Abstract
GLYCOGEN SYNTHASE KINASE3 (GSK3) is a highly conserved serine/threonine kinase involved in a variety of developmental signaling processes. The Arabidopsis (Arabidopsis thaliana) genome encodes 10 GSK3-like kinases that are clustered into four groups. Forward genetic screens have so far uncovered eight mutants, all of which carry gain-of-function mutations in BRASSINOSTEROID-INSENSITIVE2 (BIN2), one of the three members in group II. Genetic and biochemical studies have implicated a negative regulatory role for BIN2 in brassinosteroid (BR) signaling. Here, we report the identification of eight ethyl methanesulfonate-mutagenized loss-of-function bin2 alleles and one T-DNA insertional mutation each for BIN2 and its two closest homologs, BIN2-Like1 and BIN2-Like2. Our genetic, biochemical, and physiological assays revealed that despite functional redundancy, BIN2 plays a dominant role among the three group II members in regulating BR signaling. Surprisingly, the bin2bil1bil2 triple T-DNA insertional mutant still responds to BR and accumulates a more phosphorylated form of a BIN2 substrate than the wild-type plant. Using the specific GSK3 inhibitor lithium chloride, we have provided strong circumstantial evidence for the involvement of other Arabidopsis GSK3-like kinases in BR signaling. Interestingly, lithium chloride treatment was able to suppress the gain-of-function bin2-1 mutation but had a much weaker effect on a strong BR receptor mutant, suggesting the presence of a BIN2-independent regulatory step downstream of BR receptor activation.
GLYCOGEN SYNTHASE KINASE3 (GSK3) is a highly conserved Ser/Thr kinase that is implicated in a wide range of cellular and developmental processes (Woodgett, 2001). In mammals, GSK3 has only two isoforms, GSK3α and GSK3β. By contrast, many plant species contain a much larger set of GSK3-like kinases (Richard et al., 2005; Yoo et al., 2006). Arabidopsis (Arabidopsis thaliana) has 10 GSK3-like kinases, also known as AtSKs for Arabidopsis SHAGGY-like protein kinases, that can be classified into four subgroups based on phylogeny analysis (Jonak and Hirt, 2002). Genetic, transgenic, and biochemical approaches have implicated plant GSK3-like kinases in a variety of plant signaling processes, including flower development, stress/wounding responses, and hormone signaling (Jonak and Hirt, 2002). For example, transgenic Arabidopsis plants with reduced transcript levels of AtSK1-1 and AtSK1-2 contain increased numbers of floral organs and exhibit abnormal apical-basal patterning in gynoecium (Dornelas et al., 2000), whereas overexpression of mutated AtSK3-2 displayed smaller floral organs (Claisse et al., 2007). It was also known that wounding can activate an alfalfa (Medicago sativa) GSK3 kinase (Jonak et al., 2000) and that overexpression of AtSK2-2 can complement a salt stress-sensitive mutant in yeast and results in enhanced salt resistance in Arabidopsis (Piao et al., 1999, 2001).
The best studied Arabidopsis GSK3-like kinase is BIN2/UCU1/DWF12/AtSK2-1, which was believed to regulate the signal transduction of brassinosteroids (BRs; Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), a unique class of plant steroid hormones essential for plant growth and development (Clouse and Sasse, 1998). Genetic and biochemical studies in the past decade have revealed a linear signaling pathway that uses protein phosphorylation to transmit BR signals from the cell surface into the nucleus (Li, 2005; Li and Jin, 2007). In the absence of BR, BIN2 is constitutively active to phosphorylate BES1 and BZR1 (He et al., 2002; Yin et al., 2002; Zhao et al., 2002), two highly similar transcriptional factors (He et al., 2005; Yin et al., 2005), resulting in their degradation, cytoplasmic localization, and reduced DNA-binding activities (He et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Bai et al., 2007; Gampala et al., 2007; Ryu et al., 2007). BR binding to its cell surface receptor BRI1 triggers a rapid dissociation of BKI1, a putative BRI1 substrate (Wang and Chory, 2006), as well as dimerization and transphosphorylation with BAK1, a suspected BRI1 coreceptor (Li et al., 2002; Nam and Li, 2002; Russinova et al., 2004; Wang et al., 2005, 2008). These membrane events lead to BIN2 inhibition, via protein degradation (Peng et al., 2008) and/or some yet to be discovered biochemical mechanisms, thus relieving its inhibitory effects on BES1 and BZR1 that regulate the expression of BR-responsive genes (Li and Jin, 2007).
Most of our knowledge about BIN2 function came mostly from gain-of-function results. Genetic screens in Arabidopsis for BR-insensitive dwarf mutants or leaf development mutants resulted in the isolation of eight gain-of-function bin2 alleles (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), seven of which carry mutations in the highly conserved TREE (Thr-261-Arg-262-Glu-263-Glu-264) motif within the BIN2 catalytic domain (Peng and Li, 2003). In contrast, similar genetic screens identified 30 loss-of-function alleles of BRI1 (Clouse et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000; Bouquin et al., 2001; Tanaka et al., 2005; Xu et al., 2008). As a matter of fact, up to now no single loss-of-function allele of any GSK3-like kinase has been discovered in any forward genetic screen in Arabidopsis, implying that the Arabidopsis GSK3-like kinases most likely function redundantly to regulate plant development. Expression profiling results seem to support this hypothesis. Real-time PCR analysis showed that eight of the 10 AtSK genes displayed a “pseudoconstitutive” expression pattern under several different growth conditions (Charrier et al., 2002). A recent study using T-DNA insertional mutants of BIN2 and its two closest homologs concluded that the three group II AtSKs function redundantly in BR signaling (Vert and Chory, 2006). However, it remains unknown why no single gain-of-function mutation in the two BIN2 homologs was discovered in the forward genetic screens and if other Arabidopsis GSK3-like kinases might also participate in BR signaling.
In this study, we conducted a genetic screen looking for second site mutations that suppress the weak dwarf phenotype of the Arabidopsis ucu1-3 mutant and identified eight new loss-of-function bin2 alleles. Through genetic and pharmacological experiments using a well-known GSK3 inhibitor, Li+ (Klein and Melton, 1996; Stambolic et al., 1996), we discovered that BIN2 plays a major role among the group II AtSKs in regulating BR signaling and that other AtSKs can also phosphorylate BIN2 substrates. Our Li+ experiments also suggested the existence of a BIN2-independent regulatory mechanism in the BR signaling pathway.
RESULTS
Isolation of bin2 Loss-of-Function Mutants
Unlike BRI1, which was discovered through recessive loss-of-function mutants, all of the bin2 mutants uncovered from forward genetic screens carry semidominant gain-of-function alleles (Peng and Li, 2003). To test if the wild-type BIN2 plays a role in BR signaling, we carried out a suppressor screen using a weak bin2 mutant, ucu1-3 (Fig. 1A), which can produce many seeds suitable for a large-scale genetic screen. Although ucu1-3 was previously described as a weak recessive mutant (Perez-Perez et al., 2002), transformation of a genomic BIN2 construct carrying the ucu1-3 (P284S) mutation into wild-type Arabidopsis plants resulted in many dwarf plants similar to the bin2-1 mutant (Fig. 1B), indicating that ucu1-3 carries a weak gain-of-function bin2 allele. We reasoned that a suppressor screen in the ucu1-3 background could lead to identification of not only extragenic suppressors but also intragenic loss-of-function mutations that abolish the BIN2 kinase activity.
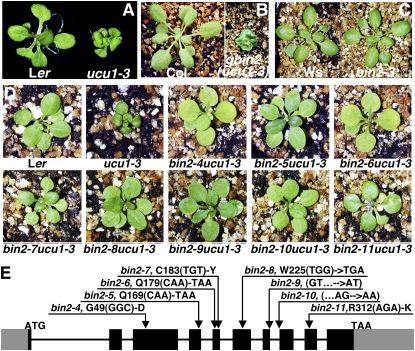
Figure 1.
Identification of bin2 loss-of-function mutants. A, ucu1-3 exhibits a weak bin2 phenotype. Shown here are 3-week-old wild-type and ucu1-3 mutant plants (both are in ecotype Ler) grown on half-strength MS medium in the light. B, ucu1-3 is a weak gain-of-function bin2 mutation. Shown here are a 5-week-old wild-type plant (left) and a transgenic plant of the same age (right) that expresses a mutated genomic gbin2 construct carrying the ucu1-3 mutation and displays the strong bin2-like phenotype (both are in ecotype Col-0). C, Phenotypic comparison between a 4-week-old wild-type plant and a 4-week-old T-DNA insertional bin2 mutant (both are in ecotype Ws). D, Screening for ucu1-3 suppressors identified eight bin2 loss-of-function mutations. Shown here from left to right in the top row are a Ler wild-type plant, ucu1-3, and bin2-4 to bin2-6 mutants and from left to right in the bottom row are bin2-7 to bin2-11 mutants (all grown in soil for 4 weeks). E, Molecular nature of eight newly discovered bin2 alleles. Black boxes represent protein-coding exons, gray boxes indicate noncoding regions, and thin lines denote introns. Arrows show the positions of the eight single nucleotide bin2 mutations, while ATG and TAA denote the start and stop codons, respectively.
Screening a total of 200,000 M2 seeds derived from 20,000 ethyl methanesulfonate-mutagenized M1 plants resulted in the identification of 50 putative suppressors. Genetic studies and PCR-based mapping of 12 putative suppressors revealed that at least eight of them are intragenic suppressors (Fig. 1D), which were confirmed by sequencing analysis of their entire BIN2 genes, each containing an additional single nucleotide change besides the ucu1-3 mutation. Therefore, we name them bin2-4 to bin2-11. As shown in Figure 1E, bin2-5, bin2-6, and bin2-8 each contain a nonsense mutation, while bin2-9 and bin2-10 each carry a single nucleotide mutation at exon/intron and intron/exon junctions of the ninth intron, respectively, most likely leading to defective RNA splicing and premature translational termination. The remaining three mutants, bin2-4, bin2-7, and bin2-11, have missense mutations at Gly-49, Cys-183, and Arg-312, respectively. Gly-49 and Arg-312 are absolutely conserved in the catalytic domain of all known Ser/Thr kinases, while Cys-183 is immediately adjacent to the absolutely conserved DFG motif of the Ser/Thr kinase activation fragment (Hanks et al., 1988). Thus, these mutations most likely lead to complete loss of the BIN2 kinase activity. All of these intragenic suppressors were morphologically indistinguishable from the corresponding wild-type plants (ecotype Landsberg erecta [Ler]; Fig. 1D), suggesting that BIN2 functions redundantly with one or more AtSKs in regulating BR signaling. We also obtained a T-DNA insertional mutant in ecotype Wassilewskija (Ws), FLAG_593C09, from the FLAGdb/FST collection (Samson et al., 2002), which was previously reported as bin2-3 (Vert and Chory, 2006). Similar to all the bin2 loss-of-function mutants of ecotype Ler, bin2-3 exhibits no obvious morphological defect compared with the corresponding Ws wild-type plant (Fig. 1C).
Analysis of bin2-3 Mutants Confirms a Regulatory Role of BIN2 in BR Signaling
Sensitized genetic backgrounds have been used widely to detect redundant gene function. To test if BIN2 plays a role in BR signaling, we used a sensitized genetic background of the bri1-5 mutation that causes endoplasmic reticulum retention of a functionally competent BR receptor (Hong et al., 2008). Because bri1-5 is in ecotype Ws, we used bin2-3 to generate the bin2-3bri1-5 double mutant to avoid any ecotype effect on individual F2 plants. As shown in Figure 2A, the bin2-3bri1-5 double mutant has a bigger rosette with visible petioles and flatter leaves than the bri1-5 mutant. The partial suppression of bri1-5 by loss of BIN2 activity confirmed that BIN2 plays a negative role in BR signaling.
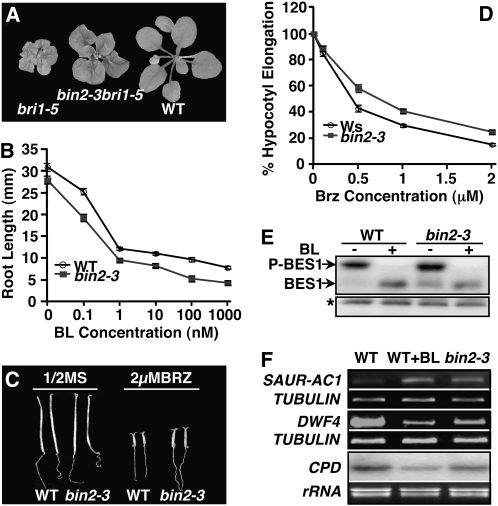
Figure 2.
Loss of BIN2 function stimulates BR signaling. A, The T-DNA insertional bin2-3 mutation partially suppresses the bri1-5 mutation. Shown from left to right are 4-week-old soil-grown plants of bri1-5, bin2-3bri1-5, and the Ws wild-type (WT) control. B, Root lengths of 2-week-old seedlings of the wild type and bin2-3 grown on half-strength MS medium containing no or increasing concentrations of BL. C, Six-day-old etiolated seedlings of the Ws wild type and bin2-3 grown on medium containing no or 2 μm Brz. D, Hypocotyl elongation of wild-type and bin2-3 seedlings grown on medium containing different concentrations of Brz relative to seedlings of the same genotype grown on regular medium (see “Materials and Methods” for details). For both B and D, each data point represents an average of 60 seedlings of two duplicate experiments. Error bars represent se. E, Western blot analysis of BES1 phosphorylation status. Eighteen-day-old seedlings were treated with 1 μm BL or mock solution for 1 h. Total proteins were extracted and analyzed by western blotting with anti-BES1 antibody. BES1-P is the phosphorylated BES1, and the asterisk indicates a nonspecific band for a loading control. F, RT-PCR and northern-blot analyses of SAUR-AC1, DWF4, and CPD gene expression in 18-d-old seedlings treated with or without 1 μm BL. See “Materials and Methods” for experimental details. β-TUBULIN was used as a control for RT-PCR analysis of both SAUR-AC1 and DWF4, while ethidium bromide staining of rRNAs served as a loading control for northern-blot analysis of CPD.
Consistent with our genetic result, bin2-3 mutants exhibit enhanced BR sensitivity measured by BR-induced root growth inhibition and resistance to brassinazole (Brz), a specific inhibitor of BR biosynthesis (Asami et al., 2000). As shown in Figure 2B, the root growth of bin2-3 mutants was slightly more inhibited than that of wild-type seedlings by brassinolide (BL), the most active member of the BR family, giving rise to a slightly down-shifted dose-response curve. Figure 2, C and D, revealed that hypocotyl elongation of dark-grown bin2-3 seedlings was less prohibited by Brz compared with wild-type seedlings, producing a slightly up-shifted dose-response curve.
Enhanced BR sensitivity of bin2-3 was also observed at both biochemical and molecular levels. Previous studies have shown that BR treatment leads to rapid dephosphorylation of BES1, a presumed BIN2 substrate (Mora-Garcia et al., 2004). As revealed by Figure 2E, wild-type plants predominantly accumulate the hyperphosphorylated form of BES1, and BL treatment results in rapid disappearance of the hyperphosphorylated BES1 accompanied by accumulation of hypophosphorylated BES1, which has a faster electrophoretic mobility on western blot. Interestingly, without BR treatment, bin2-3 mutants accumulate an easily detectable amount of hypophosphorylated BES1 compared with wild-type control plants grown under the same growth conditions (Fig. 2E), suggesting that loss of BIN2 results in enhanced BR signaling. We also measured the transcript levels of two BZR1 target genes, CPD and DWF4 (He et al., 2005), and a BES1 target gene, SAUR-AC1 (Yin et al., 2005), by northern-blot and reverse transcription (RT)-PCR analyses. Consistent with previous reports (Mathur et al., 1998; Goda et al., 2002), 1 h of BL treatment stimulated the expression of SAUR-AC1 but inhibited the accumulation of transcripts of both CPD and DWF4 genes (Fig. 2F). Interestingly, without BR treatment, bin2-3 mutants accumulated more SAUR-AC1 transcripts but lower levels of DWF4 and CPD mRNAs than wild-type plants. Taken together, our results demonstrated that the bin2-3 knockout mutant exhibits measurable physiological and molecular phenotypes despite the lack of any visible morphological defect in a BRI1+ background, confirming that BIN2 is an important negative regulator of the BR signaling pathway.
The Two Closest Homologs of BIN2 Are Also Involved in BR Signaling
Although our genetic, physiological, and biochemical assays demonstrated that BIN2 is a negative regulator of the BR signaling pathway, the lack of morphological defects of all isolated bin2 loss-of-function mutants is in sharp contrast to the severe BR-insensitive dwarf phenotype of most previously characterized bin2 gain-of-function mutants. This is likely caused by functional redundancy between BIN2 and one or more Arabidopsis GSK3-like kinases. Based on sequence similarity, BIN2 and its two closest homologs, AtSK2-2 and AtSK2-3 (hereafter named BIL1 and BIL2 for BIN2-Like1 and BIN2-Like2, respectively), belong to group II of the Arabidopsis AtSKs (Jonak and Hirt, 2002). Amino acid sequence alignment revealed that the two highly similar BILs share extensive sequence identity with BIN2, despite having extra amino acids at their N-terminal ends and different C-terminal tails (Fig. 3A), suggesting that both BIL1 and BIL2 might also be involved in BR signaling.
Figure 3.
Both BIL1 and BIL2 are capable of blocking BR signaling. A, Sequence alignment of BIN2, BIL1, and BIL2. The sequence alignment was done by the Mcoffee program at http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi, and identical/similar amino acids are shaded with black/gray backgrounds using the BoxShade Server at http://www.ch.embnet.org/software/BOX_form.html. Eight gain-of-function bin2/ucu1/dwf12 mutations are indicated by black arrows, and the conserved TREE motif is boxed in red. B, Both BIL1 and BIL2 are able to phosphorylate BES1 in vitro. The top panel shows Coomassie Brilliant Blue staining of purified MBP-AtSK and GST-BES1 fusion proteins, and the bottom panel shows an autoradiograph of 32P-labeled AtSKs and GST-BES1. C, Both BIL1 and BIL2, when carrying a mutation equivalent to bin2-1, can inhibit BR signaling. Shown clockwise are transgenic plants expressing a wild-type gBIN2 genomic construct, a mutated gbin2-1 transgene carrying the bin2-1 mutation, a genomic gBIL2 transgene with an E293K mutation, and a genomic gBIL1 construct with an E295K mutation. Percentages at bottom right indicate the percentage of T1 transgenic plants (of more than 150 lines) exhibiting each displayed morphology.
To directly test this possibility, we first expressed both BIL1 and BIL2 as fusion proteins with MALTOSE-BINDING PROTEIN (MBP) and measured their in vitro kinase activities toward BES1 expressed also in Escherichia coli as a fusion protein with glutathione-S-transferase (GST). As shown in Figure 3B, both MBP-BIL1 and MBP-BIL2 displayed similar GST-BES1 phosphorylation activity as the MBP-BIN2 fusion kinase. Since the three MBP-fused kinases failed to phosphorylate the GST tag itself (data not shown), our results demonstrated that the two BILs are biochemically capable of phosphorylating known BIN2 substrates in vitro.
We also took a gain-of-function approach and overexpressed either a mutated gbil1(E295K) or gbil2(E293K) gene driven by their native promoters in wild-type Arabidopsis plants. Both E295K and E293K mutations correspond to the gain-of-function E263K mutation responsible for the bin2-1 dwarf phenotype (Li and Nam, 2002; Fig. 3A). As shown in Figure 3C, some of the gbil1(E295K) and gbil2(E293K) transgenic plants are morphologically similar to the bin2-1 mutant or gbin2-1 transgenic plants overexpressing a mutated BIN2 gene that carries the bin2-1 mutation. It is interesting that while almost 100% of the gbin2-1 plants exhibited the bin2-1 phenotypes, only 35% of the gbil1(E295K) transgenic seedlings and 28% of the gbil2(E293K) transgenic plants (of more than 150 transgenic lines for each transgene) were bin2 like (Fig. 3C). These results suggested that both BIL1 and BIL2 are capable of blocking BR signaling, albeit with lower efficiency when the conserved acidic residue (Glu-295 in BIL1 and Glu-293 in BIL2) is changed to a basic residue.
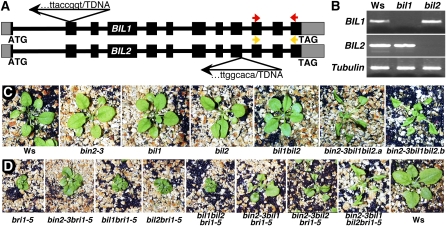
To confirm their involvement in BR signaling, we screened and obtained one T-DNA insertional mutant in the Ws ecotype for either BIL gene from the Arabidopsis Knockout Facility at the University of Wisconsin-Madison (Fig. 4A). RT-PCR analysis revealed that bil1 and bil2 mutants contained no detectable levels of BIL1 and BIL2 transcripts, respectively, suggesting that both bil1 and bil2 are likely null mutants (Fig. 4B). Similar to all of the bin2 loss-of-function mutants, neither single nor double mutants of bil1 and bil2 displayed any visible morphological/growth defect (Fig. 4C, third, fourth, and fifth plants). We also generated double and triple mutants of bil1, bil2, and bri1-5. Unlike bin2-3bri1-5, which exhibited a partial suppression phenotype, bil1bri1-5 and bil2bri1-5 double or bil1bil2bri1-5 triple mutants are morphologically indistinguishable from bri1-5 (Fig. 4D, third, fourth, and fifth plants).
Figure 4.
Genetic analysis of T-DNA insertional mutations of BIN2 and its two closest homologs. A, Schematic representation of T-DNA insertions in the BIL1 and BIL2 genes. Black boxes represent protein-encoding exon, gray boxes denote untranslated regions, and thick lines indicate introns. Arrows indicate orientations of T-DNA insertions from its left to right border, and the sequences above and below the arrows show the junction between flanking genomic DNAs and the inserted T-DNAs. The colored arrows represent the primers used for RT-PCR analysis of BIL1 or BIL2 gene expression. B, RT-PCR analysis of BIL1 and BIL2 gene expression in the wild type and bil1 and bil2 mutants. β-TUBULIN was used as a control. C, Shown from left to right are soil-grown 4-week-old Ws wild type, bin2-3, bil1, bil2, bil1bil2, and two individual bin2-3bil1bil2 mutants. D, Genetic analysis of bri1-5 with loss-of-function mutations of BIN2, BIL1, and BIL2. Shown from left to right are 4-week-old soil-grown bri1-5, bin2-3bri1-5, bil1bri1-5, bil2bri1-5, bil1bil2bri1-5, bin2-3bil1bri1-5, bin2-3bil2bri1-5, bin2-3bil1bil2bri1-5, and the Ws wild-type control.
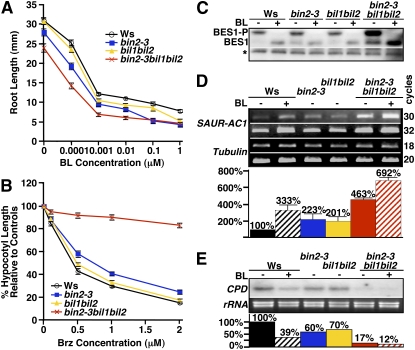
Despite the lack of any morphological effect, root growth inhibition assay and etiolated hypocotyl elongation assay revealed that simultaneous elimination of BIL1 and BIL2 had a detectable effect on BR sensitivity. Figure 5, A and B, showed that dose-responsive curves of BL-induced root growth inhibition and Brz-inhibited hypocotyl elongation of the bil1bil2 double mutant sit between those of the wild-type control and the bin2-3 mutant. The effect of the bil1bil2 double mutation on BR signaling was also examined by BES1 phosphorylation assay and SAUR-AC1 and CPD gene expression analyses. Although no detectable amount of nonphosphorylated BES1 was detected in the bil1bil2 mutant seedlings by anti-BES1 immunoblotting (Fig. 5C), RT-PCR and northern-blot analyses did show changes in transcript levels of SAUR-AC1 and CPD, respectively. The bil1bil2 mutation increases the SAUR-ACS1 transcript level by approximately 2-fold (Fig. 5D) but reduces the CPD mRNA level by approximately 30% (Fig. 5E).
Figure 5.
Physiological, biochemical, and molecular analyses of the T-DNA insertional mutations of BIN2, BIL1, and BIL2. A, Average root lengths of 2-week-old seedlings of the Ws wild type, bin2-3, bil1bil2, and bin2-3bil1bil2 mutants grown on half-strength MS medium containing no or increasing concentrations of BL. B, Hypocotyl elongation of the wild type, bin2-3, bil1bil2, and bin2-3bil1bil2 mutants grown on medium containing no or increasing concentrations of Brz. Inhibition of hypocotyl elongation by Brz is expressed relative to the average hypocotyl length of the same genotype grown on regular half-strength MS medium. For both A and B, each data point represents an average of 60 seedlings of two duplicate experiments. Error bars represent se. C, Western-blot analysis of BES1 phosphorylation status in BL- or mock-treated seedlings. Total proteins were extracted with 2× SDS buffer, separated by 10% SDS-PAGE, and analyzed by western blotting with anti-BES1 antibody. BES1-P is the phosphorylated BES1, and the asterisk indicates a nonspecific band used for a loading control. D, RT-PCR analysis of SAUR-AC1 expression. The numbers on the right represent numbers of PCR cycles. RT-PCR gel images were analyzed by ImageJ (version 1.3.7) to determine the signal intensity of individual bands. The expression level of SAUR-AC1 in the bar graph was expressed as a percentage of the ratio of the SAUR-AC1 transcript abundance versus the β-TUBULIN transcript level relative to that of the mock-treated Ws wild-type control. Each data bar represents an average of three independent RT-PCR experiments, and error bars denote se. E, Northern blot analysis of CPD gene expression. Ethidium bromide-stained rRNAs were used as a loading control. The autoradiograph was photographed and analyzed by ImageJ (version 1.3.7; http://rsb.info.nih.gov/ij/). The signal intensity of each CPD band in the bar graph below is expressed as a percentage relative to that of mock-treated Ws wild-type seedlings.
We also constructed triple mutants by crossing bil1 or bil2 single mutant with the bin2-3bri1-5 double mutant. As shown in Figure 4D (sixth and seventh plants), bin2-3bil1bri1-5 and bin2-3bil2bri1-5 mutants are larger than the parental bin2-3bri1-5 double mutant or the bil1bil2bri1-5 triple mutant with much longer petioles, revealing that suppression of the bri1-5 phenotype by bin2-3 can be significantly enhanced by elimination of either BIL1 or BIL2. Taken together, our results suggest that BIN2 functions redundantly with BIL1 or BIL2 in regulating BR signaling, with BIN2 playing a dominant role.
Elimination of BIN2, BIL1, and BIL2 Constitutively Activates BR Signaling
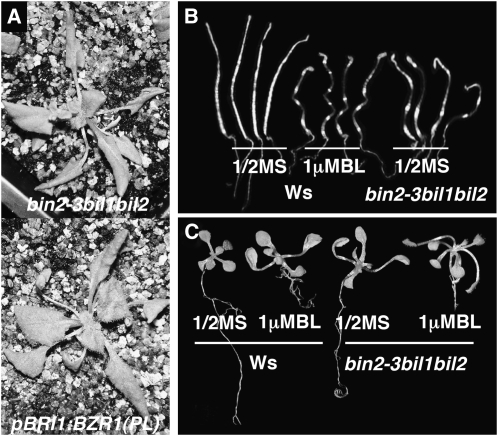
To further test functional redundancy between BIN2 and two BILs, we generated the bin2-3bil1bil2 triple mutant. Unlike bin2-3 or the bil1bil2 double mutant, the triple mutant has elongated and wavy petioles with narrow and twisted rosette leaves (Fig. 4C, sixth and seventh plants), resembling the previously characterized bes1-D and bzr1-D mutants and the BES1(P233L):GFP and BZR1(P234L):GFP transgenic plants (Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002; Fig. 6A). The P233/234L mutation is known to be responsible for a BR-hypersensitive phenotype of the bes1-D and bzr1-D mutants that accumulates high levels of both hyperphosphorylated and hypophosphorylated forms of BES1 and BZR1, respectively (He et al., 2002; Wang et al., 2002; Yin et al., 2002). The phenotypic similarity between bin2-3bil1bil2 and known mutants or transgenic plants with enhanced BR signaling outputs further supports our conclusion that BIN2 functions redundantly with BIL1 and BIL2 in regulating the BR signal transduction pathway.
Figure 6.
Simultaneous elimination of BIN2, BIL1, and BIL2 results in constitutive activation of BR signaling. A, Phenotypic comparison between the bin2-3bil1bil2 triple mutant and a transgenic line expressing a stabilized BZR1 protein grown in soil for 6 weeks. B, Etiolated 6-d-old bin2-3bil1bil2 triple mutant seedlings are morphologically similar to etiolated wild-type Ws seedlings grown on half-strength MS medium containing 1 μm BL. C, Light-grown 12-d-old seedlings of Ws and the bin2-3bil1bil2 mutant grown on half-strength MS medium supplemented with no or 1 μm BL.
To test if the phenotype of the bin2-3bil1bil2 triple mutant depends on the presence of BRI1 or active BRs, we first crossed the triple mutations into bri1-5. The resulting quadruple mutant exhibited a more or less similar morphological phenotype as the bin2-3bil1bil2 triple mutant (Fig. 4D, eighth plant). We also germinated and grew the triple mutant in the dark on Brz-containing medium. As shown in Figure 5B, while hypocotyl elongation of the wild type, bin2-3, or bil1bil2 was inhibited 85%, 73%, and 83%, respectively, by 2 μm Brz, the triple mutant was still etiolated when grown on the same concentration of Brz, with a mere 17% reduction in hypocotyl elongation.
Consistent with our genetic and physiological results, western-blot analysis of the BES1 phosphorylation status revealed that the bin2-3bil1bil2 triple mutant accumulated higher levels of both hypophosphorylated and hyperphosphorylated forms of BES1 than the wild type, bin2-3, or the bil1bil2 double mutant (Fig. 5C, lane 7). As a result, SAUR-AC1 expression was significantly enhanced in the bin2-3bil1bil3 triple mutant, with a nearly 7-fold increase compared with an approximately 3-fold BR-elicited induction in wild-type seedlings (Fig. 5D). By contrast, the transcript level of CPD was reduced by approximately 80% in the triple mutant compared with an approximately 40% reduction in BR-treated wild-type plants (Fig. 5E). Taken together, these results indicate that elimination of BIN2 and its two closest homologs leads to constitutive activation of BR signaling. Additional support for our conclusion came from our observations that both the dark- and light-grown seedlings of the triple mutants are morphologically similar to the dark- and light-grown wild-type seedlings grown in the presence of 1 μm BL, respectively (Fig. 6, B and C).
The bin2-3bil1bil2 Triple Mutant Still Responds to BR
The fact that the Arabidopsis genome encodes seven other GSK3-like kinases raises an interesting question of whether or not the BR responsiveness of the bin2-3bil1bil2 triple mutant is saturated. To answer this question, we used the BR-induced root growth inhibition assay to test if the triple mutant still exhibits a BR response. As shown in Figure 5A, roots of the triple mutant are much shorter than those of the wild type, bin2-3, or bil1bil2 without BR treatment but could be further inhibited by increasing concentrations of BL, suggesting that other GSK3 kinases might also be involved in BR signaling. Alternatively, the BR-elicited further inhibition of root growth of the bin2-3bil1bil2 triple mutant might be mediated by a GSK3-independent BR signaling pathway.
As mentioned before, our immunoblotting analysis revealed that the triple mutant accumulated not only the hypophosphorylated form but also the hyperphosphorylated form of BES1 (Fig. 5C). This observation contradicts our prediction that elimination of BIN2, BIL1, and BIL2 should significantly reduce rather than increase BES1 phosphorylation. This apparent contradiction implies that other GSK3-like kinases are able to compensate for the loss of BIN2, BIL1, and BIL2 in the triple mutant by phosphorylating BES1 in a manner that does not lead to degradation of the phosphorylated BES1. Alternatively, the increased amount of phosphorylated BES1 could be due to increased production of the BES1 protein coupled with a constant rate of phosphorylation.
In agreement with the root growth inhibition result, hyperphosphorylated BES1 disappeared while the amount of hypophosphorylated BES1 was further increased when the triple mutant was treated with BL (Fig. 5C, lanes 7 and 8). This result indicated that the kinase(s) that phosphorylate BES1 in the bin2-3bil1bil2 triple mutant could also be inhibited by a BR-activated BIN2 regulatory mechanism, thus supporting our hypothesis that BES1 in the triple mutant is likely phosphorylated by other AtSKs.
Li+ Treatment Leads to Complete Dephosphorylation of BES1
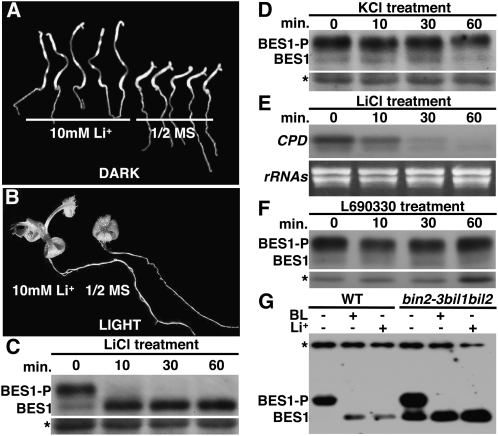
To further test whether the BES1 phosphorylation activity in the triple mutant is contributed by other Arabidopsis GSK3-like kinases, we conducted a series of pharmacological experiments using a well-studied GSK3 kinase inhibitor, lithium (Klein and Melton, 1996; Stambolic et al., 1996), which was capable of inhibiting both in vitro and in vivo phosphorylation of BES1 and BZR1 by BIN2 (Zhao et al., 2002; Peng et al., 2008). Growth of bin2-1 mutants on synthetic medium containing 10 mm LiCl rescued their short hypocotyl phenotype in the dark and their short petiole defect in the light, despite its failure to suppress their leaf phenotype (Fig. 7, A and B). Consistently, treatment with 100 mm LiCl even for 10 min resulted in complete disappearance of hyperphosphorylated BES1 accompanied by the accumulation of hypophosphorylated BES1 (Fig. 7C) and significant inhibition of CPD gene expression (Fig. 7E), whereas a similar treatment with 100 mm KCl had little effect on the BES1 phosphorylation status (Fig. 7D).
Figure 7.
BES1 phosphorylation activity in the bin2-3bil1bil2 mutant is likely catalyzed by other GSK3s. A, Li+ treatment rescued the short hypocotyl phenotype of etiolated bin2-1 seedlings. Shown are five bin2-1 seedlings grown in the dark on half-strength MS medium supplemented with or without 10 mm LiCl. B, Li+ treatment rescued the petiole length defect of the bin2-1 mutation in the light. Phenotypic comparison between 2-week-old bin2-1 mutants grown in the light on half-strength MS medium supplemented with or without 10 mm LiCl. C, Complete elimination of phosphorylated BES1 by treatment with 100 mm LiCl. D, Treatment of 100 mm KCl had no effect on the BES1 phosphorylation status. E, Li+ treatment led to rapid inhibition of CPD gene expression. Northern-blot analysis of CPD expression in Ws wild-type seedlings treated with 100 mm LiCl for 0, 10, 30, or 60 min. The top panel shows an autoradiograph of 32P-labeled CPD hybridization, and the bottom panel shows an ethidium bromide-stained RNA gel showing the amounts of rRNAs of each sample for the control of equal loading of total RNAs. F, Treatment with the inositol monophosphatase inhibitor L690330 had no effect on the in vivo BES1 phosphorylation activity. G, The BES1 phosphorylation activity in the bin2bil1bil2 mutant can be completely inhibited by treatment with BL or Li+. For C, D, F, and G, total protein crude extracts from treated seedlings were separated by 10% SDS-PAGE and analyzed by immunoblotting with an anti-BES1 antibody. BES1-P and BES1 indicate phosphorylated and nonphosphorylated forms of BES1 on western blots, and asterisks denote nonspecific bands used for loading controls.
To eliminate the possibility that the inhibitory effect of Li+ on the BES1 phosphorylation activity is mediated by another well-known Li+ target, inositol monophosphatase (Phiel and Klein, 2001), we treated Ws wild-type seedlings with a widely used inositol monophosphatase inhibitor, L690330 (Atack et al., 1993). As revealed by Figure 7F, L690330 had no measurable effect on the in vivo BES1 phosphorylation activity. We thus concluded that Li+ is quite effective at inhibiting GSK3 kinases and constitutively activates BR signaling in Arabidopsis. As shown in Figure 7G, treatment of the bin2-3bil1bil2 triple mutant with either 1 μm BL or 100 mm LiCl for 1 h completely eliminated hyperphosphorylated BES1 with a significant increase in the abundance of hypophosphorylated BES1. Taken together, these results strongly suggested that the BES1 protein in the bin2-3bil1bil2 triple mutant is phosphorylated by other GSK3 kinases that can also be inhibited by a BR-activated regulatory mechanism.
Li+ Treatment Failed to Suppress a Strong bri1 Mutation
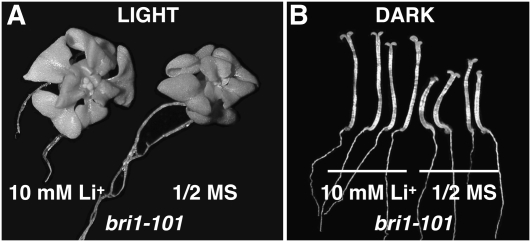
It was previously thought that BIN2 inhibition is the sole signaling output of the BR-activated BRI1 receptor complex (Vert and Chory, 2006). Our discovery that Li+ treatment was able to completely block the BES1 phosphorylation activity provided us a great opportunity to directly test this idea. We thus germinated and grew bri1-101 seedlings, carrying a mutated BR receptor with an inactive Ser/Thr kinase activity (Nam and Li, 2004), on synthetic medium containing 10 mm LiCl under both dark and light growth condition. To our surprise, the Li+ treatment only slightly enlarged the rosettes of light-grown bri1-101 seedlings (Fig. 8A) and slightly elongated the hypocotyls of dark-grown bri1-101 seedlings (Fig. 8B). The two most obvious morphological alterations that are associated with constitutive activation of BR signaling, elongated petioles in the light and twisted hypocotyls in the dark (Fig. 6, B and C), were not observed on Li+-treated bri1-101 seedlings. The different morphological responses of bin2-1 and bri1-101 mutants to the Li+ treatment thus suggested that there is a BIN2-independent regulatory mechanism downstream of BRI1 activation that is crucial for BR-regulated plant growth.
Figure 8.
LiCl treatment failed to suppress a strong BR receptor mutation. A, Shown are 3-week-old bri1-101 seedlings grown in the light on half-strength MS medium supplemented with or without 10 mm LiCl. B, Shown are 7-d-old bri1-101 seedlings grown in the dark on half-strength MS medium with or without 10 mm LiCl.
DISCUSSION
Functional Redundancy between BIN2 and Its Two Closest Homologs, with BIN2 Playing a Major Role in BR Signaling
In this study, we have identified several loss-of-function alleles of the Arabidopsis BIN2 gene, including a T-DNA insertional allele and eight intragenic suppressors of the previously characterized weak ucu1-3 mutation (Perez-Perez et al., 2002). The lack of observable morphological phenotypes in any loss-of-function bin2 mutants suggested functional redundancy between BIN2 and other AtSKs. Using T-DNA insertional mutants of BIN2 and its two closest homologs in the same Ws ecotype, we showed that simultaneous elimination of all three group II AtSKs constitutively activate BR signaling, as revealed by morphological alterations, changes in CPD and SAUR-AC1 expression, and accumulation of hypophosphorylated BES1. These results support the earlier claim that BIN2 functions redundantly with its two closest homologs in regulating BR signaling (Vert and Chory, 2006).
Our careful genetic, biochemical, and molecular analyses of several BR signaling outputs allowed us to conclude that BIN2 plays a dominant role among the three group II GSK3-like kinases despite their functional redundancy. First, while eight gain-of-function bin2 mutants were discovered as BR-insensitive dwarf mutants (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), no single gain-of-function mutant for BIL1 or BIL2 or any other AtSK has been isolated so far that exhibits the BR-insensitive dwarf phenotype. Second, loss of BIN2 function alone was able to partially suppress the bri1-5 mutation and resulted in a detectable decrease in BES1 phosphorylation, whereas single or even double mutations of bil1 and bil2 had little effect on the weak bri1-5 mutant or BES1 phosphorylation. Third, almost all transgenic plants overexpressing gbin2-1(E263K) exhibit the strong bin2-1-like phenotype, whereas only approximately one-third of gbil1(E295K) or gBIL2(E293K) transgenic plants are morphologically similar to bin2-1 mutants. Similarly, approximately 10% of transgenic plants overexpressing the wild-type BIN2 transgene exhibit the bin2-like phenotype (Li and Nam, 2002), and no single transgenic plant expressing wild-type BIL1 or BIL2 out of more than 150 independent transgenic T1 lines (per transgene) was morphologically similar to the bin2 mutant (data not shown). The E-K mutation in the highly conserved TREE motif is thought to prevent the BR-mediated BIN2 inhibition (Peng and Li, 2003), resulting in increased BIN2 stability and/or its nuclear accumulation (Vert and Chory, 2006; Peng et al., 2008), whereas overaccumulated BIN2 can saturate the BR-activated BIN2 regulatory machinery, leading to the accumulation of constitutively active GSK3 kinases in plant cells that inhibit BR signaling. One would predict that the corresponding E-K mutation in either BIL1 or BIL2 or overexpression of the two wild-type BIL genes would produce a similar morphological effect if both BILs play a significant role in regulating BR signaling. The failure to identify a single bin2-like gBIL1 or gBIL2 transgenic plant could be due to different expression profiles of the three group II AtSKs. However, a recent gene expression profiling study (Charrier et al., 2002) and a Web-based analysis of existing microarray data (Supplemental Fig. S1) showed similar expression profiles between BIN2 and the two BILs. One may argue that differential protein abundance of the three related GSK3s might be responsible for their differential effect on BR signaling. Consistent with this argument, two recent studies demonstrated that both BIN2 and a Medicago GSK3 kinase are regulated by proteasome-mediated protein degradation (Wrzaczek et al., 2007; Peng et al., 2008). Alternatively, the differential effect of the three group II AtSKs could be attributed to their primary sequences. Both BIL1 and BIL2 contain longer N-terminal ends (Fig. 5), which could affect their subcellular localizations, thus affecting their interactions with yet to be identified BIN2 regulatory proteins.
Involvement of Other GSK3-Like Kinases in Regulating BR Signaling
Our discovery that simultaneous elimination of BIN2, BIL1, and BIL2 did not completely eliminate but instead increased the phosphorylated BES1 suggested that BES1 can be phosphorylated in vivo by kinases other than the three group II AtSKs. In this study, we provided strong circumstantial evidence to suggest that the BES1-phosphorylating activity in the triple mutant is likely due to other AtSKs. Our conclusion was based on the following reasoning. First, the BES1 phosphorylation activity in the triple mutant could be completely eliminated by treatment of BL, suggesting that the BR-activated BIN2 regulatory machinery can inhibit other BES1-phosphorylating kinases. Second, the TREE motif and the Pro-284 residue critical for BIN2 regulation are almost absolutely conserved among the 10 AtSKs except AtSK3-1, which carries an Ala residue at the position of Thr (Supplemental Fig. S2), suggesting that six other AtSKs could interact with the BIN2 regulatory mechanism. Third, despite its resemblance to BR mutants or transgenic plants with constitutively activated BR signaling, the bin2-3bil1bil2 triple mutant still responds to BR in a dose-dependent manner, arguing strongly for functional redundancy between the group II AtSKs and other AtSKs. Fourth, we discovered that the BES1 phosphorylation activity in the triple mutants could be completely inhibited by treatment with Li+, which is known to inhibit in vitro and in vivo BIN2-catalyzed phosphorylation of BES1/BZR1 (Zhao et al., 2002; Peng et al., 2008). In addition, LiCl treatment resulted in phenotypic suppression of the bin2-1 mutant. Although we cannot rule out the possibility that some non-GSK3-like kinases, which can be inhibited by both BL and Li+, are responsible for phosphorylating BES1 in the bin2-3bil1bil2 triple mutant, the data presented here strongly suggested the involvement of other AtSKs in BR signaling.
The Existence of a Possible BIN2-Independent Regulatory Mechanism
Our study made a surprising discovery that Li+ treatment could suppress the gain-of-function bin2-1 mutation but had little effect on the strong BR receptor mutant bri1-101. The two most obvious morphological changes, twisted hypocotyls of dark-grown seedlings and elongated petioles of light-grown seedlings, which are associated with constitutive activation of BR signaling, were not observed on Li+-treated bri1-101 seedlings. These observations suggested that BR perception by BRI1 activates a BIN2-independent regulatory mechanism that functions together with BIN2 inhibition to regulate plant growth. Elimination of the BES1/BZR1 phosphorylation activity by Li+ without activation of such a BIN2-independent regulatory system is not able to fully activate BR signaling. It is quite possible that BR signaling behaves similarly to the signaling process of animal steroid hormones involving both genomic and nongenomic effects. The genomic effect of BR signaling is mediated by BES1 and BZR1, which are regulated at several different levels by BIN2-catalyzed protein phosphorylation, while the nongenomic effects might be mediated by known BRI1 substrates such as TTL, an Arabidopsis protein similar to vertebrate transthyretin-like proteins (Nam and Li, 2004), TRIP-1, an essential component of the translational initiation complex (Ehsan et al., 2005), or BSKs, a family of related protein kinases that are phosphorylated by BRI1 (Tang et al., 2008). It might also be possible that the activated BRI1 could regulate the proton-pumping activity of a plasma membrane or vacuolar ATPase, which is known to be critical for generating turgor pressure necessary for plant cell growth, or affect the reorganization of cortical microtubules (Peng and Li, 2003). Alternatively, the BIN2-independent regulatory mechanism might also be involved in gene regulation by inhibiting a transcriptional repressor or activating a transcriptional activator that functions together with BES1/BZR1 to control gene expression. Novel genetic screens and identification of components of a BR-activated BRI1 receptor complex could lead to complete elucidation of the suspected BIN2-independent regulatory system.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia (Col-0), Ler, and Ws-2 ecotypes were used as wild-type controls for phenotype comparison. Ws-2 was also used as the control in seedling measurement as well as in protein and RNA analysis. Col-0 was used to generate all of the transgenic plants. BR mutants bri1-5 (in Ws-2; Choe et al., 2002), ucu1-3 (in Ler; Perez-Perez et al., 2002), and all other mutants and transgenic plants were germinated on half-strength Murashige and Skoog (MS) medium and transferred into soil for continued growth at 22°C under a 16-h-light/8-h-dark photoperiod. Seed sterilization and seedling handling were carried out as described previously (Li et al., 2001).
Mutant Isolation
For isolation of loss-of-function bin2 mutants, seeds of the Arabidopsis ucu1-3 mutant (Perez-Perez et al., 2002) were treated with 0.3% ethyl methanesulfonate (Sigma-Aldrich) as described (Yin et al., 2002). About 200,000 M2 seeds derived from 20,000 M1 plants were screened on half-strength MS medium. After 4 weeks of growth in a growth chamber with a 16-h-light/8-h-dark photoperiod, putative suppressors were transferred into soil for continued growth, phenotypic analysis, and seed collection. Interesting suppressors were genotyped with the ucu13 derived cleaved amplified polymorphic sequence marker (Supplemental Table S1) to eliminate pollen/seed contaminants. The bin2-3 mutant (FLAG_593C09, Ws ecotype) was PCR screened and isolated from the FLAGdb Collection at the Versailles Genetics and Plant Biology Laboratory, Institut National de la Recherche Agronomique (http://urgv.evry.inra.fr/projects/FLAGdb++/HTML/index.shtml; Samson et al., 2002). bil1 (At2g30980) and bil2 (At1g06390) knockout mutants were obtained from the Wisconsin Arabidopsis Gene Knockout Facility at the University of Wisconsin, Madison (http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/). Both mutants are in the Ws-2 ecotype.
DNA Isolation and Analysis
Genomic DNAs of putative ucu1-3 suppressors were isolated as described previously (Li and Chory, 1998). The entire BIN2 gene was amplified using Taq polymerase (Promega) with the BIN2F/R primer set (Supplemental Table S1). The amplified BIN2 genomic DNAs pooled from four independent PCRs of each putative suppressor was gel purified using the QIAquick gel extraction kit (Qiagen) and sequenced at the University of Michigan DNA Sequencing Core Facility. The resulting BIN2 sequences were compared with those obtained from the parental ucu1-3 mutant to find single nucleotide changes.
Plasmid Construction and Plant Transformation
The gBIN2 genomic construct and its mutated version carrying the bin2-1 mutation were described previously (Li and Nam, 2002). To make gBIL1 and gBIL2 constructs, a 6.5-kb genomic fragment of At2g30980 and a 5.5-kb genomic fragment of At1g06390 were released from bacterial artificial chromosomes F7F1 and T2D23 by digestion with PstI and AflII (followed by partial filling with dTTP) and cloned into the pPZP212 binary vector (Hajdukiewicz et al., 1994) digested with PstI and EcoRI (followed by partial filling with dATP), respectively. The resulting pPZP212gBIL1 and pPZP212gBIL2 plasmids were subjected to site-directed mutagenesis using the QuickChange II XL site-directed mutagenesis kit (Stratagene) to generate gBIL1(E295K) and gBIL2(E293K) plasmids. Both the wild-type and mutated transgenes of BIN2, BIL1, and BIL2 were individually introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Col-0 wild-type Arabidopsis plants via a vacuum infiltration method (Bechtold and Pelletier, 1998).
Production of Fusion Protein and in Vitro Phosphorylation Assay
To express MBP-fused BIN2, BIL1, or BIL2 in Escherichia coli, each open reading frame of the three Arabidopsis GSK3-like kinases was individually cloned into pMAL-c1 (New England Biolabs). Induction and purification of MBP fusion proteins were carried out according to the manufacturer's recommended protocol. Expression and purification of the GST-BES1 fusion protein and in vitro phosphorylation assay using the MBP-fused GSK3-like kinases were conducted as reported previously (Zhao et al., 2002).
Hypocotyl and Root Growth Measurements
In hypocotyl growth inhibition assays, seeds were sterilized and grown on half-strength MS medium containing increasing concentrations of Brz (0.1–2 μm) or dimethyl sulfoxide (0.001% [v/v]) in the dark for 6 d. Etiolated seedlings were carefully removed from agar plates, placed on a flat surface, and photographed at a resolution of 600 pixels per inch. Hypocotyl lengths of individual seedlings were measured by ImageJ (version 1.37 of Mac OS X; http://rsb.info.nih.gov/ij/), and the results were exported into Microsoft Excel for statistical analysis. The root growth inhibition assay to analyze the effects of loss-of-function mutations of BIN2, BIL1, and BIL2 on BR sensitivity was carried out as described previously (Li et al., 2001).
Treatment with Chemicals for Protein and RNA Analyses
Eighteen-day-old seedlings grown on half-strength MS medium were gently removed from petri dishes and submerged into liquid half-strength MS medium containing different concentrations of BL, LiCl, KCl, or L690330 (Tocris). Seedlings were removed at different time points and were either directly collected into 2× SDS buffer (200 mm Tris-HCl, pH 6.8, 4% SDS, 0.2% bromphenol blue, 20% glycerol, and 0.2 m dithiothreitol) or immediately frozen in liquid nitrogen and stored in a –80°C freezer for later analysis. To analyze the effect of Li+ on plant morphology, seeds of the bin2/+ heterozygous plants were germinated and grown on half-strength MS medium with or without 10 mm LiCl at 22°C in a growth chamber under a 16-h-light/8-h-dark photoperiod. Individual seedlings were photographed for phenotype comparison and analyzed by PCR using the bin2-1 derived cleaved amplified polymorphic sequence primer set (Supplemental Table S1) to determine their genotypes.
Protein and RNA Analyses
Freshly collected or frozen samples were ground thoroughly in 2× SDS sample buffer using a plastic pestle and boiled at 100°C for 5 min. After 5 min of centrifugation to get rid of insoluble tissue debris, soluble proteins were separated on a 12% SDS-PAGE gel, transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore), and analyzed by western blot with an anti-BES1 antibody (Mora-Garcia et al., 2004). The signals on the Immobilon-P membrane were detected using the horseradish peroxidase-conjugated goat anti-rat-IgG secondary antibody (Abcam) and the Supersignal West Pico chemiluminescent substrate system (Pierce).
Total RNAs of freshly collected or frozen samples were extracted using the RNeasy Plant Mini kit (Qiagen). The resulting RNAs were separated by electrophoresis, transferred to a nylon membrane, and hybridized with a 32P-labeled CPD probe as described previously (Li et al., 2001). For RT-PCR assays, 2 to 5 μg of total RNAs was converted using the SuperScript III First-Strand cDNA synthesis kit (Invitrogen) into single-stranded cDNA templates, which were used to amplify specific transcripts with gene-specific primer sets (Supplemental Table S1). Northern blot autoradiographs and gel pictures from RT-PCR analysis were analyzed by ImageJ (version 1.37 of Mac OS X; http://rsbweb.nih.gov/ij/) to determine relative gene expression levels.
Sequence data from this article can be found in the GenBank database under the following accession numbers: BIN2, At4g18710, NP_193606; BIL1, At2g30980, NP_180655; and BIL2, At1g06390, NP_973771.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BIN2 and its two homologs display similar expression profiles.
Supplemental Figure S2. Conservation of amino acids critical for BR-mediated inhibition of GSK3-like kinase activity.
Supplemental Table S1. Primers used for genotyping, sequencing, and RT-PCR analyses.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing bacterial artificial chromosome DNA clones; Dr. J.M. Pérez-Pérez for ucu1-3 seeds; Dr. F. Tax for bri1-5 seeds; the Versailles Genetics and Plant Biology Laboratory, Institut National de la Recherche Agronomique, for bin2-3 seeds; and the Wisconsin Arabidopsis Knockout Facility for its service to screen bil1 and bil2 T-DNA insertional mutants. We thank Dr. T. Asami for his generous gift of Brz, Dr. Y. Yan for providing affinity-purified anti-BES1 antibody, and members of Li laboratory for stimulating discussion.
This work was supported by the National Institutes of Health (grant no. GM60519 to J.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jianming Li (jian@umich.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atack JR, Cook SM, Watt AP, Fletcher SR, Ragan CI (1993) In vitro and in vivo inhibition of inositol monophosphatase by the bisphosphonate L-690,330. J Neurochem 60 652–658 [DOI] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 [DOI] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127 450–458 [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3 beta-like kinase. Plant Physiol 130 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claisse G, Charrier B, Kreis M (2007) The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion. Plant Mol Biol 64 113–124 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49 427–451 [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Van Lammeren AA, Kreis M (2000) Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J 21 419–429 [DOI] [PubMed] [Google Scholar]
- Ehsan H, Ray WK, Phinney B, Wang X, Huber SC, Clouse SD (2005) Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J 43 251–261 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang YL, Li JM, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Beisteiner D, Beyerly J, Hirt H (2000) Wound-induced expression and activation of WIG, a novel glycogen synthase kinase 3. Plant Cell 12 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Hirt H (2002) Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci 7 457–461 [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA (1996) A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA 93 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J (2005) Brassinosteroid signaling: from receptor kinases to transcription factors. Curr Opin Plant Biol 8 526–531 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1998) Preparation of DNA from Arabidopsis. Methods Mol Biol 82 55–60 [DOI] [PubMed] [Google Scholar]
- Li J, Jin H (2007) Regulation of brassinosteroid signaling. Trends Plant Sci 12 37–41 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222 [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14 593–602 [DOI] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2004) The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1. Plant Cell 16 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Li JM (2003) Brassinosteroid signal transduction: a mix of conservation and novelty. J Plant Growth Regul 22 298–312 [DOI] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J (2008) Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant 1 338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242 161–173 [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Klein PS (2001) Molecular targets of lithium action. Annu Rev Pharmacol Toxicol 41 789–813 [DOI] [PubMed] [Google Scholar]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I (2001) Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J 27 305–314 [DOI] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I (1999) An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol 119 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard O, Paquet N, Haudecoeur E, Charrier B (2005) Organization and expression of the GSK3/shaggy kinase gene family in the moss Physcomitrella patens suggest early gene multiplication in land plants and an ancestral response to osmotic stress. J Mol Evol 61 99–113 [DOI] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, Ruel L, Woodgett JR (1996) Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol 6 1664–1668 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441 96–100 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15 220–235 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2 505–513 [DOI] [PubMed] [Google Scholar]
- Woodgett JR (2001) Judging a protein by more than its name: GSK-3. Sci STKE 2001 RE12. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Rozhon W, Jonak C (2007) A proteasome-regulated glycogen synthase kinase-3 modulates disease response in plants. J Biol Chem 282 5249–5255 [DOI] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18 472–478 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259 [DOI] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo MJ, Albert VA, Soltis PS, Soltis DE (2006) Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol 6 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J (2002) Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol 130 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.