Abstract
Enhancement of the carotenoid biosynthetic pathway in food crops benefits human health and adds commercial value of natural food colorants. However, predictable metabolic engineering or breeding is limited by the incomplete understanding of endogenous pathway regulation, including rate-controlling steps and timing of expression in carotenogenic tissues. The grass family (Poaceae) contains major crop staples, including maize (Zea mays), wheat (Triticum aestivum), rice (Oryza sativa), sorghum (Sorghum bicolor), and millet (Pennisetum glaucum). Maize carotenogenesis was investigated using a novel approach to discover genes encoding limiting biosynthetic steps in the nutritionally targeted seed endosperm. A combination of bioinformatics and cloning were first used to identify and map gene families encoding enzymes in maize and other grasses. These enzymes represented upstream pathways for isopentenyl diphosphate and geranylgeranyl diphosphate synthesis and the downstream carotenoid biosynthetic pathway, including conversion to abscisic acid. A maize germplasm collection was used for statistical testing of the correlation between carotenoid content and candidate gene transcript levels. Multiple pathway bottlenecks for isoprenoid biosynthesis and carotenoid biosynthesis were discovered in specific temporal windows of endosperm development. Transcript levels of paralogs encoding isoprenoid isopentenyl diphosphate and geranylgeranyl diphosphate-producing enzymes, DXS3, DXR, HDR, and GGPPS1, were found to positively correlate with endosperm carotenoid content. For carotenoid pathway enzymes, transcript levels for CrtISO inversely correlated with seed carotenoid content, as compared with positive correlation of PSY1 transcripts. Since zeaxanthin epoxidase (ZEP) depletes the carotenoid pool in subsequent conversion to abscisic acid, ZEP transcripts were examined. Carotenoid accumulation was found to be inversely associated with ZEP1 and ZEP2 transcript levels. Extension of the maize results using phylogenetic analysis identified orthologs in other grass species that may serve as potential metabolic engineering targets.
Carotenoids are a complex class of isoprenoid pigments providing nutritional value as provitamin A and nonprovitamin A compounds; their varied colors provide added commercial value as colorants in foods (for review, see Matthews and Wurtzel, 2007). Metabolic engineering of the biosynthetic pathway has been of interest to specifically address vitamin A deficiency in millions of children worldwide (for review, see Giuliano et al., 2008); this serious global health problem could be alleviated by breeding cereal crop staples in the Poaceae (grass family) for elevated levels of provitamin A carotenoids. Engineering high levels of specific carotenoid structures requires controlled enhancement of total carotenoid levels (enhancing pathway flux, minimizing degradation, and optimizing sequestration) plus controlled composition for specific pathway end products. While most of the nuclear genes for the plastid-localized pathway are available (Li et al., 2007) and/or can be identified, questions remain about the rate-controlling steps that limit the predictability of metabolic engineering in plants.
The most important cereal crop staples worldwide, such as maize (Zea mays), sorghum (Sorghum bicolor), wheat (Triticum aestivum), and rice (Oryza sativa), are all contained within the grass family. We have taken advantage of the resources available for maize, an important crop staple in sub-Saharan Africa and Latin America, to explore those factors that control endosperm carotenogenesis. Ortholog targets in related grass species can be identified using comparative genomics and thereby potentially extend maize studies for predictive metabolic engineering throughout the cereal crops.
Predictable manipulation of the seed carotenoid biosynthetic pathway in diverse maize genotypes necessitates the elucidation of biosynthetic step(s) that control carotenoid accumulation in endosperm tissue (Fig. 1). Extensive studies have implicated phytoene synthase (PSY), the first committed enzyme, as rate controlling for endosperm carotenoids (Randolph and Hand, 1940; Palaisa et al., 2003; Gallagher et al., 2004; Wong et al., 2004; Pozniak et al., 2007; Li et al., 2008a, 2008b). However, upstream precursor pathways may also positively influence carotenoid accumulation (Matthews and Wurtzel, 2000; Mahmoud and Croteau, 2001), while downstream degradative pathways may deplete the carotenoid pool (Galpaz et al., 2008). We decided to utilize natural genetic and biochemical variation in a maize germplasm collection to identify potential transcriptional control points affecting endosperm carotenoid accumulation, since transcriptional control plays a large role in carotenogenesis (Giuliano et al., 2008). An alternative approach to probe the pathway with transgenes was considered less desirable, as the resulting changes would only reflect and be predictive for the particular transformed genotype, and an ultimate goal is to derive regulatory/temporal information applicable to metabolic engineering/breeding of a broader range of genotypes by nontransgenic (Harjes et al., 2008) and/or transgenic means (Zhu et al., 2008). Association of candidate gene cis-located DNA variation with kernel carotenoid levels in a germplasm collection has been effective in identifying metabolic breeding tools for the carotenoid pathway (Harjes et al., 2008). However, association analysis may overlook a rate-controlling biosynthetic step if gene expression is regulated by another locus. Moreover, optimal timing of gene expression is not revealed. Therefore, to identify rate-controlling steps, we focused on transcript-level variation as a manifestation of DNA polymorphisms at the candidate gene or regulation by an unlinked locus. RNA profiling would also facilitate the optimization of temporal expression for transgene expression efforts.
Figure 1.
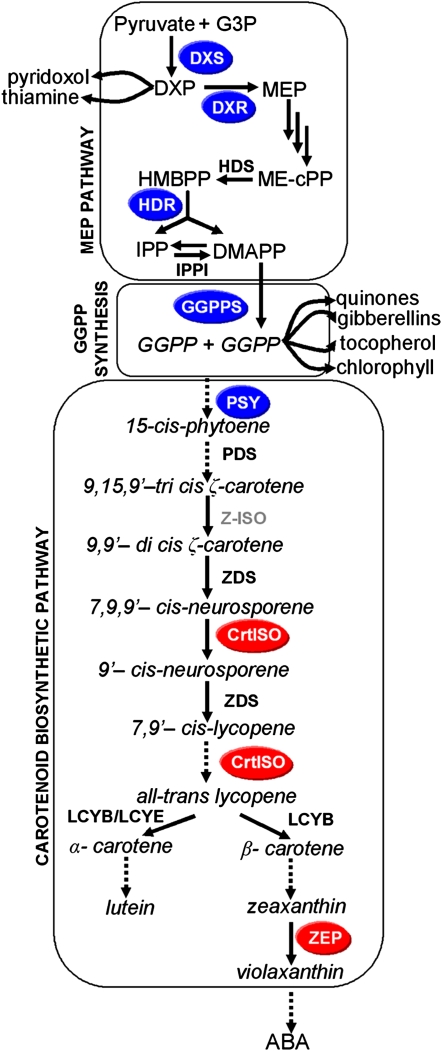
Simplified carotenoid biosynthetic pathway derived from isoprenoid precursors. Enzymatic reactions are represented by arrows. Genes encoding all enzymes shown, except Z-ISO, were tested for correlation with total accumulated carotenoids. Transcripts for enzymes in black letters had no effect; white letters on blue circles and white letters on red circles showed positive correlation and negative correlation, respectively. G3P, d-Glyceraldehyde-3-phosphate; ME-cPP, 2C-methyl-d-erythritol 2,4-cyclodiphosphate; MEP, 2C-methyl-d-erythritol-4-phosphate.
RESULTS
Pathway Gene Families and Chromosome Mapping
To be able to examine transcriptional control of pathway flux, we assembled a collection of genes from maize and other grass species to represent upstream pathways for isopentenyl diphosphate (IPP) and geranylgeranyl diphosphate (GGPP) synthesis and the downstream carotenoid biosynthetic pathway, including conversion to abscisic acid (ABA). We used a combination of bioinformatics and cloning to identify gene families for enzymes shown in Figure 1. Genes were mapped in maize to chromosome positions with no apparent linkage seen among the pathway genes (Table I; Supplemental Table S1).
Table I.
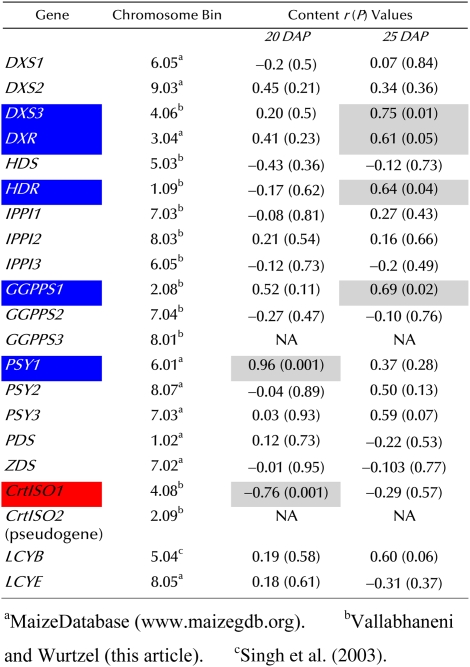
Correlation of mRNA levels to total carotenoid content in mature seeds
Pearson correlation (r) and statistical significance (P), for comparison of transcript levels and kernel carotenoid content in maize inbred lines, was performed using JMP version 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship. Letters on a white background indicate no effect; letters on blue and red backgrounds indicate positive correlation and negative correlation, respectively. Inbred lines are as listed in Figure 6. Carotenoid content correlated to mRNA levels is shown on a gray background. [See online article for color version of this table.]
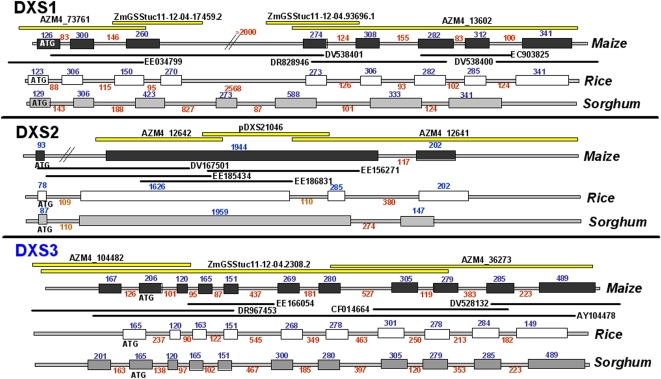
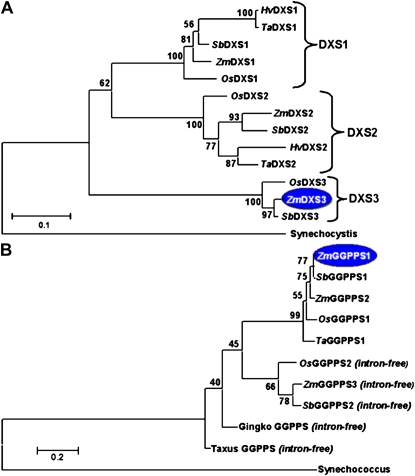
MEP pathway enzymes DXP reductoisomerase (DXR), 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (HDS), and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) were found to be encoded by single-copy genes in the maize genome. Three paralogs for maize DXS (for 1-deoxy-d-xylulose-5-phosphate synthase) were identified (Figs. 2 and 3A; Walter et al., 2002); DXS3 is a newly discovered paralog in maize and is also present in sorghum and rice, species spanning two subfamilies of the Poaceae (Figs. 2 and 3A; Supplemental Table S1; Kim et al., 2005). The ChloroP program (Emanuelsson et al., 1999) predicted a plastid localization signal in the first 51, 31, and 50 amino acids of ZmDXS1, ZmDXS2, and ZmDXS3, yielding plastid-localized proteins of 71.7, 73.5, and 73.1 kD, respectively. The three maize enzymes are similar in size, yet they share only 52.5% identity/65.8% similarity between DXS1 and DXS3 and 47.6 identity/62.4% similarity between DXS2 and DXS3 as compared with 69.7% identity/79.1% similarity between the more closely related DXS1 and DXS2 (Supplemental Fig. S1).
Figure 2.
Gene family structure of DXS genes from the Poaceae. Boxes and lines indicate exons and introns, respectively; sizes are in base pairs. Bars above maize gene structures represent the genomic counterparts used to build the gene structure; dark bars below maize gene structures represent ESTs covering the entire coding region. [See online article for color version of this figure.]
Figure 3.
Phylogenetic analysis based on translation products. A, DXS gene family. GenBank accession numbers are in parentheses: Synechocystis (sll1945, BAA17089.1), Zea mays (ZmDXS1, TC312721; ZmDXS2, TC293862; ZmDXS3, TC307233), Oryza sativa (OsDXS1, LOC_Os05g33840; OsDXS2, LOC_Os07g09190; OsDXS3, LOC_Os06g05100), Sorghum bicolor (SbDXS1, Sb09g020140; SbDXS2, Sb02g005380; SbDXS3, Sb10g002960); Hordeum vulgare (HvDXS1, TC149768; HvDXs2, TC152494); Triticum aestivum (TaDXS1, TC238788; TaDXS2, TC258088). B, GGPPS gene family. GenBank accession numbers are in parentheses: Synechococcus (crtE, ABI45773); Zea mays (ZmGGPPS1, EF417573; ZmGGPPS2, EF417574; ZmGGPPS3, EF417575); Oryza sativa (OsGGPPS1, LOC_Os07g39270; OsGGPPS2, LOC_Os01g14630); Sorghum bicolor (SbGGPPS1, Sb02g037510, SbGGPPS2, Sb03g009380); Gingko biloba (AY371321); Taxus media (AY566309). Maize enzymes are highlighted in blue if transcripts correlated with carotenoid content. [See online article for color version of this figure.]
The IPPI (for isopentenyl pyrophosphate isomerase) gene family contained three genes (Supplemental Table S1). GGPPS (for geranylgeranyl pyrophosphate synthase) was also encoded by a small gene family of three paralogs. The encoded proteins have the requisite GGPPS structural domains and chloroplast targeting signals of 52, 51 and 44 amino acids for GGPPS1, GGPPS2, and GGPPS3, yielding plastid-localized forms of 33.5, 32.9, and 32.7 kD, respectively (Supplemental Fig. S2; Supplemental Table S1; Okada et al., 2000). GGPPS3 was the only intronless gene, and its predicted peptide sequence had one variant region immediately downstream of domain III. Phylogenetic analysis showed that the intron-containing GGPPS genes and the intronless genes formed two distinct clades within the angiosperms (Fig. 3B). Maize GGPPS1 and GGPPS2 appear to be recent duplications arising after speciation. Function of the three GGPPS genes was tested by heterologous complementation in Escherichia coli (Supplemental Fig. S3) using methods described previously (Cervantes-Cervantes et al., 2006); GGPPS1 and GGPPS2 proved functional, but GGPPS3 did not (data not shown). GGPPS3 transcripts were barely detectable, as evidenced by reverse transcription (RT)-PCR and as reflected by low EST prevalence in maize and rice. Therefore, our studies limited evaluation to the two functional GGPPS genes.
For the carotenoid biosynthetic pathway, we identified two CrtISO (for carotene isomerase) genes in maize on chromosomes 2 and 4, adding to previously isolated pathway genes for PDS (for phytoene desaturase), ZDS (for zetacarotene desaturase), LCYE (for lycopene ɛ-cyclase), and LCYB (for lycopene β-cyclase), all of which are single copy in maize and other grasses (Buckner et al., 1990; Li et al., 1996, 2008a, 2008b; Matthews et al., 2003; Singh et al., 2003; Harjes et al., 2008; Supplemental Table S1). In contrast to two maize CrtISO genes, only one was found in rice, sorghum, and Arabidopsis (Arabidopsis thaliana; Supplemental Table S1; Supplemental Fig. S4). However, maize CrtISO2 proved to be a pseudogene; alignment of maize CrtISO sequences (Supplemental Fig. S4; verified from several DNA sources [see “Materials and Methods”]) revealed a stop codon in exon 1 of maize CrtISO2 that is predicted to interrupt translation. The functional CrtISO1 encodes an enzyme of 63.6 kD with a predicted plastid-targeting sequence of 43 residues, yielding a plastid-localized form of 59.3 kD. Maize CrtISO1 is located on chromosome 4 in a region syntenous with rice CrtISO and linked to marker csu704 on rice chromosome 11.
For examination of carotenoid metabolism to ABA, we identified genes for zeaxanthin epoxidase (ZEP), an enzyme catalyzing the conversion of zeaxanthin to violaxanthin, a precursor of ABA in endosperm (Fig. 1). Two ZEP genes were found in maize compared with only one in rice and sorghum (Supplemental Fig. S5; Supplemental Table S1); all showed gene and protein structural similarities, including chloroplast-targeting signals. Maize ZEP1 and ZEP2 have predicted transit peptides of 68 and 60 residues yielding 66.6-kD plastid-localized enzymes.
Transcript Profiling of Gene Family Members and Correlation with Carotenoid Content
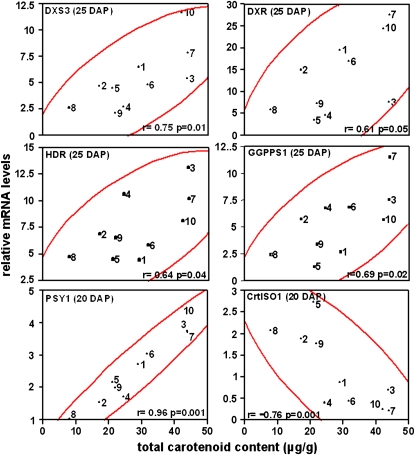
Predictive metabolic engineering/breeding of kernel carotenoid content is predicated on identifying genes and their temporal expression in developing endosperm that correlates with carotenoid content. Using the standard inbred line B73 (Li et al., 2008a), transcript levels were quantified across a panel of tissues and during endosperm development for all functional genes that were required for carotenoid accumulation (Figs. 4 and 5). Paralog-specific variation in transcript levels was observed in B73 maize. However, it is unknown how transcript levels correlate with accumulated endosperm carotenoids in genetically diverse inbreds varying in carotenoid content and composition (Islam, 2004; Harjes et al., 2008). It is also unknown how timing of gene expression during endosperm development is related to net accumulation of carotenoids in mature kernels. To investigate timing and correlation between gene expression and carotenoid content, transcript levels at specific endosperm developmental stages were compared with carotenoid content of mature kernels that varied in a genetically diverse germplasm collection. Quantitative transcript profiling was conducted in a core subset of maize lines chosen from a collection varying in kernel carotenoid content and spanning 80% of maize genetic diversity (Liu et al., 2003a; Islam, 2004; Harjes et al., 2008). The goal was to identify genes/paralogs with a statistically significant correlation between transcript levels at certain endosperm developmental stages and carotenoid content in the mature kernel. From a collection of 148 maize lines analyzed for carotenoid content (Islam, 2004), 10 lines were chosen that represented extremes of carotenoid content and composition (e.g. highest levels of zeaxanthin, lutein, etc.). These core lines spanned four major genetic diversity groups and eight subgroups (Liu et al., 2003a). To validate the utility of the core subset to yield predictive results, we used the collection to test the PSY gene family, which contains one member, PSY1, known to control flux to endosperm carotenoids. Only PSY1, of three paralogs, showed the expected correlation of transcripts and carotenoid content in endosperm (Li et al., 2008b). The validated germplasm subset was then used to assess the influence of other genes in the pathway collection described above using paralog-specific primers (Supplemental Table S2). Transcript levels were quantified in developing endosperm 10 to 25 d after pollination (DAP; Supplemental Table S3), stages when carotenoid accumulates (Li et al., 2008b). Transcript levels at each developmental stage were then statistically compared with carotenoid content of the mature kernel. Pearson correlation analysis (Table I) revealed that in addition to PSY1, several other genes showed a statistically significant correlation of transcripts with total carotenoid content (Fig. 6). Positively correlating genes upstream of the carotenoid pathway included DXS3, DXR, HDR, and GGPPS1, all at 25 DAP. Within the carotenoid pathway itself, CrtISO1 transcripts negatively correlated with carotenoid content (r = −0.76, P = 0.001) as compared with the positive correlation seen with transcripts of the carotenoid pathway gene, PSY1 (r = 0.96, P = 0.001), both at 20 DAP. The difference in r values for CrtISO1 and PSY1 is likely due to PSY1 having been under strong selection (Palaisa et al., 2003), whereas CrtISO1 has not been under any selection because it was unknown. A significant lesson drawn from these experiments is that trends in transcript accumulation patterns (Fig. 5) were not necessarily predictive of carotenoid accumulation. For example, transcripts of DXS3 and DXS1 both showed temporal increases during endosperm development, but only DXS3 showed correlative expression with carotenoids. Similarly, transcript levels of IPPI gene family members showed temporal increases during endosperm development but were not limiting as judged by the correlation analysis.
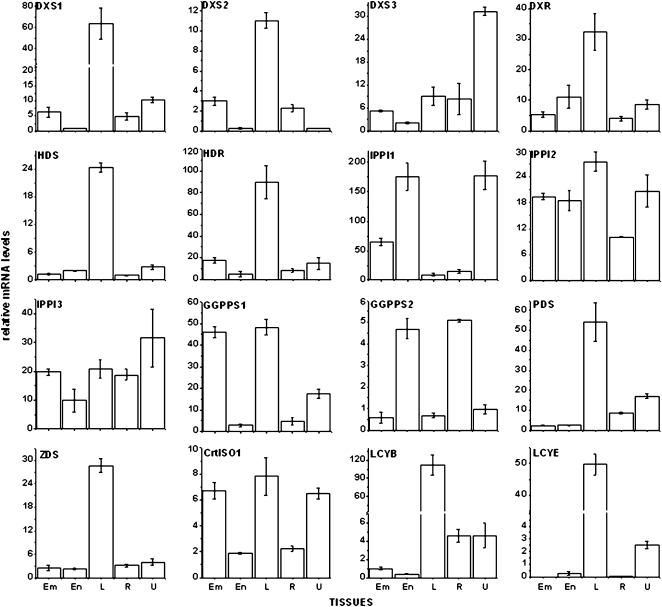
Figure 4.
Maize B73 inbred transcript profiles of genes encoding enzymes for isoprenoid and carotenoid biosynthesis. Transcript levels were expressed relative to endosperm levels at 20 DAP. Means of three replicates ± sd are shown. Embryo (Em) and endosperm (En) were collected at 20 DAP from field-grown plants; leaf (L) and root (R) samples were collected from seedlings at the six-leaf stage; U, unfertilized ovule.
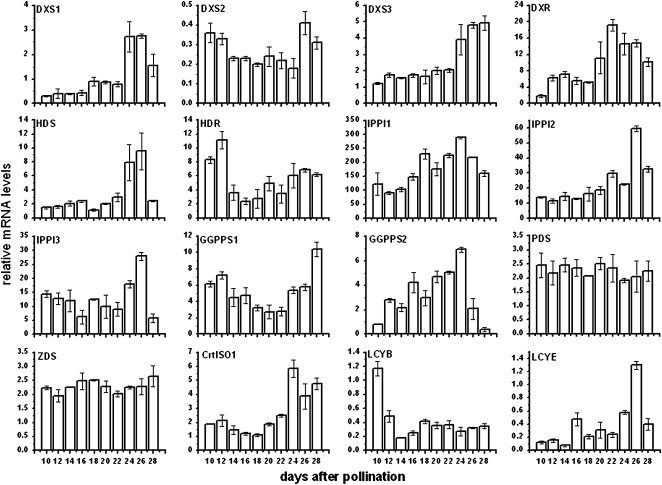
Figure 5.
Maize B73 inbred developing endosperm transcript profiles of genes encoding enzymes for isoprenoid and carotenoid biosynthesis. Means of three replicates ± sd are shown.
Figure 6.
Correlation analysis showing that endosperm carotenoid content positively or negatively correlates with mRNA levels of specific pathway enzymes. Pearson correlation (r) and statistical significance (P) for comparison of transcript levels and kernel carotenoid content in maize inbred lines were determined using JMP version 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship at a 95% confidence interval (red lines). Inbred lines are as follows: 1, A619; 2, B73; 3, B37; 4, CI.7; 5, C131A; 6, DE3; 7, KUI2007; 8, NC300; 9, SD44; 10, TZI18. PSY1 data were taken from Li et al. (2008b). [See online article for color version of this figure.]
Role of ZEP in Depleting Endosperm Carotenoids
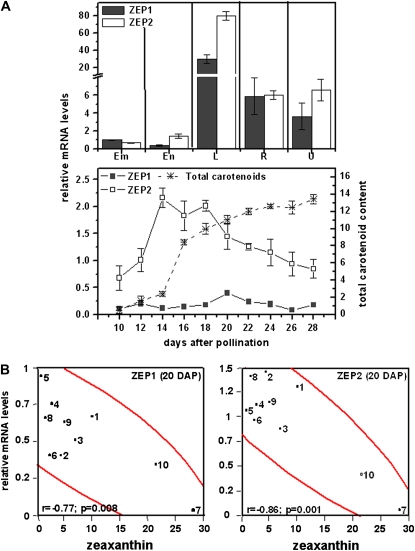
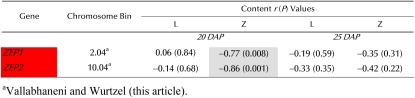
Conversion of carotenoids to ABA requires the activity of ZEP, an enzyme upstream of the rate-controlling cleavage step mediated by 9-cis-epoxycarotenoid dioxygenase (Fig. 1). To test whether ZEP expression was associated with reduced kernel carotenoids, gene-specific primers were developed for quantitative RT-PCR (Supplemental Table S1). In maize B73, ZEP genes showed similar tissue-specific transcript levels (Fig. 7A) but differed during endosperm development. ZEP transcript levels were next evaluated in developing endosperm of the 10 inbred line core subset (Supplemental Table S4) and tested for correlation with seed carotenoid composition (Table II). Both ZEP1 and ZEP2 transcript levels at 20 DAP showed statistically significant inverse correlation with seed zeaxanthin levels (Table II; ZEP1, r = −0.77, P = 0.008; ZEP2, r = −0.86, P = 0.001). This correlation matched the temporal window (20 DAP) of the earlier carotenoid pathway genes (encoding PSY and CrtISO). In contrast, no correlation was seen between ZEP transcripts and lutein levels, as predicted, since lutein is not a substrate of ZEP (Table II). The finding that ZEP expression negatively affects maize endosperm carotenoid accumulation is consistent with analysis of the tomato (Solanum lycopersicum) ZEP mutant (hp3), which showed a 30% increase in carotenoids compared with nonmutant fruits (Galpaz et al., 2008).
Figure 7.
Transcript profiling of ZEP gene family members. A, Top, tissue (see Fig. 4); bottom, developing endosperm (see Fig. 5). Transcript levels were expressed relative to endosperm levels at 20 DAP. Em, Embryo; En, endosperm; L, leaf; R, root; U, unfertilized ovule. B, Pearson correlation (r) and statistical significance (P) for comparison of transcript levels and kernel carotenoid content in maize inbred lines (see Fig. 6). Means of three replicates ± sd are shown. [See online article for color version of this figure.]
Table II.
Correlation of mRNA levels to carotenoid composition in mature seeds
Pearson correlation (r) and statistical significance (P), for comparison of transcript levels and kernel lutein and zeaxanthin content in maize inbred lines, was performed using JMP version 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship. Letters on a red background indicate negative correlation. Inbred lines are as listed in Figure 6. Carotenoid content correlated to mRNA levels is shown on a gray background. L, Lutein; Z, zeaxanthin. [See online article for color version of this table.]
DISCUSSION
Using bioinformatics and genome analysis, we produced a comprehensive identification of key genes and gene family members involved in the biosynthesis of carotenoids in maize and related grass species. Study of a genetically diverse maize germplasm helped elucidate the rate-controlling steps in seed carotenoid biosynthesis. We distinguished gene family members for which transcript levels statistically correlated with seed carotenoid content. We also provided temporal information on gene expression that will guide future breeding efforts whether transgenes or natural alleles are utilized. These data will contribute to predictive strategies to engineer the maize endosperm carotenoid pathway, to stack traits that affect multiple biosynthetic pathways competing for isoprenoid precursors, or to divert isoprenoid precursors to alternate pathways. Lastly, phylogenetic analysis was used to identify putative targets in related grasses, extending these studies to other species that may lack the resources of maize.
Upstream Isoprenoid Biosynthesis Controls Downstream Endosperm Carotenoid Content
The observation that several steps, including upstream isoprenoid biosynthesis pathways, influence flux to carotenoids indicates that PSY is not the only rate-controlling step in controlling maize endosperm carotenoid accumulation. The identification of genes encoding the isoprenoid IPP and GGPP-producing enzymes DXS3, DXR, HDR, and GGPPS1 provides new targets for metabolic engineering of carotenoids in maize across genetically diverse germplasm. DXS and GGPPS both function at branch points of metabolism, whereby several pathways must compete for enzyme products. HDR produces a 5:1 mixture of IPP and dimethallyl diphosphate isomers that must be combined in specific ratios for downstream prenyl transfer reactions of multiple pathways, including that for carotenoid biosynthesis (Rodriguez-Concepcion and Boronat, 2002). It is not surprising to have found these particular enzymes to be influential, given evidence in other species that the enzymes may be rate controlling.
Up-regulation of the upstream isoprenoid and downstream carotenoid pathways is needed to force flux to carotenoids. For example, overexpression of DXS or GGPPS in combination with a carotenoid cluster significantly enhanced carotenoids produced in an E. coli bacterial platform (Wang et al., 1999; Matthews and Wurtzel, 2000). Similar examples in other plants have shown that overexpression of genes for certain upstream precursor pathway enzymes (DXS, DXR, or HDR) plus a downstream pathway enzyme forced pathway flux to the downstream pathway (Botella-Pavia et al., 2004; Carretero-Paulet et al., 2006; Munoz-Bertomeu et al., 2006). In contrast, overexpression of HDS had no effect on enhancing carotenoid accumulation in E. coli or Arabidopsis (Flores-Perez et al., 2008), consistent with the results shown here for maize.
Manipulation of a downstream pathway, in the absence of elevating the isoprenoid pool, may have negative consequences. For example, early attempts to increase tomato fruit carotenoids by PSY overexpression caused a dwarf phenotype due to redirection of GGPP away from gibberellins (Fray et al., 1995). Similar problems in improvement of tomato volatiles by overexpression of geraniol synthase to divert isoprenoids caused reduced carotenoid accumulation (Davidovich-Rikanati et al., 2007), and overexpression of taxadiene synthase redirected GGPP from GGPPS to taxadiene but also depleted the carotenoid pool (Besumbes et al., 2004).
Given that certain isoprenoid biosynthetic enzymes play such a critical role in controlling endosperm carotenoids, breeders might consider new approaches to breeding that incorporate surveys for “biosynthetic potential.” That is, maize carrying a mutation in PSY1 is blocked in the downstream carotenoid pathway and appears as “white maize.” Such an ear would not be considered suitable for breeding for high (yellow) carotenoid content. However, this line may possess high levels of transcripts for one of the “rate-controlling” isoprenoid enzymes and hence have biosynthetic potential, once the carotenoid block is released. Conversely, ears that have high levels of ZEP transcripts may appear light yellow, masking optimal expression of other required biosynthetic steps.
Isomerase Expression Negatively Affects Endosperm Carotenoid Content
The negative correlation seen for CrtISO was surprising given its placement in the pathway. PDS and ZDS were not correlative and the Z-ISO (for 15-cis zetacarotene isomerase) gene has yet to be cloned and therefore could not be tested. The isomerase CrtISO is one of several enzymes, including the desaturases PDS and ZDS, and a second isomerase, Z-ISO, unique to plant carotenoid biosynthesis and required to mediate desaturation of 15-cis phytoene to all-trans lycopene (Li et al., 2007). In contrast, bacterial pathways carry out these conversions with only a single enzyme, CrtI, which is why bacterial CrtI has been used as a simple tool in plant carotenoid metabolic engineering (Matthews and Wurtzel, 2007; Giuliano et al., 2008). At this point, it is a mystery why the CrtISO negative regulation is operative in this part of the pathway. Given that CrtISO expression controls pathway flux in endosperm, one should question whether engineering with a bacterial gene interferes with endogenous regulatory mechanisms of isomerization, as hinted by recent evidence of epigenetic control of CrtISO in Arabidopsis (Cazzonelli et al., 2009). Evidence that an isomerase controls flux partitioning in photosynthetic tissue is suggested by reduced lutein observed in CrtISO mutants and in plants overexpressing bacterial CrtI (Misawa et al., 1994; Park et al., 2002).
Temporal Windows of Gene Expression That Influence Endosperm Carotenoid Content
Predictive metabolic breeding also requires an understanding of the timing of expression for pathway-controlling genes. Carotenoid accumulation in maize endosperm occurs continuously from 10 DAP onward as the seed matures (Li et al., 2008b). The correlations observed for upstream and downstream steps fell into two temporal categories: all genes controlling steps upstream of the carotenoid pathway showed correlation at 25 DAP, whereas the carotenoid pathway genes (PSY1 and CrtISO, ZEP1, and ZEP2) showed correlation at 20 DAP. One possible explanation for the temporal difference is that several pathways, including the carotenoid pathway, compete for isoprenoid precursors during the later phases of endosperm development.
Using Maize to Identify Putative Targets in Other Grass Species for Improvement of Seed Carotenoids
Extension of the maize results using phylogenetic analysis identified orthologs in other grass species that may serve as potential engineering targets. The DXS phylogenetic tree (Fig. 3A) showed that the DXS triplication occurred prior to evolution of the grasses, suggesting that grass orthologs may have similar subfunctionalized roles, as shown for the PSY gene family (Li et al., 2008a, 2009; Welsch et al., 2008). Therefore DXS3 orthologs in other grass species would be an attractive target for initiating investigations of rate-controlling steps for endosperm isoprenoid-derived metabolites. Similarly, the correlative single-copy genes (DXR, HDR, and CrtISO) are potential endosperm targets for other grass species having single-copy genes.
CONCLUSION
In summary, transcript levels of seven genes were found to positively or negatively correlate with endosperm carotenoid content in genetically diverse germplasm; the critical timing of expression of these seven genes fell into two temporal windows of endosperm development. Manipulation of the pathway across the diversity of maize, in cultivars worldwide, therefore, will be somewhat complicated, with timing adding another dimension to this complexity. Incorporating a DXS transgene will not overcome suboptimal alleles that limit pathway flux when the trait is introgressed into various genotypes. However, these data now allow for the development of additional tools to canvas suitable alleles for the downstream steps or to use other transgenes as necessary.
In this study, the influence of candidate gene expression on endosperm carotenoid content was examined from the perspective of synthesis. This approach can also be used to dissect those factors that control endosperm composition for specific carotenoids (e.g. provitamin A carotenoids). Future study of the degradation genes/enzymes is also needed, since carotenoid degradation (Galpaz et al., 2008; Vogel et al., 2008) may potentially reduce carotenoid pools achieved by targeting the genes reported here. Beyond candidate genes, there are likely other factors, as evidenced from genetic (mutant and quantitative trait locus) studies in maize and other plants (Liu et al., 2003b; Wurtzel, 2004).
The utilization of a genetically and biochemically diverse maize germplasm collection to infer pathway regulation is an important resource in identifying potential gene targets for controlling endosperm carotenoid content. Germplasm collections in many plant species hold untapped potential in elucidating underlying regulatory mechanisms that will guide breeding and facilitate predictive metabolic engineering. Future investigations of pathway regulation, including timing of gene expression, which underlies the global network of plant metabolism, could be accomplished by deep transcript profiling of tissues from core collections chosen by metabolite subsorting and correlations made between mRNA levels and metabolite profiles.
MATERIALS AND METHODS
Sequence Analysis and Chromosome Mapping
Rice (Oryza sativa) genes (www.gramene.org) were used as a query to identify orthologs from maize (Zea mays; www.tigr.org; www.plantgdb.org) and to decipher gene families. Sequence analysis was performed using Vector NTI Suite 9.0 (Invitrogen). ESTs and conceptual translation were used to distinguish gene paralogs. Chromosomal positions of genes in the maize B73 inbred line were mapped either by utilizing tools available in the WebAGCoL package (Pampanwar et al., 2005) or at Maize GDB (www.maizegdb.org). Prediction of the chloroplast targeting site was made using ChloroP software (Emanuelsson et al., 1999).
The presence of a stop codon in CrtISO2 was confirmed from a bacterial artificial chromosome clone (98301–103300; AC183901), EST (EE045563), genomic sequence from a methyl filtered library (AZM4_69491), and GSS contig sequence (ZmGSSTUC11-12–04.150.3) from Plant GDB (www.plantgdb.org).
Quantitative RT-PCR and Carotenoid Measurements
Plants and tissues were collected as described (Li et al., 2008b). RNA extraction and quantitative RT-PCR were performed using gene-specific primers (Supplemental Table S2) and normalized to actin, as described previously (Li et al., 2008b). Values are expressed as means of three RT-PCR replicates ± sd. The carotenoid extraction procedure was based on Kurilich and Juvik (1999) and as described previously (Li et al., 2008b). Values are expressed as means of three extraction replicates ± sd.
Plasmids and Functional Complementation
Full-length cDNAs encoding GGPPS1 and GGPPS2 were amplified using approximately 100 ng of cDNA prepared from frozen maize endosperm (20 DAP), whereas GGPPS3 was amplified using 0.5 μg of maize B73 genomic DNA by PCR. Primers were designed on available genomic sequence adapted to the pET23 series of vectors (Novagen) with EcoRI and HindIII restriction sites. The resultant PCR products were cloned in the respective sites, designated as pET23aZmGGPPS1, pET23aZmGGPPS2, and pET23cZmGGPPS3, and used to test function. These plasmids together with empty pET23a vector were transformed into Escherichia coli BL21 (DE3) cells containing pACCAR25ΔcrtE. Double transformants were grown in Luria-Bertani medium containing ampicillin and chloramphenicol and selected for their capacity to accumulate the yellow pigment zeaxanthin-β-d-glucoside. Pigments were extracted and analyzed by HPLC (Gallagher et al., 2004). The sense and antisense primers used to construct the respective plasmids are as follows: GGPPS1, 5′-GAATTCGATAAGGAGGCCGTAACCTGTC-3′ (#1000) and 5′-AAGCTTGCCTGACAAAGTCACAACAGG-3′ (#1001); GGPPS2, 5′-GAATTCATGGCGTTTCACTTCCACCCGCTGGTC-3′ (#1005) and 5′-AAGCTTAGTGAGCAAGAACAGAATAAG-3′ (#1004); and GGPPS3, 5′-GAATTCCCATGAACAAGTTGGCATCCTGCT-3′ (#1565) and 5′-AAGCTTTCAATGCTGTCTGTGCGCCATGA-3′ (#1474).
Statistical Analyses
Pearson correlation analysis of transcript and carotenoid composition from different maize inbred lines was performed using JMP version 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF417573 (ZmGGPPS1), EF417574 (ZmGGPPS2), and EF417575 (ZmGGPPS3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignment of DXS proteins.
Supplemental Figure S2. GGPPS proteins of maize.
Supplemental Figure S3. Functional complementation of GGPPS1 and GGPPS2 from maize.
Supplemental Figure S4. Multiple alignment of CrtISO protein sequences.
Supplemental Figure S5. ZEP gene family structure.
Supplemental Table S1. Gene family sizes in three species of the Poaceae.
Supplemental Table S2. Primers used in the study.
Supplemental Table S3. Transcript levels in developing endosperm from diverse maize lines.
Supplemental Table S4. Transcript levels of ZEP genes in developing endosperm from diverse maize lines.
Supplementary Material
Acknowledgments
We thank Dr. Dwight Kincaid for advice on statistical analysis, Christina Murillo for technical assistance, and members of the Wurtzel laboratory (Dr. Faqiang Li, Rena Quinlan, Oren Tzfadia, Dr. Louis Bradbury, Dr. Abby Cuttriss, Dr. Maria Shumskaya, and Dr. Yu Chen) for helpful discussions.
This work was supported by the National Institutes of Health (grant nos. S06–GM08225, 1SC1GM081160–01, and 5SC1GM081160–02 to E.T.W.), Professional Staff Congress-City University of New York, and New York State.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eleanore T. Wurtzel (wurtzel@lehman.cuny.edu).
Some figures and tables in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Besumbes O, Sauret-Gueto S, Phillips MA, Imperial S, Rodriguez-Concepcion M, Boronat A (2004) Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of taxol. Biotechnol Bioeng 88 168–175 [DOI] [PubMed] [Google Scholar]
- Botella-Pavia P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodriguez-Concepcion M (2004) Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40 188–199 [DOI] [PubMed] [Google Scholar]
- Buckner B, Kelson TL, Robertson DS (1990) Cloning of the y1 locus of maize, a gene involved in the biosynthesis of carotenoids. Plant Cell 2 867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62 683–695 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Cervantes M, Gallagher CE, Zhu C, Wurtzel ET (2006) Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol 141 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Sitrit Y, Tadmor Y, Iijima Y, Bilenko N, Bar E, Carmona B, Fallik E, Dudai N, Simon JE, et al (2007) Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat Biotechnol 25 899–901 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Perez U, Perez-Gil J, Rodriguez-Villalon A, Gil MJ, Vera P, Rodriguez-Concepcion M (2008) Contribution of hydroxymethylbutenyl diphosphate synthase to carotenoid biosynthesis in bacteria and plants. Biochem Biophys Res Commun 371 510–514 [DOI] [PubMed] [Google Scholar]
- Fray R, Wallace A, Fraser P, Valero D, Hedden P, Bramley P, Grierson D (1995) Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from gibberellin pathway. Plant J 8 693–701 [Google Scholar]
- Gallagher CE, Matthews PD, Li F, Wurtzel ET (2004) Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses (Poaceae). Plant Physiol 135 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53 717–730 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26 139–145 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, et al (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam SN (2004) Survey of carotenoid variation and quantitative trait loci mapping for carotenoid and tocopherol variation in maize. Masters thesis. University of Illinois, Urbana-Champaign, IL
- Kim BR, Kim SU, Chang YJ (2005) Differential expression of three 1-deoxy-D: -xylulose-5-phosphate synthase genes in rice. Biotechnol Lett 27 997–1001 [DOI] [PubMed] [Google Scholar]
- Kurilich A, Juvik J (1999) Quantification of carotenoid and tocopherol antioxidants in Zea mays. J Agric Food Chem 47 1948–1955 [DOI] [PubMed] [Google Scholar]
- Li F, Murillo C, Wurtzel ET (2007) Maize Y9 encodes a product essential for 15-cis zetacarotene isomerization. Plant Physiol 144 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tzfadia O, Wurtzel ET (2009) The Phytoene Synthase gene family in the grasses: Subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal Behav 4 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Wurtzel ET (2008. a) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic-stress-induced root carotenogenesis. Plant Physiol 146 1333–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008. b) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress-tolerance. Plant Physiol 147 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Matthews PD, Burr B, Wurtzel ET (1996) Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol 30 269–279 [DOI] [PubMed] [Google Scholar]
- Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003. a) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165 2117–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D (2003. b) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1 195–207 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PD, Luo R, Wurtzel ET (2003) Maize phytoene desaturase and zetacarotene desaturase catalyze a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot 54 2215–2230 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Wurtzel ET (2000) Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl Microbiol Biotechnol 53 396–400 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Wurtzel ET (2007) Biotechnology of food colorant production. In C Socaciu, ed, Food Colorants: Chemical and Functional Properties. CRC Press, Boca Raton, FL, pp 347–398
- Misawa N, Masamoto K, Hori T, Ohtani T, Boger P, Sandmann G (1994) Expression of an Erwinia phytoene desaturase gene not only confers multiple resistance to herbicides interfering with carotenoid biosynthesis but also alters xanthophyll metabolism in transgenic plants. Plant J 6 481–489 [Google Scholar]
- Munoz-Bertomeu J, Arrillaga I, Ros R, Segura J (2006) Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol 142 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa KA, Morgante M, Williams M, Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampanwar V, Engler F, Hatfield J, Blundy S, Gupta G, Soderlund C (2005) FPC Web tools for rice, maize, and distribution. Plant Physiol 138 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson B (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak CJ, Knox RE, Clarke FR, Clarke JM (2007) Identification of QTL and association of a phytoene synthase gene with endosperm colour in durum wheat. Theor Appl Genet 114 525–537 [DOI] [PubMed] [Google Scholar]
- Randolph LF, Hand DB (1940) Relation between carotenoid content and number of genes per cell in diploid and tetraploid corn. J Agric Res 60 51–64 [Google Scholar]
- Rodriguez-Concepcion M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol 130 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JKC, Mazourek M, Chomet P, Brutnell TP (2003) Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Tan BC, McCarty DR, Klee HJ (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283 11364–11373 [DOI] [PubMed] [Google Scholar]
- Walter M, Hans J, Strack D (2002) Two distantly related genes encoding 1-deoxy-D-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J 31 243–254 [DOI] [PubMed] [Google Scholar]
- Wang CW, Oh MK, Liao JC (1999) Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol Bioeng 62 235–241 [DOI] [PubMed] [Google Scholar]
- Welsch R, Wust F, Bar C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Lambert RJ, Wurtzel ET, Rocheford TR (2004) QTL and candidate genes phytoene synthase and zetacarotene desaturase associated with the accumulation of carotenoids in maize. Theor Appl Genet 108 349–359 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET (2004) Genomics, genetics, and biochemistry of maize carotenoid biosynthesis. Recent Adv Phytochem 38 85–110 [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA 105 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.