Abstract
We report here on the identification of the major plasma membrane (PM) ascorbate-reducible b-type cytochrome of bean (Phaseolus vulgaris) and soybean (Glycine max) hypocotyls as orthologs of Arabidopsis (Arabidopsis thaliana) AIR12 (for auxin induced in root cultures). Soybean AIR12, which is glycosylated and glycosylphosphatidylinositol-anchored to the external side of the PM in vivo, was expressed in Pichia pastoris in a recombinant form, lacking the glycosylphosphatidylinositol modification signal and purified from the culture medium. Recombinant AIR12 is a soluble protein predicted to fold into a β-sandwich domain and belonging to the DOMON (for dopamine β-monooxygenase N terminus) domain superfamily. It is shown to be a b-type cytochrome with a symmetrical α-band at 561 nm, fully reduced by ascorbate, and fully oxidized by monodehydroascorbate radical. AIR12 is a high-potential cytochrome b showing a wide bimodal dependence from the redox potential between +80 mV and +300 mV. Optical absorption and electron paramagnetic resonance analysis indicate that AIR12 binds a single, highly axial low-spin heme, likely coordinated by methionine-91 and histidine-76, which are strongly conserved in AIR12 sequences. Phylogenetic analyses reveal that the auxin-responsive genes AIR12 represent a new family of PM b-type cytochromes specific to flowering plants. Circumstantial evidence suggests that AIR12 may interact with other redox partners within the PM to constitute a redox link between cytoplasm and apoplast.
Complex interactions between plant cells and the environment are mediated by the apoplast. The apoplastic liquid phase permeating the cell wall contains relatively low concentrations of solutes (Dietz, 1997). Its composition, although largely determined by the protoplast, is easily perturbed by environmental challenges that can thus be perceived by the apoplast and translated into signals that trigger cell responses (Pignocchi and Foyer, 2003; Foyer and Noctor, 2005). Environmental challenges affecting the apoplast commonly result in an oxidative load, caused, for instance, by pollutants (e.g. ozone; Sandermann, 2008) or by endogenously generated reactive oxygen species (ROS). Several enzymatic and nonenzymatic systems are able to generate ROS in the apoplast (Fry, 1998; Apel and Hirt, 2004), an event that is not restricted to biotic or abiotic stresses (Torres and Dangl, 2005), but also involved in diverse physiological conditions, including stomata closure and cell growth (Foreman et al., 2003; Mori and Schroeder, 2004; Gapper and Dolan, 2006; Schopfer and Liszkay, 2006).
Apoplastic reductants not only act as an antioxidant barrier, but they could also modulate oxidative signals, thus actively contributing to plant adaptation to the environment. Ascorbate occurs at 10−4 to 10−3 m concentrations in the apoplast, where it represents the major pool of low-molecular-mass antioxidants (Dietz, 1997; Pignocchi and Foyer, 2003; Padu et al., 2005). Maintenance of the apoplastic ascorbate pool depends on transport systems of the plasma membrane (PM; Horemans et al., 2000). The redox state of the ascorbate in the apoplast is relatively flexible and typically more oxidized than in the symplast (Cordoba-Pedregosa et al., 2003, 2005; de Pinto and De Gara, 2004; Padu et al., 2005; Pignocchi et al., 2006). Ascorbate oxidation can be effected enzymatically by ascorbate oxidase or ascorbate peroxidase, and nonenzymatically by direct interaction with ROS (including ozone; Sandermann, 2008), transition metals (e.g. iron, copper; Fry, 1998), or phenolic radicals (Takahama, 1993). Oxidation of ascorbate gives rise to the monodehydroascorbate (MDA) radical, which can disproportionate into ascorbate and fully oxidized dehydroascorbate. In addition, the apoplastic MDA radical can be reduced back to ascorbate by a trans-PM redox system that uses cytosolic ascorbate as a reductant and involves a high-potential cytochrome b (Horemans et al., 1994). The latter has escaped molecular identification thus far (Trost et al., 2000; Bérczi et al., 2003; Griesen et al., 2004; Preger et al., 2005).
It was suggested (Asard et al., 2001) that the trans-PM electron transfer from cytosolic ascorbate to apoplastic MDA may be effected by a cytochrome b561, in analogy to the electron transfer of animal chromaffin vesicles (Kelley and Njus, 1986). Cytochromes b561 are high-potential, transmembrane redox proteins of about 25 kD made of six membrane-spanning α-helices, which bind two hemes b. One heme is predicted to be close to an ascorbate binding site facing the cytosol, whereas the second heme faces the opposite side of the membrane and can be oxidized by either MDA or ferrichelates (Tsubaki et al., 1997; McKie et al., 2001; Bérczi et al., 2005; Kamensky et al., 2007). Plants contain several orthologous genes to animal cytochrome b561 (Asard et al., 2000; Bashtovyy et al., 2003). Arabidopsis (Arabidopsis thaliana) contains four genes belonging to this family (Tsubaki et al., 2005) and one of these (At4g25570, CYBASC1), expressed in recombinant form, showed similar biochemical properties to animal cytochrome b561 (Bérczi et al., 2007). However, the localization in vivo of plant cytochrome b561 is controversial. Arabidopsis CYBASC1 was found to be associated with the tonoplast membrane (Griesen et al., 2004) and annotated in proteomic studies as either a tonoplast protein (Carter et al., 2004; Shimaoka et al., 2004) or a chloroplast protein (Zybailov et al., 2008). Tonoplast localization was also reported for bean (Phaseolus vulgaris) CYBASC1 in etiolated hypocotyls (Preger et al., 2005), whereas a GFP construct of CYBASC1 from wild watermelon (Citrullus lanatus) was shown to be targeted to the PM in transformed onion (Allium cepa) epidermal cells (Nanasato et al., 2005). No data are available for any other isoform of cytochrome b561 in plants.
An ascorbate-reducible cytochrome b from enriched PM preparations was purified as a glycosylated protein of 55 to 63 kD (bean hypocotyls; Trost et al., 2000) or 120 kD (Arabidopsis; Bérczi et al., 2003) in SDS-PAGE. The association to the PM of the bean hypocotyl cytochrome was confirmed by analytical Suc gradient centrifugation (Preger et al., 2005). Based on potentiometric redox titrations, both bean and Arabidopsis cytochrome b preparations were suggested to bind two hemes with distant redox potentials (Em7 +135 and +180/+200 mV). However, the nature of this high-potential cytochrome b of plant PM remained elusive, although clearly different from tonoplast cytochrome b561 (Preger et al., 2005).
In this article, we report on the purification, molecular identification, cloning, and biochemical characterization of the major ascorbate-reducible cytochrome b associated with the PM of soybean (Glycine max) etiolated hypocotyls. The coding gene, known as AIR12 (for auxin induced in root cultures), is early expressed during auxin-induced lateral root formation in Arabidopsis (Laskowski et al., 2006). We demonstrate that AIR12 is a member of a new family of ascorbate-reducible cytochromes b specific to flowering plant species. The protein is glycosylated and hydrophilic and predicted to be associated in vivo with the external face of the PM by means of a glycosylphosphatidylinositol (GPI) anchor (Borner et al., 2003). AIR12 has been found to be associated with lipid rafts together with other redox proteins (Lefebvre et al., 2007), which may act as its partners in a possible electron link between apoplast and symplast.
RESULTS
Purification of PM Ascorbate-Reducible Cytochrome b from Etiolated Soybean Hypocotyls
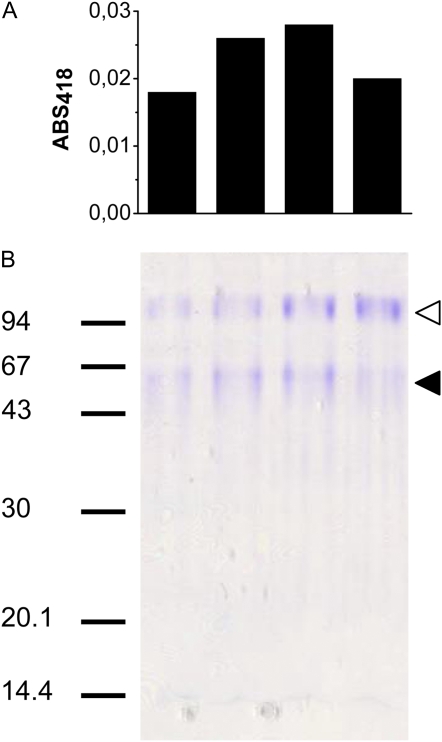
Purification of PM ascorbate-reducible cytochrome b from etiolated soybean hypocotyls was performed following the protocol described previously for bean hypocotyls (Preger et al., 2005; see “Materials and Methods” for details). This protocol leads to almost complete purification of a major ascorbate-reducible cytochrome b, previously demonstrated by isopicnic centrifugation to be associated with the PM (Preger et al., 2005). The cytochrome eluted from the last chromatographic step (Superdex 200 gel filtration chromatography; data not shown) as a peak of approximately 70 kD. This peak, analyzed by SDS-PAGE, contained two major bands. A faint and broad band around 60 kD had a distribution in the gel lanes that nicely matched the ascorbate-reduced cytochrome signals in Superdex 200 fractions (Fig. 1). An additional polypeptide of about 100 kD revealed by Coomassie Brilliant Blue staining coeluted with the cytochrome, but did not correlate with the ascorbate-reduced cytochrome signal in the eluate of the gel filtration column.
Figure 1.
SDS-PAGE of ascorbate-reducible cytochrome b-containing fractions eluted from a Superdex 200 gel filtration chromatography column. A, Histograms representing the relative amount of cytochrome in each fraction (0.4 mL) of the peak (centered at 15.05 mL of elution) of the Superdex 200 column. ABS, Absorbance. B, Fractions were concentrated 12-fold and 15 μL of each concentrated fraction were loaded on the gel. Gel was Coomassie Brilliant Blue stained. Markers are indicated on the left. On the right, black arrowhead indicates the band around 60 kD correlating with the Superdex peak. White arrowhead indicates a contaminating polypeptide. [See online article for color version of this figure.]
Protein Identification by Mass Spectrometry
Because the soybean genome is not well annotated yet, precautions were taken to avoid false identifications, and two different tandem mass spectrometry (MS/MS) search engines were used. For the 100-kD band, the X!tandem search allowed identification of a soybean EST corresponding to a predicted protein (gene no. Glyma05g04270) as revealed by a BLAST analysis against the soybean genome (http://www.phytozome.net/soybean.php). Using MASCOT, the 100-kD bands were found to correspond to a protein from grape (Vitis vinifera) annotated as a homolog of the multicopper oxidase SKU5 from Arabidopsis (gene no. At4g12420). A multiple sequence alignment with ClustalW (data not shown) revealed that these three candidate proteins share more than 76% identity, indicating that the 100-kD band likely corresponds to a SKU5 multicopper oxidase from soybean.
For the band of lower molecular mass (about 60 kD), MASCOT only detected a vegetative storage protein A from soybean (gene no. Glyma07g01730), whereas X!tandem allowed identification not only of this same protein, but also of another protein (gene no. Glyma03g22260) annotated as being similar to Arabidopsis AIR12 (gene no. At3g07390). In addition, peptide mass fingerprints were also performed with the same peptide mixtures as used for nano-liquid chromatography (LC)-MS/MS. For both bands, these analyses confirmed the identifications and increased the sequence coverage. All mass spectrometry (MS) and MS/MS results are summarized in Supplemental Table S1.
The results were further strengthened by the identification of the PM ascorbate-reducible cytochrome b of bean hypocotyls (Preger et al., 2005), having the same properties and purified by the same procedure as the soybean protein identified above. Two peptides from the tryptic digest of the 63-kD band in SDS-PAGE corresponding to the purified bean cytochrome were sequenced by Edman degradation. The two sequences (Ala-Leu-[Thr]-X-Ala-Ser-Phe-Lys and Asp-Gly-Lys-Pro-Met-Ile-His-Asp-Ser-[Lys]-Pro-Asp-Asn-Leu-Gln-Ala) perfectly match a Phaseolus coccineus complete cDNA (FJ535652) corresponding to the bean ortholog of Arabidopsis AIR12 (At3g07390). Overall, these results are compatible with the ascorbate-reducible cytochrome b of the PM being coded by AIR12 in soybean and other plants.
AIR12 Constitutes a New Protein Family Specific to Flowering Plants
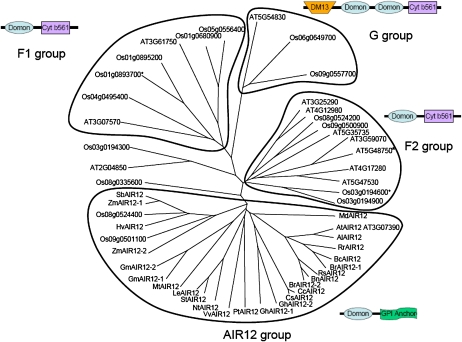
AIR12 belongs to the DOMON (for dopamine β-monooxygenase N terminus) domain superfamily, which includes proteins with very different functions, wide phyletic distribution, and different domain architectures (Aravind, 2001; Ponting, 2001; Iyer et al., 2007). The DOMON superfamily includes heme- as well as sugar-binding proteins. BLAST searches for AIR12 orthologs in public databases identified a large number of protein sequences of similar size from different flowering plant species (both monocots and dicots), but apparently no orthologs in any other group of green organisms, such as gymnosperms, ferns, mosses, or algae (green and others), or in nonphotosynthetic eukaryotes, bacteria, or archaea. Additional protein sequences, less closely related to AIR12 and generally much longer than AIR12, were also retrieved and found to contain a DOMON domain linked to a cytochrome b561 domain (Aravind, 2001; Ponting, 2001; Verelst and Asard, 2003; Tsubaki et al., 2005; Iyer et al., 2007). Extensive analysis of the complete genomes of Arabidopsis and rice (Oryza sativa) evidenced 12 and 15 DOMON-containing proteins, respectively (see “Materials and Methods” for search conditions). A phylogenetic analysis of 27 AIR12 sequences from 22 flowering plant species, together with all sequences of Arabidopsis and rice containing a DOMON domain, is presented in Figure 2. This tree reveals that all AIR12 proteins cluster together, whereas other DOMON-containing proteins cluster into three well-separated groups, corresponding to groups F1, F2, and G previously proposed by Tsubaki et al. (2005). These groups correspond to proteins containing one or two DOMON domains fused to a C-terminal cytochrome b561. Only a few sequences seem to locate outside these groups. AIR12 thus constitutes a new family of proteins within the DOMON domain superfamily, which, on the basis of public sequences, seems to be specific to flowering plants. Most species present a single AIR12-coding gene, but a few contain two (e.g. rice, maize [Zea mays], soybean, cotton [Gossypium hirsutum], and Brassica rapa). All AIR12 protein sequences were found to share similar properties, including an N-terminal signal peptide and a C-terminal GPI anchor (Fig. 3). In addition, the sequences (235–274 amino acids) contain several highly or strictly conserved residues, including Met-91 and His-175 (GmAIR12-1 numbering), which are involved in heme binding in other DOMON-containing proteins (Hallberg et al., 2000; Kloer et al., 2006; Iyer et al., 2007). This suggests that most members of the AIR12 family may represent heme-binding proteins.
Figure 2.
Phylogenetic tree of DOMON-containing proteins from Arabidopsis and rice and AIR12 sequences from diverse higher plants. The unrooted tree was constructed with the ClustalX2 program. Species abbreviations and full names: Al, Arabidopsis lyrata; At, Arabidopsis thaliana; Bc, Brassica carinata; Bn, Brassica napus (rapeseed); Br, Brassica rapa; Cc, Citrus clementina (mandarin); Cs, Citrus sinensis (orange); Gm, Glycine max (soybean); Gh, Gossypium hirsutum (cotton); Hv, Hordeum vulgare (barley); Le, Lycopersicon esculentum (tomato); Md, Malus domestica (apple); Mt, Medicago truncatula; Nt, Nicotiana tabacum (tobacco); Os, Oryza sativa ‘japonica’ group (rice); Pt, Populus trichocarpa (poplar); Rr, Raphanus raphanistrum (wild radish); Rs, Raphanus sativus (radish); Sb, Sorghum bicolor; St, Solanum tuberosum (potato); Vv, Vitis vinifera (grape vine); Zm, Zea mays (maize). For At and Os, the name of each protein in the tree corresponds to the locus number. AIR12 protein IDs from other sequenced genomes (http://genome.jgi-psf.org): Al (Araly1:478059), GmAIR12-1 (Glyma03g22260), GmAIR12-2 (Glyma16g09760), PtAIR12 (Poptr1_1:553231), and SbAIR12 (Sorbi1:4987942). AIR12 sequences accessible in NCBI entrez database (http://www.ncbi.nlm.nih.gov): Bc (ABS30419), Hv (AK248372), Vv (CAN67699), ZmAIR12-1 (ACG26288), and ZmAIR12-2 (ACG46823). AIR12 sequences translated from unigene clusters (http://www.ncbi.nlm.nih.gov/unigene): Bn (Bna14612), Br (Bra5828), Cc (Ccl16205), Cs (Csi4349), GhAIR12-1 (Ghi14385), GhAIR12-2 (Ghi16143), Le (Les20302), Md (Mdo2872), Mt (Mtr11875), Nt (Nta5372), Rr (Rra14385), Rs (Rsa16230), and St (Stu21353). A schematic representation of the organization of conserved domains in each of the four major groups is also indicated based on CD search analyses (http://www.ncbi.nlm.nih.gov/Structure/cdd) and GPI anchor prediction using the GPI-SOM program (http://gpi.unibe.ch). DM13, Domain potentially involved in redox reactions; Cyt b561, cytochrome b561. The F1, F2, and G groups have been defined based on cytochrome b561 phylogeny (Tsubaki et al., 2005). Asterisks indicate proteins belonging to the F1 and F2 groups, but containing a single DOMON domain without the cytochrome b561 domain. [See online article for color version of this figure.]
Figure 3.
Multiple sequence alignment of AIR12 from diverse higher plants. The proteins were aligned with the ClustalW program (Larkin et al., 2007). Residues boxed in black correspond to 80% identity. Residues boxed in gray correspond to 80% conservation with the PAM120 matrix. Accession numbers and abbreviations are as in Figure 1. The asterisks indicate the position of the conserved residues putatively involved in heme binding. Signal peptide sequences (SignalP 3.0 server at http://www.cbs.dtu.dk/services/SignalP) are underlined. The position of the GPI anchor (GPI-SOM prediction at http://gpi.unibe.ch) is also indicated.
Expression and Purification of Soybean AIR12 in Pichia pastoris
To get better insights into the properties of the AIR12 protein, a modified version of soybean AIR12 cDNA was introduced in the eukaryotic system P. pastoris for heterologous expression. An expression vector pPICZαB containing the soybean AIR12 cDNA downstream of the ALCOHOL OXIDASE1 (AOX1) promoter and an α-factor secretion signal sequence was constructed and used to transform P. pastoris. Transformants with multicopies of AIR12 cDNA were screened exploiting their hyper-resistance to zeocin. Positive clones were cultured and induced for production of recombinant AIR12 (recAIR12). The yields of recAIR12 were assessed by spectrophotometric analysis of the induction medium. Ascorbate-reduced minus oxidized spectra of culture solutions of recAIR12 positive clones revealed the presence of a cytochrome b starting from 24 h after induction. RecAIR12 concentrations were calculated from reduced minus oxidized spectra using a differential extinction coefficient (ɛ561–571) of 21 mm−1, as determined by the pyridine hemochrome method with the assumption that AIR12 binds a single heme (see below). The absorption maximum of the pyridine hemochrome at 557 nm strongly suggested that recAIR12 binds indeed a b-type heme (Porra and Jones, 1963). Maximum yields were obtained 48 h after induction and were estimated to be approximately 400 nmol L−1 (10 mg AIR12/L). No detectable b-type cytochromes were observed in the extracellular medium of control cultures.
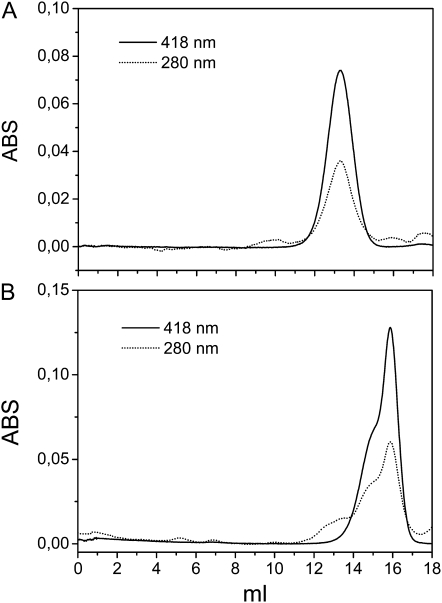
RecAIR12 was purified from the culture medium of P. pastoris by means of three chromatographic steps, including a phenyl Sepharose hydrophobic interaction column, followed by an anion-exchange column and gel filtration. The cytochrome eluted from a Superdex 200 gel filtration column as a single symmetric peak of 163 ± 25 kD (n = 6; Fig. 4A). The ratio A418:A280 of the purified protein attained a value of 2.1. By the pyridine hemochrome method, an extinction coefficient at 418 nm of 113 mm−1 could be determined for recAIR12 hemes under oxidizing conditions. On the other hand, the absorption at 280 nm was contributed both by apoprotein (ɛ280 of 29.5 mm−1 calculated on recAIR12 sequence) and heme (ɛ280 of 12 mm−1; Butt and Keilin, 1962). Based on these extinction coefficients, the ratio A418:A280 of 2.1 of purified recAIR12 could be translated into a ratio of 0.7 heme:apoprotein.
Figure 4.
Gel filtration chromatography of purified recAIR12. A, Purified recAIR12 was loaded on the Superdex 200 column. B, The peak of recAIR12 eluted from the gel filtration chromatography in A was concentrated (20-fold), deglycosylated through incubation with EndoH (2.5 μg) overnight at 22°C, and loaded again on the Superdex 200 column. ABS, Absorbance.
Upon deglycosylation of the native purified protein by endoglycosidase H, the apparent molecular mass dropped to 39 kD (Fig. 4B), closer to the theoretical molecular mass of 24 kD of the recombinant protein. The result suggests that the very high apparent molecular mass of the recombinant protein (163 kD) in comparison with the 70 kD of native AIR12, in gel filtration chromatography, may be ascribed to heavy glycosylation by the P. pastoris expression system. In addition, the presence of a 30-amino acid sequence in recAIR12, comprising the C-terminal polyhistidine tag and c-myc epitope, would also contribute to a higher apparent size in gel filtration chromatography (Supplemental Fig. S1). Accordingly, the molecular mass of the glycosylated recombinant protein determined by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS analysis was found to be only 37.3 kD.
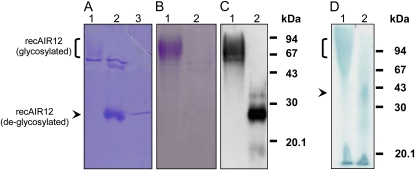
The purified recAIR12 migrates on SDS-PAGE as a high-molecular-mass broad band from 65 to 90 kD, hardly detected by Coomassie staining together with a contaminant of 50 kD (Fig. 5A). The same high-molecular-mass broad band was intensively stained when the gel was treated for glycoprotein detection (Fig. 5B). After incubation of the purified protein with endoglycosidase H, a strong band of 27 kD appeared in the Coomassie-stained gel, partially overlapping with endoglycosidase H (29 kD), whereas glycoprotein staining failed to reveal any band (Fig. 5). Antibodies directed against AIR12 recognized the diffuse, high-molecular-mass band in nondeglycosylated samples and the band at 27 kD in deglycosylated ones (Fig. 5C). Heme staining of the gel provides qualitative evidence that recAIR12 is a heme binding protein and that heme is noncovalently bound as expected for a b-type cytochrome because most of it is released by recAIR12 and recovered at the bottom of the gel under mild denaturing conditions (lithium dodecyl sulfate [LDS]-PAGE; see “Materials and Methods”).
Figure 5.
SDS-PAGE (A and B), western blotting (C), and LDS-PAGE of purified recAIR12 expressed in P. pastoris. Lane 1 (A–D), Purified recAIR12 eluted from the last gel filtration chromatography purification step (9 μg for A and B, 140 ng for C, and 9 μg for D); lane 2 (A–C), same as for lane 1, but after incubation for 1 h at 37°C with EndoH; in C, recAIR12 was 47 ng; lane 3 (A), EndoH (1 μg). Gels were stained by Coomassie Brilliant Blue (A) or glycoprotein staining (B; Leach et al., 1980). C, Western-blot analysis was performed using rabbit antisera against recAIR12 expressed in E. coli (see “Materials and Methods” for details). D, RecAIR12 was treated with EndoH overnight at 22°C prior to loading on the gel (lane 2). Gel was heme stained (Thomas et al., 1976).
For N-terminal sequencing of recAIR12, purified preparations were blotted on polyvinylidene difluoride (PVDF) membranes after SDS-PAGE and two bands were detected by Coomassie Brilliant Blue staining. Both bands were separately analyzed by Edman degradation and, in each case, two or three amino acids were detected at each cycle. However, both bands gave similar sequences corresponding to two major peptides Gln-Pro-Ala-Val-Ser-Asp-Arg-Tyr-Pro and Val-Ala-Gln-Pro-Ala-Val-Ser-Asp-Arg-Tyr-Pro and another minor peptide Gly-Ile-His-Val-Ala-Gln-Pro-Ala. This suggests that maturation of recAIR12 in P. pastoris generates a small heterogeneity at the N terminus. Nevertheless, all three peptides correspond to the N-terminal sequence of recAIR12, thereby validating the purity of recAIR12 preparations (Supplemental Fig. S1).
Characterization of AIR12 as a High-Potential, Ascorbate-Reducible Cytochrome b Binding a Single Low-Spin Heme
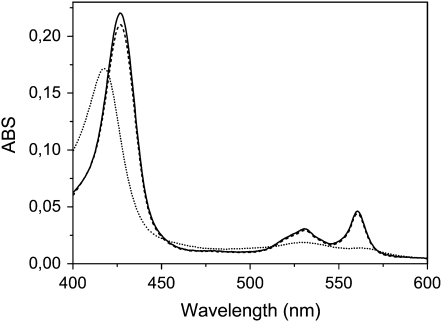
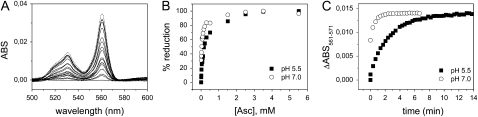
Absorption spectra of purified recAIR12 show a symmetric α-band centered at 561 nm in the reduced form together with a β-band at 530 nm and a γ-band at 427 nm (Fig. 6). Ascorbate reduction of the cytochrome reached 96% of dithionite reduction (Fig. 6). The recombinant cytochrome was extremely sensitive to ascorbate, being half reduced by 40 μm ascorbate at pH 7.0 and approximately 0.2 mm ascorbate at pH 5.5 (Fig. 7B). The α-band remained symmetric even under conditions of partial reduction (Fig. 7A). At pH 7.0, the rate of cytochrome reduction was much faster compared to pH 5.5, the cytochrome being half-reduced in a matter of seconds (Fig. 7C; 0.3 mm ascorbate). The reaction rate at pH 5.5 (half-reduction in 2 min) was not increased by changing ascorbate concentration up to 1 mm (data not shown).
Figure 6.
Optical absorption spectra of purified recAIR12 (1.6 μm). Spectra of the oxidized (dotted line), ascorbate (5 mm)-treated (dashed line), and dithionite-treated (solid line) were recorded at room temperature in 50 mm Tris-HCl, pH 7.0, 150 mm KCl. ABS, Absorbance.
Figure 7.
Reaction of ascorbate with purified recAIR12. A, Reduced minus oxidized optical spectra were recorded during titration of recAIR12 (1.6 μm) in 50 mm MES-KOH, pH 5.5, with ascorbate at the following final concentrations: 5, 10, 15, 25, 40, 55, 70, 90, 110, 160, 210, 310, 510, 1,510, 2,510, 3,510, and 5,510 μm. Finally, dithionite (a few grains) was added (dashed line). B, The percentage increase in absorbance at 561 nm is plotted as a function of ascorbate concentration for recAIR12 at pH 5.5 (same as in A; black squares) and recAIR12 (4.2 μm) equilibrated with 50 mm Tris-HCl, pH 7.0, 150 mm KCl (white circles, titrated with ascorbate at the same concentrations used for the sample at pH 5.5). C, Time courses of the absorbance change at 561 nm (relative to isosbestic point at 571 nm) upon reduction with ascorbate of purified recAIR12, at pH 5.5 (black squares) and pH 7.0 (white circles). Ascorbate (0.3 mm final concentration) was added to recAIR12 (0.7 μm) and absorption spectra were recorded approximately every 20 s. ABS, Absorbance.
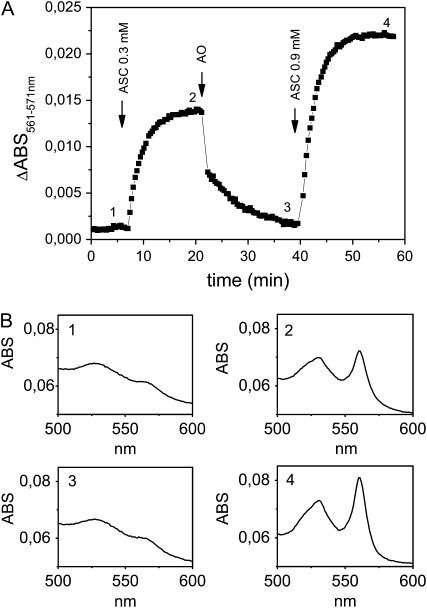
To check whether recAIR12 could be oxidized by MDA, ascorbate oxidase was added to the reduced cytochrome in the presence of ascorbate (Fig. 8). Upon MDA radical generation, a rapid decrease in α-band absorption at 561 nm (relative to the isosbestic point at 571 nm) was observed, indicating a rapid oxidation of the cytochrome. The latter could be subsequently rereduced by the addition of higher concentrations of ascorbate.
Figure 8.
A, Time course of the absorbance change at 561 nm of recAIR12 upon reduction with ascorbate (ASC; 0.3 mm) followed by oxidation through the addition of ascorbate oxidase (AO; 0.6 units) and rereduction by ascorbate (0.9 mm). RecAIR12 (1.1 μm) was in 50 mm MES-KOH, pH 5.5, 1 mm EDTA. B, Representative absorption spectra measured at points indicated in A. ABS, Absorbance.
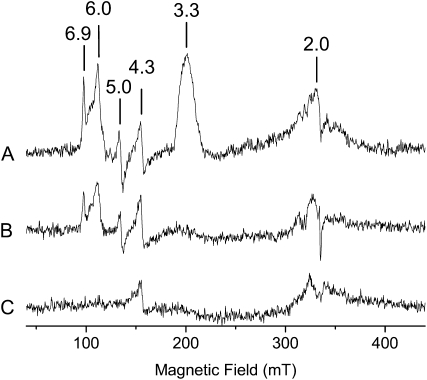
The EPR spectrum of oxidized recAIR12 shows a ramp-shaped signal with a large gmax value (3.3), and undetectable gmid and gmin (Fig. 9, trace A). This type of signal, named highly axial low spin (HALS), is commonly exhibited by low-spin hemes when the axial ligands differ from each other and is compatible with Met-91 and His-175 being the heme ligands (Salerno, 1984; Zoppellaro et al., 2006). Signals with higher g values are also evident indicating the presence of high-spin ferric heme with both axial (g = 6.0) and rhombic (g = 5.0 and 6.9) components. However, these high-spin components account for only about 3% each of total heme content, suggesting that they originate from partially degraded or misfolded protein. Ascorbate reduction led to complete disappearance of the HALS signal (Fig. 9, trace B), whereas the high-spin components were reduced by dithionite (data not shown). Apart from the g = 4.3 signal due to aspecific iron binding and to the cavity background, no other relevant signals were observed in the EPR spectrum. The signal at about g = 2.0 has not been assigned and may represent one of the components of the high-spin species. Qualitatively similar spectra were observed at both pH 5.5 (Fig. 9) and pH 7.0 (data not shown), further supporting the conclusion that recAIR12 binds a single heme.
Figure 9.
EPR spectra of recAIR12. Spectra were recorded at 10K, with a frequency of 9.4 GHz, a microwave power of 2 mW, and a modulation amplitude of 10 G. Time constant 81 ms; conversion time 163 ms. The sample contained 120 μm purified recAIR12 in 50 mm MES buffer, pH 5.5, 150 mm KCl. A, Oxidized sample. B, Sample in the presence of 5 mm ascorbate. C, Background or cavity signal.
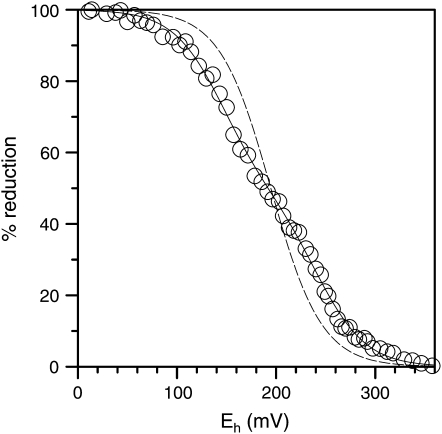
Potentiometric redox titrations of recAIR12 at pH 7.0 showed a redox response distributed over more than 200 mV, mostly comprised between +80 and +300 mV (Fig. 10). Surprisingly, experimental data could not be properly interpolated by a Nernstian curve for a single redox equilibrium, as expected for a single heme cytochrome. Better interpolations were obtained when two redox equilibria were introduced into the equation (Fig. 10). Two midpoint redox potentials were estimated (Em7 137 ± 19 and 236 ± 21 mV; n = 4), each related to a single redox couple accounting for about one-half of the total redox response (49% ± 16%; n = 4). Redox titrations performed at different pH values (5.5, 7.0, and 7.7) gave almost identical results (data not shown), indicating a very weak pH sensitivity of recAIR12 redox response within physiological pH values.
Figure 10.
Potentiometric redox titration. Purified recAIR12 (2.1 μm) was mixed with various redox mediators as described in “Materials and Methods.” After complete oxidation by ferricyanide, the redox status was monitored at pH 7.0 under anaerobic conditions following addition of reductants (ascorbate, dithionite). RecAIR12 percentage reduction, as measured by absorbance at 561–571 nm, was plotted against redox potential. Data were fitted to the Nernst equation for one (dashed line) or two (solid line) redox centers.
DISCUSSION
The capability of plant cells to reduce external electron acceptors by means of a PM redox system is a long-known phenomenon with still scarce molecular basis (Lüthje, 2008). In whole cells or intact tissues, reduction of electron acceptors was found associated with apoplast acidification, membrane depolarization, or oxidation of internal electron donors, suggesting a transmembrane electron transfer chain fed by cytosolic reductants (Vuletić et al., 2005). PM vesicles loaded with physiological electron donors [ascorbate, NAD(P)H] were found to reduce impermeable electron acceptors (MDA, ferrichelates, ferricyanide) on the apoplastic side of the membrane (Askerlund and Larsson, 1991; Asard et al., 1992; Horemans et al., 1994; Menckhoff and Lüthje, 2004). In this way, PM redox systems may influence the redox state of the apoplast and provide a redox connection between apoplast and symplast.
Search for molecular components involved in PM redox reactions led to biochemical characterization of several PM redox proteins, including flavoproteins (e.g. quinone reductases; Serrano et al., 1994; Trost et al., 1997; Schopfer et al., 2008; and MDA reductase; Bérczi and Møller, 1998), b-type cytochromes (Trost et al., 2000; Bérczi et al., 2003; Preger et al., 2005), and peroxidases (Mika et al., 2008). Moreover, molecular biology approaches led to the discovery of flavocytochromes of the FRO family that use cytosolic pyridine nucleotides to reduce apoplastic ferrichelates (Robinson et al., 1999). Structurally similar flavocytochromes (Rboh) are involved in one-electron reduction of oxygen with production of superoxide (Simon-Plas et al., 2002). However, these plant flavocytochromes apparently account for a very minor portion of b-type cytochromes associated with the PM, at least under unstressed conditions. On the other hand, PM preparations from different plant species were invariably found to contain ascorbate-reducible cytochromes b displaying a symmetric α-band around 561 nm and typically responding to high redox potentials, usually in the +50- to +300-mV range at neutral pH, with some differences among species and tissues (Asard et al., 1989; Askerlund et al., 1989). Ascorbate-reducible cytochromes b may represent up to 80% of the total cytochrome content in purified PM preparations (Asard et al., 2001).
Ascorbate-reducible cytochromes with a symmetric α-band at 561 nm were shown to be involved in trans-PM electron transport from cytosolic ascorbate to apoplastic MDA (Horemans et al., 1994). The reaction was thought to be catalyzed by a cytochrome b561 of the PM (Asard et al., 2001) in analogy with the system for ascorbate regeneration in chromaffin vesicles of mammals (Kelley and Njus, 1986). This concept needs revision, however, because true cytochrome b561 (CYBASC1) is localized in vacuolar membranes (tonoplast) in both Arabidopsis (Griesen et al., 2004) and bean (Preger et al., 2005), and chromaffin membranes are certainly closer to plant vacuolar membranes than PM. Only CYBASC1 of wild watermelon was found targeted to the PM in GFP construct experiments with onion epidermal cells (Nanasato et al., 2005). Moreover, the optical spectra of bona fide cytochrome b561 (CYBASC1 in plants; Tsubaki et al., 1997; Bérczi et al., 2007) are characterized by an asymmetric α-band with a maximum close to 561 nm and a shoulder at 557 nm, unlike the cytochrome involved in trans-PM electron transport whose α-band is definitely symmetric (Asard et al., 1992). Overall, it is unlikely that cytochrome b561 (CYBASC1) represents a significant fraction of high-potential cytochromes b of plant PM. Little is known about other, minor isoforms of cytochrome b561.
Here, we report that a major ascorbate-reducible cytochrome b of the PM, purified from either soybean or bean etiolated hypocotyls, according to previously published procedures (Preger et al., 2005), is to be identified as AIR12 (Neuteboom et al., 1999) because AIR12 is known to be one of approximately 80 Arabidopsis genes that are up-regulated in the root during the first 12 h following auxin treatment (Laskowski et al., 2006). No AIR12 protein has been characterized before. Orthologs of AIR12 are present in all flowering plant species (angiosperms) investigated, generally with one gene or, less frequently, two genes, but they appear to be absent from other groups of green plants or photosynthetic organisms, or in other eukaryotes, bacteria, or archaea. The molecular mass of native AIR12 predicted from soybean sequence (GmAIR12-1; Fig. 3) is 20 kD, after cleavage of the N-terminal (signal peptide) and the C-terminal (GPI anchor) sequences, quite different from the estimated molecular mass in SDS-PAGE (about 60 kD). The large discrepancy could be ascribed to glycosylation, however.
Bioinformatic studies predict AIR12 to belong to the DOMON (Aravind, 2001) or DoH (Ponting, 2001) domain superfamily. The DOMON domain consists of an all-β-strand fold forming a β-sandwich structure, similar to other extracellular adhesion domains (Aravind, 2001). Plant AIR12s consist of a single DOMON domain and thus appear to be largely hydrophilic proteins. Association of the cytochrome here identified as AIR12 to the PM was previously demonstrated by analytical Suc gradient centrifugation of plant microsomes (Preger et al., 2005). This result is supplemented by recent proteomic studies that identified AIR12 as a PM protein (Dunkley et al., 2006) bound to the external face of the PM by means of a GPI anchor (Borner et al., 2003) and in association with lipid rafts (Lefebvre et al., 2007). AIR12 has been reported among secreted proteins of Medicago truncatula cell cultures, suggesting that the GPI anchor is cleaved under particular conditions with release of the protein (Kusumawati et al., 2008).
Purifying adequate amounts of native AIR12 from suitable plant material is a very tedious task, so we have set up a heterologous expression system in P. pastoris. This eukaryotic expression system is also of advantage in that it produces a glycosylated recombinant protein, similar to the native one. The soybean AIR12 gene was transformed in P. pastoris cells into a form with no N-terminal signal peptide and a limited C-terminal deletion to get rid of the GPI anchor. Secreted recAIR12 was purified from culture broth and was found to be glycosylated and soluble in the absence of detergents. The expected molecular mass of the recombinant protein (24 kD) was 13 kD higher due to bound carbohydrates (MALDI-TOF MS). Enzymatic removal of N-glycans gave rise to strongly stained bands at expected positions in SDS gels (Fig. 5).
RecAIR12 shows the typical spectroscopic signature of a cytochrome b with a symmetric α-band at 561 nm and β- and γ-bands at 530 and 429 nm, respectively, following reduction by ascorbate. The cytochrome recAIR12 was sensitive to ascorbate concentrations in the 10−4 to 10−3 m range (i.e. about the same range of ascorbate concentrations found in the apoplast of different plant species; Pignocchi and Foyer, 2003). The reaction with ascorbate was reversible, MDA being an excellent electron acceptor for reduced AIR12. Besides AIR12, other members of the DOMON superfamily have been demonstrated to bind b-type heme (Iyer et al., 2007).
The best fitting of recAIR12 potentiometric redox titrations was obtained when two redox centers were introduced in the Nernst equation. RecAIR12 sensitivity to the redox potential, with two apparent midpoint redox potentials at +137 and +236 mV, is in line with that of partially purified PM cytochrome b from bean hypocotyls (+135 and +204 mV; Trost et al., 2000) and Arabidopsis leaves (+135 and +180 mV; Bérczi et al., 2003), and consistent with redox titrations of whole PM preparations (Asard et al., 1989; Askerlund and Larsson, 1991; Bérczi et al., 2003). This also indicates that AIR12 represents a major fraction of high potential and ascorbate-reducible cytochromes b of plant PM. These potentiometric results, however, are unexpected because other data point to one heme only.
The crystallographic structure of DOMON of cellobiose dehydrogenase of the fungus Phanerochaete chrysosporium (Hallberg et al., 2000) includes a single high-potential heme b (Em7 +164 mV) coordinated by a Met (Met-65) and a His (His-163). In fact, a Met-His couple (Met-91 and His-176 in GmAIR12-1; Fig. 5) is conserved in almost every plant AIR12 sequence and is likely to coordinate a heme b molecule. This observation was strengthened by EPR spectra of recAIR12 showing a HALS species with g = 3.3, compatible with His-Met coordinated heme and very similar to the HALS signal reported for cellobiose dehydrogenase (Cox et al., 1992).
The binding of a single heme per molecule is substantiated by different experimental observations, including (1) EPR spectra showing no other low-spin signal than the HALS; (2) the symmetric shape of the α-band of the optical spectrum of partially and fully reduced forms; (3) the A418/A280 ratio of the purified protein consistent with a stoichiometry of one heme per apoprotein; and, finally, (4) multiple alignment of AIR12 sequences did not show other obvious pairs of conserved residues to coordinate a second low-spin heme besides the Met-65/His-163 couple. Further studies are clearly needed to understand the bimodal redox response of AIR12.
DOMON domains are widespread in nature, usually in association with other redox domains within larger proteins (Ponting, 2001; Iyer et al., 2007). Interestingly, DOMON domains are often associated with cytochrome b561 moieties, and Arabidopsis contains 10 genes coding for hypothetical proteins with this particular domain association. Most of them have clear orthologs in other plants, including soybean (Verelst and Asard, 2003; Tsubaki et al., 2005). Animal proteins with analogous domain architecture (e.g. SDR2 of Drosophila) are able to reduce extracellular ferrichelates in transfected Xenopus oocytes, suggesting capability of transmembrane electron transport (Vargas et al., 2003). The conclusion that AIR12 is a cytochrome raises the possibility that DOMON domains bound to b561-type cytochromes (groups F1, F2, G of Fig. 2; Tsubaki et al., 2005) might contain heme themselves.
The role of AIR12 in plant PM redox reactions is still obscure, although an involvement in trans-PM electron transport, and potentially in redox signaling, seems likely. In fact, in ascorbate-loaded PM vesicles, the totality of ascorbate-reducible cytochromes could be oxidized by an external electron acceptor such as ferricyanide or MDA (Asard et al., 1992; Horemans et al., 1994), and AIR12 certainly represents a major ascorbate-reducible cytochrome of plant PM. Trans-PM electron transport involving cytosolic NAD(P)H was also demonstrated, although it is not known whether a cytochrome was involved (Askerlund and Larsson, 1991; Menckhoff and Lüthje, 2004). In any case, because AIR12 is not a transmembrane protein, interactions with other redox partners would be needed to build up an electron route across the PM (Vuletić et al., 2005; Lüthje, 2008). Notably, only a few redox proteins known to interact with the PM are up-regulated in Arabidopsis roots soon after auxin treatment and are implicated in lateral root emergence (Laskowski et al., 2006). These include AIR12, as well as flavodoxin-like quinone reductase (FQR1; Laskowski et al., 2002), multicopper oxidase SKU5, and a putative peroxidase (At5g64100), all of which could interact directly with AIR12 on the apoplastic side of the PM. SKU5, probably an ascorbate oxidase, is involved in directional growth of Arabidopsis roots (Sedbrook et al., 2002); it is also bound to the PM by means of a GPI anchor (Sedbrook et al., 2002), and it copurifies with AIR12 (see Fig. 1).
A DOMON-plus-cytochrome b561 protein (AC147406_8.1) has recently been detected in lipid rafts of M. truncatula roots together with AIR12 and other redox proteins (Lefebvre et al., 2007; i.e. orthologs of SKU5 and FQR1). Peroxidases are also part of the set of redox proteins included in lipid rafts (Lefebvre et al., 2007) and an ascorbate peroxidase was suggested to interact with ascorbate-reducible cytochrome b in bean hypocotyl PM (Preger et al., 2001). FQR1 is expected to reside on the cytosolic side of the membrane and any interaction with AIR12 would need to be mediated by a membrane-diffusible quinone for which the experimental evidence is still inconclusive (Lüthje, 2008). In any case, this assembly of PM redox proteins and compounds sequestered within lipid rafts may form a potential redox link between cells and apoplast, under the control of auxin, an intriguing hypothesis for a role of PM redox systems in lateral root formation.
MATERIALS AND METHODS
Plant Materials
Etiolated seedlings of soybean (Glycine max ‘Pacific’) were grown for 6 d in the dark on moist vermiculite at 25°C.
Chemicals
All chemicals were from Sigma-Aldrich or Merck unless otherwise stated.
Preparation of Soybean Microsomal Membranes
Microsomal membranes were prepared as described for bean (Phaseolus vulgaris) hypocotyls (Preger et al., 2005), except for minor modifications. Briefly, hypocotyls (100–200 g fresh weight) were harvested and ground on ice in 2 volumes of homogenization buffer (50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 10 mm β-mercaptoethanol, 250 mm Suc, 2 mm ascorbate, 0.5 mm phenylmethylsulfonyl fluoride) by means of a blender (Minipimer; Braun) for 20 s at full speed. The resulting slurry was filtered through a 30-μm nylon net and centrifuged at 9,000g for 20 min at 4°C. The pellet was discarded and the microsomal fraction was collected by centrifugation at 110,000g for 1 h at 4°C. Microsomes were resuspended in 20 mm Tris-HCl buffer, pH 8.0, containing 20% (v/v) glycerol, at a final concentration of 5 mg mL−1, and stored at −80°C until use.
Purification of PM Ascorbate-Reducible Cytochrome b from Soybean
Chromatographic medium and systems used for purification were from GE Healthcare. Microsomes (35 mg total protein) were solubilized for 1 h at 4°C under stirring, after the addition of 1% (w/v) octyl-β-d-glucopyranoside (OG) and 1 mm ascorbate in 7 mL of solubilization buffer (50 mm Tris-HCl [pH 8.0], 20% glycerol). Detergent-to-protein ratio was 4:1. Insoluble material was sedimented by centrifugation at 110,000g for 30 min at 4°C. Solubilized microsomal proteins (4 mL) were loaded on an anion-exchange SourceQ column (7 mL) equilibrated with 50 mm Tris-HCl (pH 8.0), 20% (v/v) glycerol, 1 mm ascorbate, 1% (w/v) OG. Proteins bound to the resin were eluted with a linear KCl gradient (from 0–0.26 m in 4.7 column volume) at a flow rate of 0.5 mL min−1. Optical absorption of the eluate was continuously monitored at 418 and 427 nm and 0.6-mL fractions were collected. PM cytochrome b-containing fractions, deriving from six anion-exchange separations at pH 8 were pooled together, concentrated to 7 mL (Amicon YM-30 and Centricon YM-30; Millipore), equilibrated with 20 mm 3-(cyclohexylamino)-1-propanesulfonic acid, pH 10.0, 20% (v/v) glycerol, 1 mm ascorbate, 1% (w/v) OG by means of HiTrap fast-desalting cartridges, and reloaded on the anion-exchange SourceQ column equilibrated with the same buffer. A linear KCl gradient (0–0.5 m in 6.3 column volume) was applied at a 0.5 mL min−1 flow rate, and 0.6-mL fractions were collected. Fractions eluted from 0.04 to 0.09 m KCl were pooled together and concentrated to 0.14 mL by Centricon. The cytochrome was finally loaded on a Superdex 200 (HR 10/30) gel filtration chromatography column equilibrated with 50 mm Tris-HCl (pH 8.0), 20% (v/v) glycerol, 1 mm ascorbate, 1% (w/v) OG. Elution was carried out at 0.5 mL min−1 and fractions of 0.2 mL were collected.
Protein Identification by MS
HPLC-grade acetonitrile (ACN), Coomassie Brilliant Blue R-250, dithiothreitol, formic acid, iodoacetamide, and trifluoroacetic acid were purchased from Sigma; Tris and Gly from Bio-Rad; and PVDF membrane from Millipore. HPLC-grade ethanol and acetic acid were purchased from VWR.
Bands of interest were excised manually and subjected to in-gel digestion by bovine trypsin (Roche Diagnostics) using the automated protein digestion system DigestPro-96 (Intavis AG). Peptides were extracted first with 1% TFA, then with 60% ACN containing 1% TFA, dried in a SpeedVac concentrator (Savant), and resuspended in 5% ACN/0.1% formic acid. Nano-LC-CHIP-IT-MS/MS experiments were performed on an Agilent 1200 nanoflow LC system coupled to a 6330 ion trap equipped with the Chip Cube orthogonal ionization system (Agilent Technologies). For MS/MS experiments, the system was operated in positive-ion mode with automatic switching between MS and MS/MS modes, whereas MS/MS scanning was performed in the ultrascan resolution mode. A total of six scans were averaged to obtain a MS spectrum, whereas three scans were averaged for the MS/MS spectrum. MS/MS data were analyzed by data analysis software, version 3.4 (Agilent Technologies). Peptide mass fingerprints were also performed by MALDI-TOF MS as described previously (Marchand et al., 2006). Mass spectra were analyzed by DataExplorer, version 5.1 (Applied Biosystems). Protein identifications were performed using two search engines: MASCOT MS/MS Ions search (http://www.matrixscience.com) and X!tandem (http://prowl.rockefeller.edu). The identifications were validated according to the established guidelines for proteomic data publication (Carr et al., 2004; Bradshaw et al., 2006) and notably protein identification was validated only if at least two different sequences were identified with high-quality mass spectra (MASCOT ion score >40). Moreover, the false discovery rate was estimated with X!tandem at less than 0.61% by searches against reversed databases. Detailed protocols and procedures are provided in Supplemental File S1.
For automated Edman degradation, peptides from in-gel tryptic digest of a purified sample were first subjected to HPLC separation on a C-18 reverse-phase column. Automated Edman degradation of two peptides was performed with an Applied Biosystems Procise 494 cLC protein sequencer at the W.M. Keck Facility.
Heme Content Analysis
Heme content of recAIR12 samples was analyzed using the pyridine hemochrome method as described in Metzger et al. (1997). Heme b content was calculated from dithionite-reduced minus ferricyanide-oxidized spectra of pyridine hemochrome using a differential extinction coefficient (557–541 nm) of 20.7 mm−1 (Porra and Jones, 1963). Using this assay, the reduced minus oxidized extinction coefficient for recAIR12 heme at 561 nm relative to the isosbestic wavelength of 571 nm was calculated to be 21 mm−1.
Assay of Protein and AIR12 Content
Total protein was assayed by the method of Bradford. RecAIR12 content was calculated from dithionite-reduced minus oxidized spectra (ɛ561−571 21 mm−1), considering a stoichiometry of one heme per AIR12 molecule.
Expression of RecAIR12 of Soybean in Pichia pastoris
An EST clone containing the full-length soybean AIR12 cDNA (GenBank accession no. FJ528079) was purchased from Biogenetic Services and used as a template for PCR. Two oligonucleotide primers were designed for ligation of a truncated form of AIR12 cDNA into the KpnI site of the expression vector pPICZαB (Invitrogen) in frame with the c-myc epitope and a polyhistidine tag. Sense and antisense primers were 5′-CATGGGTACCCTATGCTCATCTGCGCTTCCC-3′ and 5′-CATGGGTACCGGCGCGGGCGCAGTCGCGTT-3′, respectively (KpnI sites are underlined). The oligonucleotides amplified a region of the gene right after the N-terminal leader peptide encoding sequence. To recover the recombinant protein in the culture medium, avoiding GPI anchoring to the yeast membranes, the sequence coding for the C-terminal GPI signal peptide was also omitted from the amplified cDNA (Fig. 3). The PCR-amplified cDNA was cloned into the pPICZαB vector and used to transform P. pastoris X-33 cells (Invitrogen) by electroporation. Transformants were selected on YPDS medium (1% [w/v] yeast extract, 2% [w/v] peptone, 2% [w/v] dextrose, 2% [w/v] agar) containing 1.5 mg/mL zeocin. Positive clones were tested for integration by direct PCR screening. For protein expression, a single colony of P. pastoris X-33/pPICZαB-AIR12 was used to inoculate 100 mL of BMGY medium (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/v] yeast nitrogen base, 4 × 10−5% [w/v] biotin, 1% [v/v] glycerol) containing 100 μg/mL zeocin. Cells were grown overnight (28°C, 220 rpm in baffled flasks) until OD600 reached a value of 4, then pelleted (1,500g for 10 min), and resuspended in 400 mL of BMMY-inducing medium (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/v] yeast nitrogen base, 4 × 10−5% [w/v] biotin, 1% [v/v] methanol) containing 0.1 mm δ-aminolevulinic acid and 40 μm hemin. Cultures were grown (28°C, 220 rpm) for 48 h in inducing medium with the addition of 1% (v/v) methanol every 24 h.
Purification of RecAIR12
Chromatographic medium and systems used for purification were from GE Healthcare. Two days after induction, cultures of P. pastoris X-33/pPICZαB-AIR12 were spun down (10 min at 1,500g). The supernatant (220 mL) was concentrated to 10 mL (Amicon YM-10) and equilibrated to pH 5.5 with 1 m MES. The sample was then centrifuged at 35,000g for 1 h at 4°C to remove excess hemin (which precipitates at pH 5.5), then loaded on a Octyl Sepharose 4 fast-flow column (12 mL) equilibrated with 50 mm potassium phosphate, pH 7, (NH4)2SO4 1.7 m. RecAIR12-containing fractions, eluting in the column flow-through, were pooled, diluted twice with water to lower salt content, and concentrated to 7.5 mL (Amicon YM-10). Proteins were then equilibrated with 50 mm Tris-HCl, pH 7.0, 1 mm EDTA, by means of PD-10 columns and loaded on an anion-exchange SourceQ column (7 mL) equilibrated with the same buffer. A linear KCl gradient (0–0.5 m in three-column volume) was applied at a 0.5 mL min−1 flow rate, and 0.4-mL fractions were collected. RecAIR12-containing fractions were pooled, concentrated to 0.2 mL, and loaded on a Superdex 200 (HR 10/30) gel filtration chromatography column equilibrated with 50 mm Tris-HCl, pH 7.0, 2 mm EDTA, 150 mm KCl. Elution was carried out at 0.5 mL min−1 and fractions of 0.2 mL were collected.
Protein Deglycosylation
For protein deglycosylation Endo H (New England BioLabs) was used according to the manufacturer's instructions. Incubation of denatured samples with Endo H was carried out at 37°C for 1 h. For deglycosylation of the native nondenatured protein, the incubation was done overnight at 22°C.
Expression of RecAIR12 of Soybean in Escherichia coli for Polyclonal Antibody Production
A truncated form of soybean AIR12 lacking the N-terminal signal peptide and C-terminal hydrophobic signal peptide (see Fig. 3) was cloned into the expression vector pET-28a(+) (Novagen) and expressed into Escherichia coli BL-21(DE3) cells according to Sparla et al. (1999). RecAIR12 was recovered in insoluble inclusion bodies. Purified inclusion bodies were used for rabbit polyclonal antibody production at Primm.
Electrophoresis and Western Blotting
SDS-PAGE was performed on 12.5% acrylamide gels. Samples were boiled for 10 min in sample buffer prior to loading. Gels were stained with Coomassie Brilliant Blue R-250. The staining of glycoproteins was done according to Leach et al. (1980).
To visualize hemoproteins, recAIR12-containing samples were resolved on 12.5% PAGE, using LDS in the gel, sample, and running buffer instead of SDS, according to the method of Sinclair et al. (1981). Electrophoresis was conducted at a constant 90 V, 4°C, for 3 h and staining was performed as described by Thomas et al. (1976).
For western blotting, proteins were separated by SDS-PAGE and electroblotted (semidry cell; Schleicher-Schuell) onto nitrocellulose membranes. Membranes were stained with Red Ponceau before incubation with rabbit antiserum raised against AIR12 expressed in E. coli (see above), and peroxidase-conjugated secondary antibodies. Primary and secondary antibodies were diluted 1:2,000 and 1:1,000, respectively. Blots were developed by chemiluminescence according to standard procedures.
N-Terminal Sequencing and Molecular Weight Determination by MALDI-TOF MS
After deglycosylation treatment with EndoH, proteins were separated by 12% SDS-PAGE and electroblotted onto PVDF membranes (pore size 0.2 μm) using the liquid Mini Trans-Blot Cell (Bio-Rad). Bands detected by Coomassie Brilliant Blue R-250 staining were cut and subjected to automated Edman degradation using a Procise sequencer (Applied Biosystems).
The molecular weight of recAIR12 was determined by MALDI-TOF MS without the desalting step, as described previously (Zaffagnini et al., 2007).
Redox Titrations
Spectrophotometric titrations were performed essentially as described in Venturoli and Zannoni (1988), using a Jasco V-550 spectrophotometer and an anaerobic titrator (1-cm light path, 3.2-mL sample volume) equipped with a platinum electrode. Measurements were done with a pH meter using a calomel electrode as reference. The ferro-ferricyanide redox couple was used to calibrate the system according to O'Reilly (1973). An oxidized recAIR12 sample (approximately 1.2 μm) in 50 mm Tris-HCl, pH 7.0, 20 mm KCl or 50 mm MES-KOH, pH 5.5, 20 mm KCl was mixed with redox mediators: 1 μm phenazine methosulfate (Em7 = +80 mV), 1 μm phenazine ethosulfate (Em7 = +33 mV), 10 μm 1,4-naphtoquinone (Em7 = +60 mV), 10 μm 1,2-naphtoquinone (Em7 = +157 mV), 10 μm p-benzoquinone (Em7 = +280 mV). Reductive titration was performed at room temperature under N2 flow by addition of small aliquots of sodium ascorbate or sodium dithionite solution. After each addition, the sample was stirred and allowed to equilibrate before recording the visible spectrum. The fraction of maximal change in A561 as a function of redox potential was fitted to the Nernst equation for one or two redox components with unknown percentage contribution to the total response. Nonlinear regression analysis was performed with Costat (CoHort software).
Measurements of EPR Spectra
Samples containing 120 μm purified recAIR12 dissolved in 50 mm MES-KOH, pH 5.5, 150 mm KCl were withdrawn into EPR tubes. Oxygen was removed by flushing nitrogen above the solutions in the EPR tubes. The samples were flash cooled in liquid nitrogen before insertion in the EPR spectrometer at 10K. Continuous-wave EPR measurements were performed on a Bruker Elexsys E580 spectrometer at 9.4 GHz, equipped with standard TE102 cavity. The temperature was controlled with a helium flow cryostat (Oxford Instruments ESR-900) driven by a temperature controller. Reduction of the samples was obtained by adding a few microliters of a concentrated ascorbate solution (final concentration approximately 5 mm) directly in the EPR tubes. Further reduction was achieved by adding sodium dithionite (few grains). Experimental conditions were T = 10K, microwave power = 2 mW, field modulation = 100 kHz, modulation amplitude = 10 G, time constant = 82 ms, conversion time = 164 ms; g values were estimated by calibration with a diphenylpicrylhydrazyl sample. Spin quantification of the high-spin hemes relative to the HALS species was performed by simulation of the X-band EPR spectrum and double integration of the contribution of each individual species.
Sequence and Phylogenetic Analyses
All proteins containing a DOMON domain (IPR005018) from Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa ‘japonica’ group) were identified in the INTERPRO database (http://www.ebi.ac.uk/interpro). AIR12 sequences were identified by BLASTp searches in the National Center for Biotechnology Information (NCBI) protein database (http://www.ncbi.nlm.nih.gov) and in sequenced genomes in Joint Genome Institute databases (http://genome.jgi-psf.org). Additional sequences were retrieved by tBLASTn searches in NCBI GenBank and dbEST databases. For each EST identified, the corresponding Unigene cluster was retrieved (http://www.ncbi.nlm.nih.gov/unigene) and used for EST assembly with the CAP3 program (Huang and Madan, 1999). Contigs containing complete open reading frames were translated to generate AIR12 protein sequences. All protein sequences used in this study are provided in Supplemental File S2. Multiple sequence alignments were performed with the ClustalW program. The unrooted phylogenetic tree was generated with the ClustalX2 program and visualized with Treeview. Conserved domains were determined using the CD search service of NCBI conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd). The presence of signal peptides and potential cleavage sites were determined using PSORT (http://psort.ims.u-tokyo.ac.jp). GPI anchors were analyzed using the GPI-SOM program (http://gpi.unibe.ch).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ528079 and FJ535652.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Recombinant soybean AIR12 sequence expressed in P. pastoris.
Supplemental File S1. Protein identification by MS.
Supplemental File S2. Sequences used for phylogenetic analyses and sequence alignments in fasta format.
Supplemental Table S1. Protein identifications by MASCOT MS/MS ions search and X!tandem.
Supplementary Material
This work was supported by the Ministero della Pubblica Istruzione (grants FIRB 2004 and PRIN 2007).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Valeria Preger (valeria.preger@unibo.it).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Aravind L (2001) DOMON: an ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends Biochem Sci 26 524–526 [DOI] [PubMed] [Google Scholar]
- Asard H, Horemans N, Caubergs RJ (1992) Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett 306 143–146 [DOI] [PubMed] [Google Scholar]
- Asard H, Kapila J, Verelst W, Bérczi A (2001) Higher-plant plasma membrane cytochrome b561: a protein in search of a function. Protoplasma 217 77–93 [DOI] [PubMed] [Google Scholar]
- Asard H, Terol-Alcayde J, Preger V, Del Favero J, Verelst W, Sparla F, Pérez-Alonso M, Trost P (2000) Arabidopsis thaliana sequence analysis confirms the presence of cyt b-561 in plants: evidence for a novel protein family. Plant Physiol Biochem 38 905–912 [Google Scholar]
- Asard H, Venken M, Caubergs RJ, Reijnders W, Oltmann FL, De Greef JA (1989) b-Type cytochromes in higher plant plasma membranes. Plant Physiol 90 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askerlund P, Larsson C (1991) Transmembrane electron transport in plasma membrane vesicles loaded with an NADH-generating system or ascorbate. Plant Physiol 96 1178–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askerlund P, Larsson C, Widell S (1989) Cytochromes of plant plasma membranes: characterization by absorbance difference spectroscopy and redox titration. Physiol Plant 76 123–134 [Google Scholar]
- Bashtovyy D, Bérczi A, Asard H, Páli T (2003) Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma 221 31–40 [DOI] [PubMed] [Google Scholar]
- Bérczi A, Caubergs RJ, Asard H (2003) Partial purification and characterization of an ascorbate-reducible b-type cytochrome from the plasma membrane of Arabidopsis thaliana leaves. Protoplasma 221 47–56 [DOI] [PubMed] [Google Scholar]
- Bérczi A, Møller IM (1998) NADH-Monodehydroascorbate oxidoreductase is one of the redox enzymes in spinach leaf plasma membranes. Plant Physiol 116 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérczi A, Su D, Asard H (2007) An Arabidopsis cytochrome b561 with trans-membrane ferrireductase capability. FEBS Lett 81 1505–1508 [DOI] [PubMed] [Google Scholar]
- Bérczi A, Su D, Lakshminarasimhan M, Vargas A, Asard H (2005) Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Arch Biochem Biophys 443 82–92 [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw RA, Burlingame AL, Carr S, Aebersold R (2006) Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics 5 787–788 [DOI] [PubMed] [Google Scholar]
- Butt WD, Keilin D (1962) Absorption spectra and some other properties of cytochrome c and of its compounds with ligands. Proc R Soc Lond B Biol Sci 156 429–458 [DOI] [PubMed] [Google Scholar]
- Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A (2004) The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol Cell Proteomics 3 531–533 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba-Pedregosa MdC, Cordoba F, Villalba JM, Gonzalez-Reyes JA (2003) Zonal changes in ascorbate and hydrogen peroxide contents, peroxidase and ascorbate activities in onion roots. Plant Physiol 131 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba-Pedregosa MdC, Villalba JM, Cordoba F, Gonzalez-Reyes JA (2005) Changes in intracellular and apoplastic peroxidase activity, ascorbate redox status, and root elongation induced by enhanced ascorbate content in Allium cepa L. J Exp Bot 56 685–694 [DOI] [PubMed] [Google Scholar]
- Cox MC, Rogers MS, Cheesman M, Jones GD, Thomson AJ, Wilson MT, Moore GR (1992) Spectroscopic identification of the haem ligands of cellobiose oxidase. FEBS Lett 307 233–236 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, De Gara L (2004) Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J Exp Bot 55 2559–2569 [DOI] [PubMed] [Google Scholar]
- Dietz KJ (1997) Functional responses of the leaf apoplast under stress. Prog Bot 58 221–254 [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC (1998) Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochem J 332 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper C, Dolan L (2006) Control of plant development by reactive oxygen species. Plant Physiol 141 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesen D, Su D, Bérczi A, Asard H (2004) Localization of an ascorbate-reducible cytochrome b-561 in the plant tonoplast. Plant Physiol 134 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg BM, Bergfors T, Bäckbro K, Pettersson G, Henriksson G, Divne C (2000) A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Structure 8 79–88 [DOI] [PubMed] [Google Scholar]
- Horemans N, Asard H, Caubergs RJ (1994) The role of the ascorbate free radical as an electron acceptor to cytochrome b-mediated trans-plasma membrane electron transport in higher plants. Plant Physiol 104 1455–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horemans N, Foyer CH, Asard H (2000) Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci 5 263–267 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: A DNA Sequence Assembly Program. Genome Res 9 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Aravind L (2007) The DOMON domains are involved in heme and sugar recognition. Bioinformatics 23 2660–2664 [DOI] [PubMed] [Google Scholar]
- Kamensky Y, Liu W, Tsai AL, Kulmacz RJ, Palmer G (2007) Axial ligation and stoichiometry of heme centers in adrenal cytochrome b561. Biochemistry 46 8647–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley PM, Njus D (1986) Cytochrome b561 spectral changes associated with electron transfer in chromaffin-vesicle ghosts. J Biol Chem 261 6429–6432 [PubMed] [Google Scholar]
- Kloer DP, Hagel C, Heider J, Schulz GE (2006) Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 14 1377–1388 [DOI] [PubMed] [Google Scholar]
- Kusumawati L, Imin N, Djordjevic MA (2008) Characterization of the secretome of suspension cultures of Medicago species reveals proteins important for defense and development. J Proteome Res 7 4508–4520 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al (2007) ClustalW2 and ClustalX version 2. Bioinformatics 23 2947–2948 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R (2006) Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol 47 788–792 [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Dreher KA, Gehring MA, Abel S, Gensler AL, Sussex IM (2002) FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol 128 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach BS, Collawn JF, Fish WW (1980) Behavior of glycopolypeptides with empirical molecular weight estimation methods. 1. In sodium dodecyl sulfate. Biochemistry 19 5734–5741 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, Rossignol M, Napier JA, Cullimore J, Bessoule JJ, et al (2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje S (2008) Plasma membrane redox systems: lipid rafts and protein assemblies. In UE Lüttge, W Beyschlag, J Murata, eds, Progress in Botany, Vol 69. Springer-Verlag, Berlin, pp 169–200
- Marchand C, Le Maréchal P, Meyer Y, Decottignies P (2006) Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics 24 6528–6537 [DOI] [PubMed] [Google Scholar]
- McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, et al (2001) An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291 1755–1759 [DOI] [PubMed] [Google Scholar]
- Menckhoff M, Lüthje S (2004) Transmembrane electron transport in sealed and NAD(P)H-loaded right-side-out plasma membrane vesicles isolated from maize (Zea mays L.) roots. J Exp Bot 55 1343–1349 [DOI] [PubMed] [Google Scholar]
- Metzger SU, Cramer WA, Whitmarsh J (1997) Critical analysis of the extinction coefficient of chloroplast cytochrome f. Biochim Biophys Acta 1319 233–241 [DOI] [PubMed] [Google Scholar]
- Mika A, Buck F, Lüthje S (2008) Membrane-bound class III peroxidases: identification, biochemical properties and sequence analysis of isoenzymes purified from maize (Zea mays L.) roots. J Proteomics 71 412–424 [DOI] [PubMed] [Google Scholar]
- Mori IC, Schroeder JI (2004) Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol 135 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanasato Y, Akashi K, Yokota A (2005) Co-expression of cytochrome b561 and ascorbate oxidase in leaves of wild watermelon under drought and high light conditions. Plant Cell Physiol 46 1515–1524 [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JM, Kuyper M, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ (1999) Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol 39 273–287 [DOI] [PubMed] [Google Scholar]
- O'Reilly JE (1973) Oxidation-reduction potential of the ferro-ferricyanide system in buffer solutions. Biochim Biophys Acta 292 509–515 [DOI] [PubMed] [Google Scholar]
- Padu E, Kollist H, Tulva I, Oksanen E, Moldau H (2005) Components of apoplastic ascorbate use in Betula pendula leaves exposed to CO2 and O3 enrichment. New Phytol 165 131–141 [DOI] [PubMed] [Google Scholar]
- Pignocchi C, Foyer CH (2003) Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr Opin Plant Biol 6 379–389 [DOI] [PubMed] [Google Scholar]
- Pignocchi C, Kiddle G, Hernández I, Foster SJ, Asensi A, Taybi T, Barnes J, Foyer CH (2006) Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in tobacco. Plant Physiol 141 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP (2001) Domain homologues of dopamine beta-hydroxylase and ferric reductase: roles for iron metabolism in neurodegenerative disorders? Hum Mol Genet 10 1853–1858 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Jones OTG (1963) An investigation of the role of ferrochelatase in the biosynthesis of various haem prosthetic groups. Biochem J 87 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preger V, Pesaresi A, Pupillo P, Trost P (2001) Ascorbate-independent electron transfer between cytochrome b-561 and a 27-kD ascorbate peroxidase of bean hypocotyls. Protoplasma 217 137–145 [DOI] [PubMed] [Google Scholar]
- Preger V, Scagliarini S, Pupillo P, Trost P (2005) Identification of an ascorbate-dependent cytochrome b of the tonoplast membrane sharing biochemical features with members of the cytochrome b561 family. Planta 220 365–375 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Salerno JC (1984) Cytochrome electron spin resonance line shapes, ligand fields, and components stoichiometry in ubiquinol-cytochrome c oxidoreductase. J Biol Chem 259 2331–2336 [PubMed] [Google Scholar]
- Sandermann H (2008) Ecotoxicology of ozone: bioactivation of extracellular ascorbate. Biochem Biophys Res Commun 366 271–274 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Heyno E, Drepper F, Krieger-Liszkay A (2008) Naphthoquinone-dependent generation of superoxide radicals by quinone reductase isolated from the plasma membrane of soybean. Plant Physiol 147 864–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A (2006) Plasma membrane-generated reactive oxygen intermediates and their role in cell growth of plants. Biofactors 28 73–81 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano A, Cordoba F, Gonzales-Reyes JA, Navas P, Villalba JM (1994) Purification and characterization of two distinct NAD(P)H dehydrogenases from onion (Allium cepa L.) root plasma membrane. Plant Physiol 106 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K, Maeshima M, Yokota A, Tomizawa K, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Simon-Plas F, Elmayan T, Blein JP (2002) The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J 31 137–147 [DOI] [PubMed] [Google Scholar]
- Sinclair JF, Healey JF, McAllister R, Bonkowsky HL, Sinclair PR (1981) Improved retention of heme with increased resolution of microsomal proteins in polyacrylamide gel electrophoresis. Anal Biochem 114 316–321 [DOI] [PubMed] [Google Scholar]
- Sparla S, Tedeschi G, Pupillo P, Trost P (1999) Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett 463 382–386 [DOI] [PubMed] [Google Scholar]
- Takahama U (1993) Redox state of ascorbic acid in the apoplast of stems of Kalanchoë daigremontiana. Physiol Plant 89 791–798 [Google Scholar]
- Thomas PE, Ryan D, Levin W (1976) An improved procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem 75 168–176 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8 397–403 [DOI] [PubMed] [Google Scholar]
- Trost P, Bérczi A, Sparla F, Sponza G, Marzadori B, Asard H, Pupillo P (2000) Purification of cytochrome b-561 from bean hypocotyls plasma membrane. Evidence for the presence of two heme centers. Biochim Biophys Acta 1468 1–5 [DOI] [PubMed] [Google Scholar]
- Trost P, Foscarini S, Preger V, Bonora P, Vitale L, Pupillo P (1997) Dissecting the diphenylene iodonium-sensitive NAD(P)H: quinone oxidoreductase of Cucurbita plasma membrane. Plant Physiol 114 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki M, Nakayama M, Okuyama E, Ichikawa Y, Hori H (1997) Existence of two heme B centers in cytochrome b561 from bovine adrenal chromaffin vesicles as revealed by a new purification procedure and EPR spectroscopy. J Biol Chem 272 23206–23210 [DOI] [PubMed] [Google Scholar]
- Tsubaki M, Takeuchi F, Nakanishi N (2005) Cytochrome b561 protein family: expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta 1753 174–190 [DOI] [PubMed] [Google Scholar]
- Vargas JD, Herpers B, McKie AT, Gledhill S, McDonnell J, van den Heuvel M, Davies KE, Ponting CP (2003) Stromal cell-derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim Biophys Acta 1651 116–123 [DOI] [PubMed] [Google Scholar]
- Venturoli G, Zannoni D (1988) Oxidation-reduction thermodynamics of the acceptor quinone complex in whole-membrane fragments from Chloroflexus aurantiacus. Eur J Biochem 178 503–509 [DOI] [PubMed] [Google Scholar]
- Verelst W, Asard H (2003) A phylogenetic study of cytochrome b561 proteins. Genome Biol 4 R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuletić M, Sukalović VH, Vucinić Z (2005) The coexistence of the oxidative and reductive systems in roots: the role of plasma membranes. Ann N Y Acad Sci 1048 244–258 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, Le Maréchal P, Miginiac-Maslow M, Noctor G, Trost P, Lemaire SD (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J 274 212–226 [DOI] [PubMed] [Google Scholar]
- Zoppellaro G, Teschner T, Harbitz E, Schuenemann V, Karlsen S, Arciero DM, Ciurli S, Trautwein AX, Hooper AB, Andersson KK (2006) Low-temperature EPR and Mossbauer spectroscopy of two cytochromes with His-Met axial coordination exhibiting HALS signals. Chemphyschem 7 1258–1267 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3 e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.