Abstract
Adventitious root formation at the base of plant cuttings is an innate de novo organogenesis process that allows massive vegetative propagation of many economically and ecologically important species. The early molecular events following shoot excision are not well understood. Using whole-genome microarrays, we detected significant transcriptome remodeling during 48 h following shoot removal in Populus tremula × Populus alba softwood cuttings in the absence of exogenous auxin, with 27% and 36% of the gene models showing differential abundance between 0 and 6 h and between 6 and 24 h, respectively. During these two time intervals, gene networks involved in protein turnover, protein phosphorylation, molecular transport, and translation were among the most significantly regulated. Transgenic lines expressing a constitutively active form of the Populus type-B cytokinin response regulator PtRR13 (ΔDDKPtRR13) have a delayed rooting phenotype and cause misregulation of CONTINUOUS VASCULAR RING1, a negative regulator of vascularization; PLEIOTROPIC DRUG RESISTANCE TRANSPORTER9, an auxin efflux transporter; and two APETALA2/ETHYLENE RESPONSE FACTOR genes with sequence similarity to TINY. Inappropriate cytokinin action via ΔDDKPtRR13 expression appeared to disrupt adventitious root development 24 h after shoot excision, when root founder cells are hypothesized to be sensitive to the negative effects of cytokinin. Our results are consistent with PtRR13 acting downstream of cytokinin to repress adventitious root formation in intact plants, and that reduced cytokinin signaling after shoot excision enables coordinated expression of ethylene, auxin, and vascularization pathways leading to adventitious root development.
Adventitious rooting is an ecologically and economically important developmental process. Populus and other perennial species adapted to riparian ecosystems naturally propagate clonally when stems or branches detached by a natural disturbance are carried downstream and lodge in a moist environment conducive to rooting (Rood et al., 2003). For many species, therefore, clonal propagation via adventitious root formation is a natural complement to sexual propagation by seeds. The stimulation of adventitious rooting to facilitate clonal propagation is a cornerstone of the ornamental horticulture and forest products industries. Treatment of cuttings with synthetic auxins has been used for more than 60 years to induce and accelerate rooting in hard-to-root species (Kevers et al., 1997). Genetic improvement programs for forest trees including Eucalyptus species, Populus species, and Pinus radiata are almost exclusively based on the production of rooted cuttings for breeding and deployment in operational plantations (Davis and Becwar, 2007).
In woody plants, adventitious root primordia primarily arise from ray cells adjacent to the vascular cambium, from buds or leaf gaps, or from callus formed at the base of cuttings (Lovel and White, 1986). Ectopic formation of roots is regulated by both endogenous (e.g. hormones, sugars, phenolic compounds) and exogenous (e.g. temperature, light) factors (Eliasson, 1978; Davies and Hartmann, 1988; Kevers et al., 1997; De Klerk et al., 1999). Among the plant hormones, it is well established that basipetal transport and accumulation of auxin at the base of cuttings precedes adventitious root formation (Liu and Reid, 1992; Hausman et al., 1995; Guerrero et al., 1999). Auxin synthesized in shoot tips is important during adventitious rooting, since removal of the shoot apex decreases both the level of endogenous auxin in the basal portion of a cutting and the number of adventitious roots produced (Nordstrom and Eliasson, 1991). Elevated auxin concentrations give rise to new root primordia by activating the differentiation and elongation of phloem parenchyma cells adjacent to vascular bundles in the stem (Lund et al., 1996; De Klerk et al., 1999).
Adventitious root formation can be seen as a three-stage process: (1) activation, where the cells originating the root primordia become competent to respond to the rhizogenic action of auxin; (2) induction, comprising the determination of root primordia and initial cell divisions; and (3) outgrowth, where root primordia elongate and vascular connections are established to preexisting vasculature within the stem. Auxin and cytokinin appear to play antagonistic roles in more than one stage of the adventitious rooting process (De Klerk et al., 1997). Quantification of the endogenous levels of auxin and cytokinins in the basal region of cuttings from diverse woody and herbaceous plants, including Populus, Malus, Solanum, and Phaseolus, reveals that the concentrations of these two hormones follow opposite patterns during the initial 48 h of rooting. Auxin concentrations are high during activation and induction stages and low during outgrowth, while cytokinin levels are low during activation and induction stages and high during outgrowth (Blakesley et al., 1985; Bollmark and Eliasson, 1986; Maldiney et al., 1986; Hausman et al., 1997; Kevers et al., 1997). In addition, exogenous application of cytokinin to cuttings during the induction phase strongly inhibited root formation (De Klerk et al., 1999). A naturally occurring cytokinin in cucumber root xylem sap (trans-zeatin riboside) was identified as a major suppressor of adventitious root formation in hypocotyls (Kuroha et al., 2002). Histological studies indicate that cytokinin inhibits the differentiation of primordia at an early stage in development (Bollmark and Eliasson, 1986).
Cytokinin signaling resembles two-component systems first identified in bacteria and yeast. In two-component systems, an extracellular cue is sensed by a plasma membrane-localized His kinase (HK), which transfers the signal to a response regulator (RR) in the form of a phosphoryl group (Mizuno, 1998; West and Stock, 2001). In plants, these signaling pathways include a third component, a His phosphotransfer protein, which functions as a carrier of the phosphoryl group from the HK to the RR (Mok and Mok, 2001; Kakimoto, 2003; Ferreira and Kieber, 2005). The RRs are the final step of this phosphorelay and have been traditionally divided into two groups: type A and type B. The type-A RRs are cytokinin primary response genes whose proteins consist mostly of a receiver domain with conserved D-D-K residues (Brandstatter and Kieber, 1998; Taniguchi et al., 1998; D'Agostino et al., 2000). The type-B RRs are more complex proteins that contain, in addition to the receiver domain, a DNA-binding motif (GARP domain) that resembles a domain originally found in the mammalian oncoprotein c-Myb (Imamura et al., 1998; Sakai et al., 1998). While phosphorylated type-B RRs act as transcriptional activators of cytokinin-regulated genes, the type-A RRs down-regulate cytokinin signaling by competing with the type-B RRs for phosphoryl groups (for review, see To and Kieber, 2008). Genetic analyses to define the function of RRs in Arabidopsis (Arabidopsis thaliana) suggest functional redundancy among family members, since phenotypes are not obvious unless mutant lines contain null alleles from several loci (To et al., 2004; Argyros et al., 2008; Ishida et al., 2008). Because of their potential functions as transcriptional activators, type-B RRs are thought to represent key elements orchestrating transcriptome changes triggered by cytokinin. We previously identified 22 genes in Populus that exhibit the typical features of plant RRs (Ramirez-Carvajal et al., 2008).
Although the physiological and anatomical processes associated with adventitious rooting have been described in a variety of genera, including Eucalyptus, Pinus, and Malus (De Klerk et al., 1999; Fett-Neto et al., 2001), the molecular mechanisms involved in determining the competence of cells to generate adventitious roots as well as the development of adventitious roots per se are not well defined. Studies in Arabidopsis and tobacco (Nicotiana tabacum) have provided compelling evidence linking cytokinin signaling to adventitious root formation. Arabidopsis mutants lacking the cytokinin HK receptors AHK2, AHK3, and AHK4 exhibit enhanced adventitious root growth, whereas the elongation of primary and lateral roots is virtually eliminated (Higuchi et al., 2004; Nishimura et al., 2004). Similar phenotypes have been observed in cytokinin-deficient tobacco plants, where overexpression of the cytokinin-degrading enzyme cytokinin oxidase results in abundant adventitious roots (Werner et al., 2001, 2003). The Arabidopsis triple mutant arr1 arr10 arr12 lacks the function of three type-B RRs, shows almost complete insensitivity to exogenously applied cytokinin, and spontaneously produces adventitious roots in hypocotyls (Argyros et al., 2008). While these genetic studies imply a potential role of cytokinin in adventitious rooting, they do not illuminate when or where these effects occur. Analyses of shifts in gene expression in conjunction with adventitious rooting (Kohler et al., 2003; Brinker et al., 2004) examined relatively small sets of genes or targeted relatively late stages of root formation. Thus, the potential role of cytokinin in regulating adventitious root development remains obscure, particularly the activation and induction phases that constitute the very early stages of adventitious root development.
As part of a comprehensive analysis of the primary molecular mechanisms orchestrating de novo adventitious root formation in Populus cuttings, we report transcriptome remodeling involving about half of the transcriptional units annotated in the poplar (Populus tremula × Populus alba) genome within 24 h of shoot excision. We investigated the role of cytokinin signaling by contrasting whole-genome expression data acquired from nontransgenic (NT) poplar, with transgenic lines delayed in rooting due to ectopic expression of a constitutively active form of PtRR13, referred here as ΔDDKPtRR13. Perturbation of rooting due to alteration of cytokinin signaling through the expression of ΔDDKPtRR13 appears to be physiologically relevant at 24 h, where a large set of signaling, developmental, and metabolic networks are differentially regulated in the transgenic and NT lines. Promoter sequence examination of differentially regulated genes across all time points between NT and ΔDDKPtRR13 lines reveals putative direct and indirect targets of PtRR13. These putative targets include a negative regulator of vascularization, CONTINUOUS VASCULAR RING1 (COV1), an auxin efflux transporter, PLEIOTROPIC DRUG RESISTANCE TRANSPORTER9 (PDR9), and two APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) proteins similar to TINY.
RESULTS
The Poplar Transcriptome Is Remodeled in the Base of Excised Shoots
Populus softwood cuttings of the genotype used in this study form an adventitious root system that is sufficiently developed within 10 to 14 d so that plants can be removed from periodic mist treatments. No exogenous auxin treatment is required; the lack of a requirement for auxin treatment may be due to high rates of auxin synthesis and/or transport in the actively growing shoots used to generate cuttings. Observations in Malus and Populus microcuttings reveal cell divisions as early as 48 h after auxin exposure and the presence of organized meristemoids by 96 h (De Klerk et al., 1995; Wu, 2004). Because early physiological and biochemical evidence indicates that there are significant changes in endogenous hormone pools, including ethylene, auxin, and cytokinin, during the first 48 h after excision (Maldiney et al., 1986; Selby et al., 1992; Hausman, 1993; De Klerk et al., 1997), we predicted that changes in gene expression during this time frame would provide information about the early and poorly defined molecular networks involved in adventitious root formation.
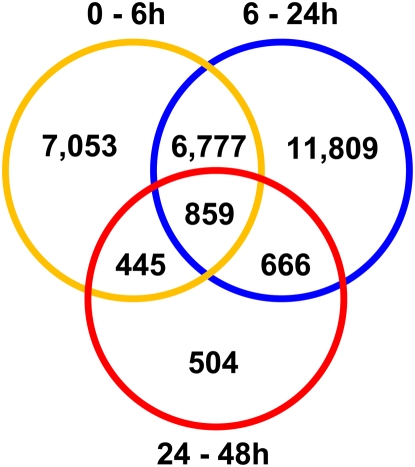
To analyze transcriptome changes during adventitious root formation in Populus, the basal 5 mm of softwood cuttings, where adventitious roots originate, were harvested at 0, 6, 24, and 48 h after excision. RNA extracted from these samples was used for microarray analyses with a custom-designed NimbleGen array containing features representing 55,793 annotated gene models from the sequenced genome of Populus trichocarpa, including 45,555 predicted gene models reported previously (Tuskan et al., 2006) plus an additional 10,238 gene models showing evidence of transcription (Quesada et al., 2008). Transcripts showing significant differences in abundance at a false discovery rate (FDR; Storey and Tibshirani, 2003) of 5% between adjacent time points (0–6 h, 6–24 h, and 24–48 h) were identified. A total of 15,134 transcripts (representing 27% of the predicted gene models used in this study) were differentially regulated between 0 and 6 h, 20,111 (36%) were differentially regulated between 6 and 24 h, and 2,474 (4%) were differentially regulated between 24 and 48 h (Fig. 1). This high proportion of differentially regulated genes is consistent with massive transcriptome remodeling after shoot excision. Gene set enrichment analysis of these genes suggests differential regulation of functional networks with potential roles in protein degradation, protein phosphorylation, and RNA metabolism (Supplemental Table S1; Supplemental Fig. S1, A and B).
Figure 1.
Differentially expressed genes during early stages of adventitious root formation. Microarray expression data were contrasted between 0 and 6 h, 6 and 24 h, and 24 and 48 h after shoot excision. Genes whose contrasts were significant at FDR < 5% are displayed in the Venn diagram. [See online article for color version of this figure.]
Gene Clustering Reveals Diverse Patterns of Gene Expression and Reflects Contrasting Roles of Ethylene, Auxin, and Cytokinin
To discern global patterns of differential transcript abundance over the time course, transcripts with contrasts significant at FDR < 5% were further filtered to include only those with greater than 2-fold changes in transcript abundance at adjacent time points (0–6 h, 6–24, and 24–48 h). Transcripts meeting these criteria were clustered using the Gaussian Clustering application of ArrayMiner5 (Optimal Designs; Fig. 2). We chose to present n = 7 clusters since the number of transcripts not included in any cluster (Fig. 2, Unc.) was similar with clusters of n ≥ 7. Cluster size ranged from 3,796 to 1,330 transcripts. Current research indicates that the early (activation and induction) stages of adventitious root development probably involve complex interactions among ethylene, auxin, and cytokinin. Therefore, we evaluated the clusters to determine if and how these hypothesized hormone interactions might be reflected in the transcript data. For clarity, we distinguish clusters reflecting general trends toward increasing transcript abundance (red; clusters 1, 4, and 7) and decreasing transcript abundance (green; clusters 2, 3, 5, and 6) over time.
Figure 2.
Clusters of differentially regulated genes during early stages of adventitious root formation. Microarray expression data were contrasted between 0 and 6 h, 6 and 24 h, and 24 and 48 h after shoot excision. Genes whose contrasts were significant at 5% FDR and 2-fold regulation at any of the three contrasts were clustered using the Gaussian Clustering application from ArrayMiner5. Clusters of genes showing overall down-regulation are shown in green, while up-regulated clusters are shown in red. The number of genes in each cluster is displayed. Selected gene families involved in cytokinin, auxin, and ethylene signaling/biosynthesis are displayed. Unc, Unclustered genes.
Ethylene biosynthesis is predicted to increase after shoot excision (Bollmark et al., 1988; O'Donnell et al., 1996; Van Loon et al., 2006). We observed an increase in transcript abundance for 9 of the 12 members of the aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase gene families that were placed in clusters (Fig. 2). Seven of these nine transcripts were located in either cluster 4 or 7 (both clusters with the lowest transcript abundance at 0 h). These data are consistent with an increase in ethylene biosynthesis following shoot excision.
Two families of transcriptional regulators (IAAs and ARFs) are involved in auxin signaling. IAAs negatively modulate auxin signaling by repressing the transcriptional activity of ARFs through heterodimerization (Ulmasov et al., 1999). Auxin can induce the transcription of specific IAAs, which is thought to establish a negative feedback loop and ensure a transient response (Abel and Theologis, 1996; Liscum and Reed, 2002). Poplar IAAs and ARFs have been annotated and evaluated for auxin inducibility (Kalluri et al., 2007). We found that transcripts for IAA gene family members increased (four of 11 clustered genes) in concert with transcripts for ethylene biosynthesis or decreased (seven of 11; Fig. 2). In contrast, all ARF gene family members that were placed in clusters (all eight of the clustered genes) decreased in transcript abundance.
Physiologically relevant quantities of cytokinin are exported from roots to aboveground organs (Hirose et al., 2008). Since type-A RRs are postulated to be transcriptionally regulated by cytokinin, changes in the transcript abundance of these genes may operate as reporters for cytokinin availability. Two subfamilies of RRs (type A and type B) have been annotated in the poplar genome and evaluated for cytokinin inducibility (Ramirez-Carvajal et al., 2008). Of the eight RR family members that occurred in clusters, all of them decreased in transcript abundance over time (Fig. 2, clusters 2 and 3). While this general pattern coincides with the ARFs in that the greatest transcript abundance was at time 0, the transcript abundance for most of the RRs decreased within 6 h (seven of the eight RR transcripts grouped in clusters 3 and 5). In contrast, most of the ARFs decreased at 24 h (five of the eight ARF transcripts grouped in cluster 2).
PtRR13 Lacking a Receiver Domain Generates a Phenotype, But Not Full-Length or RNA Interference Constructs
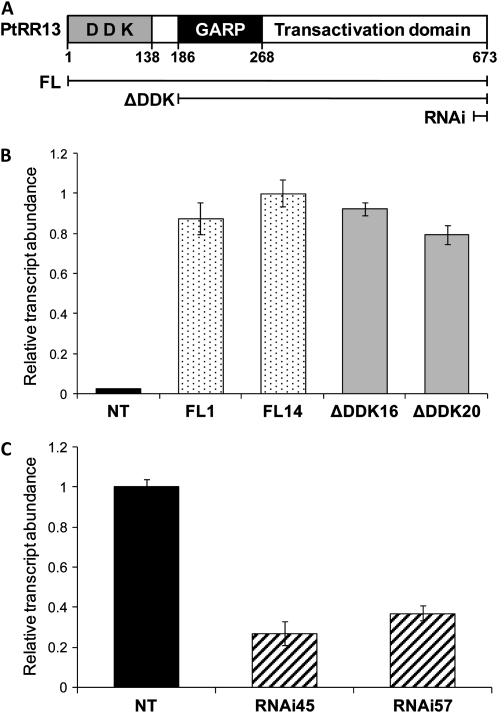
PtRR13 displays the typical domain organization of type-B RRs: an N-terminal receiver domain with invariable D-D-K residues, a GARP domain, and a long C-terminal transactivation domain (Fig. 3A). Sequence similarity and neighboring gene colinearity suggest that PtRR13 is an ortholog of Arabidopsis ARR1 (Supplemental Fig. S2). To develop knowledge about the in vivo roles of PtRR13, three different constructs were designed to direct gain or loss of PtRR13 function. Two different overexpression constructs, one for production of full-length PtRR13 (FLPtRR13) and a second for production of a truncated version in which the receiver domain was deleted (ΔDDKPtRR13), were engineered. Since the type-B receiver domain is proposed to inhibit the DNA-binding activity of the GARP domain in the absence of the phosphorylated Asp residue associated with cytokinin signaling (Sakai et al., 2000, 2001), engineering a construct in which this domain is missing is predicted to create a constitutively active version of PtRR13. The third construct, consisting of a transitive RNA interference (RNAi) vector containing approximately 200 bp of the last exon of PtRR13, was generated for a loss-of-function approach. Transgenic P. tremula × P. alba plants were generated for all three constructs via Agrobacterium tumefaciens-mediated transformation, and the two strongest lines from each construct were chosen for detailed phenotyping. To identify these, 60 lines (20 independent transgenic events per construct) were regenerated, hardened off to greenhouse conditions, propagated as rooted cuttings, and then screened for transgene expression levels. Transgene transcript abundance for the FLPtRR13 and ΔDDKPtRR13 lines was assayed by real-time quantitative PCR, and the two lines with highest expression for each construct were chosen for further phenotyping (FLPtRR13-1 and FLPtRR13-14 for the full length and ΔDDKPtRR13-16 and ΔDDKPtRR13-20 for the truncated; Fig. 3B). The two RNAi lines with the greatest reductions in endogenous PtRR13 expression compared with the nontransgenic controls were also selected for further phenotyping (lines RNAi45 and RNAi57, with 73% and 62% reduction, respectively; Fig. 3C).
Figure 3.
PtRR13 domain organization and screening of transgenic lines. A, Domain organization of PtRR13 was deduced by amino acid sequence similarity with Arabidopsis RRs (Ramirez-Carvajal et al., 2008). The three coding regions used for construct synthesis included the full-length protein (FL), a truncated version missing the receiver domain (ΔDDK), and a short region (60 amino acids) in the last exon (RNAi). B and C, Transgenic line screening. Transgenic lines carrying the FL or ΔDDK (B) and RNAi (C) constructs were screened for transgene/endogenous PtRR13 expression by real-time PCR and compared with the nontransgenic control NT. For all lines, RNA extracted from shoots was reversed transcribed and used as a template in real-time PCR. Fluorescence intensities were normalized to the intensity of the actin control and are presented relative to the NT. Error bars represent se where n = 3.
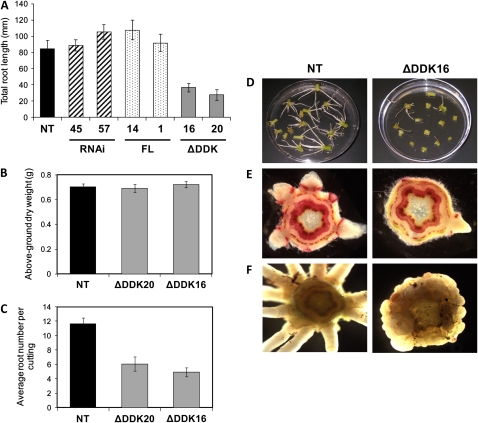
A perturbation in adventitious rooting during propagation was also observed. Measuring total root length of apical cuttings 10 d after shoot excision revealed that the two ΔDDKPtRR13 lines had significantly lower total root lengths than the nontransgenic line (36.3 ± 5.2 and 27.3 ± 6.5 mm for ΔDDKPtRR13-16 and ΔDDKPtRR13-20, respectively, versus 84.2 ± 10.5 mm for the NT; Fig. 4A). No significant differences were found between the FLPtRR13 and RNAi and the NT lines. The absence of phenotypes for the FLPtRR13 lines may be because ectopically expressed PtRR13 does not alter cytokinin signaling, which may be driven by shifts in RR phosphorylation status. The lack of phenotypes in the PtRR13 RNAi lines could be a consequence of incomplete gene silencing, since the line with the most severe reduction may retain normal cytokinin signaling. Alternatively, PtRR13 may be functionally redundant with another RR. Based on the observed delay in adventitious rooting, subsequent analyses focused on the ΔDDKPtRR13 lines.
Figure 4.
PtRR13 is a negative regulator of adventitious root formation. A, Total root length was measured in 14-cm, 10-d-old cuttings for two independent transgenic lines per construct (FL, ΔDDK, and RNAi) and in NT. B and C, The ΔDDKPtRR13 lines exhibit fewer roots than the NT (B), and the phenotype is specific to roots (C). D, Overview of the rooting phenotypes. E, Phloroglucinol-HCl-stained stem sections at rooting sites. F, Abnormal callus formation at the base in ΔDDK lines. The rooting experiments described here were repeated a minimum of five times to confirm the phenotypes. A typical experiment is shown. Error bars represent se where n = 15.
PtRR13 Is a Negative Regulator of Adventitious Root Formation
The differences in total root length observed above were a consequence of decreased numbers of roots in the ΔDDKPtRR13 lines. While no differences were observed in plant growth, cuttings of equivalent biomass (Fig. 4B) revealed consistent differences in root number between the control NT line, which formed, on average, 11.66 ± 0.84 (se) roots per cutting, and the ΔDDKPtRR13-16 and ΔDDKPtRR13-20 lines, which formed 4.92 ± 0.65 and 6.07 ± 0.99 roots per cutting, respectively. This indicates that the differences observed in rooting were not driven by disparities in plant size or shoot biomass. The reduction in root number in the ΔDDKPtRR13 lines (Fig. 4, D and E) was typically accompanied by callus formation at the base of the cutting adjacent to the wound site (Fig. 4F), implying a discrete disruption in adventitious root development.
To evaluate whether the proliferation of unorganized cells at the base of the cuttings reflected an altered response to hormones, stem explants from the ΔDDKPtRR13 lines were cultured under six different auxin:cytokinin ratios (0:0, 10:0, 100:0, 0:10, 10:10, and 10:100 μm indole-butyric acid:μm zeatin, respectively; Supplemental Fig. S3). The ΔDDKPtRR13 lines showed greater tissue growth in the absence of exogenous cytokinins, implying an up-regulation of cytokinin signaling in the transgenic lines. Collectively, the results obtained in these experiments imply a perturbation of cytokinin signaling in transgenic poplar expressing ΔDDKPtRR13, with an apparent negative role in adventitious root formation.
ΔDDKPtRR13 Expression Has Minor Pleiotropic Effects on the Transcriptome
To gain insights into the molecular mechanisms altered by a constitutively active version of PtRR13, whole-transcriptome monitoring data were generated to detect shifts in transcript abundance. Line ΔDDKPtRR13-16 was arbitrarily selected, since both ΔDDKPtRR13-16 and ΔDDKPtRR13-20 lines displayed identical phenotypes with respect to the adventitious rooting and the tissue culture response to cytokinin. Expression data at 0, 6, 24, and 48 h after shoot excision for line ΔDDKPtRR13-16 was contrasted with that for NT, and transcripts significantly altered across all four time points (transgene effect in the ANOVA model) were identified. Consistent with our whole-plant studies, which collectively implied a lack of pleiotropic effects of the transgene, we identified only 11 transcripts that showed differential abundance across all time points at a 10% FDR and a 2-fold regulation threshold. Of the 11 genes differentially regulated across all time points, five were up-regulated (including transgene ΔDDKPtRR13) and six were down-regulated (Table I). The two transcripts most increased were COV1, a negative regulator of vascular formation in stems (Parker et al., 2003) and PDR9, encoding a protein involved in auxin efflux (Ito and Gray, 2006). The other genes up-regulated by ΔDDKPtRR13 were FIMBRIN-LIKE2 (FIM2), encoding an actin-binding protein, and MECHANOSENSITIVE CHANNEL OF SMALL CONDUCTANCE-LIKE10 (MSL10), encoding an ion channel protein. The transcripts with lower abundance in the ΔDDKPtRR13 line (or increased in NT) included two TINY-like transcription factors, the cytochrome P450 CYP94B3, a caffeic acid O-methyltransferase family 2 protein, a metal-nicotinamide transporter YELLOW STRIPE-LIKE7 (YSL7), and a homeodomain protein, BELL1.
Table I.
Differentially regulated genes between NT and ΔDDKPtRR13 from 0 to 48 h
Transgene effects on gene expression during 48 h following shoot excision were determined by contrasting expression estimates of the ΔDDKPtRR13 line relative to NT across all time points. Genes with contrasts significant at FDR < 10% and 2-fold regulation are displayed. Significantly regulated Populus gene models were annotated by identifying putative orthologs from GenBank databases (closest hit). Positive fold changes indicate higher transcript abundance in ΔDDKPtRR13, whereas negative fold changes indicate higher transcript abundance in NT.
| Description | Populus Gene Model | Closest Hit | Fold Regulation | P |
|---|---|---|---|---|
| ΔDDKPtRR13 | estExt_Genewise1_v1.C_LG_X3573 | At3g16857.1 | 11.00 | 7.0E-16 |
| Membrane protein COV1 | estExt_Genewise1_v1.C_570139 | At2g20120.1 | 4.41 | 3.0E-11 |
| Pleiotropic drug resistance transporter PDR9 | fgenesh4_pm.C_LG_X000603 | At3G53480.1 | 3.92 | 8.0E-06 |
| Actin-binding protein FIM2 | estExt_fgenesh4_pm.C_LG_XI0146 | At4g26700.1 | 2.45 | 3.2E-07 |
| Mechanosensitive ion channel MSL10 | gw1.28.2.1 | At5g12080.1 | 2.30 | 2.5E-07 |
| ERF/AP2 transcription factor TINY-like1 | fgenesh4_pm.C_scaffold_41000071 | At1g71450.1 | −4.44 | 8.2E-08 |
| Cytochrome P450 CYP94B3 | fgenesh4_pg.C_LG_XII000950 | At3g48520.1 | −3.58 | 7.7E-07 |
| Caffeic acid O-methyltransferase | fgenesh4_pg.C_LG_XIX000854 | At4g35160.1 | −2.48 | 2.1E-06 |
| ERF/AP2 transcription factor TINY-like2 | gw1.XIX.1847.1 | At1g71450.1 | −2.43 | 1.3E-05 |
| Metal-nicotianamine transporter YSL7 | fgenesh4_pg.C_LG_IV000291 | At1g65730.1 | −2.28 | 1.7E-05 |
| Homeodomain protein BELL1 | gw1.III.246.1 | At5g41410.1 | −2.08 | 1.1E-05 |
Arabidopsis ARR1 binds DNA in a sequence-specific manner to the semipalindromic motif AGATC (Sakai et al., 2001). This motif has been detected in the promoters of several Arabidopsis type-A RRs and in several cytokinin-regulated genes (Rashotte et al., 2003). A manual search for this ARR1 motif in the promoters of the 10 genes (less the ΔDDKPtRR13 transgene) regulated across all time points revealed that the COV1 promoter had the highest number of AGATC motifs (five motifs) that all occurred in a 374-bp segment approximately 680 bp upstream of the presumed translation start (Supplemental Fig. S4). The number of ARR motifs in the other gene promoters ranged from one to four. Because Arabidopsis TINY has been demonstrated to bind the dehydration-responsive element with a core sequence of A/GCCGAC as well as the ethylene-responsive element with a core sequence of AGCCCGCC (Sun et al., 2008), we manually searched for these motifs but found no indication of enrichment.
Cytokinin Action May Interfere with Adventitious Rooting at 24 h
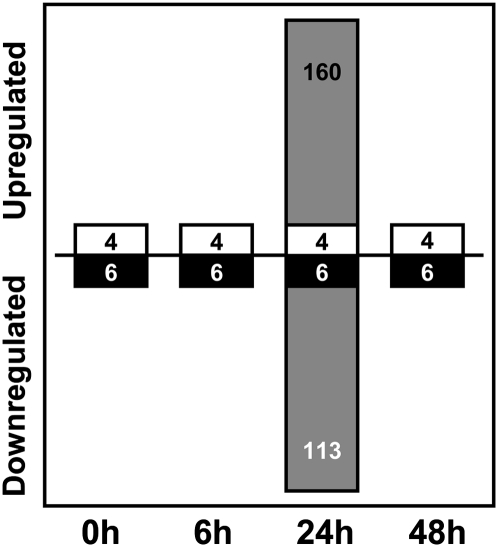
To gain insights into the molecular mechanisms altered during early adventitious root formation in transgenic plants, we identified genes significantly regulated at each time point (transgene × time interaction terms in the ANOVA model). These analyses revealed no significant gene regulation differences at 6 and 48 h (above and beyond that incited by the transgene alone), although a significant transcriptional response at 24 h was identified (273 genes at 1% FDR and 2-fold abundance threshold, or 5,111 genes at 5% FDR and 2-fold abundance threshold; Fig. 5). Of the 273 genes differentially regulated at 24 h at 1% FDR (Supplemental Table S2), 160 (59%) had significantly greater expression in ΔDDKPtRR13-16 than in NT, while the remaining 113 (41%) were more abundantly expressed in NT. The top five most significantly up-regulated transcripts and the top five most significantly down-regulated transcripts were identified (Table II). These transcripts encode genes involved in diverse biological processes such as auxin homeostasis (IAA-conjugating enzyme GH3-8), brassinosteroid signaling (BEE transcription factor), transcriptional regulation (Myb102), calcium signaling (IQD33), defense responses (SOBER1), and transport (CAT7 and MATE efflux proteins).
Figure 5.
Overall effects of ΔDDKPtRR13 overexpression on gene expression. In addition to the transgene, 10 genes were differentially regulated between NT and ΔDDKPtRR13 across all time points (white and black bars) at a 10% FDR and 2-fold regulation threshold; four were up-regulated and six were down-regulated in ΔDDKPtRR13. An additional 273 genes were differentially regulated between NT and ΔDDKPtRR13 at 24 h at a 1% FDR and 2-fold regulation threshold (gray bars); 160 were up-regulated and 113 were down-regulated in ΔDDKPtRR13.
Table II.
Top 10 differentially regulated genes between NT and ΔDDKPtRR13 at 24 h
The genotype × time point interaction was significant only at 24 h after shoot excision. The top five up- and down-regulated genes at 1% FDR and 2-fold abundance threshold are displayed. Positive fold changes indicate higher transcript abundance in ΔDDKPtRR13, whereas negative fold changes indicate higher transcript abundance in NT.
| Description | Populus Gene Model | Closest Hit | Fold Regulation | P |
|---|---|---|---|---|
| MATE efflux family protein | fgenesh4_pg.C_LG_XI000017 | At3g21690.1 | 8.01 | 1.5E-05 |
| IAA-conjugating enzyme GH3-8 | gw1.III.363.1 | At4g27260.1 | 7.91 | 1.6E-07 |
| Transcription factor BEE3 | eugene3.01230094 | At1g73830.1 | 7.48 | 2.0E-05 |
| Cationic amino acid transporter CAT7 | grail3.0090011402 | At3g10600.1 | 7.11 | 7.5E-06 |
| Cytochrome P450 CYP81D2 | eugene3.00640122 | At4g37360.1 | 6.62 | 1.0E-04 |
| Calmodulin-binding protein IQD33 | grail3.0018026401 | At4g23060.1 | −8.56 | 1.0E-04 |
| Defense-related carboxylesterase SOBER1 | gw1.166.165.1 | At4g22300.1 | −8.23 | 1.0E-04 |
| Myb transcription factor MYB102 | fgenesh4_pg.C_LG_XI001038 | At4g21440.1 | −6.39 | 1.0E-04 |
| Abscisic acid stress ripening-like protein | grail3.0002027201 | gi16588758 | −6.27 | 1.0E-04 |
| Extensin-like protein | fgenesh4_pm.C_LG_X000304 | At3g22800.1 | −6.19 | 1.0E-04 |
To determine if there was significant enrichment of any particular cis element(s) in these genes differentially regulated at 24 h, we adapted the overrepresentation computational analysis described by Nemhauser et al. (2004) to Populus. In genes up-regulated by ΔDDKPtRR13, a significant enrichment of the element OSE2ROOTNODULE (CTCTT; Fehlberg et al., 2005), originally found in promoters of genes activated during root nodule formation, was detected (Z score = 3.44, P < 0.001; data not shown). For genes down-regulated by ΔDDKPtRR13, we detected enrichment of the root-hair-specific cis element RHERPATEXPA7 (KCACGW; Z score = 2.37, P = 0.009; Kim et al., 2006). Interestingly, cytokinin receptors are required to activate cell divisions during nodule organogenesis (Murray et al., 2007), implying potential biological significance of these promoter motifs.
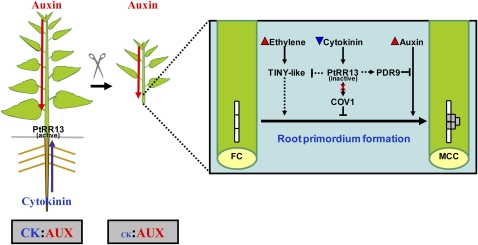
We developed a model (Fig. 6) to organize the interpretation of these results and to serve as a set of working hypotheses to guide further dissection of adventitious root development.
Figure 6.
Hypothesized role of PtRR13 in adventitious root development. In intact plants, cytokinin-auxin ratios regulate growth and development. Under these circumstances, a continuous supply of root-borne cytokinins maintains RRs, such as PtRR13, in a phosphorylated (active) state. Shoot excision alters the normal hormone balance by stimulating ethylene- and auxin-dependent pathways (red triangles) and decreasing cytokinin-auxin ratios (blue triangle). Abolition of cytokinin supply from the roots down-regulates cytokinin signaling, leading to inactivation of PtRR13. Adventitious root formation requires that a small group of root founder cells (FC) become competent to form a root meristematic cell cluster (MCC). Active PtRR13 (phosphorylated or constitutively active ΔDDKPtRR13) plays a negative role by activating transcription of COV1, a negative regulator of vascularization. At the same time, PtRR13 also potentially perturbs root primordia formation by interfering with auxin gradient establishment by stimulating transcription of the auxin efflux pump PDR9 and also by inhibiting stress/ethylene-inducible expression of TINY-like transcription factors.
DISCUSSION
Adventitious rooting is a complex process that encompasses signaling networks involved in physiological responses to mechanical injury, wound repair, and organ development. In this study, we monitored the Populus transcriptome during early stages of adventitious rooting and gained new insights into the regulation of endogenous hormone signaling cascades during this developmental process. We investigated the poorly understood effect of cytokinin on adventitious root formation by altering the expression of the type-B response regulator PtRR13. Significant transcriptional alterations at discrete times may imply when cytokinin antagonizes adventitious root development and identifies potential points at which cytokinin may interact with ethylene and auxin pathways.
A Molecular Genetic Roadmap: From Intact Plants to Adventitious Roots
Our results are consistent with a model in which cytokinin, synthesized in the roots and transported throughout the plant body, acts through PtRR13 to repress adventitious root development in intact Populus plants. Although cytokinins can be synthesized in various plant organs, significant amounts of root-borne cytokinins are continuously exported to aboveground organs in the xylem sap (Kuroha et al., 2002; Matsumoto-Kitano et al., 2008). Such root-to-shoot transport is thought to signal the nitrogen and nutrient status of the soil and thus coordinate soil nutrient availability with metabolism and development (Samuelson et al., 1992; Hirose et al., 2008). Our finding that constitutive cytokinin activity, implemented via ΔDDKPtRR13 expression, disrupts adventitious root development is consistent with observations that Arabidopsis mutant lines lacking cytokinin receptors develop numerous adventitious roots and that the cytokinin-insensitive triple mutant arr1 arr10 arr12 spontaneously develops adventitious roots from the hypocotyl (Werner et al., 2003; Higuchi et al., 2004; Nishimura et al., 2004; Argyros et al., 2008). While an impact on adventitious root development was not reported, transgene-directed cytokinin biosynthesis in lateral root founder cells disrupted their development from primary roots (Laplaze et al., 2007). We speculate that in intact plants the supply of cytokinin might maintain PtRR13 in an active, phosphorylated state. We note that ΔDDKPtRR13 plants lack a visible phenotype, perhaps because the transgene-encoded protein mimics the predominant form of endogenous PtRR13 in intact plants. In the phosphorylated state, PtRR13 would be expected to transcriptionally regulate downstream targets that collectively repress adventitious root development.
Shoot excision caused a significant reduction in type-A RR transcripts in the stem base, and one possible explanation is a disruption in cytokinin supply from the roots. Cytokinin progressively declines within basal portions of cuttings during the first 24 h after shoot excision, a time period known to be critical for adventitious root development (Blakesley et al., 1985; Bollmark and Eliasson, 1986; Maldiney et al., 1986; Hausman et al., 1997). The results of gene set analysis suggested potential roles for protein phosphorylation (known to alter global protein activity or degradation processes associated with hormone action; Jonak et al., 2002; Ouaked et al., 2003; Nakagami et al., 2005; Yoo et al., 2008) and protein turnover (a mechanism for rapid removal of protein inhibitors and effectors of diverse hormone signaling and stress responses; Dreher and Callis, 2007), which, if they were to involve PtRR13 itself, may target phosphorylated PtRR13 for proteasomal degradation. Accompanied by de novo translation of PtRR13 transcripts, the PtRR13 pool might be quickly shifted to a dephosphorylated (i.e. inactive as a transcription factor) form. Accompanying the removal of cytokinin-mediated repression, adventitious root development could then proceed. The phosphorylation status of RRs affects the extent to which they are proteasomally degraded in plants (Murray et al., 2007) as well as in yeast (Sato et al., 2003), where it is thought to be a mechanism for rapid response to acute changes in osmolarity of the growth medium. Rapid acquisition of tissue capacity for adventitious root development could be similarly adaptive in woody plants, since the acquisition of competence for rooting would occur shortly after shoot detachment in coordination with cellular adaptation to desiccation and tissue repair.
Interactions between Cytokinin and Other Factors
The two most significantly up-regulated genes in the ΔDDKPtRR13 transgenic lines, COV1 and PDR9, may imply effects of cytokinin on vascular tissue formation and cross talk with auxin signaling, respectively. COV1 is an integral membrane protein that negatively regulates vascular tissue differentiation in stems (Parker et al., 2003). In Arabidopsis, COV1 mutants lack appropriate definition of vascular bundles in the stem, resulting in a continuous ring-like pattern of xylem and phloem with little interfascicular tissue (Parker et al., 2003). The phenotypes of these mutants are independent of auxin and were attributed to the action of an unknown inhibitory factor. If our hypothesis is correct, then the unknown inhibitory factor proposed by Parker et al. (2003) may be cytokinin or cytokinin dependent. We hypothesize that ΔDDKPtRR13 transcriptionally activates the Populus COV1 homolog, which interferes with the establishment of vascular continuity between root primordia and stem vascular tissue.
ΔDDKPtRR13 up-regulation of PDR9 could suggest direct effects on auxin distribution. Arabidopsis PDR9 is exclusively expressed in roots, and the protein is predicted to act as a 2,4-dichlorophenoxyacetic acid efflux pump (Ito and Gray, 2006). Recent studies by Delker et al. (2008) indicate that PDR9 is a genetic suppressor of the SCF complex mutant tir1-1 that is able to restore wild-type root growth when supplied with auxin. Similarly, in Arabidopsis, transgene-directed cytokinin accumulation in root founder cells caused disorganized cell divisions and disrupted PIN-mediated auxin gradient establishment (Laplaze et al., 2007). Our results suggest that the Populus PDR9 homolog is expressed in stems of ΔDDKPtRR13 lines and may perturb an auxin gradient required for adventitious root initiation or organization.
Ethylene synthesis and action coordinate early responses to tissue injury (Adie et al., 2007; Jackson, 2008), which are likely reflected in the up-regulation of the ethylene biosynthetic enzymes ACC synthase and ACC oxidase. A clue to how cytokinin interacts with ethylene may be found in the observation that ΔDDKPtRR13 expression reduced the transcript abundance of two TINY-like transcription factors. TINYs belong to the DREB (for dehydration-responsive element-binding protein) subfamily of AP2/ERF transcription factors. Among ERF/AP2 family members, the DREB and ERF (for ethylene-responsive element-binding factor) subfamilies participate in responses to abiotic stress, such as mechanical injury and reduced water availability (Shinozaki and Yamaguchi-Shinozaki, 2000; Lorenzo et al., 2003). TINY appears to be a positive effector of ethylene action based on its transcript induction in response to ethylene treatment and the triple response phenotype observed when TINY is overexpressed in seedlings (Wilson et al., 1996; Sun et al., 2008). Genes encoding other members of the AP2/ERF family, PLETHORA1 and PLETHORA2, and BABY BOOM are up-regulated during the actual formation of adventitious roots in culture (Imin et al., 2007). The mechanism by which ΔDDKPtRR13 or other type-B RRs reduce transcript abundance in the presence of cytokinin is not known, although Arabidopsis lines that lack type-B RRs are impaired in their normal responses to cytokinin treatments, including cytokinin down-regulated genes (Argyros et al., 2008). Our model proposes that shoot excision induces the expression of TINY-like genes (due to rapid loss of active PtRR13), reflecting a possible mechanism for integrating ethylene and cytokinin signaling.
Stages of Adventitious Root Development
Early responses to shoot excision are complex, as revealed by the differential regulation of about half of the nuclear genes within 24 h. Adventitious rooting is generally thought to progress in a stepwise manner, with discrete stages identified using a combination of hormone measurements and treatments. In vitro rooting experiments of apple (Malus domestica) microcuttings revealed that following a lag period during the initial 24 h after excision, founder cells acquired a transient state of higher sensitivity to auxin and cytokinin, since exogenous applications of auxin or cytokinin at this time strongly stimulate or inhibit root formation, respectively (De Klerk et al., 1999). Likewise, cytokinin effects on lateral root formation have been shown to be stage specific, with lateral root founder cells sensitive to cytokinin and lateral root primordia not sensitive (Laplaze et al., 2007). Consistent with these observations, we found that a significant (time point × transgene) interaction for gene expression was specific to the 24-h time point. We hypothesize that these significant effects reveal the ability of adventitious root founder cells to respond to the rhizogenic stimulus of auxin and that a low cytokinin-auxin ratio might be required for normal induction of rooting. High cytokinin availability mimicked by the constitutively active PtRR13 at 24 h after excision might disrupt this ratio. The transient effects of ΔDDKPtRR13 on downstream gene expression reinforce the stage-specific nature of cytokinin inhibition of adventitious root development.
The discrete effect of the ΔDDKPtRR13 construct on the timing of adventitious root development merits attention, because it stands in contrast to the pleiotropic effects of ARR1ΔDDK expression in Arabidopsis that includes reduced plant growth, disordered cellular proliferation around the shoot apex, and ectopic shoots on cotyledons (Sakai et al., 2001). Distinct phenotypes may reflect functional divergence of Populus PtRR13 and Arabidopsis ARR1 or reveal differences in the transformation/regeneration procedures that facilitate the recovery of lines with stronger “alleles” with the Arabidopsis floral dip method compared with the Populus tissue culture-based regeneration method. The effect of ΔDDKPtRR13 expression in delaying, but not abolishing, adventitious root development may also reflect selection of comparatively weak transgenic events that could be propagated. Alternatively, ΔDDKPtRR13 may completely disrupt one developmental pathway that is independent of alternative pathways, potentially involving diverse cell type precursors of adventitious roots (Lovel and White, 1986). If this hypothesis is correct, then this study has illuminated only one of several distinct pathways for adventitious root development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Root induction experiments were conducted in a greenhouse at ambient temperature between the months of September and December in Gainesville, Florida. Populus tremula × Populus alba INRA-clone 717-1-B4 plants were given 12 to 14 h of natural light, supplemented in the winter with artificial illumination to maintain indeterminate growth. Plants were grown in 11.4-L pots on flood benches subirrigated once daily with a nutrient solution containing Peters Professional Blend 20-10-20 fertilizer solution (adjusted to 4 mm nitrogen). When plants reached 60 to 80 cm tall, single cuttings per plant were collected, planted in 25-cm2 pots containing Fafard's-4 mix potting media, and placed under mist until harvested.
Real-Time PCR
Total RNA was isolated using a cetyltrimethylammonium bromide method (Chang et al., 1993). RNA samples were subjected to DNase treatment with RQ1 RNase-free DNase (Promega) and purified using the RNeasy Mini Kit (Qiagen). One microgram of DNA-free RNA was used to synthesize first-strand cDNAs using oligo(dT) primers and M-MLV reverse transcriptase (Promega). Gene-specific primers were designed using the Joint Genome Institute assembly for P. trichocarpa version 1.1 and are given in Supplemental Table S3. To avoid nonspecific PCR amplification, primers were designed against the most variable regions in the coding sequences using NetPrimer (Premier Biosoft International), followed by melt curve analysis to verify single products. For each biological sample (i.e. RNA extracted from an individual plant), three technical replications were performed, each using 1 μL of a 20-μL reverse transcription reaction on that template RNA. The number of biological replications (samples derived from individual plants) varied from n = 3 to n = 5 depending on the experiment, and these values are indicated in the figure legends showing the results. Gene expression was quantified using the SYBR Green kit (Stratagene) and the Mx3000P thermocycler (Stratagene) according to the manufacturer's instructions. The obtained expression values were scaled to the mean expression of the ACTIN2 and UBIQUITIN control genes.
Tissue Culture Experiments
Stems were harvested from plants grown as described to a height of 60 cm. Leaves were removed and stems were surface sterilized by four consecutive washes of 70% (v/v) 95% ethanol, 30% (v/v) bleach (5.25% sodium hypochlorite), and two washes with autoclaved deionized water. Stems were cut into 0.5-cm sections and placed on MS agar plates (0.443% [w/v] Murashige and Skoog medium, 0.01% [w/v] myoinositol, 3% [w/v] Suc, and 0.65% [w/v] phytoagar) containing the indicated cytokinin-auxin concentrations. Plates were placed in a Percival growth chamber under 12-h light/dark cycles at a constant temperature of 25°C and 75% relative humidity.
PtRR13 Plasmid Construction and Transgenic Line Generation
cDNAs encoding the full-length protein (FLPtRR13), the DDK truncation (ΔDDKPtRR13), and the RNAi fragment were amplified by PCR and cloned into the TOPO entry vector (Invitrogen). Cloned sequences were transferred to destination vectors using Gateway technology (Invitrogen) with the FLPtRR13 and ΔDDKPtRR13 cDNA sequences cloned into pZKY1 Overexpress and the RNAi sequence cloned into pZKY2 Direct. Vectors were kindly provided by Oak Ridge National Laboratories, and sequence information can be found at http://www.esd.ornl.gov/PGG/foo_vectors.htm#35 s%20Overexpression. Poplar INRA-clone 717-1-B4 was transformed via an Agrobacterium tumefaciens-mediated protocol developed by Han et al. (2000). Twenty independent transformations (lines) per construct were obtained, and explants were grown on kanamycin-selection medium. Transgenic lines were screened using PtRR13-specific primers and primers directed against the 35S promoter and octopine synthase inverted repeats (for full-length and DDK truncated lines) or the PIV2 intron (for RNAi lines).
Microarray Analysis
For these analyses, apical cuttings 14 cm in length were collected from NT and ΔDDKPtRR13 plants 60 cm in height. Cuttings were placed in 25-cm2 pots containing Fafard's-4 mix potting medium and placed on a bench with intermittent mist to prevent shoot desiccation. Samples consisting of a 5-mm section measured up from the base of the cutting (one sample per cutting) were collected at the indicated time points. Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and DNase treated in-column with the RNase-Free DNase set (Qiagen). Double-stranded cDNA was synthesized using the SuperScript Double Strand cDNA Synthesis Kit (Invitrogen) with oligo(dT) primers following the manufacturer's protocol, except that the synthesis step was extended to 16 h. Cy-3 labeling and hybridization steps were performed by NimbleGen using standard procedures. A custom-designed microarray platform was used comprising single 60-mer probes designed against 55,793 annotated gene models from the sequenced genome of P. trichocarpa. Each 60-mer probe was chosen from a group of six to seven nonoverlapping probes designed against different parts of the gene model. The probe whose value was most similar to the average of the six to seven nonoverlapping experimental probes was assumed to be the most reliable for transcript level estimation. A total of 40 microarray chips were used in these experiments: 40 chips = 2 genotypes (NT and ΔDDK) × 4 time points (0, 6, 24, and 48 h) × 5 biological replications. Signal intensities were log2 transformed and quantile normalized (Bolstad et al., 2003). Normalized signals were analyzed in SAS 9.1 (SAS Institute) using a mixed-model ANOVA with genotype and genotype × time interactions as fixed effects and biological replication as a random effect. Data are available through the Gene Expression Omnibus (accession no. GSE15049). Microarray expression data were verified for a subset of significantly regulated genes by real-time PCR, obtaining similar gene expression patterns with both analysis methods (Supplemental Fig. S5).
Gene Set Analysis
Gene set analysis was performed using the overrepresentation analysis application of the ErmineJ software (version 2.1.16), which uses the binomial approximation to the hypergeometric distribution to find gene set enrichment. The corresponding Gene Ontology (GO) annotations of the Populus genes were obtained by querying the GO annotation tool of The Arabidopsis Information Resource (http://www.arabidopsis.org/tools/bulk/go/index.jsp) with the closest Arabidopsis hit for each Populus gene. GO annotations were downloaded from the GO Web site (http://www.geneontology.org/GO.downloads.ontology.shtml). Only biological processes-related GO categories were considered in the analysis. Genes with contrasts significant at FDR < 0.5% were used for gene set enrichment analysis. Gene sets were determined significant at FDR < 10%.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Top 10 genes in the highly significant gene set categories enriched during the initial 24 h after shoot excision.
Supplemental Figure S2. Arabidopsis and Populus PtRR13 closest relatives.
Supplemental Figure S3. ΔDDKPtRR13 overexpression stimulates stem growth in culture.
Supplemental Figure S4. ARR1-, DRE-, and ERF-binding motifs in promoters of genes regulated across all time points.
Supplemental Figure S5. Microarray data verification by real-time PCR.
Supplemental Table S1. Gene set analysis of differentially regulated genes in NT during early stages of adventitious root formation.
Supplemental Table S2. Differentially regulated genes in ΔDDKPtRR13 at 24 h.
Supplemental Table S3. Primer list and gene models.
Supplementary Material
Acknowledgments
We thank Ms. Elizabeth Etherington in the laboratory of Steve Strauss for transgenic lines, Matias Kirst and Derek Drost for microarray chip design, Carolina Novaes Boaventura for help with cDNA labeling, Catherine Benedict for assistance with promoter analysis, and Barry Goldfarb, Harry Klee, and Mark Settles for helpful discussions.
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (grant no. DE–AC05–00OR22725).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John M. Davis (jmdavis@ufl.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie B, Chico JM, Rubio-Somoza I, Solano R (2007) Modulation of plant defenses by ethylene. J Plant Growth Regul 26 160–177 [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakesley D, Hall JF, Weston GD, Elliott MC (1985) Endogenous plant growth substances and the rooting of Phaseolus aureus cuttings. In M Bopp, M Knoop, BW Rademaeher, eds, Abstracts of the 12th International Conference on Plant Growth Substances. IPGSA, Heidelberg, p 87
- Bollmark M, Eliasson L (1986) Effects of exogenous cytokinins on root-formation in pea cuttings. Physiol Plant 68 662–666 [Google Scholar]
- Bollmark M, Kubat B, Eliasson L (1988) Variation in endogenous cytokinin content during adventitious root-formation in pea cuttings. J Plant Physiol 132 262–265 [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193 [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker M, van Zyl L, Liu WB, Craig D, Sederoff RR, Clapham DH, von Arnold S (2004) Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol 135 1526–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies FT, Hartmann HT (1988) The physiological basis of adventitious root formation. Acta Hortic 227 113–120 [Google Scholar]
- Davis JM, Becwar MR (2007) Developments in tree cloning. In Developments in Fibres and Fibre Treatment Series. PIRA International, Surrey, UK, p 69
- Delker C, Anja R, Gray WM, Quint M (2008) PDR9 functions as a genetic suppressor of the SCF complex mutant tir1-1. 19th International Conference on Arabidopsis Research. http://plantconferences.org/Arabidopsis2008/abstracts/finaljustabstracts.cfm?abcategory=Cell%20Biology (May 7, 2009)
- De Klerk GJ, Arnholdt-Schmitt B, Lieberei R, Neumann KH (1997) Regeneration of roots, shoots and embryos: physiological, biochemical and molecular aspects. Biol Plant 39 53–66 [Google Scholar]
- De Klerk GJ, Keppel M, Terbrugge J, Meekes H (1995) Timing of the phases in adventitious root-formation in apple microcuttings. J Exp Bot 46 965–972 [Google Scholar]
- De Klerk GJ, Van Der Krieken W, De Jong JC (1999) The formation of adventitious roots: new concepts, new possibilities. In Vitro Cell Dev Biol Plant 35 189–199 [Google Scholar]
- Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson L (1978) Effects of nutrients and light on growth and root-formation in Pisum sativum cuttings. Physiol Plant 43 13–18 [Google Scholar]
- Fehlberg V, Vieweg MF, Dohmann EMN, Hohnjec N, Puhler A, Perlick AM, Kuster H (2005) The promoter of the leghaemoglobin gene VfLb29: functional analysis and identification of modules necessary for its activation in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots. J Exp Bot 56 799–806 [DOI] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ (2005) Cytokinin signaling. Curr Opin Plant Biol 8 518–525 [DOI] [PubMed] [Google Scholar]
- Fett-Neto AG, Fett JP, Goulart LWV, Pasquali G, Termignon RR, Ferreira AG (2001) Distinct effects of auxin and light on adventitious root development in Eucalyptus saligna and Eucalyptus globulus. Tree Physiol 21 457–464 [DOI] [PubMed] [Google Scholar]
- Guerrero JR, Garrido G, Acosta M, Sanchez-Bravo J (1999) Influence of 2,3,5-triiodobenzoic acid and 1-N-naphthylphthalamic acid on indoleacetic acid transport in carnation cuttings: relationship with rooting. J Plant Growth Regul 18 183–190 [DOI] [PubMed] [Google Scholar]
- Han KH, Meilan R, Ma C, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19 315–320 [DOI] [PubMed] [Google Scholar]
- Hausman JF (1993) Changes in peroxidase-activity, auxin level and ethylene production during root-formation by poplar shoots raised in vitro. Plant Growth Regul 13 263–268 [Google Scholar]
- Hausman JF, Evers D, Kevers C, Gaspar T (1997) Internal controls of root induction in poplar shoots raised in vitro. Angew Bot 71 104–107 [Google Scholar]
- Hausman JF, Kevers C, Gaspar T (1995) Auxin-polyamine interaction in the control of the rooting inductive phase of poplar shoots in vitro. Plant Sci 110 63–71 [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59 75–83 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T (1998) Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA 95 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Wu T, Rolfe BG (2007) Factors involved in root formation in Medicago truncatula. J Exp Bot 58 439–451 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49 47–57 [DOI] [PubMed] [Google Scholar]
- Ito H, Gray WM (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB (2008) Ethylene-promoted elongation: an adaptation to submergence stress. Ann Bot (Lond) 101 229–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Okresz L, Bogre L, Hirt H (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5 415–424 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54 605–627 [DOI] [PubMed] [Google Scholar]
- Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevers C, Hausman JF, Faivre-Rampant O, Evers D, Gaspar T (1997) Hormonal control of adventitious rooting: progress and questions. J Appl Bot/Angew Bot 71 71–79 [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Delaruelle C, Martin D, Encelot N, Martin F (2003) The poplar root transcriptome: analysis of 7000 expressed sequence tags. FEBS Lett 542 37–41 [DOI] [PubMed] [Google Scholar]
- Kuroha T, Kato H, Asami T, Yoshida S, Kamada H, Satoh S (2002) A trans-zeatin riboside in root xylem sap negatively regulates adventitious root formation on cucumber hypocotyls. J Exp Bot 53 2193–2200 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49 387–400 [PubMed] [Google Scholar]
- Liu JH, Reid DM (1992) Adventitious rooting in hypocotyls of sunflower (Helianthus annuus) seedlings. IV. The role of changes in endogenous free and conjugated indole-3-acetic-acid. Physiol Plant 86 285–292 [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovel PH, White J (1986) Anatomical changes during adventitious root formation. In MB Jackson, ed, New Root Formation in Plants and Cuttings. Martinus Nijhoff Publishers, Dordrecht, The Netherlands, pp 111–140
- Lund ST, Smith AG, Hackett WP (1996) Cuttings of a tobacco mutant, rac, undergo cell divisions but do not initiate adventitious roots in response to exogenous auxin. Physiol Plant 97 372–380 [Google Scholar]
- Maldiney R, Pelese F, Pilate G, Sotta B, Sossountzov L, Miginiac E (1986) Endogenous levels of abscisic-acid, indole-3-acetic-acid, zeatin and zeatin-riboside during the course of adventitious root-formation in cuttings of Craigella and Craigella lateral suppressor tomatoes. Physiol Plant 68 426–430 [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Vaclavikova K, Miyawaki K, Kakimoto T (2008) Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci USA 105 20027–20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T (1998) His-Asp phosphotransfer signal transduction. J Biochem (Tokyo) 123 555–563 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52 89–118 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10 339–346 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom AC, Eliasson L (1991) Levels of endogenous indole-3-acetic-acid and indole-3-acetylaspartic acid during adventitious root-formation in pea cuttings. Physiol Plant 82 599–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917 [DOI] [PubMed] [Google Scholar]
- Ouaked F, Rozhon W, Lecourieux D, Hirt H (2003) A MAPK pathway mediates ethylene signaling in plants. EMBO J 22 1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G, Schofield R, Sundberg B, Turner S (2003) Isolation of COV1, a gene involved in the regulation of vascular patterning in the stem of Arabidopsis. Development 130 2139–2148 [DOI] [PubMed] [Google Scholar]
- Quesada T, Li Z, Dervinis C, Li Y, Bocock PN, Tuskan GA, Casella G, Davis JM, Kirst M (2008) Comparative analysis of the transcriptomes of Populus trichocarpa and Arabidopsis thaliana suggests extensive evolution of gene expression regulation in angiosperms. New Phytol 180 408–420 [DOI] [PubMed] [Google Scholar]
- Ramirez-Carvajal GA, Morse AM, Davis JM (2008) Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol 177 77–89 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood SB, Kalischuk AR, Polzin ML, Braatne JH (2003) Branch propagation, not cladoptosis, permits dispersive, clonal reproduction of riparian cottonwoods. For Ecol Manage 186 227–242 [Google Scholar]
- Sakai H, Aoyama T, Bono H, Oka A (1998) Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol 39 1232–1239 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294 1519–1521 [DOI] [PubMed] [Google Scholar]
- Samuelson ME, Eliasson L, Larsson CM (1992) Nitrate-regulated growth and cytokinin responses in seminal roots of barley. Plant Physiol 98 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Kawahara H, Toh-e A, Maeda T (2003) Phosphorelay-regulated degradation of the yeast Ssk1p response regulator by the ubiquitin-proteasome system. Mol Cell Biol 23 6662–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C, Kennedy SJ, Harvey BMR (1992) Adventitious root formation in hypocotyl cuttings of Picea sitchensis (Bong) Carr.: the influence of plant growth regulators. New Phytol 120 453–457 [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3 217–223 [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yu JP, Chen F, Zhao TJ, Fang XH, Li YQ, Sui SF (2008) TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J Biol Chem 283 6261–6271 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T (1998) Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett 429 259–262 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Kieber JJ (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13 85–92 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11 184–191 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmulling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26 369–376 [DOI] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G (1996) A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q (2004) Isolation of genes associated with adventitious root development in Populus. Master's thesis. North Carolina State University, Raleigh, NC
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.