Abstract
As an adaptation to life in a world with predictable daily changes, most eukaryotes and some prokaryotes have endogenous circadian (approximately 24 h) clocks. In plants, the circadian clock regulates a diverse range of cellular and physiological events from gene expression and protein phosphorylation to cellular calcium oscillations, hypocotyl growth, leaf movements, and photoperiod-dependent flowering. In Arabidopsis (Arabidopsis thaliana), as in other model organisms, such as Drosophila (Drosophila melanogaster) and mice, circadian rhythms are generated by molecular oscillators that consist of interlocking feedback loops involving a number of elements. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYLS (LHY) are closely related single myb transcription factors that have been identified as key elements in the Arabidopsis oscillator. Research in other model organisms has shown that posttranslational regulation of oscillator components plays a critical role in the generation of the approximately 24-h cycles. To examine the role of posttranslational regulation of CCA1 and LHY in the Arabidopsis oscillator, we generated transgenic plants with tagged CCA1 and LHY under the control of their own promoters. We have shown that these tagged proteins are functional and can restore normal circadian rhythms to CCA1- and LHY-null plants. Using the tagged proteins, we demonstrate that CCA1 can form both homodimers and heterodimers with LHY. Furthermore, we also show that CCA1 is localized to the nucleus in vivo and that there is no significant delay between the translation of CCA1 and its translocation to the nucleus. We discuss our findings in the context of the functioning of the Arabidopsis oscillator.
The circadian, approximately 24 h, clock has an enormous influence on the biology of plants and controls a plethora of processes, including hypocotyl growth, shade avoidance, leaf movements, scent production, and stomatal opening (Yakir et al., 2007). Consistent with the role of the circadian clock in the regulation of a wide range of activities, transcription from approximately one-third of the genome (including noncoding genes) of the model plant Arabidopsis (Arabidopsis thaliana) is under circadian control (Michael and McClung, 2003; Covington et al., 2008; Michael et al., 2008; Hazen et al., 2009). In addition, the circadian clock serves as a timekeeper to regulate daylength-dependent processes, such as flowering time and tuberization. The circadian systems responsible for generating circadian rhythms are ubiquitous in eukaryotes and have also been found in some prokaryotes (Dunlap, 1999). Conceptually, a circadian system can be divided into three parts: the oscillator mechanism, input pathways, and output pathways. Interestingly, although their components differ, the basic oscillator mechanism appears to be well conserved in all the eukaryotic model organisms that have been studied (Dunlap, 2006). Oscillators are comprised of interlocking positive/negative feedback loops made from clock proteins that control their own rhythms. The oscillators can be entrained by signals from the environment, such as temperature and light changes. Output pathways from the oscillating clock proteins in turn convey circadian rhythms to the various physiological and molecular processes.
The model of the Arabidopsis oscillator consists of interlocking feedback loops of several components, including CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1 (TOC1). CCA1 and LHY are homologous myb-related transcription factors that show robust circadian oscillations in their RNA and protein levels (Schaffer et al., 1998; Wang and Tobin, 1998). Overexpression of CCA1 or LHY in transgenic Arabidopsis plants abrogates the circadian rhythmicity of clock-controlled processes, including gene expression and leaf movements (Schaffer et al., 1998; Wang and Tobin, 1998; Thain et al., 2004). Mutations in CCA1 or LHY result in a shorter period of circadian-controlled gene expression and leaf movements than wild-type plants, while plants with mutations in both genes appear to be unable to maintain sustained oscillations (Green and Tobin, 1999; Alabadi et al., 2002; Mizoguchi et al., 2002). However, despite their homology, CCA1 and LHY may have somewhat different roles in the oscillator mechanism (Gould et al., 2006; Zeilinger et al., 2006). TOC1 (also known as PSEUDO-RESPONSE REGULATOR1 [PRR1]) belongs to the PRR/TOC1 family of genes (Makino et al., 2000; Strayer et al., 2000) that encode nuclear proteins with a region that is common to several transcription factors. Mutations in TOC1 also cause a short period phenotype (Somers et al., 1998). Other genes, such as GIGANTEA (GI), EARLY FLOWERING4 (ELF4), LUX ARRHYTHMO, TIME FOR COFFEE, LIGHT INSENSITIVE PERIOD1, PRR3/5/7/9, LIGHT-REGULATED WD1 (LWD1), LWD2, and FIONA1, might operate in the oscillator or close to it (Doyle et al., 2002; Hazen et al., 2005; Locke et al., 2005; Nakamichi et al., 2005; Edwards et al., 2006; Gould et al., 2006; Ding et al., 2007; Kevei et al., 2007; McWatters et al., 2007; Para et al., 2007; Kim et al., 2008; Wu et al., 2008).
Considerable progress has been made in determining the mechanisms by which the circadian elements described above interact to generate circadian rhythms in Arabidopsis. In the least complex model of the main oscillator, CCA1 and LHY expression rises before dawn and suppresses the expression of TOC1 by binding to its promoter. In the evening, when CCA1 and LHY levels decrease, TOC1 expression rises. TOC1 then activates CCA1 and LHY expression by an as yet unknown mechanism (Alabadi et al., 2001). Recently, a combination of experimental and mathematical modeling techniques (Locke et al., 2006; Zeilinger et al., 2006) have supported the idea that other morning and evening genes, such as PRR7, PRR9, and GI, are involved in the oscillator, possibly by forming two other additional feedback loops interacting with the CCA1/LHY/TOC1 loop.
However, although the CCA1/LHY/TOC1 transcription/translation feedback loop is clearly central to the mechanism of the Arabidopsis oscillator, it is probably insufficient to create a cycle that takes approximately 24 h. Thus, there are likely to be additional steps, including posttranslational modifications, built in to the cycling of oscillator components to ensure that the loop takes about a day to complete. Consistent with the idea of posttranslation modifications having a role in the oscillator, in the oscillators of two of the best-studied organisms, mice and Drosophila (Drosophila melanogaster), phosphorylation, protein interactions, and cellular localization play a crucial role in the regulation of the oscillator (Kwon et al., 2006; Meyer et al., 2006).
Posttranslational modifications also play a role in the Arabidopsis oscillator. There is evidence for the role of posttranslational regulation of TOC1. ZEITLUPE (ZTL), an F-box protein, has been shown to regulate TOC1 degradation, probably through a CULLIN1-containing SCF complex, and ZTL itself is stabilized by interaction with GI (Mas et al., 2003; Kim et al., 2007; Harmon et al., 2008). GI stability is regulated by CONSTITUTIVE PHOTOMORPHOGENIC1 and ELF3 (Yu et al., 2008). In addition, a phosphorylated form of TOC1 can interact with a phosphorylated form of PRR3. The TOC1/PRR3 interaction competes with the TOC1/ZTL interaction and possibly prevents TOC1 degradation in the vascular tissues (Para et al., 2007; Fujiwara et al., 2008). Some work, mostly in vitro, has also started to show how CCA1 and LHY are modified. In western blots of total cellular protein extracts, CCA1 and LHY levels oscillate with a similar pattern, peaking around dawn with no detectable protein late in the day (Wang and Tobin, 1998; Kim et al., 2003). In addition, LHY may be degraded through the proteasomal pathway, and DE-ETIOLATED1 inhibits LHY turnover (Wang and Tobin, 1998; Song and Carre, 2005). In vitro, both CCA1 and LHY are phosphorylated by CK2 (Sugano et al., 1998), and when the CK2 phosphorylation sites in CCA1 are mutated, CCA1 activity is altered (Daniel et al., 2004). Moreover, the DNA-binding activity of CCA1 from plant extracts requires phosphorylation by CK2 (Portolés and Más, 2007). CCA1 and LHY have been shown to be localized to the nucleus (Wang et al., 1997; Carre and Kim, 2002; Perales and Mas, 2007; Gutierrez et al., 2008), although there are no data about the timing and control of this localization.
In general, however, little is known about the crucial posttranslational localization and interactions of CCA1 and LHY in the Arabidopsis oscillator in vivo. To study the posttranslational regulation of CCA1 and LHY, we generated transgenic plants with tagged CCA1 and LHY proteins and have shown that these tagged proteins are fully functional. Using the tagged proteins, we demonstrate that in vivo CCA1 can interact with both itself and with LHY; moreover, after translation CCA1 moves rapidly into the nucleus. Thus, posttranslational delays built into the oscillators of other model organisms appear not to be present in the Arabidopsis circadian oscillator.
RESULTS
CCA1 Moves Rapidly into the Nucleus after Translation
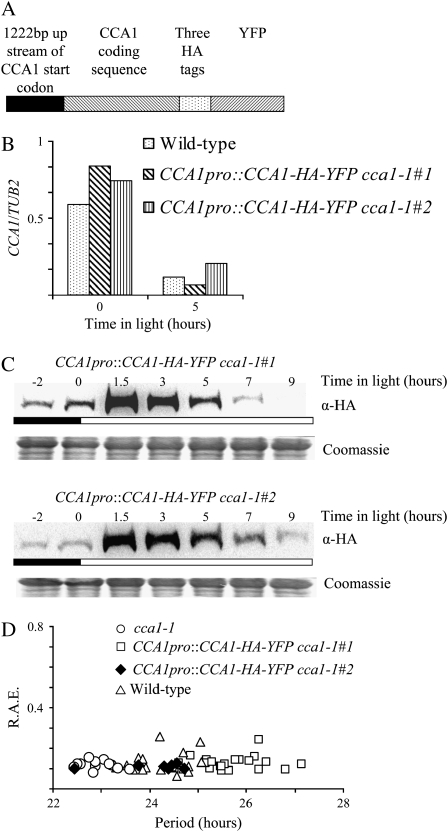
To be able to follow the subcellular localization and interactions of CCA1 protein, we generated transgenic plants with tagged CCA1. We made a construct of the CCA1 cDNA fused to three HA-tags and yellow fluorescent protein (YFP) under the control of the CCA1 promoter (1,222 bp upstream of the start codon) shown in Figure 1A. The resulting construct was used to transform CCA1-null plants (cca1-1; Green and Tobin, 1999). Two transgenic lines, CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2, were chosen for further studies.
Figure 1.
CCA1 is correctly regulated in CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 transgenic plants. A, The CCA1-HA-YFP construct used to transformed cca1-1 plants. B, CCA1pro∷CCA1-HA-YFP cca1-1#1, CCA1pro∷CCA1-HA-YFP cca1-1#2, and wild-type plants were grown in LD for 2 weeks before harvested and the levels of CCA1-HA-YFP mRNA and CCA1 mRNA determined by quantitative PCR and plotted on a graph relative to TUB2 mRNA levels. C, CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants were grown in LD for 2 weeks before harvesting, and the levels of CCA1-HA-YFP protein were determined by western analysis. The Coomassie-stained loading control is shown below. The white and black bars represent light and dark periods, respectively. D, One-week-old CCA1pro∷CCA1-HA-YFP cca1-1#1, CCA1pro∷CCA1-HA-YFP cca1-1#2, cca1-1, and wild-type plants were transferred to LL after entrainment in LD. Leaf movements were recorded every 20 min over 7 d and analyzed by FFT-NLLS. The RAE of the rhythms is plotted against period.
To determine whether CCA1 levels in the CCA1pro∷CCA1-HA-YFP cca1-1 lines are comparable to the levels in wild-type plants, CCA1pro∷CCA1-HA-YFP cca1-1#1, CCA1pro∷CCA1-HA-YFP cca1-1#2, and wild-type plants were grown for 2 weeks in 14 h light:10 h dark (LD). Tissue was harvested at zero and 5 h after lights-on and mRNA extracted. CCA1-HA-YFP mRNA levels in the transgenic plants and CCA1 mRNA levels in the wild type were determined by real-time quantitative PCR. Figure 1B shows that the levels of CCA1-HA-YFP mRNA in both of the transgenic lines are comparable to CCA1 levels in wild-type plants, with higher levels at lights-on than 5 h later. We then examined the accumulation of the CCA1-HA-YFP protein. CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants were grown for 2 weeks in 14 h light:10 h dark (LD). Tissue was harvested at intervals and protein extracted and determined by western analysis of total protein extracts using an anti-HA antibody. Our results show that the levels of CCA1-HA-YFP protein in both transgenic lines reach a maximum at 1.5 h after lights-on (Fig. 1C). Thus, the peak of CCA1-HA-YFP is similar to that previously reported for CCA1 in wild-type plants (Wang and Tobin, 1998). Taken together, our results suggest that CCA1-HA-YFP is expressed at both the transcriptional and translational levels in the CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants in a similar way to CCA1 in wild-type plants.
Our next goal was to check if, in addition to showing the correct pattern of accumulation in LD, the CCA1-HA-YFP protein could also mimic the activity of wild-type CCA1. As a test for CCA1-HA-YFP activity, we examined whether CCA1-HA-YFP could lengthen the period of circadian-regulated leaf movement in the cca1-1 plants that have been shown previously to have a shorter circadian period than wild-type plants (Mizoguchi et al., 2002). CCA1pro∷CCA1-HA-YFP cca1-1#1, CCA1pro∷CCA1-HA-YFP cca1-1#2, cca1-1, and wild-type plants were grown for 7 d in LD before being transferred to continuous light (LL). Figure 1D shows that the periods of leaf movement of the CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants are longer (25.6 ± 0.2 se and 24.7 ± 0.6 se, respectively) than those of cca1-1 plants (22.9 ± 0.2 se) and resemble the wild type (24.3 ± 0.1 se). The average traces for leaf movements of the transgenic and control lines also showed that both transgenic lines were rhythmic with, especially CCA1pro∷CCA1-HA-YFP cca1-1#1, a pattern of rhythmicity close to the wild type (Supplemental Fig. S1A). Thus, our results show that the CCA1-HA-YFP in the CCA1pro∷CCA1-HA-YFP cca1-1 plants has not only a similar pattern of expression to CCA1 in wild-type plants but also a similar circadian activity.
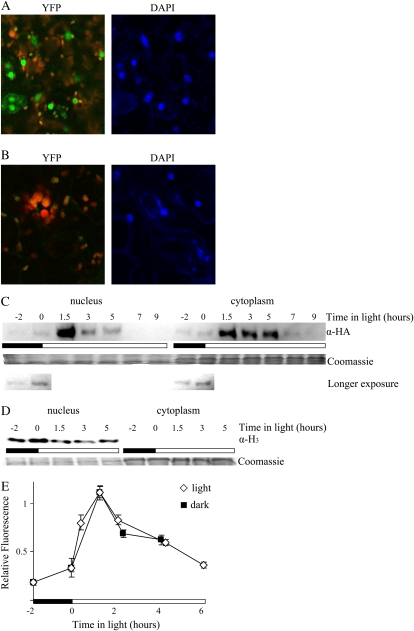
CCA1 has been reported in the nuclei of wild-type plants at dawn (Gutierrez et al., 2008), and we examined whether CCA1-HA-YFP could also be detected in the nuclei of CCA1pro∷CCA1-HA-YFP cca1-1 plants by examining the subcellular localization of fluorescence from YFP. CCA1pro∷CCA1-HA-YFP cca1-1#1, CCA1pro∷CCA1-HA-YFP cca1-1#2, and wild-type plants were grown for 2 weeks in LD and then examined by confocal microscopy. Figure 2A and Supplemental Figure S2A shows that 3 h after lights-on there is a fluorescence signal in both the epidermal and mesophyll cells of CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants. Staining the cells with 4′,6-diamino-phenylindole (DAPI) indicated that the majority of the fluorescence in each cell was localized to the nucleus. We could not detect any fluorescence signals in the nuclei of wild-type plants (Fig. 2B). Thus, CCA1-HA-YFP is found in the nuclei of the CCA1pro∷CCA1-HA-YFP cca1-1 plants.
Figure 2.
CCA1-HA-YFP protein can move into the nucleus almost immediately after translation. A and B, Two-week-old CCA1pro∷CCA1-HA-YFP cca1-1#1 (A) and wild-type (B) plants were examined for yellow fluorescence by confocal microscopy 3 h after lights-on. Right panels, DAPI staining (blue) of nuclei; left panels, YFP (green) and chloroplast autofluorescence (red). C and D, Two-week-old CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD. Tissue was harvested at intervals starting 2 h before lights-on and the nuclear and cytoplasmic fractions of the cells separated as described in “Materials and Methods.” The levels of CCA1-HA-YFP protein were determined by western analysis with anti-HA antibodies. The Coomassie-stained loading control and a longer exposure of the first two time points (showing that CCA1-HA-YFP is in both the nucleus and cytoplasm 2 h before lights-on) are shown below (C). The levels of histone 3 protein were determined by western analysis with antihistone 3 antibodies (D). E, Two-week-old CCA1pro∷CCA1-HA-YFP cca1-1#1 plants transferred to light or kept in dark were examined at intervals for yellow fluorescence by confocal microscopy. The white and black bars represent light and dark periods, respectively.
Since in Drosophila the timing of PERIOD (PER) and TIMELESS (TIM) movement into the nucleus is an important part of the mechanism of the oscillator that allows it to cycle with an approximately 24-h period, we examined the rate of CCA1-HA-YFP movement into the nucleus. CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants were grown for 2 weeks in LD and harvested at intervals. The nuclear and cytoplasmic fractions of the tissue were isolated (as described in “Materials and Methods”) and examined by western analysis using an anti-HA antibody. Figure 2C and Supplemental Figure S2B show that in both lines even at zero hours (lights-on) when total protein levels are still very low, CCA1-HA-YFP is already in the nucleus at levels that are similar to those in the cytoplasm. CCA-HA-YFP continues to accumulate in the nucleus before declining. Figure 2C and Supplemental Figure S2B also show that in both lines, the levels of CCA1-HA-YFP protein remain higher in the cytoplasm for longer. The fact that we observed high levels of CCA1-HA-YFP protein in the cytoplasmic extracts at a time when the nuclear CCA1-HA-YFP levels have declined suggests that our nuclear extracts were free of significant amounts of cytoplasmic contamination. To verify the purity of the nuclear extracts, we used antibodies against histone 3 (Fig. 2D), histone 4 (data not shown), and RNA polymerase II (Supplemental Fig. S2C) on western blots with the cytoplasmic and nuclear fractions. Figure 2D and Supplemental Figure S2C show that the cytoplasmic fraction was free from detectable contamination by nuclear material.
To confirm our findings that CCA1-HA-YFP moves into the nucleus soon after translation and to examine the effects of light on its translocation, we also examined the subcellular localization of fluorescence from YFP. CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#2 plants were grown for 2 weeks in LD and then, at the end of the last dark period, transferred to light or kept in the dark. YFP fluorescence was examined by confocal microscopy. The levels of fluorescence in the nuclei of epidermis, stomata, and mesophyll cells were calculated and plotted as a function of time. As can be seen in Figure 2E and Supplemental Figure S2D, at lights-on, there were detectable levels of fluorescence in the nuclei. Fluorescence levels reached a maximum around 1.5 h after lights-on and then started to decrease. A comparison between the changes in CCA1-HA-YFP protein levels (Fig. 1C) and the fluorescence in the nucleus (Fig. 2E; Supplemental Figure S2D) indicates that CCA1-HA-YFP protein can move into the nucleus very soon after translation. The fact that we detected CCA1-HA-YFP in the cytoplasm on western blots but little fluorescence from the YFP in the cytoplasm is probably because the CCA1-HA-YFP in the cytoplasm is more diffuse resulting in a weaker signal. Figure 2E also shows that the translocation of CCA1-HA-YFP protein into the nucleus is unaffected by dark. This finding is consistent with the results shown in Figure 2C and Supplemental Figure S2B that there are already detectable levels of CCA1-HA-YFP in the nucleus at lights-on. Taken together, our results show that CCA1-HA-YFP is able to move rapidly in to the nucleus after translation and that this translocation is unaffected by light and dark.
CCA1 and LHY Interact in Vitro
Several structural studies have shown that in proteins containing two myb domains, both are necessary for DNA binding, and it has been suggested that proteins containing only one myb domain might bind DNA as dimers (Jin and Martin, 1999). Moreover, other proteins containing single myb domains have been shown to dimerize (Bianchi et al., 1997). We therefore hypothesized that CCA1 and LHY might function as heterodimers.
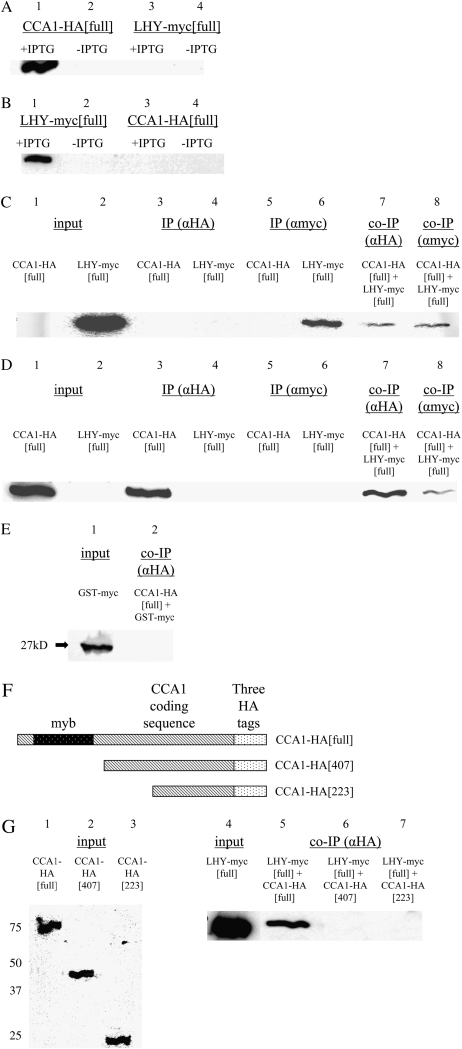
As a preliminary step to check whether CCA1 and LHY might function as heterodimers, we determined whether CCA1 and LHY could form heterodimers in vitro. We expressed the full-length CCA1 cDNA fused to three HA-tags (CCA1-HA[full]) and the full-length LHY cDNA fused to a myc-tag (LHY-myc[full]) in Escherichia coli. Extracts from the bacterial cultures were separated by SDS-PAGE and CCA1-HA and LHY-myc identified by western blots with anti-HA and anti-myc antibodies. Figure 3, A and B, shows that CCA1-HA and LHY-myc are expressed and recognized correctly by the antibodies. The extracts were then used in reciprocal coimmunoprecipitation (co-IP) experiments (as described in “Materials and Methods”). The co-IPs were done on a mixture of CCA1-HA and LHY-myc protein extracts with either anti-HA antibody (Fig. 3, C and D, lane 7) or anti-myc antibody (Fig. 3, C and D, lane 8). The immunoprecipitated proteins were separated by SDS-PAGE and CCA1-HA and LHY-myc identified by western blots using the anti-myc (Fig. 3C) or anti-HA (Fig. 3D) antibodies. Our results show that CCA1-HA was immunoprecipitated by LHY-myc (Fig. 3D, lane 8) and that LHY-myc was immunoprecipitated by CCA1-HA (Fig. 3C, lane 7). Thus, CCA1 and LHY form heterodimers in vitro. As a control for the specificity of the antibodies, we showed that anti-HA precipitates CCA1-HA[full] and not LHY-myc[full] (Fig. 3D, lane 3, compared to Fig. 3C, lane 4) and that anti-myc precipitates LHY-myc[full] and not CCA1-HA[full] (Fig. 3C, lane 6, compared to Fig. 3D, lane 5). As a further control, to check that there is a specific interaction between CCA1-HA[full] and LHY rather than with the myc tag, we coimmunoprecipitated a mixture of CCA1-HA[full] and glutathione S-transferase (GST)-myc protein extract with anti-HA antibodies. No GST-myc band was detected (Fig. 3E, lane 2).
Figure 3.
CCA1 and LHY interact in vitro. A and B, E. coli transformed with CCA1-HA[full] or with LHY-myc[full] were incubated for 3 h at 37°C with or without IPTG. Proteins were extracted and equal amounts of protein were loaded into SDS-PAGE gels and analyzed by western blots with anti-HA (A) or anti-myc (B). C and D, E. coli transformed with CCA1-HA[full] or with LHY-myc[full] were incubated for 3 h at 37°C with IPTG. Extracts were immunoprecipitated with anti-HA (lanes 3 and 4) or anti-myc (lanes 5 and 6), or extracts were mixed and then coimmunoprecipitated with anti-HA (lane 7) or anti-myc (lane 8). Proteins were analyzed by western blots with anti-myc (C) or anti-HA (D). E, Protein extracts from cells of E. coli transformed with CCA1-HA[full] and cells of E. coli transformed with GST-myc were mixed and used in co-IPs with anti-HA. The proteins pulled down were analyzed on western blots with anti-myc. F, CCA1-HA[full], CCA1-HA[407], and CCA1-HA[223] constructs. G, Protein extracts from cells of E. coli transformed with CCA1-HA[full] (lane 5), CCA1-HA[407] (lane 6), or with CCA1-HA[223] (lane 7) and cells of E. coli transformed with LHY-myc[full] were mixed and coimmunoprecipitated with anti-HA. Input extract of CCA1-HA[full], CCA1-HA[407], and CCA1-HA[223] constructs analyzed by western blots using anti-HA antibodies (lanes 1–3). Input extract of LHY-myc[full] construct analyzed by western blots using anti-myc antibodies (lane 4). Co-IPs were analyzed by western blots using anti-myc antibodies (lanes 5–7).
To start to determine the domains in CCA1 that are necessary for its interactions with LHY, we made two truncated CCA1-HA proteins and expressed them in E. coli. The constructs, CCA1-HA[407] and CCA1-HA[223], contained the last 407 or 223 amino acids of CCA1, respectively, with three HA-tags (Fig. 3F). Bacterial extracts of CCA1-HA[full], CCA1-HA[407], and CCA1-HA[223] were used in co-IP experiments with LHY-myc[full] (as described in “Materials and Methods”). The immunoprecipitated proteins were separated by SDS-PAGE and LHY-myc[full] identified by western blots using the anti-myc antibodies. Figure 3G shows that only CCA1-HA[full] immunoprecipitated LHY-myc[full] (lane 5 compared to lanes 6 and 7). Since LHY-myc[full] was not immunoprecipitated by either of the truncated CCA1-HA proteins, it is likely that the region of CCA1 responsible for CCA1/LHY interactions is on the N terminus, perhaps near the myb domain. However, we cannot rule out the possibility that the truncated CCA1-HA proteins are misfolded and that the correct folding of CCA1 is necessary for its interaction with LHY. Taken together, our results show that CCA1 and LHY interact in vitro and that this interaction probably requires the N terminus of CCA1.
CCA1 and LHY Interact in Vivo
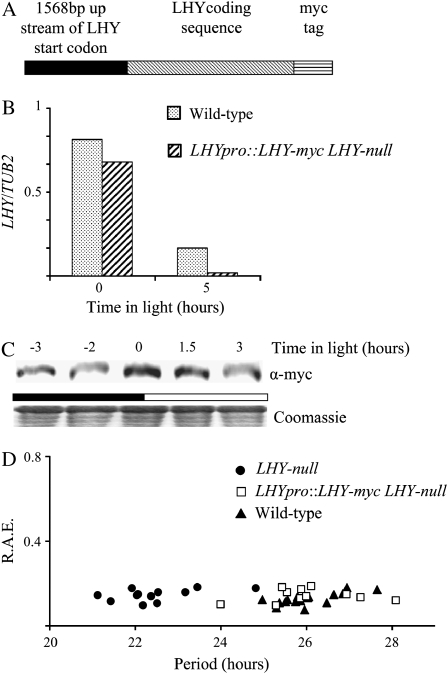
To be able to examine whether CCA1 and LHY also interact in vivo, we generated transgenic plants with tagged LHY. We made a construct (LHY-myc) of the LHY cDNA fused to a myc-tag under the control of the LHY promoter (1,568 bp upstream of the start codon) shown in Figure 4A. A line of plants with a T-DNA insertion in the LHY sequence was obtained from the SALK institute. From this T-DNA line, we isolated a homozygous plant (LHY-null) that did not express LHY (Supplemental Fig. S3). We then transformed LHY-null with the LHY-myc construct to generate LHYpro∷LHY-myc LHY-null.
Figure 4.
LHY is correctly regulated in LHYpro∷LHY-myc LHY-null transgenic plants. A, The LHY-myc construct used to transform LHY-null plants. B, LHYpro∷LHY-myc LHY-null and wild-type plants were grown in LD for 2 weeks before being harvested, and the levels of LHY-myc mRNA and LHY mRNA were determined by quantitative PCR and plotted on a graph relative to TUB2 mRNA levels. C, LHYpro∷LHY-myc LHY-null plants were grown in LD for 2 weeks before being harvested, and the levels of LHY-myc protein were determined by western analysis. The Coomassie-stained loading control is shown below. The white and black bars represent light and dark periods, respectively. D, One-week-old LHYpro∷LHY-myc LHY-null, LHY-null, and wild-type plants were transferred to LL. Leaf movements were recorded every 20 min over 7 d and analyzed by FFT-NLLS. The RAE for the rhythms was plotted against the period.
To determine whether LHY-myc levels in the transgenic line are comparable to the levels of LHY in wild-type plants, LHYpro∷LHY-myc LHY-null and wild-type plants were grown for 2 weeks in 14 h light:10 h dark (LD). Tissue was harvested at zero and 5 h after lights-on and mRNA extracted. LHY-myc mRNA levels in the transgenic plants and LHY mRNA levels in the wild type were determined by real-time quantitative PCR. Figure 4B shows that the levels of LHY-myc mRNA from the LHYpro∷LHY-myc construct in the transgenic line are comparable to LHY levels in wild-type plants. We then examined the accumulation of the LHY-myc protein. LHYpro∷LHY-myc LHY-null plants were grown for 2 weeks in 14 h light:10 h dark (LD). Tissue was harvested in intervals and protein extracted and examined by western analysis of total protein extracts using an anti-myc antibody. The LHY-myc protein levels peak around dawn in the LHYpro∷LHY-myc LHY-null plants (Fig. 4C). Thus, the peak of LHY-myc is close to that previously reported for LHY in wild-type plants (Kim et al., 2003)
To verify that the LHY-myc activity in plants is also similar to the endogenous LHY activity, we checked whether LHYpro∷LHY-myc could restore normal circadian rhythms of leaf movements to LHY-null plants. Plants with mutated LHY have been shown to have a shorter circadian period than wild-type plants (Mizoguchi et al., 2002). LHYpro∷LHY-myc LHY-null, LHY-null, and wild-type plants were grown for 7 d in LD before being transferred to LL. Figure 4D demonstrates that the leaf movement period of LHYpro∷LHY-myc LHY-null plants (24 ± 0.3 se) is longer than that of the LHY-null plants (20.5 ± 0.4 se) and close to that of wild-type plants (24 ± 0.2 se). We also observed that the average traces for leaf movements of the LHYpro∷LHY-myc LHY-null closely matched those of the wild-type control (Supplemental Fig. S1B). Thus, our results show that the LHY-myc in the LHYpro∷LHY-myc LHY-null plants has a similar circadian activity to LHY in wild-type plants.
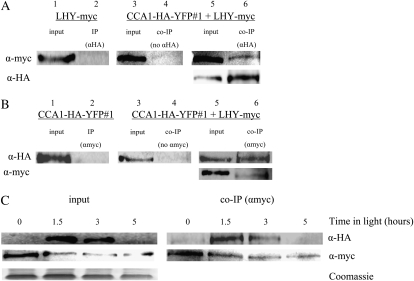
Our next goal was to examine whether CCA1 and LHY could interact in vivo. The LHYpro∷LHY-myc LHY-null plants were crossed into CCA1pro∷CCA1-HA-YFP cca1-1#1 plants. The resulting LHYpro∷LHY-myc LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD for 4 weeks. Tissue was harvested 2 h after lights-on and used for co-IP assays with anti-HA antibodies. The immunoprecipitated proteins were separated by SDS-PAGE and LHY-myc identified by western analysis using anti-myc. Our results show that the co-IP resulted in a band the size of LHY-myc (Fig. 5A, lane 6), suggesting that CCA1-HA-YFP and LHY-myc interact in vivo. In a control co-IP carried out without the anti-HA antibody, no significant LHY-myc could be detected (Fig. 5A, lane 4). As a further control to show that the anti-HA antibody interacted specifically with CCA1-HA-YFP and not with LHY-myc, the immunoprecipitation assay was also performed on LHYpro∷LHY-myc LHY-null extracts. Figure 5A, lane 2, shows that in the absence of CCA1-HA-YFP, no LHY-myc was precipitated. We also performed the reciprocal assay using the anti-myc antibody to co-IP CCA1-HA-YFP. Figure 5B shows that CCA1-HA-YFP was precipitated only when the anti-myc antibody was used (Fig. 5B, lane 4 compared to lane 6). By contrast, the anti-myc antibody could not pull down CCA1-HA-YFP from CCA1pro∷CCA1-HA-YFP cca1-1#1 plants (Fig. 5B, lane 2). Our results clearly show that CCA1 and LHY interact in vivo.
Figure 5.
CCA1 and LHY interact in vivo. A, LHYpro∷LHY-myc LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 and LHYpro∷LHY-myc LHY-null plants were grown in LD for 4 weeks. Tissue was harvested and used for co-IPs with anti-HA (αHA). As a control, the co-IP experiments were also carried out without anti-HA antibody (no αHA). Proteins were analyzed by western blots with anti-myc (top row) or anti-HA (bottom row). B, LHYpro∷LHY-myc LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD for 4 weeks. Tissue was harvested and used for co-IPs with anti-myc (αmyc). As a control, the co-IP experiments were also carried out without anti-myc antibody (no αmyc). Proteins were analyzed by western blots with anti-HA (top row) or anti-myc (bottom row). C, LHYpro∷LHY-myc LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD for 2 weeks. Tissue was harvested at intervals and used in a co-IP experiment with anti-myc. The proteins were analyzed by western blots with anti-HA (top row) or anti-myc (bottom row). Input, Levels of CCA1-HA-YFP and LHY-myc in extracts. Co-IP (αmyc), CCA1-HA-YFP and LHY-myc pulled down by anti-myc antibodies. The Coomassie-stained loading control is shown below.
Since our results (Fig. 1C) showed that the levels of CCA1-HA-YFP protein changes during the day, we examined whether the in vivo interactions between CCA1-HA-YFP and LHY-myc also show changes over time. We collected leaf samples from 2-week-old LD-grown LHYpro∷LHY-myc LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants at intervals as described (Fig. 5C). The samples were subjected to co-IP with the anti-myc antibody, the immunoprecipitated proteins were separated by SDS-PAGE, and CCA1-HA-YFP was identified using anti-HA. Figure 5C shows that there is correlation between the levels of CCA1-HA-YFP and the amount of CCA1-HA-YFP interacting with LHY-myc. Our results suggest that the interaction between CCA1 and LHY depends mostly on their abundance and that they may form heterodimers whenever they are present together in the cell.
CCA1 Can Form Homodimers in Vivo
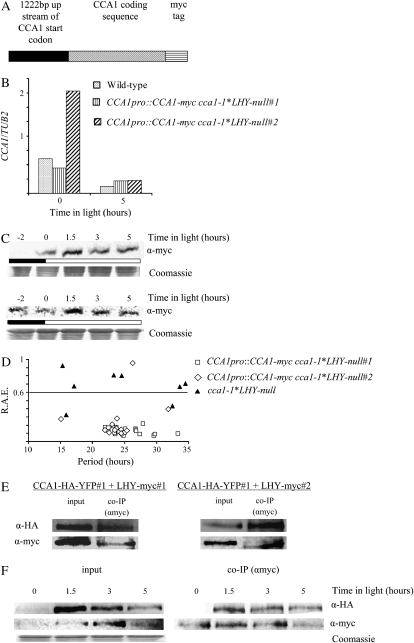
Previous work in several labs has established that CCA1 and LHY are partially redundant and that in the absence of LHY, CCA1 might function as a substitute for LHY in the circadian system (Green and Tobin, 1999; Alabadi et al., 2002; Mizoguchi et al., 2002). To examine whether CCA1 might also be able to form homodimers, we made a construct of the CCA1 cDNA fused to a myc-tag under the control of the CCA1 promoter (1,222 bp upstream of the start codon; Fig. 6A). The resulting construct was used to transform cca1-1*LHY-null plants (CCA1pro∷CCA1-myc cca1-1*LHY-null). Figure 6B shows that the levels of CCA1-myc mRNA in CCA1pro∷CCA1-myc cca1-1*LHY-null#1 are comparable to the levels of CCA1 mRNA in the wild type. In the CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants, CCA1-myc mRNA levels are higher at zero hours than CCA1 mRNA in the wild type but similar at 5 h. Figure 6C shows that, like CCA1-HA-YFP in the CCA1pro∷CCA1-HA-YFP cca1-1#1 plants, the level of CCA1-myc in the CCA1pro∷CCA1-myc cca1-1*LHY-null#1 and CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants is highest 1.5 h after lights-on, although the levels continue to remain fairly high throughout the time course of the experiment.
Figure 6.
CCA1 forms homodimers in vivo. A, The CCA1-myc construct used to transform cca1-1*LHY-null plants. B, CCA1pro∷CCA1-myc cca1-1*LHY-null#1, CCA1pro∷CCA1-myc cca1-1*LHY-null#2, and wild-type plants were grown in LD for 2 weeks before being harvested, and the levels of CCA1-myc mRNA and CCA1 mRNA were determined by quantitative PCR and plotted on a graph relative to TUB2 mRNA levels. C, Two-week-old CCA1pro∷CCA1-myc cca1-1*LHY-null#1 and CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants were grown in LD. The levels of CCA1-myc protein were determined by western analysis. The Coomassie-stained loading control is shown below. The white and black bars represent light and dark periods, respectively. D, One-week-old CCA1pro∷CCA1-myc cca1-1*LHY-null#1, CCA1pro∷CCA1-myc cca1-1*LHY-null#2, and cca1-1*LHY-null plants were transferred to LL after grown in LD. Leaf movements were recorded every 20 min over 7 d and analyzed by FFT-NLLS. The RAE was plotted against the period. E, Four-week-old CCA1pro∷CCA1-myc cca1-1*LHY-null#1;CCA1pro∷CCA1-HA-YFP cca1-1#1 and CCA1pro∷CCA1-myc cca1-1*LHY null#2;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD. Tissue was harvested and used for co-IPs carried out with anti-myc. Proteins were analyzed by western blots with anti-HA (top row) or anti-myc (bottom row). F, CCA1pro∷CCA1-myc cca1-1*LHY null#1;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown in LD for 2 weeks. Tissue was harvested at intervals and used in a co-IP experiment with anti-myc. The proteins were analyzed by western blots with anti-HA (top row) or anti-myc (bottom row). Input, Levels of CCA1-HA-YFP and CCA1-myc in extracts. Co-IP (αmyc), CCA1-HA-YFP and CCA1-myc pulled down by anti-myc antibodies. The Coomassie-stained loading control is shown below.
We also examined whether CCA1-myc could restore circadian rhythms of leaf movements to cca1-1*LHY-null plants. Consistent with previous reports that plants with mutated CCA1 and LHY have weak circadian rhythms (Mizoguchi et al., 2002), Figure 6D shows that cca1-1*LHY-null plants do not have robust rhythms. The relative amplitude error (RAE) is a measurement of the robustness of a circadian rhythm (Somers et al., 2004) and <15% of the cca1-1*LHY-null plants have an RAE of <0.6, i.e. are significantly rhythmic. By contrast, Figure 6D shows that both CCA1pro∷CCA1-myc cca1-1*LHY-null#1 and CCA1pro∷CCA1-myc cca1-1*LHY-null#2 have robust circadian rhythms of leaf movements. More than 85% of the CCA1pro∷CCA1-myc cca1-1*LHY-null#1 plants and >80% of the CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants showed an RAE of <0.6. In addition, the average traces for leaf movements of the CCA1pro∷CCA1-myc cca1-1*LHY-null lines showed rhythmicity, while the trace for cca1-1*LHY-null plants was arrhythmic (Supplemental Fig. S1C). Thus, CCA1-myc in the CCA1pro∷CCA1-myc cca1-1*LHY-null plants not only has a similar pattern of expression to endogenous CCA1 in wild-type plants but also has similar circadian activity.
To determine whether CCA1 can interact with itself, CCA1pro∷CCA1-myc cca1-1*LHY-null#1 and CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants were crossed into CCA1pro∷CCA1-HA-YFP cca1-1#1 plants. The resulting CCA1pro∷CCA1-myc cca1-1*LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown for 4 weeks in LD. Tissue was harvested 2 h after lights-on and used for co-IP assays with anti-myc antibodies. The immunoprecipitated proteins were separated by SDS-PAGE and CCA1-HA-YFP identified by western analysis using anti-HA antibodies. Our previous results (Fig. 5B, lane 2) showed that CCA1-HA-YFP is not precipitated by anti-myc antibodies in plants that do not express CCA1-myc. Figure 6E shows that, in the CCA1pro∷CCA1-myc cca1-1*LHY-null#1 and CCA1pro∷CCA1-myc cca1-1*LHY-null#2 plants that express both CCA1-HA-YFP and CCA1-myc, the anti-myc antibodies precipitated a protein that could be detected on a western blot with anti-HA. Thus, it appears that CCA1 can interact with itself in vivo.
We also examined the in vivo interactions between CCA1-HA-YFP and CCA1-myc over time. CCA1pro∷CCA1-myc cca1-1 LHY-null;CCA1pro∷CCA1-HA-YFP cca1-1#1 plants were grown for 2 weeks in LD. Tissue was harvested at intervals and used for co-IP assays with anti-myc antibodies. Figure 6F shows that there is correlation between the levels of CCA1-HA-YFP and CCA1-myc and the amount of CCA1-HA-YFP interacting with CCA1-myc.
DISCUSSION
In other model organisms, such as mice and Drosophila, it has been established that posttranslation events, such as dimerization, phosphorylation, and cellular localization, have important roles in generating and maintaining the approximately 24-h rhythms. PER and TIM, the two main components of the negative feedback loop of the Drosophila oscillator, accumulate slowly, forming heterodimers in the cytoplasm before dissociating and moving separately to the nucleus (Meyer et al., 2006). In both the nucleus and the cytoplasm, phosphorylation by kinases, such as CASEIN KINASE1 (CK1) and CK2, controls the activity and stability of PER (Bae and Edery, 2006). Similarly, in mouse cells, BMAL1 shuttles between the cytoplasm and the nucleus where it regulates the accumulation of another element of the oscillator loop, CLOCK (Kwon et al., 2006). In the nucleus, CLOCK/BMAL1 heterodimers control expression of the clock genes mPER1, mPER2, mPER3, and CHRYPTOCHROME1 (CRY1) and CRY2. These mPER and CRY proteins form multimeric complexes that reenter the nucleus and repress the transcriptional activity of CLOCK and BMAL (Ko and Takahashi, 2006; Kwon et al., 2006). Phosphorylation by kinases, especially CK1ɛ and CK1δ, is critical for regulating the stability and nuclear translocation of mPER and CRY.
While the basic model proposed for the Arabidopsis oscillator (Alabadi et al., 2001; Locke et al., 2005) shows clear parallels with those of other model organisms, for example, mice and Drosophila, it is clear that there are also differences in the mechanisms. The aim of the experiments described in this article was to examine posttranslational regulation of CCA1 and LHY, the negative elements in the Arabidopsis oscillator. In particular, we wanted to examine if, like PER and TIM in Drosophila and CLOCK and BMAL1, CCA1 and LHY form dimers and whether the timing of their movement into the nucleus may add a significant delay to the oscillator mechanism to ensure that it takes approximately 24 h.
To examine the posttranslational regulation of CCA1, we used the endogenous sequence of CCA1 (including its promoter) attached to three HA-tags and YFP in transgenic plants. We first demonstrated that this construct could mimic the activity of endogenous CCA1. We then followed the rate of CCA1-HA-YFP translocation to the nucleus by comparing the changes in total levels of CCA1-HA-YFP protein during the course of the day (Fig. 1C) to the levels of CCA1-HA-YFP protein in the nucleus (Fig. 2, C and E; Supplemental Fig. S2B and S2D). We examined the levels of CCA1-HA-YFP protein in the nucleus both biochemically (Fig. 2C; Supplemental Fig. S2B) and using microscopy (Fig. 2E; Supplemental Fig. S2D). Both approaches to examining the subcellular localization of CCA1-HA-YFP gave essentially similar results and showed that the rate of increase in CCA1-HA-YFP levels in the nucleus is almost identical to the rate of increase in total CCA1-HA-YFP levels in the cell. Furthermore, CCA1 protein can be detected in the nucleus even before lights-on (Fig. 2C). Thus, it appears that, unlike the Drosophila oscillator, in the Arabidopsis oscillator there is not necessarily a significant delay between the translation of a key negative element and its initial translocation to the nucleus. We also observed that the levels of CCA1-HA-YFP protein in the nucleus decrease more rapidly than the levels of protein in the cytoplasm. This slightly different pattern of protein accumulation might indicate a time-dependent mechanism for regulating CCA1 translocation or a difference in subcellular degradation rates of CCA1.
In further experiments, we made transgenic plants harboring the endogenous sequence of LHY (including its promoter) attached to a myc tag to examine whether CCA1 and LHY could dimerize. Our results show that CCA1 and LHY do indeed interact and that this interaction appears to occur whenever they are both present in the cell (Fig. 5). Since CCA1 and LHY both have single myb DNA binding domains and two myb domains may be required for their DNA binding, it is possible that CCA1 and LHY bind DNA as dimers. However, further experiments will be required to determine the significance of the CCA1/LHY interaction and its importance for subcellular localization and transcriptional regulation. Moreover, a recent report has shown that the oscillator mechanism in roots involves both CCA1 and LHY but is otherwise a simplified version of the oscillator found in the aerial parts of the plant (James et al., 2008). It will be interesting to see how CCA1 and LHY interactions and localization are affected in the root.
We have also shown that CCA1 can dimerize with itself in vivo. There is clear evidence for different biochemical activities of CCA1 and LHY in the circadian system (Gould et al., 2006). Moreover, loss of either CCA1 or LHY shortens the period of circadian rhythms, so there is only partial redundancy of CCA1 and LHY. It is possible that the different interactions of CCA1, with itself or with LHY, affect its function as a transcriptional regulator. Thus, different interactions of CCA1 may regulate the expression of different genes, different expression from the same gene, or the same gene differently under different conditions. In future experiments, it will be interesting to determine when and under what conditions CCA1 and LHY form homo- and heterodimers.
In conclusion, while there are clearly some similarities between the Arabidopsis circadian oscillator and that of other model organisms in the regulation of the negative elements, for example, their dimerization, there are also differences, for example, in the timing of their entry into the nucleus.
MATERIALS AND METHODS
Plant Materials and Growth
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used for all experiments unless stated otherwise. CCA1-null plants are cca1-1 plants (Green and Tobin, 1999) originally in Wassilewskija but backcrossed six times into the Columbia-0 background. The LHY-null is Salk_031092 obtained from the Salk Institute Genome Analysis Laboratory with a T-DNA insertion in the third intron of LHY.
All seeds were imbibed and cold treated at 4°C for 4 d to optimize germination. Plants for transformation and for some of the co-IP assays were grown on soil. For other purposes, plants were grown in petri dishes on Murashige and Skoog (MS; Weigel and Glazebrook, 2002) medium from Duchefa Biochemie supplemented with 0% Suc (w/v) (leaf movement assay), 1% Suc (RNA and protein extractions), or 3% Suc (confocal microscopy). Unless otherwise stated, plants were grown for 2 weeks under 14:10 light:dark (125 μE m−2 s−1) at a constant 23°C. Philips fluorescent lights TLD 18W/29 and TLD18W/33CW provided lighting for plant growth.
RNA Analysis
RNA extractions were carried out as previously described (Green and Tobin, 1999). RNA samples (3.3 μL of 1.5 μg/μL) were treated with DNase (DNA-free from Ambion) according to the manufacturer's instructions. From each DNA-free RNA sample, 5-μL aliquots were used as a template to produce cDNA, using the Reverse-iT Max 1st Strand Synth Kit from Abgene with random-hexamer primers according to the manufacturer's instructions. cDNA samples were diluted 5-fold and used as templates for the quantitative real-time PCR reaction using ABsolute SYBR Green ROX Mix from ABgene according to the manufacturer's instructions. Reactions were performed in a Rotagene real-time PCR machine. The primers for quantitative real-time PCR were as follows: CCA1 forward, 5′-TCCAGATAAGAAGTCACGCTCA-3′, and CCA1 reverse, 5′-TCTAGCGCTTGACCCATAGC-3′; LHY forward, 5′-GCTAAGGCAAGAAAGCCATA-3′, and LHY reverse, 5′-TGCCAAGCTCTTCCATAAAG-3′; and TUB2 forward, 5′-GGTTGAGCCTTACAACGCTACTCT-3′, and TUB2 reverse, 5′-GTGGTTCAAATCACCAAAGCTGGG-3′.
Protein Analysis
Protein extractions from plants were carried out as previously described (Somers et al., 2004). Briefly, Arabidopsis plants were ground in liquid nitrogen before resuspending in extraction buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, 3 mm DTT, 1:100 protease inhibitor cocktail [P9599 from Sigma-Aldrich], and 1 mm phenylmethylsulfonyl fluoride) by gentle vortexing. The extracts were clarified by centrifugation at 14,000g for 10 min at 4°C. Protein concentrations were determined using a commercial protein assay reagent (Rc/Dc reagent; Bio-Rad). Protein (100 μg) was loaded onto each lane. For the protein localization experiments, nuclei were separated from cytoplasm with CELLYTPN1 CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich), crude preparation. To be able to determine the levels of protein in each subcellular compartment, nuclei and cytoplasm fractions were loaded onto the western gel in proportion to their amounts in the total protein extract calculated by their relative volumes in the original extractions.
For protein extractions from Escherichia coli, cells were incubated with 1 mm isopropylthio-β-galactoside (IPTG) for 3 h for protein induction. The cells were collected by centrifugation and then lysed by incubation in 1 mm EDTA, 1 mm PMSF, 1:200 protease inhibitor cocktail (Sigma-Aldrich), 50 μg/mL DNaseI (Sigma-Aldrich), 10 mm MgCl2, and 10 mg/mL lysozyme for 30 min on ice followed by disruption in a French press. Proteins were collected from the extract by centrifugation at 3,000g for 10 min. Protein concentrations were determined using a commercial protein assay reagent (Rc/Dc reagent; Bio-Rad).
For the western blots, proteins were fractionated by SDS-PAGE (7% or 8%) and transferred to nitrocellulose membrane (Amersham). Immunoblotting was performed using 1:500 monoclonal mouse anti-HA and anti-myc first antibodies from Santa Cruz (sc-7392 and sc-40, respectively) and 1:5,000 goat anti-mouse second antibody from KPL (474–1806) or 1:1,000 polyclonal rabbit anti-H3 first antibodies from Abcam (ab1791) or 1:1,000 Phospho RNA Polymerase II (S2) first antibodies from Bethyl (A300-654A) and 1:5,000 goat anti-rabbit second antibody from Sigma-Aldrich (A0545). Peroxidase chemoluminance reactions were carried out on the membranes using SuperSignal reagents from Pierce and the membranes photographed using a chemoluminance sensitive camera Fuji-Film LAS-3000. Levels of protein were calculated by ImageJ using the Mean Gray Value option. To verify the linearity of the detection system, aliquots of CCA1-HA-YFP were run on a western blot and the antigen load plotted against the detected signal (Supplemental Fig. S4).
Leaf Movement Analysis
One-week-old Arabidopsis plants grown on MS medium (Plant Materials and Growth) in LD were transferred to 24-well cell culture plates (Greiner Labortechnik), one plant per well. The plates were transferred to continuous white light (30–40 μmol m−2 s−1), and leaf movement was recorded every 20 min over 7 d by Panasonic CCTV cameras, model WV-BP120 (Matsushita Communications Industrial). Post-run analysis was performed using the Kujata software program (Millar et al., 1995), and traces were analyzed by FFT-NLLS (Plautz et al., 1997).
Preparation of the LHY and CCA1 Fusion Constructs
pBluescript KS− plasmids containing the CCA1 cDNA and the LHY cDNA were donated by Elaine Tobin. The endogenous stop codons of both CCA1 and LHY were mutated using a site-directed mutagenesis kit (Intron Biotechnology; MutaDirect kit). The 3HA tag sequence or the myc tag sequence was introduced into CCA1 cDNA containing pBluescript KS− downstream of the CCA1 cDNA between HindIII and BamHI to create CCA1-HA[full] or CCA1-myc, respectively. The myc tag sequence was also introduced into LHY cDNA containing pBluescript KS− downstream of the LHY cDNA between BamHI and NotI to create LHY-myc[full]. To express CCA1-HA[full] in E. coli cells, the CCA1-HA construct was cloned into pHIS-parallel1 plasmid (pET22b, Dr. Peter Sheffield, University of Virginia) downstream of the ribosomal binding site. To express LHY-myc[full] in E. coli, the LHY-myc construct was cloned into pACYCDuet-1 (Novagen) plasmid downsteam of the ribosomal binding site.
The CCA1-HA[407] construct was made by digesting CCA1-HA[full] with NdeI and SacI and followed by ligation with an NdeI_SacI_linker_FP, 5′-TATGGTGCTAGTGCCATTGGGGAGCT-3′, and NdeI_SacI_linker_RP, 5′-CCCCAATGGCACTAGCACCA-3′. The CCA1-HA[223] constructs was made by digesting CCA1-HA[full] with NdeI and AgeI and ligation with NdeI_AgeI_linker_FP, 5′-TATGGGAGACAGAAAACAAGTTGA-3′, and NdeI_AgeI_linker_RP, 5′-CCGGTCAACTTGTTTTCTGTCTCCCA-3′.
The GST-myc control was created by cloning the myc tag into the EcoRI site in the pGST-parallel1 plasmid (Dr. Peter Sheffield, University of Virginia) downstream of the GST and in frame.
CCA1pro∷CCA1-HA-YFP was made by cloning the CCA1 promoter (1,222 bp upstream of the start codon) into pBluescript KS− with CCA1-HA with XhoI and BglII before the CCA1. The stop codon was mutated by a site-directed mutagenesis kit (Intron Biotechnology MutaDirect kit). The resulting CCA1-HA with the CCA1 promoter and without a stop codon was cloned between the XhoI and BamHI sites in the vector 10 OP CE-YFP containing YFP (CLONTECH) given by Yuval Eshed, and the CCA1pro∷CCA1-HA-YFP construct was cloned into the NotI site in the binary vector pMLBART also obtained from Yuval Eshed (Goldshmidt et al., 2008). The direction of the CCA1pro∷CCA1-HA-YFP construct in the pMLBART vector was checked by PCR. LHYpro∷LHY-myc was made by cloning the LHY promoter (1,568 bp upstream of the start codon) into pBluescript KS− with LHY-myc with BstEII and NcoI before the LHY. The LHY with LHY promoter was cloned into binary vector pMLBART. CCA1pro∷CCA1-myc was made by cloning the CCA1 promoter (1,222 bp upstream of the start codon) into pBluescript KS− with CCA1-myc with XhoI and BglII before the CCA1. The CCA1-myc with CCA1 promoter was cloned into binary vector pMLBART.
Plant Transformation
GV3101∷pMP90RK Agrobacterium tumefaciens containing the binary vectors described above was cultured in Luria-Bertani medium at 28°C with agitation until OD600 = 1. Three-week-old flowering Arabidopsis plants were dipped in floral dip medium for 5 min (Weigel and Glazebrook, 2002). Plants were left horizontally in the dark for 24 h and then grown for three more weeks until the seeds were ready for harvesting. Transformed plants were identified by their resistant to 1% Basta (glufosinate ammonium).
Co-IP
Co-IP experiments in plants were carried out as described in Weigel and Glazebrook (2002). Briefly, plant tissue samples were collected and ground in liquid nitrogen, then resuspended in grinding buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitors [Roche]), and clarified by centrifugation twice at 16,000g for 10 min at 4°C. Protein concentrations were determined using a commercial protein assay reagent (Rc/Dc reagent; Bio-Rad). Aliquots of 300 μL of protein extracts were incubated with 10 μL antibody for 2 h at 4°C. Protein A beads (20 μL) were added to each sample and then incubated for a further 24 h at 4°C. The samples were then washed with grinding buffer followed by centrifugation three times at 1,500g for 5 min at 4°C. The precipitated proteins were fractionated by SDS-PAGE (Protein Analysis) and analyzed by western blots as described above.
For co-IPs from E. coli extracts, cells were incubated with 1 mm IPTG for 3 h for protein induction. Cells were collected by centrifugation and then lysed by incubation in 1 mm EDTA, 1 mm PMSF, 1:200 protease inhibitor cocktail (Sigma-Aldrich), 50 μg/mL DNaseI (Sigma-Aldrich), 10 mm MgCl2, and 10 mg/mL lysozyme for 30 min on ice. The cells were disrupted using a French press and centrifuged at 3,000g for 10 min at 4°C. The supernatant was than incubated for with 1/500 (v/v) antibody at 4°C overnight. Protein A/G agarose beads (20 μL) were added to the supernatant, and after 1 h of incubation, the samples were centrifuged for 5 min at 1,000g in 4°C and the resulting pellets washed with cold PBS (8 g/L NaCl, 1.15 g/L Na2HPO4, 0.2 g/L KCl, and 0.2 g/L KH2PO4, pH 7.4) four times. Proteins were fractionated by SDS-PAGE and analyzed by western blots, as described above.
Confocal Microscopy
Two-week-old Arabidopsis plants grown on MS medium (Plant Materials and Growth) were examined using an FV-1000 Olympus Confocal Microscope with a ×40/1.3 oil immersion objective. Excitation and emission for DAPI-stained cells were 405 nm and 370 to 430 nm; excitation and emission for YFP were 515 nm and 535 to 565 nm; and excitation and emission for chloroplast autofluorescence was 633 nm and 655 to 755 nm. Levels of fluorescence were calculated by ImageJ using the Mean Gray Value option.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf movement in the transgenic and control lines.
Supplemental Figure S2. CCA1-HA-YFP is localized to the nucleus in CCA1pro∷CCA1-HA-YFP cca1-1#2 plants.
Supplemental Figure S3. LHY-null plants do not express LHY mRNA.
Supplemental Figure S4. The system used for protein detection is linear for the concentrations of protein used in our experiments.
Supplementary Material
Acknowledgments
We thank Elaine Tobin, Steve Knowles, Yuval Eshed, and Alexander Goldshmidt for their generous gifts of plasmids and seeds, Nir Ohad and Aviva Katz for technical advice, and Shai Yerushalmi for his critical reading of the manuscript.
This work was supported by the Binational Science Foundation (grant no. 0378415) and the Deutsch-Israelische Projektkooperation (grant no. 0307712).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rachel M. Green (rgreen@vms.huji.ac.il).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12 757–761 [DOI] [PubMed] [Google Scholar]
- Bae K, Edery I (2006) Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem 140 609–617 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J 16 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre IA, Kim JY (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53 1551–1557 [DOI] [PubMed] [Google Scholar]
- Covington M, Maloof J, Straume M, Kay S, Harmer S (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9 R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96 271–290 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (2006) Physiology. Running a clock requires quality time together. Science 311 184–186 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JCW, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283 23073–23083 [DOI] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y (2008) Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S, Naef F, Quisel T, Gendron J, Chen H, Ecker J, Borevitz J, Kay S (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10 R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322 1832–1835 [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Feher B, Toth R, Viczian A, Kircher S, Rea D, Dorjgotov D, Schafer E, Millar AJ, et al (2007) Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol 17 1456–1464 [DOI] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carre IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360 [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 2 R271–277 [DOI] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH (2006) BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol 26 7318–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1 E1–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41 791–803 [DOI] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW (2006) PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311 226–229 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, Kay SA (1995) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46 686–698 [DOI] [PubMed] [Google Scholar]
- Para A, Farre EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Mas P (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12 204–217 [DOI] [PubMed] [Google Scholar]
- Portolés S, Más P (2007) Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J 51 966–977 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125 485–494 [DOI] [PubMed] [Google Scholar]
- Song HR, Carre IA (2005) DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol Biol 57 761–771 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771 [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Vandenbussche F, Laarhoven LJ, Dowson-Day MJ, Wang ZY, Tobin EM, Harren FJ, Millar AJ, Van Der Straeten D (2004) Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol 136 3751–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Harir Y, Green RM (2007) Regulation of output from the plant circadian clock. FEBS J 274 335–345 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ III (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.