Abstract
Cotton (Gossypium hirsutum) provides the world's dominant renewable textile fiber, and cotton fiber is valued as a research model because of its extensive elongation and secondary wall thickening. Previously, it was assumed that fibers elongated as individual cells. In contrast, observation by cryo-field emission-scanning electron microscopy of cotton fibers developing in situ within the boll demonstrated that fibers elongate within tissue-like bundles. These bundles were entrained by twisting fiber tips and consolidated by adhesion of a cotton fiber middle lamella (CFML). The fiber bundles consolidated via the CFML ultimately formed a packet of fiber around each seed, which helps explain how thousands of cotton fibers achieve their great length within a confined space. The cell wall nature of the CFML was characterized using transmission electron microscopy, including polymer epitope labeling. Toward the end of elongation, up-regulation occurred in gene expression and enzyme activities related to cell wall hydrolysis, and targeted breakdown of the CFML restored fiber individuality. At the same time, losses occurred in certain cell wall polymer epitopes (as revealed by comprehensive microarray polymer profiling) and sugars within noncellulosic matrix components (as revealed by gas chromatography-mass spectrometry analysis of derivatized neutral and acidic glycosyl residues). Broadly, these data show that adhesion modulated by an outer layer of the primary wall can coordinate the extensive growth of a large group of cells and illustrate dynamic changes in primary wall structure and composition occurring during the differentiation of one cell type that spends only part of its life as a tissue.
Fiber from cotton (Gossypium hirsutum) is the world's dominant natural textile fiber (Chen et al., 2007). This unique single cell is also a valuable research model because of its extreme length (≥2.25 cm) and thick (3–6 μm) cellulosic secondary wall (Kim and Triplett, 2001). Cotton fiber morphogenesis initiates with the bulging of selected seed epidermal cells near the day of flowering (anthesis) and is described temporally by days post anthesis (DPA). Seed and fiber development occurs within (typically) four individual carpels (locules) within the ovary (or cotton boll). A typical locule of Deltapine 90 cotton contains eight to nine developing seeds each with approximately 14,500 fibers (Bowman et al., 2001). Therefore, approximately 123,000 fibers reach a final length of approximately 2.7 cm in the confined space of one locule, which expands to only approximately 5 cm3 internal volume (inclusive of seed volume). Previously, there has been little consideration of how the extreme elongation of so many cells could be optimized within a confined space. Because cotton fibers initiate as individuals and form a fluffy mass after boll opening, it was previously assumed without direct evidence that they also progressed through the entire elongation stage as individuals.
Similar to other expanding plant cells, the primary walls of 10-DPA cotton fibers contain 23% cellulose fibrils and 22% protein as well as noncrystalline matrix polysaccharides including xyloglucan (XG) and pectin (Meinert and Delmer, 1977). Matrix polysaccharides are chemically complex. Briefly, XG has a β-1,4-glucan backbone with frequent side groups of α-1,6-linked Xyl, which may be further modified by the addition of Gal and Fuc (Pauly and Keegstra, 2008). Fucosylated xyloglucan (Fuc-XG), with α-Fuc-(1,2)-β-Gal side chains, exists in cotton fiber walls (Hayashi and Delmer, 1988). Pectins are GalUA-rich polysaccharides including, among others, homogalacturonan (HG), substituted HGs (including xylogalacturonan), and rhamnogalacturonan I (RG-1) with a GalA-Rha disaccharide repeating unit in its backbone. HG may have varying amounts of methyl and/or acetyl side groups, whereas RG-1 has a wide variety of sugar side groups and simple or branched oligosaccharide side chains, including arabinan and galactan (Mohnen, 2008).
Between elongation and secondary wall thickening in cotton fiber, there is a “transition stage” of development when elongation slows and the wall thickening begins via deposition of the “winding” layer. Like the S1 layer in wood fiber, the winding layer has intermediary cellulose content and microfibril angle (relative to the longitudinal fiber axis) compared with the primary and secondary walls. At the end of the transition (typically at approximately 21 DPA), elongation and winding layer deposition cease while approximately 95% cellulose deposition in the secondary wall begins (Meinert and Delmer, 1977). Finally, the boll opens and the fiber and seed dry at approximately 45 to 60 DPA (Basra, 1999).
Since cotton fibers can be easily obtained in bulk as a single cell type, their staged cell wall synthesis offers unique opportunities for understanding the mechanisms of construction and deconstruction of cell walls. This is foundational knowledge to support the optimization of plant biomass as feedstocks for biofuels production as well as the improvement of fiber crops as renewable materials (Pauly and Keegstra, 2008). Previously, there was little specific knowledge about cotton fiber cell wall structure or understanding of how cell walls might control fiber differentiation beyond traditional concepts of thin, extensible, primary walls versus thick, cellulose-rich, secondary walls (Basra, 1999). A notable exception was the demonstration of biphasic distribution of wall polymers in 1- to 2-DPA fibers (Vaughn and Turley, 1999), but functional consequences of this wall structure were unknown. Similarly, the transition stage involves up-regulation of genes putatively encoding cell wall hydrolases as well as the loss of certain neutral cell wall sugars and reduction in pectin and XG mass (Meinert and Delmer, 1977; Shimizu et al., 1997; Tokumoto et al., 2002, 2003; Guo et al., 2007), but a cell biological explanation for these developmental events was lacking.
This paper reports the surprising discovery that a specialized cell wall layer, which we called the cotton fiber middle lamella (CFML), fuses elongating cotton fibers into tissue-like bundles during elongation. The CFML was visualized by both cryo-field emission-scanning electron microscopy (cryo-FE-SEM) and transmission electron microscopy (TEM) and shown, through TEM immunolabeling, to contain cell wall polymers including HG and XG. The means of forming and consolidating cotton fiber bundles via the CFML, as well as the highly organized packing of fiber bundles within the boll, were revealed by life-like views of elongating cotton fibers by cryo-FE-SEM. Near the end of elongation in the transition stage, the CFML was degraded under developmental control. Complementary changes in gene expression, enzyme activity, and cell wall structure were demonstrated by microarray analysis and quantitative real-time reverse transcription-PCR (qPCR), in vitro enzyme assays, analysis of both neutral and acidic glycosyl residues in noncellulosic cell wall matrix polymers, characterization of epitopes within extractable cell wall polymers, and TEM immunolabeling. Cumulatively, the data show that (1) a specialized outer layer of the primary wall can coordinate the extensive growth of a large group of cells, and (2) dynamic changes in primary wall structure and composition occur in one cell type to facilitate its oscillation between differentiation as an individual, a simple tissue, and then an individual once more.
RESULTS
Time Course of Fiber Development
Fiber harvested from Deltapine 90 cotton was used for the biochemical and gene expression experiments. Deltapine 90 is an advanced cultivar of upland cotton (the type most commonly grown worldwide), and it was the source of a unique set of ESTs biased toward genes expressed during early secondary wall deposition (Haigler et al., 2005). Because the temporal progression of cotton fiber development varies somewhat with genotype and is particularly sensitive to temperature differences, we determined the time course of fiber length and weight gain, as well as cellulose content, during growth in an air-conditioned greenhouse with a 26°C/22°C day/night cycle (Supplemental Fig. S1). Under these highly controlled temperature conditions, late elongation and the first low-level rise in cellulose percentage within the winding cell wall layer occurred concurrently at 17 to 22 DPA, representing the transition stage. At 22 DPA, secondary wall deposition via approximately 95% cellulose deposition began, as shown by the subsequent rapid increases in fiber weight, fiber weight per unit length, and cellulose content, with elongation slowing greatly after that time (Supplemental Fig. S1).
Microscopic Evidence for a Transient, Tissue-Like Phase of Cotton Fiber Development
In contrast to previous conceptions of elongation of cotton fibers as individual cells, cross-sections of elongating cotton fibers observed with the light microscope showed bundles of tightly packed cells (Fig. 1A) reminiscent of a simple plant tissue (a tissue composed of only one cell type; Evert, 2006). Higher magnification views by TEM were also consistent with the existence of tissue-like aggregates of fiber (Fig. 1B). Hundreds of fiber cross-sections have been observed in one bundle, and Supplemental Figure S2 proves that these bundles of cells do represent fiber cross-sections.
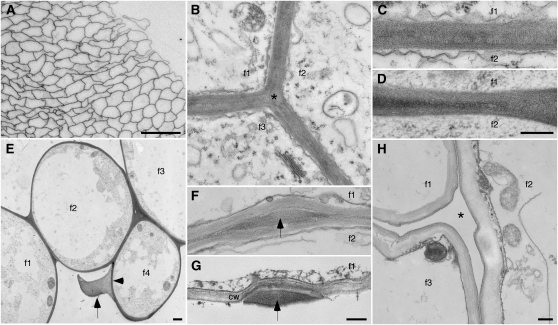
Figure 1.
Micrographs showing the existence of cotton fiber bundles. f1 to f4 indicate interiors of individual fibers, where applicable. A, Light micrograph of a cross-section of part of a fiber bundle. B to H, TEM images prepared by chemical (B, C, F, and H) or cryo (D, E, and G) fixation. During elongation, thin coherent walls (B–E) join adjacent fibers, including filling corners between three fibers (asterisk in B). E shows the distinction between the inner primary wall (arrowhead) and material between adjacent fibers (arrow) in a case where a missing fiber (from space at bottom center) likely separated from the forming bundle during specimen preparation. Occasionally, the thin joined walls are interrupted by material-filled bulges (F and G, arrows). In G, the lower fiber is missing, and the bulge material is clearly distinct from the traditional primary cell wall (cw) of the upper fiber. H shows a cleared space (asterisk) between three fibers at the onset of secondary wall thickening after the interfacial material had broken down (see below). Ages of fiber (DPA) are as follows: A, 15; B, C, and G, 10; D and E, 5; F, 20; and H, 25. Bars = 50 μm (A), 300 nm (D, for C and D; G, for F and G; and H, for B and H), and 1 μm (E).
Both chemical and cryo fixation prior to TEM observations revealed a thin (approximately 300–400 nm), apparently coherent (double) wall between adjacent fibers in many locations (Fig. 1, B–D). There were no discernible boundaries between adjacent fibers over large regions, proving that these bundles were not merely aggregates of coaligned individual fibers. In addition, we sometimes saw material (Fig. 1E, arrow) that appeared distinct from the inner primary wall surrounding the protoplast (Fig. 1E, arrowhead). In this specimen, the outer material was probably attached to only one fiber, because there was no lower fiber to keep it compressed within a fiber bundle. Occasionally, 1- to 2-μm-wide material-filled bulges occurred between adjacent fibers after both chemical and cryo fixation (Fig. 1, F and G). In longitudinal sections, the bulges were approximately 4 to 12 μm long and randomly located along the fiber length between regions of coherent (double) primary walls (Supplemental Fig. S3). The tissue-like bundles of fiber persisted during elongation, but the material between fibers disappeared and fiber individuality was restored as secondary wall deposition commenced (Fig. 1H; the asterisk indicates the cleared interfiber space).
Cryo-FE-SEM Revealed How Fiber Bundles Formed and Their Packing within the Boll
Using cryo-FE-SEM, we determined how the fiber bundles originated and their implications in situ within the cotton boll. After cutting a window in the boll wall, followed by rapid freezing and mounting on the ultracold cryo-FE-SEM stage, cotton fibers were observed in their life-like state within the boll. By this method, the outermost layer of fibers of each seed was observed. Shortly after initiation, the tips of elongating fibers began to intertwine, which appeared to nucleate and entrain fiber bundles (Fig. 2A). Starting at 2 DPA (data not shown), a distinct outer layer of material was observed to fuse adjacent fibers (Fig. 2B, double arrow), and this material was present even at the fiber tips (Fig. 2C, arrow). During early elongation, the forming bundles also engaged in directional, curved elongation (Fig. 2D) as a precursor to forming wave-like paths as elongation progressed (see below). Until 3 DPA, many fiber tips were visible on the outer surface of the fiber mass, but afterward, only a few tips were visible, apparently because most of them turned inward toward the seed (Fig. 2E).
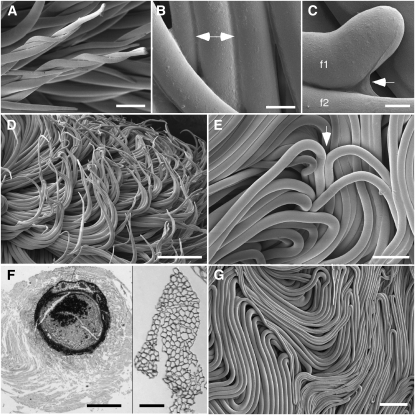
Figure 2.
Micrographs showing the onset and effects of fiber bundling. A to E and G, Cryo-FE-SEM observations of fiber within the boll in the “as living” state, showing twisting fiber tips (A), material fusing adjacent fibers (double arrow; B), material (arrow) connecting the tip of one fiber to another fiber below (C), fiber bundles adopting curving paths (D), and a site on the outside of the fiber mass (arrow) where fiber tips have turned inward toward the seed (E). F, Specialized paraffin embedding of intact locules revealed organized fiber around each developing ovule. The left panel shows a cross-section of a 10-DPA ovule (densely stained) surrounded by a packet of its own fiber (gray halo). The distinct outer boundary of the “fiber packet” was consistently observed. The right panel shows a higher magnification view of a cross-sectioned fiber bundle within the fiber packet, even though the extended specimen preparation for paraffin embedding resulted in some fiber separation (see Supplemental Fig. S4 for additional views). G, From 5 DPA onward, a mass of tightly packed fiber bundles in meandering patterns was closely appressed to the boll wall. Ages of fiber (DPA) are as follows: A and D, 2; B and C, 6; E, 4; F, 10; and G, 5. Bars = 50 μm (A), 9 μm (B), 5 μm (C), 250 μm (D and G), 100 μm (E and F, right), and 750 μm (F, left).
After specialized cryogenic processing for paraffin sectioning, sections of developing ovules with attached fiber showed the ovule (dark circle) surrounded by a packet of fiber with a distinct, relatively smooth surface, because fiber edges, not tips, dominated the surface (Fig. 2F, left; see Supplemental Fig. S4A for a higher magnification view). Throughout the depth of the fiber packet, fiber bundles were running at various angles relative to the plane of the section, but where they traveled perpendicularly, bundled fiber cross-sections were once again observed (Fig. 2F, right; Supplemental Fig. S4B). The upper surface of the fiber mass (appressed to the inner boll wall) showed tightly packed bundles of fiber in meandering, wave-like patterns (Fig. 2G), and organized fibers filled the locule (Supplemental Fig. S4C). Because of the constrained space within the locule, fibers did not change their overall packing pattern even after they once more became separate individuals during secondary wall deposition (data not shown).
TEM Immunolabeling Demonstrated the Cell Wall Nature of the CFML
Higher magnification views of the CFML and its degradation products were consistent with its existence as a specialized layer of adhesive cell wall polymers, distinct from the inner primary wall. In cryo-FE-SEM, the CFML appeared sheet-like as it stretched between two fibers at a separation point (Fig. 3A), which is suggestive of its adhesive, or glue-like, properties. After cryo fixation for TEM, the fibrillar nature of the CFML was evident in a “fringed” outer layer of the primary wall, probably where fibers had separated during processing (Fig. 3B). The fibrillar degradation products of the CFML at the transition stage resembled cell wall components, and they cleanly separated from the inner primary wall (Fig. 3C, arrowhead) before they finally disappeared from interfiber spaces (Fig. 1H).
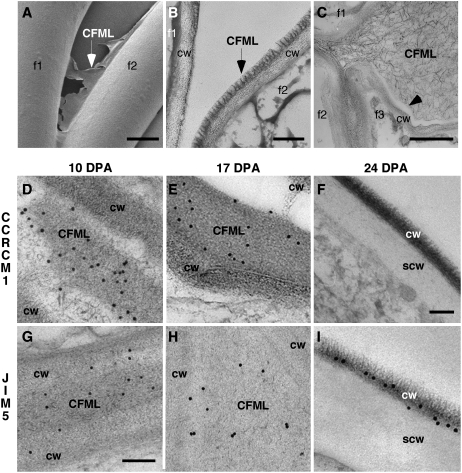
Figure 3.
Evidence from cryo-FE-SEM and TEM analyses that the CFML is a special cell wall layer that breaks down in transition-stage fiber. Individual fibers are indicated in A to C by f1 to f3. The traditional primary cell wall of an individual fiber is indicated by cw, and the secondary wall at 24 DPA is indicated by scw. TEM immunolabeling was performed at 10 DPA (D and G), 17 DPA (E and H), and 24 DPA (F and I). In contrast to other panels including parts of two or three fibers, only one fiber is shown at 24 DPA in F and I, because CMFL degradation occurring previously led to spaces between fibers. Antibodies used were CCRC-M1 (D–F) and JIM5 (G–I). CCRC-M1 recognizes fucosylated side chains of XG, and JIM5 recognizes HG with a relatively low degree of methylesterification. A, Cryo-FE-SEM at 4 DPA showed a torn sheet of CFML material (arrow) between the inner primary walls of two adjacent fibers at a separation point. B, Similarly, TEM after cryo fixation at 10 DPA showed a distinct fringed layer (arrow) as the outer layer of the primary wall. Probably, this fiber separated from the one on the left during specimen preparation, with CFML material remaining attached to only one fiber. C, At 20 DPA during the transition, CFML polymers dispersed between fibers before they disappeared entirely (compare with Fig. 1H). The arrowhead shows a sharp separation zone between CFML polymers and the inner primary cell wall. D to F, CCRC-M1 labeled the CFML at 10 and 17 DPA, but labeling was undetectable by 24 DPA. In E, a gap was present between CFML in a bulged area and the primary cell wall of the adjacent fiber (top right corner), again indicating a distinction between these primary wall layers. G to I, JIM5 labeled the CFML at 10 and 17 DPA and the inner primary wall more lightly. However, at 24 DPA during secondary wall deposition, labeling on the primary wall that remained after CFML degradation was more intense than at earlier DPA. Bars = 5 μm (A), 200 nm (B), 1 μm (C), 300 nm (F, for D–F and I), and 150 nm (G, for G and H).
Twelve monoclonal antibodies (mAbs) recognizing epitopes in cell wall polymers (and one polyclonal antibody) gave positive results in TEM or fluorescence immunolabeling to show that the CFML contained cell wall polymers. Supplemental Table S1 summarizes the probes used and all results. Supplemental Figure S5 shows the larger cellular context of the TEM immunolabeling results shown in Figure 3 and demonstrates that immunolabeling was specific for cell wall regions of the samples. Antibodies that labeled CFML material at 10 and 17 DPA included CCRC-M1 (Fig. 3, D and E), which recognizes the α-Fuc-(1,2)-β-Gal side chain of Fuc-XG. In addition, nine of 11 other XG antibodies tested labeled positively, including CCRC-M106 with specific affinity for the XXFG epitope [containing α-Fuc-(1,2)-β-Gal] and a polyclonal anti-XG that does not depend on a fucosylated side chain for binding and likely recognizes the XG backbone (Lynch and Staehelin, 1992; Supplemental Table S1). Positive labeling of the CFML was also obtained with JIM5, which recognizes short stretches of HG pectin, typically with a relatively low degree of methyl esterification (Fig. 3, G and H). CCRC-M38, which is able to bind fully deesterified HG, also positively labeled the CFML (Supplemental Table S1; data not shown). Immunolabeling was negative at 10 and 17 DPA for four antibodies recognizing the RG-1 backbone and five antibodies recognizing possible RG-1 side chains (Supplemental Table S1).
At 24 DPA, the CFML had been degraded so that the space between fibers was cleared (Fig. 3, F and I). Interestingly, however, the density of JIM5 labeling in the residual primary wall was higher at 24 DPA during secondary wall deposition than it was during elongation (compare the numerous colloidal gold particles on the primary wall, labeled cw in Fig. 3I, versus much lighter labeling of cw areas in elongating fibers in Fig. 3, G and H). (Note that at 24 DPA, the primary wall [cw] had been pushed to the outside due to deposition of the secondary wall [scw; Fig. 3, F and I].) No such increase of primary wall labeling at 24 DPA was observed with CCRC-M1 (Fig. 3F), pointing to pectin-specific changes.
Similar to results from other systems (Derbyshire et al., 2007), the increase in JIM5 labeling of the residual primary wall (Fig. 3I) could reflect the deesterification of HG at the end of cotton fiber elongation. Initial attempts to use JIM7, which typically recognizes more highly esterified HG, in TEM experiments in order to probe further into changing pectin esterification did not result in convincing positive labeling (data not shown). Since the ester groups on the HG backbone that are required for JIM7 labeling may be oxidized in air, JIM7 and JIM5 were retested in parallel by fluorescence immunolabeling of freshly cut semithin sections (Fig. 4). At 10 and 17 DPA, when fibers were bundled, JIM5 labeling was weak (Fig. 4, A and B), whereas JIM7 labeling was strong (Fig. 4, G and H). At 24 DPA, when the CFML had been degraded to release individual fibers, JIM5 labeling increased (Fig. 4C), while JIM7 labeling decreased (Fig. 4I). After treatment of sections with Na2CO3 as a deesterifying agent, the JIM5 signal became markedly brighter as expected at all DPA tested (Fig. 4, D to F), whereas the JIM7 signal dimmed but did not disappear (data not shown). These results are consistent with an overall relatively high level of HG esterification in the cotton fiber primary wall during elongation followed by deesterification as elongation ends. However, there was evidence of zonation in the degree of HG esterification within the wall during elongation, as shown by superimposing the images of 10-DPA fiber labeled with JIM5 before (Fig. 4A, signal changed to red) and after Na2CO3 treatment (Fig. 4D, signal retained as green). In the composite image (Fig. 4J), the red JIM5 signal from the untreated sections occurred frequently in the middle of the double primary walls between paired fibers as well as in corners between three fibers. These data are consistent with the TEM results for JIM5 labeling (Fig. 3, G and H), and cumulatively, the data indicate that the CFML is enriched in HG with a relatively low degree of esterification.
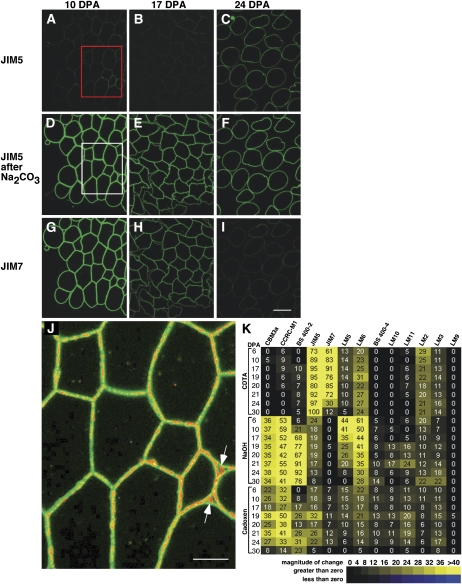
Figure 4.
Fluorescence analysis of cotton fiber cell wall epitopes illustrates changes correlated temporally with the end of elongation as well as transition-stage CFML breakdown. A to I, Fluorescence micrographs of semithin sections of 10-, 17-, and 24-DPA cotton fiber cross-sections immunolabeled with JIM5 or JIM7. Sections were optionally pretreated with 100 mm Na2CO3 as a deesterifying agent prior to labeling. Successive sections of the same sample are shown for each DPA. The exposure time was equivalent, and no image processing was performed, which allowed comparison of labeling intensity between samples. The boxed areas in A and D were superimposed to produce J (see below). At 10 and 17 DPA (D, E, G, and H), strong signals arose from HG distributed throughout the primary wall following labeling with JIM5 (after Na2CO3) and JIM7. These images show that the fibers were bundled into a simple tissue during elongation. In contrast, by 24 DPA, CFML degradation had occurred to release individual fibers (C and F). Comparison of C with A and B shows that HG in the main primary wall was less esterified after elongation ended, a conclusion that was reinforced by the strong decrease in the JIM7 signal at 24 DPA in I compared with G and H. Bar in I = 20 μm for A to I. J, The dim positive signal for JIM5 untreated (boxed area in A; signal changed to red) was superimposed onto the bright signal after Na2CO3 pretreatment (boxed area in D; signal retained as green). The red signal aligned with the middle of joined primary walls between adjacent fibers in many locations and filled larger spaces between three fibers (arrows), indicating that fiber junctions were enriched in HG with a lower degree of esterification. Bar = 10 μm. K, Heat map arising from CoMPP analysis of 6- to 30-DPA fiber extracted with CDTA, NaOH, and cadoxen. The 24 extracts (listed on the y axis) were printed and exposed to 13 cell wall polymer probes (listed on the top x axis). Supplemental Table S1 shows binding properties of the probes, and all intensity values are relative to 100. In the CDTA fraction, the inverse changes in the JIM5 (increasing) and JIM7 (decreasing) signals after 20 DPA correlate with fluorescence immunolabeling (compare C and I). In the cadoxen fraction, the CCRC-M1 and JIM5 epitopes (but not the JIM7 epitopes) had elevated signals at 19 to 21 DPA, which correlated with the timing of release of these epitopes from the CFML as observed by TEM.
Transition-Stage CFML Breakdown Was Correlated with Changes in Cell Wall Polysaccharides
Given that the CFML existed only during cotton fiber elongation, we sought evidence for biochemical changes in the cell wall correlated with its breakdown at the transition stage. One technique used was comprehensive microarray polymer profiling (CoMPP), which involves sequentially extracting cell wall polymers. The cell wall extracts are then spotted as arrays and probed with mAbs or carbohydrate binding modules (CBMs; Supplemental Table S1). Quantified spot signals provide semiquantitative information about epitope or polymer occurrence (Moller et al., 2007; Fig. 4K). Three extractants (cyclohexane diamine tetraacetic acid [CDTA; a Ca2+ chelator], NaOH, and cadoxen) were used in succession to extract wall polymers that were less and then more tightly bound within cell walls. The consistency of CoMPP data with known aspects of cotton fiber differentiation is shown by the following: (1) strong signals from 6 to 30 DPA for CDTA-soluble HG (JIM5) and NaOH-soluble XG (CCRC-M1), which likely reflect the matrix components of the persistent inner primary wall; and (2) a high signal for NaOH-extractable β-1,3-glucan (callose; detected by BS 400-2) beginning at 17 DPA. Other analyses have shown that callose is deposited just outside the cotton fiber at the transition stage (Maltby et al., 1979; Salnikov et al., 2003). In addition, HG deesterification at the end of elongation detected by fluorescence immunolabeling was paralleled by inverse changes for the signals from CDTA-soluble JIM5 (increased) and JIM7 (decreased) after 21 DPA.
We examined the CoMPP results for patterns of change that correlated temporally with the transition-stage degradation of the CFML. Such changes were evident in the cadoxen fraction: both the CCRC-M1 and JIM5 signals were approximately constant during elongation, approximately doubled between 17 and 19 DPA, and then declined with similar patterns thereafter. Since CCRC-M1 and JIM5 did not label the thickening cell wall (see lack of colloidal gold on scw areas in Fig. 3, F and I), their maximum signals in the cadoxen fraction at 19 DPA in CoMPP profiling could reflect a breakdown of the CFML (see “Discussion”). Notably, this pattern was not shared by JIM7 in the cadoxen fraction, which is consistent with evidence that low esterified HG is enriched in the CFML (Figs. 3, G and H, and 4J). In a second trial of CoMPP profiling, LM8 recognizing an RG-1 side chain, xylogalacturonan, was tested because it labels walls of some cells that undergo complete detachment, such as those in the root cap (Willats et al., 2004). No signals above 5 (the threshold for interpretation) were detected in cotton fiber extracts, and LM8 was also negative in immunolabeling experiments (data not shown). Similarly, the antibody 2F4 that recognizes a Ca2+-dependent epitope in HG (Bush and McCann, 1999, and refs. therein) did not yield positive results when tested with optimum procedures in TEM or fluorescence analyses (Supplemental Table S1; Supplemental Protocol S2).
Analysis of sugars in hot water-soluble versus insoluble cell wall matrix polymers (excluding cellulose) further documented changes consistent with CFML degradation at the transition stage. Gas chromatography-mass spectrometry analysis of trimethylsilyl (TMS)-methylglycoside derivatives allowed quantitation of both neutral and acidic glycosyl residues (Fig. 5A; complete data are shown in Supplemental Tables S2 and S3). Expressing the data as nanomoles of sugar per milligram of dried fiber revealed changes over developmental time within a unit of fiber cell wall mass. A 56% decrease in the total amount of noncellulosic sugars in the hot water-insoluble fraction occurred between 17 and 19 DPA. The amounts of Ara, Fuc, Gal, GalA, and Rha showed significant decreases by 19 and/or 21 DPA compared with 17 DPA (P ≤ 0.05). For Ara, Gal, and Rha, these results paralleled other cotton fiber neutral sugar analyses that showed decreases near the time of onset of secondary wall deposition as well as low levels of Fuc throughout fiber development (Meinert and Delmer, 1977). Notably, these decreases were reflected in the lack of increase (trend downward) in fiber weight/length between 17 and 21 DPA even though cellulose content within the newly deposited winding layer was increasing simultaneously (Supplemental Fig. S1). Such an effect could arise from CFML degradation. By 30 DPA, the amounts of Ara and Rha in the hot water-soluble fraction cell wall fraction had increased significantly compared with 17 DPA (P ≤ 0.05), and Gal and GalA showed the same trend (Supplemental Table S3). The CFML had physically disappeared several days earlier, but remnant sugars or oligomers could have persisted within the closed boll compartment and been hot water extractable, a possibility that is supported by the increase in transition-stage fiber of soluble carbohydrate oligomers detected by HPLC (Murray et al., 2001).
Figure 5.
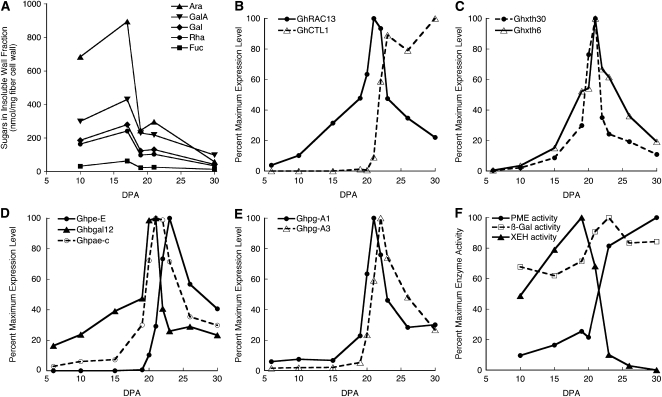
Biochemical and molecular analyses showing changes in cell wall composition and capacity for cell wall hydrolysis in transition-stage fiber. A, In the hot water-insoluble fraction of fiber cell wall matrix polymers, the sugars Ara, GalA, Gal, Rha, and Fuc showed significant decreases (P ≤ 0.05) at 19 and/or 21 DPA. See Supplemental Tables S2 and S3 for complete results for sugar analysis in the insoluble and soluble fractions, respectively. B to F, Percentage of the maximum observed for that measurement type; nonnormalized data are shown in Supplemental Tables S5 and S6. Transcriptional profiles in 6- to 30-DPA fiber are shown for genes related to the following developmental stages: GhRAC13 with peak expression in the transition and GhCTL1 with peak expression during secondary wall deposition (B); XG modification (Ghxth30 and Ghxth6; XG endotransglycosylase/hydrolase type; C); pectin modification (Ghpe-E and Ghpae-c; pectin esterase type) and galactosidase (Ghbgal12; D); and pectin hydrolysis (Ghpg-A1 and Ghpg-A3; PG type; E). F shows transition-stage peaks and/or up-regulation of enzyme activities: PME, β-galactosidase (β-Gal), and XEH.
Transition-Stage Fibers Showed Genetic and Enzymatic Competence for Cell Wall Modifications
Microarray analysis was performed to compare gene expression at 6 and 10 DPA (elongation stage), 24 DPA (early secondary wall stage), and 20 DPA (transition stage) with the aim of revealing genes that were up-regulated at the transition stage to support secondary wall deposition. The cDNAs spotted on the array originated from 20 DPA (Haigler et al., 2005), which therefore became the common reference sample for the experiment (Smyth, 2005). The data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE12202 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12202). This site includes a tab-delimited text file, “Cotton_Microarray_Summary_Annotated,” showing (1) gene expression ratios (6:20, 10:20, and 24:20 DPA) and P values, and (2) best plant protein matches and E-values for the cotton cDNAs on the microarray as well as annotations as found in public databases. Preliminary analysis of this microarray experiment showed that (1) homologs of many secondary wall-related genes required for xylem sclerenchyma differentiation were up-regulated in cotton fiber at 20 and 24 DPA (transition and early secondary wall stages) compared with 6 and 10 DPA (elongation/primary wall stage), and (2) there was a high (approximately 90%) agreement between the microarray results and qPCR analysis (Haigler et al., 2009).
In the microarray data, the group of genes with significant up-regulation at 20 DPA compared with 6 and 10 DPA (P ≤ 0.05) included sequences apparently encoding cell wall-modifying enzymes. Note that in the “Cotton_Microarray_Summary_Annotated” file posted at NCBI, genes that are up-regulated at 20 DPA have negative 6:20 and 10:20 log2 ratios. These included genes encoding two putative pectin-modifying enzymes: (1) a pectin methylesterase (PME; NCBI EST identifier CO495780; Ghpe-E) with 10:20 DPA log2 ratio = −2.2 (4.6-fold up-regulation at 20 versus 10 DPA), and (2) a polygalacturonase (PG; NCBI EST identifier CO492632; Ghpg-A3) with 10:20 DPA log2 ratio = −2.48 (5.7-fold up-regulation at 20 versus 10 DPA). These microarray results were confirmed by qPCR between 6 and 30 DPA of fiber development (Supplemental Table S4 shows qPCR primers). Corresponding to its higher dynamic range (Busch and Lohmann, 2007), qPCR showed 4,229-fold and 12-fold up-regulation at 20 versus 10 DPA for Ghpe-E and Ghpg-A3, respectively (Supplemental Table S5).
The qPCR analyses were expanded to include a total of 31 genes encoding representatives of several families of cell wall-modifying proteins (Fig. 5, C–E; Supplemental Table S5). Fiber developmental marker genes were also included: (1) GhRAC13, with peak expression at 21 to 22 DPA during the transition (Kim and Triplett, 2004), and (2) GhCTL1, with peak expression from 23 DPA onward during secondary wall deposition (Zhang et al., 2004; Fig. 5B). GhRAC13 encodes a small GTPase that can modulate hydrogen peroxide production (Delmer et al., 1995), and GhCTL1 encodes a protein with an obligatory but unknown role in secondary wall cellulose synthesis (Brown et al., 2005). In order to emphasize developmental patterns of expression, the graphs show percentages of the maximum expression observed for each gene, and nonnormalized data are shown in Supplemental Table S5. The graphs show genes with transition-stage expression that was increased by a factor of at least 4 compared with 10 DPA (during rapid elongation) in four sequence families related to modification or hydrolysis of cell wall HG or pectin: (1) XG endotransglucosylase/hydrolase (xth; Fig. 5C); (2) β-galactosidase (β-gal; Fig. 5D); (3) pectin esterases, which include pectin methylesterase (pme) and pectin acetylesterase (pae; Fig. 5D); and (4) polygalacturonase-like (pg; Fig. 5E). In Figure 5, C to E, all but Ghpe-E reached their peak expression at 20 to 22 DPA, toward the end of the transition when the CFML was degraded, similar to GhRAC13 (Fig. 5B). The up-regulation of Ghpe-E was later and similar to that of GhCTL1, although Ghpe-E expression peaked at 23 DPA and GhCTL1 expression was sustained as secondary wall deposition proceeded (Fig. 5, compare B and D).
The enzymes encoded by cell wall hydrolase genes could help to degrade the CFML, and relevant enzyme activities were demonstrated in fiber extracts. As before, percentage maximum activity is graphed in each case (Fig. 5F), and nonnormalized data are shown in Supplemental Table S6. XTH enzymes with an oxygen acceptor may act as XG endohydrolases (XEHs), degrading the XG backbone (Vicente et al., 2007). XEH activity peaked at 19 DPA, which correlates with the timing in CoMPP of the transitory increase in the cadoxen-soluble CCRC-M1 epitope within Fuc-XG (Fig. 4K). β-Galactosidases hydrolyze terminal, nonreducing, β-d-galactosyl residues from β-d-galactosides, including pectin side chains, and this enzyme activity was elevated between 19 and 23 DPA in fiber extracts. β-Galactosidase activity could account for the posttransition disappearance of the LM5 epitope, putatively in galactan, in the CoMPP NaOH fraction (Fig. 4K). NaOH-extractable LM5 epitopes may arise from a subset of pectin galactan domains that are covalently linked to other cell wall polymers (Moller et al., 2007). However, LM5 did not label CFML in situ in TEM immunolabeling experiments (data not shown), perhaps because the LM5 epitope was rendered inaccessible by interaction with other polymers.
DISCUSSION
These data show that an outer layer of the cotton fiber primary wall, the CFML, functions as an adhesive middle lamella, bundling fibers together and aiding their organized, extensive elongation within the confined locule space. Numerous types of data collected from two cultivars of cotton (Deltapine 90, a modern elite line, and Coker 312, an older commercial cultivar) growing in three different greenhouses consistently converged to reveal this new aspect of cotton fiber development. The typical middle lamella between cells of the plant body, together with cell wall specializations at tricellular junctions, connect adjacent cells during normal cell expansion. Covalent or noncovalent cross-links between cell wall polymers in the middle lamella and the inner primary walls of two adjacent cells generate intercellular adhesion (Jarvis et al., 2003; Waldron and Brett, 2007). In contrast to the typical persistence of the middle lamella during growth of tissues in the plant body, cotton fiber exerts developmental control over the synthesis and degradation of the middle lamella that joins adjacent fibers (the CFML). After initiation as individual cells, cotton fiber bundles are entrained by twisting fiber tips during early elongation. In the same time frame as the development of extremely pointed fiber tips at 2 DPA, the synthesis of the CFML begins to generate adhesion between large bundles of adjacent fibers, forming a simple cotton fiber tissue during elongation. This picture of fiber elongation contrasts markedly with prior assumptions that cotton fibers elongated as individuals. The formation and packing of fiber bundles caused each developing ovule to become surrounded by an organized packet of its own fiber. We now realize that previously published images reflected the ability of the CFML to keep fiber closely packed around a developing ovule even when single ovule/fiber units were removed from the boll during elongation and processed for detection of GUS activity (see figure 7A in Zhang et al., 2004; compare the packed fiber at 17 DPA with the dispersed fiber at 21 DPA). The properties of the CFML, its developmentally controlled degradation at the onset of secondary wall deposition, and comparison with other cell detachment processes are discussed further below.
The Material Properties of the CFML
The existence of fiber bundles and apparently coherent “double” walls between adjacent fibers supports the adhesive properties of the CFML under physiological conditions. In contrast, as occurs for many synthetic adhesives (Bascom and Cottington, 1976), the CFML tended to crack upon exposure to ultracold temperatures during cryo fixation for cryo-FE-SEM or TEM freeze substitution. When this occurred, joined fibers separated to reveal distinct CFML material as the outer layer of the primary wall (Figs. 1E and 3, A and B). Separated fibers were less frequent in cryo-FE-SEM views, probably because largely immobilized fibers still within the boll were observed immediately. In contrast, fibers on isolated seeds had many opportunities to separate during the prolonged freeze-substitution protocol, with its multiple solvent exchanges prior to resin embedding for sectioning. Nevertheless, the observation of some joined fibers in sections after cryofixation and freeze substitution (Fig. 1, D and E) provided important confirmation of the reality of fiber bundling via an adhesive CFML in vivo because cryo fixation instantly immobilizes cellular structure (Dahl and Staehelin, 1989). Because of frequent cracking of the CFML upon freezing, chemical fixation, which provided similar images of the cell wall (Fig. 1, B, C, and F), was usually preferred for TEM immunolabeling experiments.
Other middle lamellae often contain HG and may include RG-1 pectin and/or XG (Ishii, 1981; Moore et al., 1986; Moore and Staehelin, 1988; Bush and McCann, 1999), whereas the CFML clearly contains HG and XG. The middle lamella within tension wood of sweetgum (Liquidambar styraciflua) trees, like the CFML, contains HG (labeling strongly with JIM5 and weakly with JIM7) and XG (labeling strongly with CCRC-M1 and CCRC-M38; Bowling and Vaughn, 2008). Cumulatively, the positive immunolabeling results (Figs. 3 and 4; Supplemental Table S1) support the possibility that the XG in the CFML includes regions with and without fucosylated side chains, while part of the HG has relatively low and/or no esterification. Low-ester HG pectin has been commonly found in the middle lamella of dicotyledonous plants (Marry et al., 2006, and refs. therein). Possibly related to the earliest stages of CFML synthesis, initiating cotton fibers have bilayered primary walls with a thin outer sheath enriched in low-ester, JIM5-reactive HG, whereas the inner layer was enriched in JIM7-reactive HG. The predominant occurrence of JIM5 epitopes in the outer sheath of these young fibers (Vaughn and Turley, 1999) could indicate initial synthesis of the CFML. Alternatively, the outer sheath could be an initiation-specific fiber wall layer, since, unlike the CFML, it was not enriched in XG (Vaughn and Turley, 1999).
The CMFL is also likely to contain special molecules that confer its adhesive properties. Like the CMFL, the inner primary wall labeled with HG antibodies (JIM5, JIM7, and CCRC-M38) and some XG antibodies (anti-XG, CCRC-M1, and CCRC-M88). In contrast, several XG antibodies labeled the CFML but not the inner primary wall (Supplemental Table S1), suggesting that the CFML may contain specific forms of XG. In addition, although the list of cell wall probes is expanding, unusual variants of cell wall molecules are not likely to be comprehensively targeted. As yet unknown constituents of the CFML, including cell wall proteins or phenolics, could help to confer its adhesive properties. Other supporting evidence for special wall polymers in the CFML will be discussed in the context of cell wall changes during its breakdown in the transition stage (see below).
CFML Breakdown Is Correlated with Up-Regulation of Gene Expression and Enzyme Activities Related to Cell Wall Modification and Hydrolysis
Targeted cell wall hydrolysis occurs at the transition stage, because the inner primary wall persists while the CFML disappears (along with its CCRC-M1- and JIM5/JIM7-reactive polymers), thereby restoring fiber individuality (Fig. 3, C, F, and I). As the CFML was degraded, (1) Fuc and Gal (plausibly in the XG side chain recognized by CCRC-M1) as well as GalA (plausibly in HG recognized by JIM5) within hot water-insoluble matrix polymers diminished, and (2) cadoxen-soluble CCRC-M1 and JIM5 epitopes (but not JIM7 epitopes) showed a transient increase in the CoMPP profile. Since CCRC-M1 and JIM5 did not label the thickening cell wall (Fig. 3, F and I), the latter CoMPP pattern could reflect breakdown of the interfacial CFML. As observed here in the CoMPP profile (Fig. 4K), substantial amounts of HG and XG in primary walls are typically CDTA and NaOH soluble, respectively, and the corresponding signals were strong and relatively consistent in polymers extracted from cotton fibers at 6 to 30 DPA. In contrast, transition-stage changes in the JIM5 and CCRC-M1 epitopes were observed only in the strong solvent, cadoxen. Therefore, we propose that a special polymer network that imparts the adhesive nature of the CFML also confers resistance to extraction in CDTA, NaOH, and even cadoxen until hydrolytic changes begin to occur during the transition. In such a case, the transition-stage increase in the CoMPP signals for CCRC-M1 and JIM5 epitopes in the cadoxen fraction could reflect increasing extractability of CFML polymers at the onset of CFML degradation. Similarly, JIM5-reactive antigens in the middle lamella of beet (Beta vulgaris) roots, but not in the inner primary wall, were resistant to extraction by a Ca2+ chelator (Marry et al., 2006). We could not immunolabel polymers that typically exist as RG-1 side chains in situ, but LM5- and LM6-reative antigens, typically galactan and arabinan, were detected in CoMPP profiling. Because of the sharp decline of polymer(s) containing the extractable LM5 epitope during the transition stage (Fig. 4K), it will be worthwhile to explore further a possible role of galactans in cotton fiber adhesion.
Detailed analysis by qPCR over a finely resolved developmental time course showed transition-stage peaks in expression of genes in several classes required for XG or pectin modification or hydrolysis. Pectin esterases (e.g. PME that catalyzes pectin demethylesterification) and PG-like enzymes degrade HG, with prior removal of methyl or other esters (by pectin esterases) making the backbone more susceptible to hydrolysis (by PGs; Vicente et al., 2007). Therefore, PME activity up-regulated at the transition stage could facilitate CFML degradation and cotton fiber separation in a manner similar to the PME-mediated separation of pea (Pisum sativum) root border cells (Wen et al., 1999) and Arabidopsis (Arabidopsis thaliana) pollen tetrads (Francis et al., 2006). In contrast to positive results for PME activity, neither PG nor pectin lyase activity was detected in vitro when commercial polygalacturonic acid from oranges (Citrus sinensis) was used as a substrate (data not shown). Perhaps the PG-like genes up-regulated in transition-stage fibers encode pectin-degrading enzymes that are active against different pectin substrates, such as specialized CFML polymers. Supplemental Table S5 also shows different transcriptional profiles for other genes within the same families, which shows that specific, developmentally regulated hydrolase isoforms carry out fiber wall remodeling during the transition.
It is also possible that the cell wall-related genes and enzyme activities up-regulated at the transition stage have additional or alternative roles in other processes. For example, after CFML degradation occurred, both immunolabeling and the CoMPP signals in the CDTA-soluble fraction showed that the degree of HG esterification in the primary wall was lower after the transition stage. Correspondingly, PME activity was up-regulated after 20 DPA. As normal elongation of Arabidopsis hypocotyls requires a degree of pectin esterification of approximately 60% or greater (Derbyshire et al., 2007), these cell wall changes may be negative regulators helping to end fiber elongation at 22 DPA.
Relationship between the Cotton Fiber Tissue and Cuticle
How can an elongation-stage cotton fiber tissue be reconciled with the cotton fiber cuticle given that the plant cuticle prevents cell and organ fusion (for review, see Jarvis et al., 2003)? The existence of a hydrophobic “cuticle” has only been proven for mature cotton fiber. This strongly OsO4-positive, thin (approximately 250 nm wide, approximately 1.3% of fiber weight), morphologically homogeneous outer layer contains an approximately 1:1 mixture of primary wall polysaccharides and wax/cutin molecules, and it is sharply distinct from the underlying cellulosic secondary wall (Yatsu et al., 1983; Cui et al., 2002; Degani et al., 2002; Wakelyn et al., 2007). This structure differs from the bilayered, cuticulated surface of vegetative, epidermal, trichomes, which mixes gradually into an underlying cell wall containing several components (Marks et al., 2008). Within the cotton boll, elongating fibers would not require protection from a cuticle, and its absence during cotton fiber elongation would allow the fiber tissue to form. The absence of a cotton fiber cuticle during elongation, although contrary to common assumptions, is consistent with existing data. First, we did not observe any continuous cuticle-like demarcation between primary cell walls of adjacent fibers in bundles (Figs. 1, B–D, and 3G). Second, 2-DPA cotton fibers lacked the distinct cuticle layer that was easily seen in adjacent regular epidermal cells (Vaughn and Turley, 1999), and other images are consistent with an elongating cotton fiber leaving the ovule epidermal cuticle behind at its base (see Fig. 9 in Weis et al., 1999). Third, images from freeze-fracture transmission electron microscopy were interpreted in terms of a thinning of the cotton fiber cuticle during elongation. A multilamellate structure typical for the cuticle was shown only at 2 DPA (Willison and Brown, 1977), and this could have existed at the fiber base. In contrast, at 10 DPA, only a relatively smooth, thin layer existed between primary walls of adjacent fibers (Willison and Brown, 1977). We propose that this was not cuticle but instead the CFML described here. It is most likely that the wax and cutin components may be intercalated into (or attain their mature form within) the residual, inner cotton fiber primary wall only after the CFML breaks down. Consistent with this possibility, the “hairy,” OsO4-positive surface of 24-DPA cotton fiber (Fig. 3, F and I; Salnikov et al., 2003) resembles the amorphous cuticle of Arabidopsis stem epidermis (see Fig. 2.10e in Jeffree, 2006). The array data presented here do not provide further insight because cotton genes with best homology to three genes required for wax biosynthesis in Arabidopsis (CER3 [At5g57800], CER6 [At1g68530], and CER10 [At3g55630]) showed no significant change between 6 and 24 DPA in cotton fiber. How cuticle and wax are deposited remains largely unknown for all plant cells (Pollard et al., 2008; Samuels et al., 2008), and future research on single-celled cotton fibers will be worthwhile.
Other Implications of the CFML for Cotton Fiber Development
In addition to promoting fiber packing within the boll, the existence of the CFML is likely to have other implications for fiber development. For example, a fiber tissue may better withstand the high turgor that drives the rapid phase of elongation (Ruan, 2007), and the CFML may have mechanical properties that facilitate elongation (e.g. possibly allowing elongating fibers to slip past each other while still reinforced within a tissue-like bundle). Similar arguments based on a consideration of biomechanical principles and the structure of parenchyma tissue have been made previously (Niklas, 1992).
The demonstration of cell wall-mediated fiber adhesion also explains and unifies disparate prior observations about cotton fiber. We can now understand that twisting fiber tips on cultured ovules (Stewart, 1975) reflect a developmentally controlled mechanism that nucleates and entrains fiber bundles. Honeycomb-like aggregates of cross-sections of primary wall-stage fibers observed after deembedding prior to traditional SEM (Goynes et al., 1995) are now shown to reflect fiber bundles that existed in vivo. Fragmentary views of orderly fibers in replicas of the fiber surface inside the boll (Paiziev and Krakhmalev, 2004) are now demonstrated to exist on a large scale and to arise from the orderly packing of fiber bundles around each seed. Because the CFML breaks down before secondary wall deposition begins, a strong rationale now exists for changes in cell wall polymers and cell wall hydrolytic activity at the transition stage (Meinert and Delmer, 1977; Shimizu et al., 1997; Tokumoto et al., 2002, 2003; Guo et al., 2007; this study). CFML hydrolysis may also explain the increasing extractability of glycan oligomers near the onset of secondary wall deposition in cotton fiber (Murray et al., 2001).
Comparisons with Other Cell Adhesion and Detachment Events
There are several examples of separation of plant cells, including aerenchyma formation, sloughing of root border cells, pollen maturation, fruit ripening, dehiscence, and abscission (Roberts and Gonzalez-Carranza, 2007). However, none of these processes serves to remove a middle lamella that previously coordinated the extensive elongation of a large group of cells, implying that the degradation of the CFML is developmentally unique. Interestingly, several cotton genes with peak expression at the transition stage, including Ghbgal12, Ghpe-E, Ghxth30, and Ghxth6, have close homologs that are up-regulated during ripening-associated fruit softening (BAE72073 in pear [Pyrus communis], AAB57671 in citrus, AAS46240 in tomato [Solanum lycopersicum], and AAS46243 [XTH7] in tomato, respectively). Fruit texture depends on primary wall strength and cell-to-cell adhesion via the middle lamella, and softening occurs as newly expressed proteins modulate changes in cell wall structure, including (1) removal of pectin side groups and branches (e.g. arabinan and galactan), and (2) depolymerization and/or solubilization of XG, HG, and possibly RG-I (Martel and Giovannoni, 2007; Vicente et al., 2007). Since cotton fiber differentiation occurs within the developing boll (fruit), similarities between CFML degradation and edible fruit ripening are reasonable.
FUTURE RESEARCH
The highly defined cotton fiber system will facilitate future research to determine (1) linkage structure of CFML polymers, (2) how this defined cell wall polymer network develops adhesive properties, (3) activities of purified enzymes that hydrolyze particular CFML polymers, (4) mechanisms of synthesis and spatial localization of the CFML as the outer primary wall layer, (5) possible signaling effects of small molecules released by degradation of CFML polymers, (6) commonalities and differences of CFML-related mechanisms in various Gossypium species, and (7) effects on cotton fiber quality of genetic experiments to modify CFML-related mechanisms. Currently, we are analyzing Gossypium barbadense, which has longer fiber, a longer period of fiber elongation, and greater overlap of elongation with secondary wall deposition, in order to gain additional insight into how the CFML modulates fiber development.
MATERIALS AND METHODS
Cotton Plant Growth
Cotton (Gossypium hirsutum) flowers were tagged on the day of anthesis to allow harvesting of bolls of known age. All qPCR gene expression assays, enzyme assays, chemical fixation for TEM, growth curves, CoMPP profiling, and sugar analyses were performed on fiber of Deltapine 90 plants grown in an air-conditioned Phytotron greenhouse at North Carolina State University. Plants (one plant per 6-L pot) were grown in a 1:2 (v/v) mixture of Redi-earth Plug and Seedling Mix (product no. F1153; Sun Gro Horticulture Canada) and gravel (no. 16, construction grade) and watered twice a day with a solution containing macronutrients and micronutrients (http://www.ncsu.edu/phytotron/manual.pdf). The greenhouse had a 26°C/22°C (12-h/12-h) day/night temperature cycle and natural lighting, expect the night was interrupted for 3 h to regulate flowering (from 11 pm to 2 am, light was provided from incandescent bulbs at 11–12 μmol m−2 s−1). Supplemental Figure S1 shows the time course of fiber development during July under these conditions, which was typical for April to October, when bolls were harvested for these experiments. The slightly cool day temperature for cotton plant growth avoided any high-temperature stress effects and slowed fiber development somewhat, which allowed relatively transient changes in transition-stage gene expression to be captured reliably.
Microarray analysis was performed with RNA extracted from greenhouse-grown Deltapine 90 cotton at Michigan State University; fiber development occurred similarly to that in Supplemental Figure S1, and 20 DPA was during the transition (Haigler et al., 2005). Cryo-FE-SEM observations, cryo fixation for TEM, and paraffin histology were performed on Coker 312 cotton grown in June in a Texas Tech University greenhouse (approximately 28°C/22°C day/night cycle, with other conditions as described [Singh et al., 2005]). Because of the higher day temperature in the less controlled Texas greenhouse and, possibly, cultivar variation, fiber development was somewhat accelerated in the cryo-FE-SEM experiments.
Determination of Fiber Growth Parameters
For fiber differentiating in the 26°C/22°C Phytotron greenhouse, length and weight were measured from five bolls of different plants collected on each DPA. For weight determination, fiber from two locules of each boll was separated from the counted seeds (except 30 DPA, for which only one locule was used), dried (48 h, 80°C), and weighed. For length measurements, fibers of at least five seeds per boll were relaxed by boiling in 0.025 n HCl. Fiber still attached to the seed was straightened by a flow of water on the convex side of a watch glass, and the length of the bulk of the fiber mass was measured with a millimeter ruler. se values were determined for each mean, but some are not visible when plotted in Supplemental Figure S1. The sharp increase in the ratio of fiber weight to length beginning at 22 DPA (Supplemental Fig. S1) showed this to be the time of onset of secondary wall thickening.
Microscopy
For cryo-FE-SEM, fibers that had been snap frozen in slushed liquid nitrogen were observed through a window cut in the boll wall using a cryo-FE-SEM apparatus coupled with a cryogenic preparation unit. Additional details for cryo-FE-SEM are listed in Supplemental Protocol S1.
Prior to thin sectioning, cryo fixation and freeze substitution were accomplished as described previously (Salnikov et al., 2003). For ultrastructural analysis of cell walls and CFML, a laboratory microwave oven was used to process samples from the steps of secondary fixation onward. TEM immunolabeling was performed similarly to published methods (Ruan et al., 2004). Additional details for TEM structural analysis and immunolabeling prior to TEM or fluorescence analysis are listed in Supplemental Protocol S2.
For histology, developing ovules with attached fiber were snap frozen, freeze substituted, and then embedded in paraffin prior to sectioning and staining by combination and modification of published methods (Ruzin, 1999; Grimson and Blanton, 2006). Additional details are listed in Supplemental Protocol S3.
Analysis of Cell Wall Cellulose and Sugars
Frozen fiber at 10, 17, 19, 21, and 30 DPA was pulverized under liquid nitrogen, lyophilized, and determined to be starch free. Crystalline cellulose content was determined by a modification of the Updegraff (1969) assay. Sugars were analyzed as TMS-methylglycoside derivatives, which quantifies both neutral and acidic glycosyl residues in noncellulosic cell wall matrix polymers (York et al., 1985). Additional details are listed in Supplemental Protocol S4.
Relative composition for each sugar was expressed as nanomoles per milligram of starting material. One sample for each DPA contained lyophilized pooled fiber from three to five individual bolls from several plants. For insoluble sugars, means and sd values were based on two separate samples of pooled fiber, each assayed in triplicate so that five to six independent TMS procedures contributed to each mean (in only a few cases, outlier values due to experimental error were discarded). For soluble sugars, means and sd values were based on two independent TMS procedures (one assay each for two separate pooled fiber samples). Significant differences (P ≤ 0.05) between means from the two pooled fiber samples at 17 DPA versus other DPA were assessed by t test (one-tailed test with unequal variance).
CoMPP Profiling
Following published methods (Moller et al., 2007), cotton fiber cell wall extracts were sequentially extracted in three solvents: (1) 50 mm CDTA, pH 7.5, a Ca2+ chelator, typically extracting pectin; (2) 4 m NaOH with 1% (v/v) NaBH4, typically extracting other noncellulosic wall matrix components; and (3) a strong base, cadoxen (31% [v/v] 1,2-diaminoethane with 0.78 m, or 10% [w/v], CdO to create a saturated solution). Cadoxen in this range of formulation swells the cotton fiber secondary wall and solubilizes a small percentage of the fiber weight (Evans and Jeffries, 1970). After CDTA and NaOH extraction, cadoxen-solubilized polymers must represent tightly bound matrix components and/or low-crystallinity cellulose.
For each of three replicates of fiber at each DPA (100 mg fresh weight of fiber, previously liquid nitrogen pulverized, then freeze dried), the fiber was further ground in a mixer mill and sequentially incubated in the three solvents (250 μL of solvent, shaking, 2 h) with retention of the supernatant and pellet at each extraction step. The supernatants from the three sample replicates were pooled, and each pooled sample was printed in six replicates and three dilutions (undiluted and 1:4 and 1:24 [v/v] dilutions), giving a total of 18 spots representing each DPA for each solvent. Arrays were probed with mAbs or CBMs and developed similarly to a dot blot. Arrays were scanned, and the spot intensities (signal mean minus local background median) were quantified using microarray analysis software (ImaGene 6.0; BioDiscovery). A heat map was generated to display the relative intensity of each signal to the maximum signal observed in the experiment, which occurred for JIM5 in the CDTA fraction at 30 DPA (brightest yellow; value of 100). Although CoMPP signal intensities reflect the quantity of a particular epitope within a sample, the technique is not fully quantitative because mAb and CBM probes bind with different avidities and epitopes are not necessarily extracted from samples with equal efficiency.
Microarray Analysis
Based on the G.h.fbr-sw ESTs, which were biased toward the early secondary wall stage of cotton fiber development (Haigler et al., 2005), a spotted cDNA microarray of 3,185 cDNAs was hybridized with Cy3/Cy5 probes prepared from RNA isolated from fiber at 6, 10, 20, and 24 DPA. In the microarray analysis, all time points were compared with the 20-DPA sample as a common reference (Smyth, 2005) because (1) this time point was expected to show measurable expression given that the spotted cDNAs derived from 20-DPA RNA (Haigler et al., 2005), and (2) our major interest was to explore genes that were up-regulated for secondary wall deposition. Two copies of each amplified cDNA were spotted on each slide, and three biological replications and two technical replications (dye swaps) were performed for each data point, followed by quality control and statistical analysis of the results (Smyth and Speed, 2003; Smyth, 2004, 2005; R Development Core Team, 2005; Smyth et al., 2005). Briefly, RNA was labeled using the amino-allyl procedure (http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml). Hybridization was performed using SlideHyb #1 buffer (Ambion). Slides were scanned using an Affymetrix 428 Array Scanner (Affymetrix) and analyzed using the GenePix Pro 3.0 software (MDS Analytical Technologies). Array normalization and statistical analysis were performed using the “limma: Linear Models for Microarray Data” library module (version 2.2.0) of the R statistical package (version 2.2.0). Slide intensity data were normalized using the global Loess method. The least squares method was used for the linear model fit utilizing the Benjamini and Hochberg method to control the false discovery rate.
A summary file of the microarray data was created, “Cotton_Microarray_Summary_Annotated,” including expression ratios (log2) and P values (for averaged duplicate spots of each cDNA on each slide) for the following DPA comparisons: 6:20, 10:20, and 24:20. Note that following the convention of placing the value for the common reference in the denominator resulted in negative 6:20 and 10:20 ratios for genes that were up-regulated at 20 DPA during the transition stage. To provide annotations, the cotton cDNAs on the array were compared by BLASTx (August 5, 2008) with the Arabidopsis protein database (TAIR8_pep_20080412) and with the Green Plant protein database (minus Arabidopsis proteins; PLANTallAA), both downloaded from ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/. A value of E-3 was used as the cutoff to include a matching gene identifier and annotation; otherwise “no hits found” is indicated. From information automatically retrieved by BLAST, the file was parsed to generate a brief, informative annotation for inclusion in the summary file.
qPCR
Details of gene selection, primer design, RNA isolation, and qPCR procedures are listed in Supplemental Protocol S5. In qPCR, Gheif5 (for eukaryotic translation initiation factor 5) was used as an endogenous normalizing gene; it had nearly equal expression in 6- to 30-DPA fiber (Haigler et al., 2009). The qPCR procedure was repeated three times, beginning with RNA isolation from independent samples at each DPA. Average gene expression patterns were calculated relative to a time of high elongation rate at 10 DPA, which showed clearly any potential up-regulation during the shift to secondary wall deposition at the transition stage (Supplemental Table S5). Selected gene expression profiles were graphed as percentage maximum increase for each gene in order to allow comparison of developmental patterns of expression regardless of possibly different expression amplitudes (nonnormalized data are shown in Supplemental Table S5).
Enzyme Activity Assays
Additional details of assay methods are listed in Supplemental Protocol S6. Liquid nitrogen-ground fiber samples from 10 to 30 DPA were assayed, with results normalized to total soluble protein (Bradford assay; Thermo Fisher Scientific). The reported incubation time was determined to be in the linear range of each assay. Fiber extract was replaced with equal volumes of the extraction buffer in blanks. For each data point, an average for four biological replications was determined ± se.
β-Galactosidase activity was measured by hydrolysis of β-d-galactopyranoside (Pressey, 1983). Activity of XEH was determined as described with minor modifications (Kallas et al., 2005). PME activity in cotton fibers was quantified as described (Jiang et al., 2005).
Using published methods and polygalacturonic acid (P-3889; Sigma-Aldrich) as substrate, negative results for cotton fiber extracts were obtained for PG activity (Zhang et al., 1999) and pectate lyase activity (Payasi et al., 2004). Similarly, negative results for cotton fiber extracts were obtained in gel diffusion assays (Taylor and Secor, 1988) in which Rhizopus PG (P-2401; Sigma-Aldrich) yielded positive results.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fiber growth and cellulose accumulation from 6 to 30 DPA for Deltapine 90 cotton growing with a 26°C/22°C day/night cycle.
Supplemental Figure S2. Micrographs of 15-DPA fiber showing how fiber bundle cross-sections were generated.
Supplemental Figure S3. TEM views of CFML-filled bulges between 15-DPA fibers observed in longitudinal orientation.
Supplemental Figure S4. Images of fiber packing around an ovule at high magnification and fiber packing within a locule.
Supplemental Figure S5. Overviews of fibers immunolabeled in Figure 3.
Supplemental Table S1. Probes used in CoMPP or TEM immunolabeling and qualitative results of immunolabeling intensity.
Supplemental Table S2. Sugar analysis of insoluble fiber wall matrix fraction.
Supplemental Table S3. Sugar analysis of soluble fiber wall matrix fraction.
Supplemental Table S4. Primers used to amplify cotton genes by qPCR.
Supplemental Table S5. Expression patterns for cotton genes in 6- to 30-DPA fiber determined by qPCR.
Supplemental Table S6. Activities of cell wall-modifying enzymes in cotton fiber between 10 and 30 DPA.
Supplemental Protocol S1. Cryo-FE-SEM.
Supplemental Protocol S2. Ultrastructural analysis and immunolabeling.
Supplemental Protocol S3. Paraffin histology using cryogenic methods.
Supplemental Protocol S4. Cell wall cellulose and sugar analysis.
Supplemental Protocol S5. qPCR methods.
Supplemental Protocol S6. Enzyme activity assays.
Supplementary Material
Acknowledgments
We thank P. Knox for the gift of CBM3a and L.A. Staehelin for the gift of anti-XG polyclonal antibody. We thank M. Hahn for contributing a prototype large-scale testing kit of mAbs raised against cell wall polymers and for providing information on epitope characterization in progress. We thank two anonymous reviewers for constructive suggestions.
This work was supported by Cotton, Inc., of Cary, North Carolina, by the National Science Foundation (Plant Genome grant nos. DBI–0211797, R98RA1829, and DBI–0110173), and by the National Research Initiative, Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture (grant no. 2006–35318–17301). Development and distribution of some antibodies (from the CarboSource Services at the Complex Carbohydrate Research Center, University of Georgia) were supported by the National Science Foundation (grant nos. RCN0090281 and DBI0421683).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Candace H. Haigler (candace_haigler@ncsu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bascom WD, Cottington RL (1976) Effect of temperature on the adhesive fracture behavior of an elastomer-epoxy resin. J Adhes 7 333–346 [Google Scholar]
- Basra AS, editor (1999) Cotton Fibers, Developmental Biology, Quality Improvement, and Textile Processing. Food Products Press, New York
- Bowling AJ, Vaughn KC (2008) Immunocytochemical characterization of tension wood: gelatinous fibers contain more than just cellulose. Am J Bot 95 655–663 [DOI] [PubMed] [Google Scholar]
- Bowman DT, Van Esbroeck GA, Van't Hof J, Jividen GM (2001) Ovule fiber cell numbers in modern upland cottons. J Cotton Sci 5 81–83 [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Lohmann JU (2007) Profiling a plant: expression analysis in Arabidopsis. Curr Opin Plant Biol 10 136–141 [DOI] [PubMed] [Google Scholar]
- Bush MS, McCann MC (1999) Pectin epitopes are differentially distributed in the cell walls of potato (Solanum tuberosum) tubers. Physiol Plant 107 201–213 [Google Scholar]
- Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, et al (2007) Towards sequencing cotton (Gossypium) genomes. Plant Physiol 145 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XL, Price JB, Calamari TA Jr, Hemstreet JM, Meredith W (2002) Cotton wax and its relationship with fiber and yarn properties. I. Wax content and fiber properties. Text Res J 72 399–404 [Google Scholar]
- Dahl R, Staehelin LA (1989) High-pressure freezing for the preservation of biological structure: theory and practice. J Electron Microsc Tech 13 165–174 [DOI] [PubMed] [Google Scholar]
- Degani O, Gepstein S, Dosoretz CG (2002) Potential use of cutinase in enzymatic scouring of cotton fiber cuticle. Appl Biochem Biotechnol 102-103 277–289 [DOI] [PubMed] [Google Scholar]
- Delmer DP, Pear JR, Andrawis A, Stalker DM (1995) Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol Gen Genet 248 43–51 [DOI] [PubMed] [Google Scholar]
- Derbyshire P, McCann MC, Roberts K (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol 7 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GM, Jeffries R (1970) The swelling of cotton in cadoxen, ethylenediamine, and cuprammonium hydroxide. J Appl Polym Sci 14 633–653 [Google Scholar]
- Evert RF (2006) Esau's Plant Anatomy, Ed 3. Wiley Interscience, Hoboken, NJ
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goynes WR, Ingber BF, Triplett BA (1995) Cotton fiber secondary wall development: time vs. thickness. Text Res J 65 400–408 [Google Scholar]
- Grimson MJ, Blanton RL (2006) Cryofixation methods for ultrastructural studies of Dictyostelium discoideum. Methods Mol Biol 346 339–366 [DOI] [PubMed] [Google Scholar]
- Guo JY, Wang LJ, Chen SP, Hu WL, Chen XY (2007) Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary wall synthesis. Cell Res 2007 1–13 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Singh B, Wang G, Zhang D (2009) Genomics of cotton fiber secondary wall deposition and cellulose biogenesis. In AH Paterson, ed, Genetics and Genomics of Cotton. Plant Genetics and Genomics: Crops and Models 3. Springer Science + Business Media, New York, pp 385–417
- Haigler CH, Zhang D, Wilkerson CG (2005) Biotechnological improvement of cotton fiber maturity. Physiol Plant 124 285–294 [Google Scholar]
- Hayashi T, Delmer DP (1988) Xyloglucan in the cell walls of cotton fiber. Carbohydr Res 181 273–277 [Google Scholar]
- Ishii S (1981) Isolation and characterization of cell-wall pectic substances from potato tuber. Phytochemistry 20 2329–2333 [Google Scholar]
- Jarvis MC, Briggs SPH, Knox JP (2003) Intercellular adhesion and cell separation in plants. Plant Cell Environ 26 977–989 [Google Scholar]
- Jeffree CE (2006) The fine structure of the plant cuticle. In M Riederer, C Müller, eds, Biology of the Plant Cuticle. Blackwell Publishing, Oxford, pp 11–125
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan C, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas AM, Pines K, Denman SE, Henriksson H, Faldt J, Johansson P, Brumer H, Teeri TT (2005) Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula × tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA (2004) Characterization of GhRac GTPase expressed in developing cotton (G. hirsutum L.) fibers. Biochim Biophys Acta 1679 214–221 [DOI] [PubMed] [Google Scholar]
- Lynch MA, Staehelin LA (1992) Domain-specific and cell type-specific localization of two types of cell wall matrix polysaccharides in the clover root tip. J Cell Biol 118 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby D, Carpita NC, Montezinos D, Kulow C, Delmer DP (1979) β-1,3-Glucan in developing cotton fibers. Plant Physiol 63 1158–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger J, Dixon RA, Haigler CH (2008) A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J 56 483–492 [DOI] [PubMed] [Google Scholar]
- Marry M, Roberts K, Jopson SJ, Huxham M, Jarvis MC, Corsar J, Robertson E, McCann MC (2006) Cell-cell adhesion in fresh sugar-beet root parenchyma requires both pectin esters and calcium cross links. Physiol Plant 126 243–256 [Google Scholar]
- Martel C, Giovannoni J (2007) Fruit ripening. In JA Roberts, Z Gonzalez-Carranza, eds, Plant Cell Separation and Adhesion. Blackwell Publishing, Oxford, pp 164–176
- Meinert MC, Delmer DP (1977) Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol 59 1088–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11 266–277 [DOI] [PubMed] [Google Scholar]
- Moller I, Sorensen I, Bernal AJ, Blaukopf C, Lee K, Obro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, et al (2007) High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J 50 1118–1128 [DOI] [PubMed] [Google Scholar]
- Moore PJ, Darvill AG, Albersheim P, Staehelin LA (1986) Immunogold localization of xyloglucan and rhamnogalacturonan I in the cell walls of suspension-cultured sycamore cells. Plant Physiol 82 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PJ, Staehelin LA (1988) Immunogold localization of the cell-wall-matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.: implications for secretory pathways. Planta 174 433–445 [DOI] [PubMed] [Google Scholar]
- Murray AK, Nichols RL, Sassenrath-Cole GF (2001) Cell wall biosynthesis: glycan containing oligomers in developing cotton fibers, cotton fabric, wood, and paper. Phytochemistry 57 975–986 [DOI] [PubMed] [Google Scholar]
- Niklas KJ (1992) Plant Biomechanics: An Engineering Approach to Plant Form and Function. University of Chicago Press, Chicago
- Paiziev AA, Krakhmalev VA (2004) Self-organization phenomena during development of cotton fibers. Curr Opin Solid State Mater Sci 8 127–133 [Google Scholar]
- Pauly M, Keegstra K (2008) Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 54 559–568 [DOI] [PubMed] [Google Scholar]
- Payasi A, Misra PC, Sanwal GG (2004) Effect of phytohormones on pectate lyase activity in ripening Musa acuminate. Plant Physiol Biochem 42 861–865 [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JG (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13 236–246 [DOI] [PubMed] [Google Scholar]
- Pressey R (1983) β-Galactosidases in ripening tomato. Plant Physiol 71 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2005) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
- Roberts JA, Gonzalez-Carranza Z, editors (2007) Plant Cell Separation and Adhesion. Blackwell Publishing, Oxford
- Ruan YL (2007) Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fiber. Funct Plant Biol 34 1–10 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Xu SM, White R, Furbank RT (2004) Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol 136 4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin SE (1999) Plant Microtechnique and Microscopy. Oxford University Press, Oxford
- Salnikov V, Grimson MJ, Seagull RW, Haigler CH (2003) Localization of sucrose synthase and callose in freeze substituted, secondary wall stage, cotton fibers. Protoplasma 221 175–184 [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59 683–707 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T (1997) Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol 38 375–378 [DOI] [PubMed] [Google Scholar]
- Singh B, Haley L, Nightengale J, Kang WH, Haigler CH, Holaday AS (2005) Long-term night chilling of cotton, Gossypium hirsutum, does not result in reduced CO2 assimilation. Funct Plant Biol 32 655–666 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes method for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article 3 [DOI] [PubMed]
- Smyth GK (2005) Limma: linear models for microarray data. In R Gentleman, V Carey, S Dudoit, R Irizarry, W Huber, eds, Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, pp 397–420
- Smyth GK, Michaud J, Scott H (2005) The use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21 2067–2075 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP (2003) Normalization of cDNA microarray data. Methods 31 265–273 [DOI] [PubMed] [Google Scholar]
- Stewart JMcD (1975) Fiber initiation on the cotton ovule (Gossypium hirsutum). Am J Bot 62 723–730 [Google Scholar]
- Taylor RJ, Secor GA (1988) An improved diffusion assay for quantifying the polygalacturonase content of Erwinia culture filtrates. Phytopathology 78 1101–1103 [Google Scholar]
- Tokumoto H, Wakabayashi K, Kamisaka S, Hoson T (2002) Changes in the sugar composition and molecular mass distribution of matrix polysaccharides during cotton fiber development. Plant Cell Physiol 43 411–418 [DOI] [PubMed] [Google Scholar]
- Tokumoto H, Wakabayashi K, Kamisaka S, Hoson T (2003) Xyloglucan breakdown during cotton fiber development. J Plant Physiol 160 1411–1414 [DOI] [PubMed] [Google Scholar]
- Updegraff DM (1969) Semi-micro determination of cellulose in biological materials. Anal Biochem 32 420–424 [DOI] [PubMed] [Google Scholar]
- Vaughn KC, Turley RB (1999) The primary walls of cotton fibers contain an ensheathing pectin layer. Protoplasma 209 226–237 [Google Scholar]
- Vicente AR, Saladie M, Rose JKC, Labavitch JM (2007) The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric 87 1435–1448 [Google Scholar]
- Wakelyn PJ, Bertoniere NR, French AD, Thibodeaux DP, Triplett BA, Rousselle MA, Goynes WR Jr, Edwards JV, Hunter L, McAlister DD, et al (2007) Cotton Fiber Chemistry and Technology. CRC Press, New York
- Waldron KW, Brett CT (2007) The role of polymer cross-linking in intercellular adhesion. In JA Roberts, Z Gonzalez-Carranza, eds, Plant Cell Separation and Adhesion. Blackwell Publishing, Oxford, pp 183–204
- Weis KG, Jacobsen KR, Jernstedt JA (1999) Cytochemistry of developing cotton fibers: a hypothesized relationship between motes and non-dyeing fibers. Field Crops Res 62 107–117 [Google Scholar]
- Wen F, Yanmin Z, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Steele-King CG, Marcus SE, Mort A, Huisman M, van Alebeek GJ, Schols HA, Voragen AG, Le Goff A, et al (2004) A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta 218 673–681 [DOI] [PubMed] [Google Scholar]
- Willison JHM, Brown RM Jr (1977) An examination of developing cotton fiber: wall and plasmalemma. Protoplasma 92 21–41 [Google Scholar]
- Yatsu LY, Espelie KE, Kolattukudy PE (1983) Ultrastructural and chemical evidence that the cell wall of green cotton fiber is suberized. Plant Physiol 73 521–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P (1985) Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 118 3–40 [Google Scholar]
- Zhang D, Hrmova M, Wan CH, Wu C, Balzen J, Cai W, Wang J, Densmore LD, Fincher GB, Zhang H, et al (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54 353–372 [DOI] [PubMed] [Google Scholar]
- Zhang J, Bruton BD, Biles CL (1999) Purification and characterisation of a prominent polygalacturonase isozyme produced by Phomopsis cucurbitae in decayed muskmelon fruit. Mycol Res 103 21–27 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.