelets are converted from rachis branch meristems. Three factors, timing of IM abortion, conversion of the rachis branch meristem to the terminal spikelet meristem, and program shift of lateral meristem identity along a basal-apical direction on a rachis branch, determine the overall architecture of the rice inflorescence. The key event in all three factors is the transition of meristem state, in particular, acquisition of the spikelet meristem identity.
Several genes that are crucial for determining meristem fates in the grass inflorescence have been reported (Bommert et al., 2005a; Bortiri and Hake, 2007). Rice FRIZZY PANICLE (FZP) gene and its maize (Zea mays) ortholog branched silkless1 (bd1) are regulators of spikelet meristem initiation (Chuck et al., 2002; Komatsu et al., 2003). In fzp and bd1 mutants, spikelet meristem identity is not established and, instead, ectopic branching continues infinitely. The SUPERNUMERARY BRACT gene of rice and indeterminate spikelet mutation (ids) and sister of ids genes in maize are involved in the control of the next step, transition from spikelet meristem to the floral meristem (Lee et al., 2007; Chuck et al., 2008). Recently, it was revealed that accumulation of the ids mRNA is fine tuned by miRNA-mediated regulation to establish the correct spatial arrangement of floral meristems (Chuck et al., 2007a, 2007b). Timing of the shift of meristem identity along a basal-apical direction is also under strict genetic control. Three genes, ramosa1 (ra1) and ra2, encoding transcription factors, and ra3, encoding a putative signal transduction component, regulate the program shift from long branch initiation to short branch initiation in maize inflorescence development (Vollbrecht et al., 2005; Bortiri et al., 2006; Satoh-Nagasawa et al., 2006). Interestingly, there is no cognate homolog of ra1 whereas ra2 and ra3 orthologs are found in the rice genome, although their functions are yet to be determined.
In addition to genes controlling establishment of meristem identity, genes regulating meristem size are critical for normal inflorescence architecture. Rice floral organ number1 (fon1), fon2, and maize fascinated ear and thick tassel dwarf1, all defective in the CLAVATA signaling pathway, contain enlarged SAMs and exhibit abnormal branching patterns (Taguchi-Shiobara et al., 2001; Suzaki et al., 2004, 2006; Bommert et al., 2005b; Chu et al., 2006). In contrast, the lonely guy (log) mutant that has a defect in synthesis of active cytokinins (CKs) has a small SAM and produces an inflorescence markedly reduced in size (Kurakawa et al., 2007). This demonstrated that CKs are critical in the control of inflorescence form in rice. The role of CKs in the control of inflorescence branching has also been demonstrated through the analysis of GRAIN NUMBER1, a quantitative trait locus that affects rice panicle size (Ashikari et al., 2005).
A number of studies showed that the molecular basis of floral development is conserved among a wide range of plant species (Shepard and Purugganan, 2002). In Arabidopsis, key regulators of floral meristem identity have been reported (for review, see Sablowski, 2007). Among them, LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO) are positive regulators of floral meristem identity and work in the same genetic pathway (Weigel et al., 1992; Simon, et al., 1994; Ingram et al., 1995; Lee et al., 1997; Chae et al., 2008). Basically, conserved systems driven by LFY/UFO orthologs control floral meristem identity in Antirrhinum and petunia (Petunia hybrida) although minor divergences in their mode of operation are observed. For example, DOUBLE TOP (DOT), a UFO ortholog, is the major determinant of floral meristem identity in petunia, whereas, in Arabidopsis and Antirrhinum, LFY and FLORICAULA (FLO) are major players that determine floral meristem identity (Weigel et al., 1992; Simon et al., 1994; Souer et al., 2008). Mutations in UFO or FIMBRIATA (FIM), an Antirrhinum UFO ortholog, confer only subtle effects on floral development.
Surprisingly, UFO and all its orthologs in dicot species reported so far including FIM, DOT, Proliferating Floral Organs, ANANTHA (AN), and Stamina pistillaoida promote floral fate, whereas ABERRANT PANICLE ORGANIZATION1 (APO1), a rice UFO ortholog, suppresses floral fate (Simon et al., 1994; Levin and Meyerowitz 1995; Souer et al., 1998, 2008; Taylor et al., 2001; Zhang et al., 2003; Ikeda et al., 2007; Lippman et al., 2008). apo1 loss-of-function mutant plants exhibit precocious transition of the IM and rachis branch meristems into spikelet meristems, leading to formation of small inflorescences (Ikeda et al., 2005, 2007). Although no complete loss-of-function mutants for RICE FLORICAULA LEAFY (RFL), a rice LFY ortholog, have been reported, the knock-down phenotypes that resemble apo1 suggest that RFL could act in the same genetic pathway as APO1, and negatively regulate floral fate (Kyozuka et al., 1998; Prasad et al., 2003; Rao et al., 2008). In contrast to the divergence of the APO1 and RFL functions in rice from that of their eudicot orthologs, the function of TERMINAL FLOWER1 orthologs as repressors of floral fate seems to be conserved in rice (Nakagawa et al., 2002).
Elucidation of the molecular basis for APO1 function is apparently one of the critical steps in understanding molecular mechanisms controlling rice inflorescence development. Here, we analyzed dominant gain-of-function mutant alleles of APO1 and transgenic plants overexpressing APO1. Increases in APO1 expression level led to an accelerated proliferation of cells in the IM and an enhanced inflorescence branching phenotype. We propose that APO1 controls the transition of meristem phases via control of cell proliferation in the meristem.
RESULTS
Isolation of Mutants with Increased Inflorescence Branching
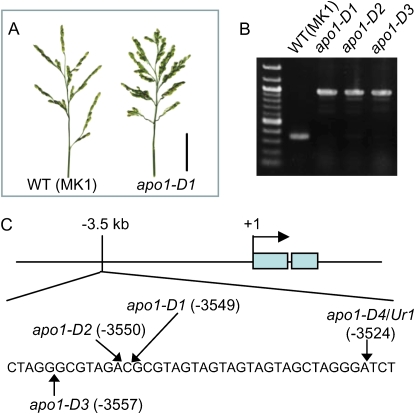
To obtain further insights into the genetic mechanisms controlling inflorescence development, we conducted a screening for mutants that show an increased inflorescence branching phenotype using a population in which nDart1, a recently identified transposon, actively transposes (Tsugane et al., 2006). From the screening of 5,781 independent lines, three lines of mutants with increased inflorescence branching were identified (Fig. 1A). Considering the activity of nDart1 transposition, we anticipated that the mutant phenotypes are likely caused by nDart1, and thus, carried out inverse PCR to isolate the genomic sequences flanking nDart1 from each mutant. Numerous DNA fragments were amplified by inverse PCR, reflecting the active transposition of nDart1. In total, 17 different DNA sequences that flank nDart1-0 were successfully obtained and among them was a promoter sequence for APO1. The presence of nDart1-0 in the APO1 promoter was confirmed by PCR and DNA sequence analysis (Fig. 1B). Because the inflorescence phenotypes were quite similar in the three mutants, we predicted that all three lines had a mutation in the APO1 gene. Further analysis indicated that nDart1-0 was present in the promoter region of the APO1 gene in all three identified mutants (Fig. 1, B and C), thus, the mutants were designated as apo1-D1, -D2, and -D3. The nDart1-0 inserts are in close but distinct positions of the APO1 promoter between 3,549 and 3,557 bp upstream of the translation start site (Fig. 1C). All three alleles contained an 8-bp duplication of the genomic sequence at the site of the insertion that is presumably generated during nDart1-0 insertion (data not shown). No other differences were seen in the coding sequence or 5-kb upstream of the translation start site of the APO1 sequence in the three mutants.
Figure 1.
Isolation of dominant mutants showing the inflorescence with increased branching. A, Inflorescence of wild type (WT; left) and apo1-D1 (right); bar = 5 cm. B, PCR amplification of APO1 promoter region in three apo1-D mutants and wild-type (‘MK1’) plants. C, Schematic representation of the APO1 gene. Positions of nDart1-0 insertion in the apo1-D mutants about 3.5 kb from the translation initiation site of APO1 are shown. The shaded boxes indicate the translated regions of APO1 starting at +1.
nDart1 Is Present in the Promoter Region of the APO1 Gene in the Undulate rachis1 Mutant
The Undulate rachis1 (Ur1) mutant (Nagao et al., 1958) has been reported to carry an increased number of secondary rachis branches per panicle and spikelets per secondary rachis branch, resulting in an increased number of spikelets (Fig. 2A; Nagao et al., 1958; Murai and Iizawa, 1994). Because Ur1 is a dominant mutant located on the distal region of the long arm of chromosome 6 (Nagao and Takahashi, 1963) where APO1 resides, we hypothesized that Ur1 might also be a dominant allele of APO1. To test this notion, we sequenced the APO1 gene including the promoter region and the coding sequence in the Ur1 mutant and found an insertion of nDart1-0 in the APO1 promoter (Figs. 1C and 2, B and C). The complete cosegregation of the nDart1-0 insertion and the mutant phenotype strongly supported the supposition that the nDart1-0 insertion was the cause of the mutant phenotype of Ur1 (Fig. 2D). To further confirm this notion, we screened for revertants in which nDart1-0 excised from the APO1 promoter. Through PCR-based screening of 400 homozygous Ur1 plants, five independent siblings were found that amplified a lower molecular mass band (Fig. 2B). Sequence analysis demonstrated that nDart1-0 had indeed excised from the APO1 promoter of all five plants, leaving a footprint of 8 bp (Fig. 2C). These plants had inflorescences of normal appearance (Fig. 2A). These results demonstrate that the Ur1 phenotype was indeed caused by an nDart1-0 insertion into the APO1 promoter, thus, Ur1 is also a dominant mutant allele of APO1 and was renamed apo1-D4. In summary, the three newly identified mutants and Ur1 are dominant mutant alleles of the APO1 gene.
Figure 2.
Analysis of apo1-D4/Ur1 mutant and revertants. A, Inflorescences of Apo1-D4/Ur1 (left) and the revertant line Ur1:3 (center) and the wild type (WT; ‘Bozu2’, right); bar = 5 cm. B, PCR analysis of the APO1 promoter in wild type (‘Bozu2’) and the apo1-D4/Ur1 mutant. C, Nucleotide sequences of the APO1 promoter around 3,520 bp from the translation initiation site. An 8-bp target site duplication, GCTAGGGA, is found in apo1-D4/Ur1-1 (shown in italic font and underlined). The footprint of 8 bp found in the revertant plant is marked with a box. D, Cosegregation of the nDart1-0 insertion and mutant phenotype. m, Mutant phenotype; W, wild-type phenotype.
Quantitative But Not Spatial Changes in APO1 Expression in the nDart1-0 Insertion Mutants
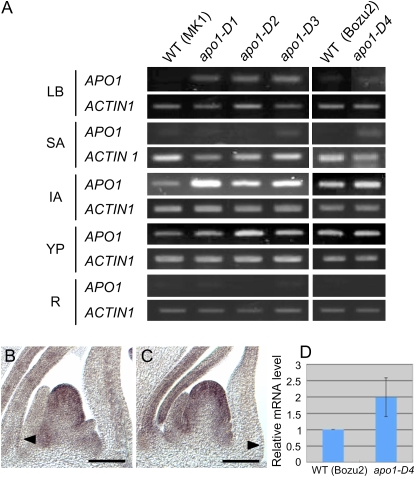
The phenotypes of apo1-D mutants, which are opposite to what is seen in apo1 loss-of-function mutants, suggested a possibility that the insertion of nDart1-0 in the promoter region of APO1 caused an increase in APO1 expression levels. In wild-type rice plants, APO1 transcripts localize to the leaf primordia, and the epidermis of young leaves during the vegetative phase (Ikeda et al., 2007). After the transition to the reproductive phase, APO1 transcripts accumulate in the IM and branch meristems. We compared APO1 mRNA accumulation in different organs of APO1 dominant alleles by semiquantitative reverse transcription (RT)-PCR analysis. Significant increases in APO1 expression were observed in leaf blades and inflorescence apices, the tissues where APO1 mRNA localizes in wild-type plants (Fig. 3A). Tissue specificity of APO1 expression was not significantly changed in apo1-D mutants, indicating that the nDart1-0 insertion did not alter the spatial distribution of APO1 expression. This result was confirmed by in situ hybridization analysis; APO1 expression was detected in the leaf margin and in the inflorescence branch primordia in both wild-type and apo1-D plants (Fig. 3, B and C). Quantitative RT-PCR showed that approximately twice as much APO1 mRNA accumulated in shoot apices in apo1-D4 than wild type (Fig. 3D).
Figure 3.
Expression levels of APO1 are increased in apo1-D mutants but tissue specificity of APO1 mRNA accumulation is not altered. A, Tissue specificity of APO1 expression analyzed by RT-PCR. RNA was isolated from leaf blades (LB), shoot apices of 4-week-old plants (SA), inflorescence apex (IA) at the secondary panicle branch initiation stage, young panicles at about 1 cm in height (YP), and roots (R). apo1-D1 to apo1-D3 were derived from ‘MK1’, and apo1-D4 was derived from ‘Bozu2’. PCR cycles performed on ACTIN1 and APO1 genes were 22 and 35, respectively. WT, Wild type. B and C, In situ analysis of APO1 expression. Longitudinal sections of the wild-type (B) and apo1-D1 (C) inflorescences at PB initiation stage. Arrowheads, APO1 signal in the leaf margin; bar = 100 μm. D, Quantitative RT-PCR analysis of the APO1 mRNA expression in whole seedlings of wild type and apo1-D4 at 10 d after germination (Bar: sem). ACTIN1 was used as a reference for normalization.
Inflorescence Morphology of apo1-D Dominant Mutants
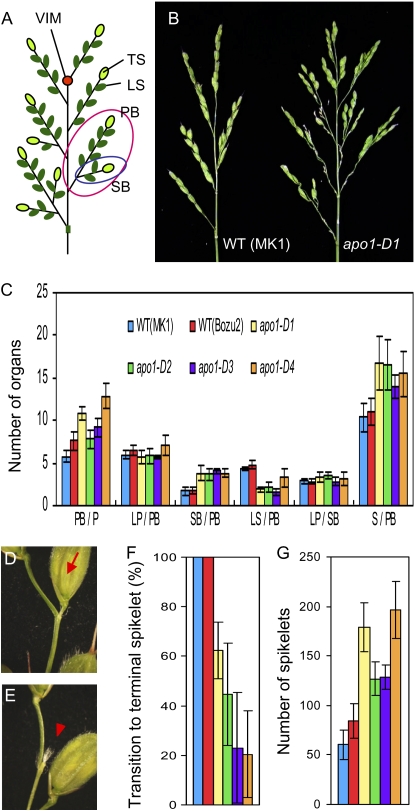
To reveal the effects of enhanced APO1 activity on inflorescence development, we analyzed the inflorescence phenotypes of apo1-D gain-of-function alleles in detail. During rice inflorescence development, the IM loses its activity and stops rachis branch meristem production, leaving a vestige of the meristem. Thus, timing of the IM abortion determines the number of PBs (Fig. 4A). In apo1-D mutants, the number of PBs increased up to 50%, implying that the timing of IM abortion was delayed by increased APO1 expression (Fig. 4, B and C).
Figure 4.
Inflorescence phenotypes of apo1-D mutants. A, A schematic representation of the rice inflorescence. VIM, Vestige of IM; TS, terminal spikelet; LS, lateral spikelet; SB, secondary rachis branch. Red and blue open circles indicate a primary branch and a secondary branch, respectively. B, Inflorescences of wild-type (WT; ‘MK1’, left) and apo1-D1 (right). C, Number of lateral organs in the inflorescence. apo1-D1, apo1-D2, and apo1-D3 were derived from ‘MK1’ and apo1-D4/Ur1 was from ‘Bozu2’. P, Panicle; LP, lateral primordium; SB, secondary rachis branch; LS, lateral spikelet; and S, total spikelet. D and E, Close-up view of branch apex. A terminal spikelet (arrow) is produced at the top of PBs in wild-type inflorescence (D) but a vestige of the rachis branch meristem (arrowhead) is observed in apo1-D mutants (E). F, Frequency of terminal spikelet formation on the PB. G, Total number of spikelets in an inflorescence. Vertical bars in C, F, and G indicate sem; n = 10 for wild type, apo1-D1, -D2, -D3, and -D4.
In wild-type rice inflorescences, growth of the rachis branches is determinate, that is to say, the rachis branch meristem transforms into a terminal spikelet meristem and terminates its activity (Fig. 4A). The timing of conversion of rachis branch meristem identity to the spikelet determines the number of lateral primordia produced on each branch (Fig. 4C, LP/PB, LP/SB). Although significant differences in the number of lateral primordia produced on each primary or secondary branch (LP/PB, LP/SB) were not observed in the apo1-D mutants (Fig. 4C), 40% to 80% of rachis branch meristems did not transform into a terminal spikelet and, instead, were left as a pedicel with panicle hair (Fig. 4, D–F). This finding implies that conversion of the meristem identity from a rachis branch meristem to a terminal spikelet meristem is retarded or suppressed in apo1-D mutants.
Among the meristems generated by a rachis branch meristem, the first few grow as next order rachis branches, whereas the rest directly acquire spikelet meristem identity and grow as lateral spikelets (Fig. 4A). Thus, the timing of the program shift in lateral meristem identity along a basal-apical direction, from next order rachis branch formation to spikelet formation, is also critical for determining the branching pattern and the inflorescence form. In apo1-D mutants, there was a clear tendency of a delayed program shift, and more lateral meristems grew to secondary branches rather than to lateral spikelets, resulting in an increase of the secondary rachis branches (SB/PB) and an decrease of lateral spikelets on a PB (LS/PB; Fig. 4C). The increase in the number of primary and secondary rachis branches was a main reason for increases in the total number of spikelets per branch as well as per inflorescence in the apo1-D mutants (Fig. 4, C and G). In summary, in apo1-D mutants, timing of all three major determinants of rice inflorescence architecture, namely, abortion of the IM activity, conversion from the rachis branch meristem to a terminal spikelet meristem, and the shift from next order rachis branch production to lateral spikelet production, is delayed.
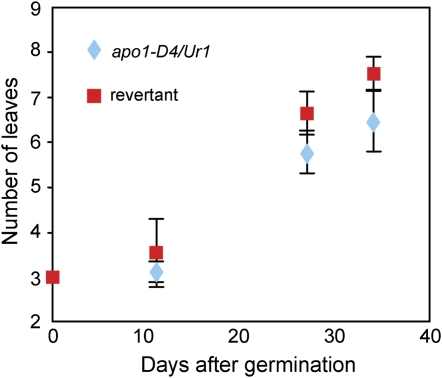
The Plastochron Is Prolonged in apo1-D Mutants
In addition to the pronounced phenotypes in inflorescence development, modest alterations of development were observed during the vegetative phase. We previously reported that loss of APO1 function causes the acceleration of leaf emergence (Ikeda et al., 2007). Contrary to this result, leaf number was slightly but significantly lower in apo1-D4 than that in the reverted plants at 35 d after germination (Fig. 5). These results indicated that the increase in APO1 transcripts confers a prolonged plastochron, the opposite phenotype of apo1. Moreover, the width of the flag leaf and the inflorescence axis of apo1-D4 were significantly larger than those of the wild type (data not shown).
Figure 5.
Prolonged plastochron in apo1-D4 mutants. A significant decrease in the plastochron length was observed in apo1-D4/Ur1 from very early stages of vegetative development.
Constitutive Overexpression of APO1 Caused a Significant Increase in Inflorescence Branching
To confirm that the increased level of APO1 expression was responsible for increases in inflorescence branching, we generated transgenic rice plants constitutively expressing APO1 under the control of the cauliflower mosaic virus 35S or maize UBIQUITIN promoters. Both promoters conferred essentially the same phenotypes. The most prominent phenotype observed in the transgenic plants was a significant increase in inflorescence branching. All four independent UBQ∷APO1 plants and 14 out of 28 independent 35S∷APO1 plants showed this strong hyperbranched phenotype. The inflorescence of transgenic plants with the severe phenotype remained enclosed by leaves due to suppressed stem elongation; removal of surrounding leaves revealed the presence of an extremely branched inflorescence (Fig. 6A). Close observation of the hyperbranched inflorescence revealed a large number of higher order branches (Fig. 6, B and C). Because the entire inflorescence remained undeveloped and packed so tightly it was not possible to count branch number accurately. To get around this problem, we made transverse sections of an internode immediately below the neck node and counted the number of vascular bundles that roughly reflect the number of PBs (Hoshikawa, 1989). apo1-D1 stem had more vascular bundles than wild type, indicating an increase in primary branches (Fig. 6, D–G). The stems of the UBQ∷APO1 plants were obviously thicker than apo1-D1 and wild-type stems and contained a highly increased number of vascular bundles (Fig. 6, H and I). Although at low frequency, plants with a mild phenotype that had tertiary and even higher order branches and more vascular bundles were also obtained (Fig. 6, J–L). The expression levels of APO1 are consistent with the observed degree of severity in the phenotypes (Fig. 6M).
Figure 6.
Ectopic over expression of APO1 caused an extreme increase in inflorescence branching. A to C, Different views of an inflorescence of 35S∷APO1. B and C, Higher magnification of the inflorescence shown in A; bar = 1 cm. D to I, Cross sections of inflorescence stems of wild type (‘MK1’, D and E), apo1-D1 (F and G), and 35S∷APO1 with severe phenotype (H and I). E, G, and I, Closer views of the stem enclosed with white boxes in D, F, and H, respectively. Vascular bundles are indicated with asterisks; bar = 0.5 mm. J, An inflorescence of 35S∷APO1 showing a mild phenotype. K and L, Vascular bundles of the 35S∷APO1 with mild phenotype are indicated with asterisks; bar = 0.5 mm. L, The close-up of the stem region enclosed with a white box in K. M, Expression level of APO1 analyzed by RT-PCR. The number indicates the numbers of PCR cycles. N, Young plants of a control containing an empty vector (‘Nipponbare’) and transgenic 35S∷APO1 and UBQ∷APO1; bar = 10 cm. O to Q, Tiller growth in the plants shown in N.
In addition to the inflorescence phenotype, ectopic APO1 expression affected other aspects of rice development. Suppression of tiller outgrowth was the next most prominent phenotype observed (Fig. 6, N–Q). With regard to this phenotype, the 35S∷APO1 plants showed moderate effects, whereas the UBQ∷APO1 plants allowed no or a few tillers to outgrow. Lumpy and wavy leaves were also observed in transgenic plants (Supplemental Fig. S1).
APO1 Promotes Cell Proliferation in the Meristem
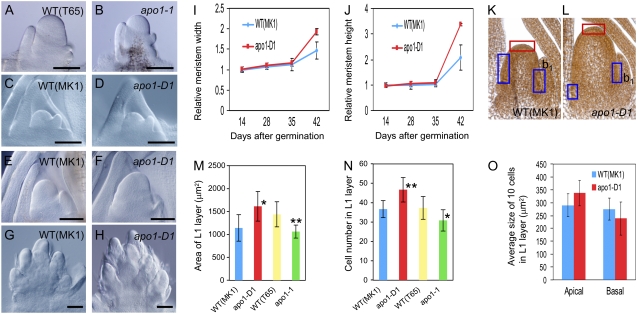
To further understand how APO1 controls meristem activity, we examined the IM of mutants in more detail. In wild-type plants, the SAM gradually increases in size after germination, accelerating growth at the late vegetative stage (Ito et al., 2005). Upon transition from the vegetative SAM to the reproductive IM, the meristem begins to enlarge rapidly in height and width, and starts to differentiate PB primordia. The apo1 SAM increases in size during the early vegetative phase, similar to wild type; however, enlargement of the SAM at the late vegetative stage is less vigorous than that in the wild type (Ikeda et al., 2005; Fig. 7, A and B). An opposite phenotype was observed in apo1-D mutants. The size of the SAM in apo1-D1 was comparable to that in the wild type during the vegetative phase, showing a gradual increase in height and width (Fig. 7, C, D, I, and J). apo1-D1 SAM started to increase in size more rapidly than the wild type upon the onset of reproductive development (Fig. 7, E, F, I, and J). The IM of apo1-D1 at the PB initiation stage was clearly larger both longitudinally and horizontally than that of wild type (Fig. 7, E–J). These observations implied that the level of APO1 activity was positively correlated with cell proliferation in the meristem.
Figure 7.
Influence of APO1 activity on cell proliferation in the IM. A to H, Cleared views of the vegetative SAM (C and D), IM (A, B, E, and F), and young inflorescences at the rachis branch initiation stage (G and H) of wild-type (WT; A, C, E, and G) apo1-1 (B) and apo1-D1 mutants (D–F); bars = 100 μm. I and J, Growth of the apical meristem. The width and height of the meristem were measured and expressed as a value relative to that of the meristem in 14-d-old seedlings (n = 7 to approximately 12). K and L, Sections of wild-type (K) and apo1-D1 (L) IM. Stage of the IM was estimated as the development of the panicle bract (b1). M, Comparison of areas of L1. N, Comparison of cell number in L1. O, Comparison of cell size in the apical region (red box in K and L) and basal region (blue boxes in K and L). Average of 10 cells is shown. M to O, n = 6 for wild type (MK1) and apo1-D1, n = 8 for wild type (T65) and apo1-1. Asterisks indicate significantly different from wild type at *, P < 0.05; **, P < 0.01; bars = 100 μm.
The difference in IMs between apo1, wild-type, and apo1-D mutants was examined more carefully to obtain clues for understanding how APO1 is involved in the control of inflorescence size. Growth of an inflorescence bract (b1 in Fig. 7, K and L) was used to check the correspondence of developmental stages of IMs used in this analysis. The area and number of cells in the L1 layer of the IM at the early primary branch initiation stage were significantly higher in apo1-D1 than in wild type (Fig. 7, K–N). Conversely, significant differences were not observed in cell sizes of both apical and basal regions of the IM between wild type and apo1-D1 (Fig. 7O).
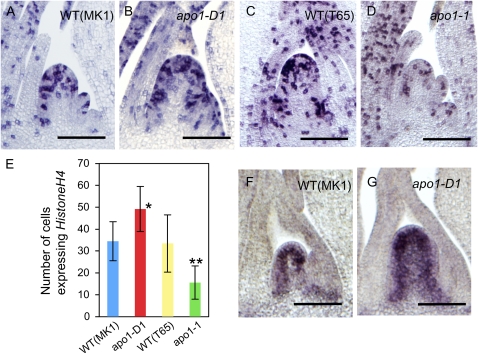
Patterns of cell division and distribution of meristematic cells were examined by in situ hybridization analysis using Histone H4 and OSH1 as probes (Fig. 8). The number of cells expressing Histone H4 was higher in apo1-D1 than in wild type, although there were fewer cells in apo1-1 than in wild type in accordance with the differences in IM size (Fig. 8, A–E). Gross patterns of Histone H4 expression were similar among wild type and the mutants. Likewise, in apo1-D1, more cells expressed OSH1 compared with that in wild-type IM, whereas the spatial pattern of OSH1, a marker for meristematic cells, was comparable (Fig. 8, F and G). Taken together, these analyses indicated that accelerated cell division mainly accounted for the increase in IM size in apo1-D.
Figure 8.
Pattern of cell proliferation in the IM. A to D, In situ hybridization analysis of Histone H4 expression in wild type (WT; ‘MK1’; A), apo1-D1 (B), wild type (T65; C), and apo1-1 (D). E, Number of cells expressing Histone H4. n = 6 for wild type (‘MK1’) and apo1-D1, n = 8 for wild type (T65) and apo1-1. F and G, In situ hybridization analysis of OSH1 expression in wild type (MK1; F) and apo1-D1 (G). Asterisks indicate significantly different from wild type at *, P < 0.05; **, P < 0.01; bars = 100 μm.
DISCUSSION
Here, we identified dominant gain-of-function mutant alleles of APO1. Analyses of the four dominant apo1-D alleles and transgenic plants expressing APO1 under the control of strong constitutive promoters have shown that the identity of a meristem as a spikelet negatively depends on APO1 expression level. A positive relationship between APO1 expression level and meristem size, especially cell number, suggested that APO1 controls the transition of meristem phases via control of cell proliferation in the meristem.
Genetic Networks Controlling Rice Inflorescence Branching
The rice inflorescence is classified as a panicle. The panicle is a branched raceme in which the branches are themselves racemes. Transition of meristem state to the spikelet meristem, which is a key event in determining the overall branching pattern of rice inflorescence, is delayed in apo1-D mutants. Furthermore, overexpression of APO1 severely repressed the transition to spikelet meristem. These observations are opposite to the phenotypes in the apo1 loss-of-function mutant, implying that the identity of a meristem as a spikelet negatively depends on the level of APO1 expression. Intriguingly, the function of APO1, suppression of spikelet (i.e. floral) identity in the meristem, is opposite to that of APO1 counterparts. All UFO orthologs in dicot species reported so far promote floral fates (Simon et al., 1994; Levin and Meyerowitz, 1995; Taylor et al., 2001; Zhang et al., 2003; Lippman et al., 2008; Souer et al., 2008).
Genetic analyses have shown that UFO acts with LFY (Lee et al., 1997). Recently, physical interactions between UFO and LFY in Arabidopsis and between DOT and ABERRANT LEAF AND FLOWER (ALF), the petunia LFY, in petunia have been demonstrated (Chae et al., 2008; Souer et al., 2008). The UFO/LFY complex is recruited to the promoter of the LFY target genes to induce transcription. Considering the high sequence similarities between RFL and LFY and between UFO and APO1, and a resemblance of inflorescence defects between the apo1 mutants and RFL knock-down plants (Kyozuka et al., 1998; Rao et al., 2008), we considered the possibility that RFL and APO1 also work together at least for inflorescence development. Based on the analogy of the interaction between UFO/LFY and DOT/ALF, it is likely that RFL function is required for APO1 to work. RFL mRNA accumulation is not observed in the SAM during the vegetative stage, but starts to localize in the SAM immediately after the transition to the reproductive phase that is coincident with the onset of excessive SAM proliferation in apo1-D mutants and 35S∷APO1 plants. Together, these results suggest a possible scenario of APO1, stimulating cell proliferation in the meristem with RFL, and because APO1 requires RFL to exert its effect, the impact of APO1 overexpression on the meristem is limited to the reproductive meristem.
In addition to the role as regulators of floral meristem identity, LFY/FLO act as direct activators of MADS-box genes required for stamen and carpel development (Blazquez and Weigel, 2000). The function as a positive regulators of class B and C functions is conserved in APO1/RFL (Chujo et al., 2003; Ikeda et al., 2007). This implies that the divergence in the roles that APO1/RFL and UFO/LFY play in the control of meristem identity may be achieved by differences in regulatory sequences in the downstream target genes rather than in the activity of RFL as a transcription factor. To reveal the molecular basis of the divergence in the function of APO1/RFL from that of UFO/LFY and their homologs, analysis of genes working downstream of APO1/RFL and comparison with downstream components of LFY pathway is needed (Busch et al., 1999; William et al., 2004; Saddic et al., 2006). It is also necessary to study interactions between APO1/RFL and other putative orthologs of known meristem identity genes to gain a comprehensive view of mechanisms controlling rice inflorescence development.
APO1 Promoter and Rice Inflorescence Form
Here we demonstrated that an increased level of APO1 expression in apo1-D alleles was caused by the insertion of nDart1-0 in the APO1 promoter. This suggests that a cis-element(s) involved in the negative regulation of APO1 expression might exist in the 3.5-kb upstream region of APO1. The finding that the nDart1-0 insertion locates within a short region of 35 bp in the dominant Apo1-D alleles suggests two possibilities: (1) this region might be a hot spot for nDart1-0 insertion or (2) this is the only region where insertion of nDart1-0 causes an enhancement of APO1 expression. Because the nucleotide sequence around the −3.5-kb position of APO1 gene does not share significant similarities with that of other nDart1-0 insertions (Tsugane et al., 2006), the first hypothesis is unlikely. We favor the second explanation, implying that the insertion of nDart1-0 to this particular sequence was obligatory to achieve the enhancement of APO1 expression that can lead to increases in inflorescence size.
Recent works have shown that alterations in the spatial and temporal expression patterns of a few regulatory genes generate a dramatic divergence of inflorescence form (Prusinkiewicz et al., 2007; Lippman et al., 2008; Rebocho et al., 2008). In petunia and tomato (Solanum lycopersicum), which form cymose inflorescences, fine tuning of spatial and temporal distribution of EVERGREEN (EVG) and Compound inflorescence (S) expressions is crucial to form the cymose inflorescence. EVG and S exert their functions through the control of expression patterns of DOT and AN, UFO orthologs. It will be interesting to test if alterations in APO1 expression levels through the modification of this sequence have generated diversity in grass inflorescence morphology (Malcomber et al., 2006). Future analysis of APO1 function in other grass species will provide additional insight into the involvement of APO1 in the evolution of inflorescence form in grass species.
Molecular Function of APO1
In addition to the negative control of spikelet meristem identity, APO1 likely plays a positive role to maintain the IM activity. In normal rice inflorescence development, the IM ceases its activity before acquiring the next identity, thus, the number of PBs mostly depends on the timing of the IM abortion. In apo1 loss-of-function mutants, the IM is converted to a spikelet identity (Ikeda et al., 2005, 2007). This supports the notion that the rice IM is genetically programmed to be finally converted to the spikelet meristem; however, this program is not completed due to the abortion that takes place prior to the transition in apo1 loss-of-function mutants. In spite of a marked increase in the number of primary branches, the vestige of the IM was observed in apo1-D mutants. This indicates that increased levels of APO1 expression caused the delay of meristem abortion in apo1-D mutants. This finding raises the attractive possibility that APO1 not only prevents the transition of meristem phase to spikelet meristem, but also plays a more general role that is concerned with control of meristem activity.
There is a clear positive relationship between APO1 expression levels and meristem size at the reproductive phase. This result raised the possibility that the increase in PB number in apo1-D mutants likely reflects the increased IM size. Similar relationships between meristem size and inflorescence branching have been described in several mutants including knotted1 in maize and log in rice, both exhibit reduced meristem size and inflorescence branching (Kerstetter et al., 1997; Vollbrecht et al., 2000; Kurakawa et al., 2007). A fascinating question that needs to be answered is how APO1 regulates the meristem size at the early stage of inflorescence development.
Meristem Size and Plastochron
In contrast to the positive relationship between APO1 expression level and meristem size, we showed a negative correlation between the length of the plastochron and APO1 level. A shortened plastochron was observed in apo1, whereas the plastochron was slightly lengthened in apo1-D mutants. This suggests that the timing of leaf emergence depends on the level of APO1 expression. A consequence of meristem size and plastochron is often discussed because meristem size tends to be altered in plastochron mutants (Wang et al., 2008); however, the relationship is complex and there is no general rule. In higher plants, cells in the peripheral zone of meristems undergo differentiation and become incorporated into the leaf primordium on the meristem flanks. One possible explanation might be that more frequent recruitment of leaf founder cells in apo1 loss-of-function mutants than in wild type due to accelerated leaf initiation resulted in a smaller number of cells in the apo1 SAM. However, although the altered plastochron was observed from the early stage of development, the alteration of meristem size became evident from the late stage of vegetative development or after the transition to reproductive phase. Therefore, it is unlikely that the change in meristem size is a direct consequence of the altered plastochron in apo1 mutants. Nevertheless, we cannot completely rule out the possibility that the effect of the shortage or surplus of cells becomes evident only at later stages of development.
MATERIALS AND METHODS
Plant Material and Genetics
Plants were grown in pots under natural conditions or in a growth chamber set at 13 L (28°C)/11 D (25°C). An nDart1-0-promoted mutant collection derived from MK1 selected in the cross between the japonica variety ‘Matsumoto-mochi’ and ‘Shiokari’ variegated pyl NIL carrying nDart1-0 and an active autonomous transposable element was used for the mutant screening. The primers used for amplification to identify the revertant plants were APOPF (5′-GCCTGCTATCGTGTCACTCA-3′) and APOPR (5′-GAGCTCGTGGGACTTAC-3′).
Amplification of nDart1-0-Flanking Sequences by Inverse PCR
Inverse PCR (Hartl and Ochman, 1996) was employed to obtain sequences flanking the transposed nDart1-0 in ‘MK1’. Five micrograms of genomic DNA isolated from a single plant were digested overnight with AluI whose restriction site at base position 173 is specific for nDart1-0. One-hundred nanograms of the purified, digested DNA was self ligated with T4 DNA ligase (Promega) in a 20 μL reaction volume at 16°C for 60 min. One microliter of the self-ligated DNA was used as a template for a first round of PCR with a set of primers, inv3 (5′-GGCCGTGCCGGCTACAGGTTCC-3′) and inv5 (5′-CGGGCCGGCACGGCACGGCACA-3′). The PCR reaction was performed at 94°C for 2 min, 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 90 s, with a final cycle of 72°C for 5 min. The product of the first round of PCR amplification was diluted 100 times with water and used for the second PCR. The second PCR was performed using 1 μL of the diluted product with inv4 (5′-TACAGGTTCCGCCGTGCCTCGGG-3′) and inv6 (5′-CGGCACGGCACGGCACAGCT-3′). The PCR conditions were identical to those used for the first round PCR except for decreasing the cycle number to 24 cycles. Amplified DNA fragments were run on a gel, purified, and sequenced.
Identification of Reverted Plants
Germinal revertants were screened from about 400 apo1-D4 homozygous siblings by PCR. The primers used for amplification were APK3F (5′-GCCTGCTATCGTGTCACTCA-3′) and APK3-1R (5′-CATGTCTGCACTCTGCACCT-3′), spanning the nDart1-0 inserted region. For revertant candidate plants, further PCR was performed using another set of primers, APK3F (5′-GCCTGCTATCGTGTCACTCA-3′) and APK3R (5′-GAGCTCGTGGGACTTAC-3′) to confirm that the amplified bands are derived from the APO1 promoter region and nDart1-0 was deleted. The genotype and phenotype of the revertant plants were further confirmed in the next generation.
Histological Analysis
For paraffin sectioning, samples were fixed with formalin:glacial acetic acid:70% ethanol (1:1:18) at 4°C overnight, and dehydrated in a graded ethanol series. Following substitution with xylene, samples were embedded in Paraplast Plus (Oxford Labware) and sectioned 8-μm thick using a rotary microtome. Sections were stained with hematoxylin and observed under a light microscope.
Samples were fixed overnight in formalin:glacial acetic acid:70% ethanol (1:1:18) for clearing of seedlings and young inflorescences. After dehydration in a graded ethanol series, samples were transferred into BB4-1/2 clearing fluid (Herr, 1982). The cleared samples were observed using a microscope (IX70; Olympus Optical) equipped with Nomarski differential interference contrast optics. The width of the shoot apex was measured above the P1 primordium insertion, and the height was the shortest distance from the line used for measuring the width to the tip of the apex.
In Situ Hybridization
Tissues were fixed with 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 m sodium phosphate buffer, dehydrated through a series of butanol extractions, and embedded in Paraplast Plus. Microtome sections were applied to glass slides treated with Vectabond (Vector Laboratories). Digoxygenin-labeled antisense probes were prepared as described previously (Ikeda et al., 2007). Hybridization and immunological detection with alkaline phosphatase were performed as described in Kouchi and Hata (1993).
Quantitative RT-PCR
RNA extraction and cDNA synthesis were performed as described previously (Ikeda et al., 2007). The primer sets used to amplify APO1 and ACTIN1 cDNAs were, APO1F (5′-GTCATCTGAGTTGGTAGTGTG-3′) and APO1R (5′-CAACAGATCTCATGGCAAG-3′), ACTF (5′-CATTCCAGCAGATGTGGATTG-3′) and ACTR (5′-TCTTGGCTTAGCATTCTTGG-3′), respectively. The relative abundances were estimated with a LightCycler system (Roche Applied Science) using ACTIN1 gene expression as a reference for normalization.
Generation of Transgenic Plants
A genomic region of the APO1 gene containing the entire coding region was amplified from genomic DNA of ‘Nipponbare’ using a set of primers, APO1-150F (5′-GCCTCACTCCACTCCACTTC-3′) and APO1+1437R (5′-CCCCATTGCTGAAACAACTT-3′). The amplified fragment was cloned into pGEM-T Easy (Promega) and sequenced. Subsequently, an approximately 1.6-kb fragment was cut out with EcoRI and cloned into pMSH1, a binary vector carrying a cauliflower mosaic virus 35 S promoter (Kawasaki et al., 1999). Rice (Oryza sativa) transformation was carried out as described (Nakagawa et al., 2002).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB292777 (APO1) and AB116151 (nDart1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Wide and waving leaves in plants ectopically expressing APO1.
Supplementary Material
Acknowledgments
We thank H. Nishimura for his technical assistance.
This work was supported by a grant from the Promotion of Basic Research Activities for Innovative Biosciences (to M.M.), by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation grant no. IPG–0001 to J.K.), and by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Scientific Research on Priority Areas to J.K.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Junko Kyozuka (akyozuka@mail.ecc.u-tokyo.ac.jp).
The online version of this article contains Web-only data.
References
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309 741–745 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D (2000) Integration of floral inductive signals in Arabidopsis. Nature 404 889–892 [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005. a) Genetic and evolution of grass inflorescence and flower development. Plant Cell Physiol 46 69–78 [DOI] [PubMed] [Google Scholar]
- Bommert PB, Lunde C, Nardmann J, Vollbrecht E, Running MP, Jackson D, Hake S, Werr W (2005. b) thick tassel dwarf1 encodes a putative maize orthologue of the Arabidopsis CLAVATA1 leucine-rich receptor-like kinase. Development 132 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rochefold T, Martienssen R, Hake S (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristem of maize. Plant Cell 18 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E, Hake S (2007) Flowering and determinacy in maize. J Exp Bot 58 909–916 [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of floral homeotic gene in Arabidopsis. Science 285 585–587 [DOI] [PubMed] [Google Scholar]
- Chae E, Tan QKG, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135 1235–1245 [DOI] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al (2006) The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S (2007. a) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39 544–549 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135 3013–3019 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S (2007. b) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet. Nat Genet 39 1517–1521 [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298 1238–1241 [DOI] [PubMed] [Google Scholar]
- Chujo A, Chu Y, Kishino H, Shimamoto K, Kyozuka J (2003) Partial conservation of LFY function between rice and Arabidopsis. Plant Cell Physiol 44 1311–1319 [DOI] [PubMed] [Google Scholar]
- Hartl DL, Ochman H (1996) Inverse polymerase chain reaction. Methods Mol Biol 58 293–301 [DOI] [PubMed] [Google Scholar]
- Herr JM (1982) An analysis of methods for permanently mounting ovules cleared in four-and-a-half type clearing fluids. Stain Technol 57 161–169 [DOI] [PubMed] [Google Scholar]
- Hoshikawa K (1989) The Growing Rice Plant. Nosan Gyoson Bunka Kyokai, Tokyo
- Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51 1030–1040 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y (2005) ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol 282 349–360 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn GW, Coen ES (1995) Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish EE (1997) Experimental analysis of tassel development in the maise mutant Tassel Seed 6. Plant Physiol 114 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46 23–47 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Laudensia D, Smith L, Hake S (1997) Loss-of-function mutants in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124 3045–3054 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130 3841–3850 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238 106–119 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin activating enzyme. Nature 455 652–655 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K (1998) Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci USA 95 1979–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Lee J, Moon S, Park SY, An G (2007) The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49 64–78 [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7 95–104 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44 425–481 [Google Scholar]
- McSteen P, Laudencia-Chingucuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5 61–66 [DOI] [PubMed] [Google Scholar]
- Murai M, Iizawa M (1994) Effects of major genes-controlling morphology of panicle in rice. Breed Sci 44 247–255 [Google Scholar]
- Nagao S, Takahashi M (1963) Trial construction of twelve linkage groups in Japanese rice: genetical studies on rice plants, XXVII. Mem Fac Agr Hokkaido Univ 53 72–130 [Google Scholar]
- Nagao S, Takahashi M, Kinoshita T (1958) Inheritance on a certain ear type in rice: genetical studies on plant, XXIII. Mem Fac Agr Hokkaido Univ. 3 38–47 [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29 743–750 [DOI] [PubMed] [Google Scholar]
- Prasad K, Kushalappa K, Vijayraghavan U (2003) Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence. Mech Dev 120 491–502 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007) Evolution and development of inflorescence architectures. Science 316 1452–1456 [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105 3646–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebocho AB, Bliek M, Kusters E, Castel R, Procissi A, Roobeek I, Souer E, Koes R (2008) Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev Cell 15 437–447 [DOI] [PubMed] [Google Scholar]
- Sablowski R (2007) Flowering and determinacy in Arabidopsis. J Exp Bot 58 899–907 [DOI] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D (2006) The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133 1673–1682 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441 227–230 [DOI] [PubMed] [Google Scholar]
- Shepard K, Purugganan MD (2002) The genetics of plant morphogenesis evolution. Curr Opin Plant Biol 5 49–55 [DOI] [PubMed] [Google Scholar]
- Simon R, Carpenter R, Doyle S, Coen E (1994) Fimbriata controls development by mediating between meristem and organ identity genes. Cell 78 99–107 [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Blieck M, Kusters E, de Bruin RAM, Koes R (2008) Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20 2033–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van der Krol AR, Kloos D, Spelt C, Bliek M, Mol J, Koes R (1998) Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125 733–742 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131 5649–5657 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY (2006) Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol 47 1591–1602 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Hofer J, Murfet I (2001) Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane K, Maekawa M, Takagi K, Takahara H, Qian Q, Eun CH, Iida S (2006) An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J 45 46–57 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 3161–3172 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen RM (2005) Architecture of floral branch. Nature 436 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59 253–279 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859 [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sandal N, Polowick PL, Stiller J, Stougaard J, Fobert PR (2003) Proliferating Floral Organs (Pfo), a Lotus japonicas gene required for specifying floral meristem determinacy and organ identity, encodes an F-box protein. Plant J 33 607–619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.