Abstract
Drug-resistant bacteria have caused serious medical problems in recent years, and the need for new antibacterial agents is undisputed. Transglycosylase, a multidomain membrane protein essential for cell wall synthesis, is an excellent target for the development of new antibiotics. Here, we determined the X-ray crystal structure of the bifunctional transglycosylase penicillin-binding protein 1b (PBP1b) from Escherichia coli in complex with its inhibitor moenomycin to 2.16-Å resolution. In addition to the transglycosylase and transpeptidase domains, our structure provides a complete visualization of this important antibacterial target, and reveals a domain for protein–protein interaction and a transmembrane helix domain essential for substrate binding, enzymatic activity, and membrane orientation.
Keywords: antibacterial development, antibiotic resistance, membrane protein structure, peptidoglycan synthesis, protein–protein interaction
In the last decade, the prevalence and occasional outbreaks of drug-resistant bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE), have posed appalling hurdles in the treatment of bacterial infections (1, 2). New antibacterial agents are, as a result, in desperate demand to combat these pernicious antibiotic-resistant problems that can otherwise cause life-or-death struggles.
Bacteria cell wall is a mesh-like structure of cross-linked peptidoglycan, which is essential to scaffold the cytoplasmic membrane and to maintain structural integrity of the cell (3). Cell wall synthesis at the membrane surface is mainly carried out by the membrane-bound enzymes, transpeptidases and transglycosylases, and inhibitors of the transpeptidase are among the most popular antibiotics in clinical use today (3).
Escherichia coli PBP1b is a bifunctional transglycosylase, also known as peptidoglycan glycosyltransferase or murein synthase. It contains a transmembrane (TM) helix, 2 enzymatic domains [transglycosylase (TG) and transpeptidase (TP)] (4), and a domain composed of ≈100 aa residues between TM and TG with unknown structure and functionality (Fig. 1B). For >50 years, TP has been the main target for 2 most important classes of antibiotics: β-lactams (e.g., penicillin and methicillin) and glycopeptides (e.g., vancomycin). Not too long after they were introduced, resistant bacteria had emerged rapidly and caused serious medical problems. In contrast, resistant strains against moenomycin, the only natural inhibitor to TG from Streptomyces, have rarely been found. The development of new antibiotics against TG has been highly anticipated (5), and not until recently have the molecular structures of TG been available, even with the TM structure undefined.
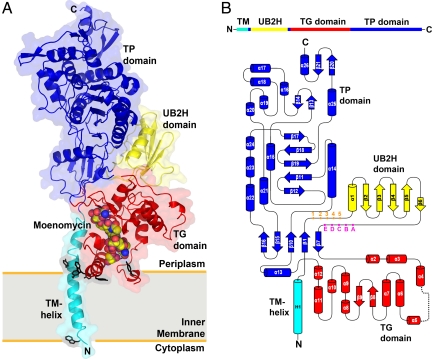
Fig. 1.
Overall structure and topology of E. coli PBP1b. (A) The crystal structure of PBP1b is represented as a ribbon diagram. The TM, UB2H, TG, and TP domains are color coded in cyan, yellow, red, and blue, respectively. Moenomycin is represented as van der Waals spheres. Tryptophan and tyrosine residues located near the water–membrane interfaces are shown in black sticks. The proposed membrane location is indicated by a gray rectangle. All figures of 3D structural representations were made with PyMOL (www.pymol.org). (B) The 1D and 2D topology of E. coli PBP1b are color-coded as in A. The numbering (1–5) at the N terminus of UB2H domain and the alphabet (A–E) at the C terminus of UB2H domain are markers for the locations used in the combinatorial domain deletion strategy (see SI Materials and Methods).
Two crystal structures of transglycosylase, a bifunctional transglycosylase from S. aureus (referred to as SaPBP2) and a transglycosylase domain from Aquifex aeolicus (referred to as AaPGT), have been determined recently with their TM domain or TM and TP domains removed, respectively (6–8). These structures revealed critical interactions between protein and moenomycin and served as good platforms for antibiotic development. We have previously demonstrated that the TM helix domain is important for the binding between E. coli PBP1b and moenomycin (9). In addition, we found that the full-length PBP1b also showed a substantially higher TG enzymatic activity than a TM truncated counterpart [supporting information (SI) Fig. S1]. Therefore, in this study, our purified full-length PBP1b possessing a similar enzymatic activity [kcat is 3.14 ± 0.236 s−1, Km is 18.3 ± 4.05 μM and kcat/Km is (1.74 ± 0.3) × 105 M−1s−1] to previous studies (10) was chosen for structure determination by X-ray crystallography.
Results and Discussion
Overall Structure of PBP1b–Moenomycin Complex.
The crystal structure of E. coli PBP1b in complex with moenomycin was solved at 2.16-Å resolution (Fig. 1A). Our protein construct includes amino acid residues 58–804, containing TM, an unknown domain, TG, and TP domains. In the process to obtain protein crystals with good X-ray diffracting quality, the solubilization, purification, and crystallization of the PBP1b required the use of multiple detergents, including N-dodecyl-β-d-maltopyranoside, N-decyl-β-d-maltopyranoside, and N-dodecyl-N,N-dimethylamine-N-oxide. By using a multiwavelength anomalous dispersion (MAD) approach with crystals from selenomethionine-labeled proteins, the phase information was obtained to generate a protein electron density map. The structure was built from residues 66–800, except 2 loop regions with absent electron density (residues 249–267 and 399–406), and was refined to good quality with Rwork and Rfree values of 20.6% and 25.1%, respectively (Table S1).
At the amino terminus, the TM domain consists of a single long helix, encompassing residues 66–96. The residues 83–88 in the TM helix are in close vicinity to residues 292–296 in the TG domain (Fig. S2A). Further examination of the corresponding residues in the TM helix and TG domains among homologous transglycosylases revealed a moderate conservation of hydrophobic amino acid residues, suggesting that similar interactions between the TM and TG domains in other transglycosylases are possible (Fig. S2B).
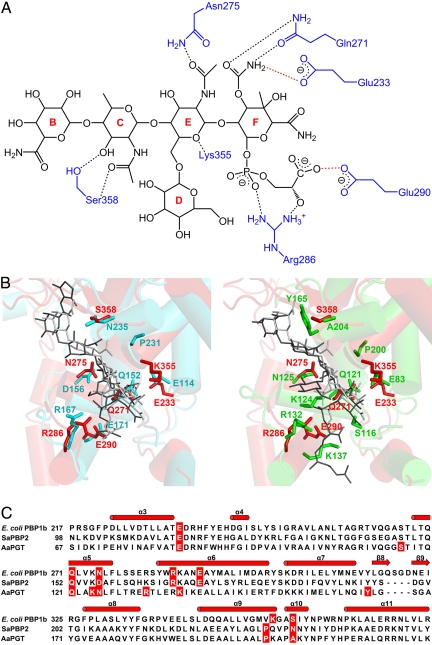
The overall fold of the TG domain in our structure, in complex with moenomycin, is highly similar to the 2 available transglycosylase structures from SaPBP2 and AaPGT (Fig. S3). The RMSD is 1.53 Å for 145 Cα atoms between TG domains from E. coli PBP1b and SaPBP2, and 1.46 Å for 143 Cα atoms between E. coli PBP1b and AaPGT. However, the residues involved in potential interactions with moenomycin (defined with distance cutoff of 3.2 Å) showed similarities and differences in these transglycosylase structures (Fig. 2 B and C). The resemblance between our structure and SaPBP2 may explain the observation that transglycosylases from E. coli and S. aureus share comparable binding affinity to moenomycin (9). In addition, the interacting residues of the TG domain around the E ring, the F ring, the phosphate group, and the carboxylate group of moenomycin are more conserved than the interacting residues with the remaining parts (Fig. 2A and Fig. S4). The conserved interacting residues in the binding pocket of transglycosylases can be considered as the most critical region to be studied in the process of antibiotic design. Our result is in agreement with the previous findings to define the minimal pharmacophore in moenomycin, in which the EF-ring phosphoglycerate portion together with either the C or the D ring forms critical interactions with proteins (8). Although A. aeolicus (11), like E. coli, was classified as Gram-negative bacterium, the interaction pattern with moenomycin in AaPGT showed differences from our structure and SaPBP2 (Fig. 2 B and C). It is noted that a positively charged Lys-137 residue in AaPGT can form an interaction with the carboxylate group of the phosphoglycerate in moenomycin, the corresponding interacting residue is, however, a negatively charged residue glutamic acid in both our structure and that of SaPBP2. The mutagenesis study also confirmed that the activity of AaPGT was nearly abolished after mutating Lys-137 to alanine (8). However, Lys-287 of E. coli PBP1b (corresponding to Lys-137 in AaPGT) seems to be less critical for the activity of peptidoglycan synthesis in E. coli, because the Lys-287 to alanine mutant still possessed 63% of wild-type activity, whereas the Glu-290-to-glutamine mutant displayed only 2% of wild-type activity (12). The fact that the corresponding lysine residues act differently in AaPGT and E. coli PBP1b is interesting. It will be worthwhile to conduct a thorough study to understand the role of this residue and how it affects the activity of the homologous proteins from 2 Gram-negative bacteria.
Fig. 2.
Residues interacting with moenomycin and structure-based sequence alignment of transglycosylases. (A) The potential hydrogen-bonding interactions (distance cutoff 3.2 Å) between E. coli PBP1b and moenomycin are shown as dashed lines in black. The interactions between the putative active sites (E233 and E290) and moenomycin proposed in ref. 6 are shown as dashed lines in red. (B) Comparison of moenomycin-binding modes between E. coli PBP1b and SaPBP2 (Left); between E. coli PBP1b and AaPGT (Right). TG and residues of E. coli PBP1b, SaPBP2, and AaPGT are shown in red, cyan, and green, respectively. Moenomycin are shown in light gray (for E. coli PBP1b) and dark gray (for SaPBP2 and AaPGT). (C) Sequences of TG from E. coli PBP1b, SaPBP2, and AaPGT are aligned according to their secondary structure elements. Residues forming the potential interaction with moenomycin are shaded in red.
The crystal structure of E. coli PBP1b represents a structural platform of transglycosylase, in particular for Gram-negative bacterial pathogens, for the development of antibiotics. Together with the 2 structures of transglycosylases from Gram-positive (SaPBP2) and thermophilic bacteria (AaPGT), addition of our structure completes the structural scope of transglycosylases across the bacterial spectrum.
Several compounds with molecular weights smaller than moenomycin have been reported to compete with the moenomycin and bind directly to the transglycosylase domain (9, 13). Although the inhibition efficiencies of these compounds to bacteria are lower than that of moenomycin, the structural information between these compounds and transglycosylase can be studied via molecular modeling or by X-ray crystallography using current E. coli PBP1b structure as a template for structure-based drug design.
Structure and Function of UvrB Domain 2 Homolog.
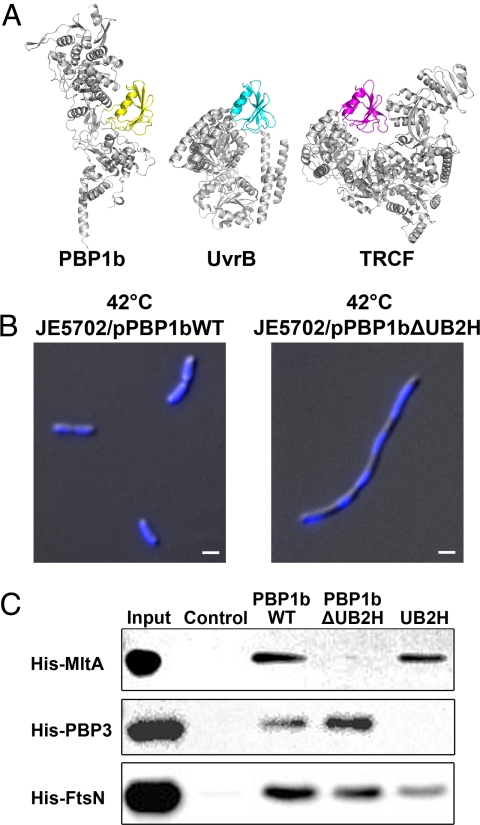
In addition to the TM, TG, and TP domains that are commonly found in bifunctional transglycosylases, an unexpected domain was observed in our crystal structure (Fig. 1A). This domain, comprising residues 109–200, folds with a 5-antiparallel-stranded β-sheet (β2-β6) and 1 α-helix (α1) and forms more interactions with the TP domain (with buried surface area of 630.17 Å2) and less interactions with the TG domain (313.01 Å2). In comparison with the structure of SaPBP2, which shows no direct interactions between the TG and TP domain, addition of this extra domain makes E. coli PBP1b a more compact structure. By using Dali search (14), this domain was found to be structurally homologous to domains in UvrB (RMSD is 1.8 Å for 82 Cα atoms, with 24% sequence identity) and TRCF (transcription-coupled repair factor) (RMSD is 1.6 Å for 82 Cα atoms, with 14% sequence identity) (Fig. 3A). UvrB and TRCF are critical components of nucleotide excision repair (NER) system in DNA damage repairs (15, 16). The corresponding homologous domains in UvrB and TRCF specifically bind to a domain in UvrA in a competitive manner to coordinate the functionality of bacterial NER system. Based on the highly similar fold, we referred to this domain as UB2H (UvrB domain 2 homolog) domain.
Fig. 3.
UB2H domain, its deletion phenotype and pull-down assay. (A) The structurally homologous domains from PBP1b, UvrB (PDB ID code 2NMV), and TRCF (PDB ID code 2EYQ) are shown in yellow, cyan, and magenta, respectively. The nonhomologous parts of these proteins are colored in gray. (B) Morphological differences and DNA segregation between the wild type and UB2H-truncated (PBP1bΔUB2H) strains are shown in differential interference contrast microscopy combined with DAPI staining images. (Scale bar, 1 μm.) (C) Wild-type PBP1b, deletion mutant PBP1bΔUB2H, and UB2H domain only were coupled onto the CNBr-activated Sepharose, and their binding ability to MltA, PBP3, and FtsN was examined.
UB2H deletion mutants of PBP1b were then constructed to gain insights into their functions (Fig. 1B). The mutant PBP1bΔUB2H (Glu-114 to Gln-191 were deleted) provided on a plasmid was found to be able to rescue a PBP1b-deletion/PBP1a-temperature-sensitive E. coli host strain, JE5702, at 42 °C (see Tables S2 and S3). At this temperature, both PBP1b and PBP1a are not functional; thus, the survival of JE5702 indicated that PBP1bΔUB2H can complement the enzymatic functions of TG and TP. However, we observed that UB2H deletion caused an aberrant growth rate (Fig. S5) and an elongated cell shape containing multiple copies of DNA in JE5702 (Fig. 3B). In E. coli, PBP1b interacts with different proteins during the course of cellular growth and division. For example, MltA, the membrane-bound lytic transglycosylase, interacts with PBP1b and participates in the peptidoglycan processing during cell elongation and cell division (17); PBP3, a transpeptidase catalyzing the formation of cross-linked peptidoglycan, interacts with PBP1b for peptidoglycan synthesis during cell division (18); FtsN, the essential cell division protein that can interact with PBP1b, may play a role in stabilizing the divisome during cell division (19). We hence tested the idea that whether UB2H serves as the binding domain in PBP1b for the interaction with different binding partners. Pull-down assay was performed, and the result showed that PBP1bΔUB2H lost the binding ability with protein MltA, but not PBP3 or FtsN (Fig. 3C). In addition, the UB2H domain alone possessed binding ability similar to wild-type PBP1b, indicating that the UB2H domain participates in the interaction with MltA.
The UB2H domain exists only in bifunctional transglycosylases of some Gram-negative bacteria (183 of 988 bacterial genomes in the National Center for Biotechnology Information database as of November 2008). The protein–protein interaction between PBP1b and MltA established by the pull-down assay can be via a third protein MipA involved in bacterial cell-wall synthesis (17). A previous study, however, has reported that a mltA deletion did not affect the morphology of E. coli (20). Thus, the aberrant morphology caused by UB2H deletion may not have a direct correlation to MltA. Other UB2H-interacting proteins can be involved in this morphological change.
It is surprising that PBP1b, like UvrB and TRCF, can also interact with UvrA in the pull-down assay (Fig. S6). Whether the UB2H domain participates in the regulation between DNA repair and/or synthesis and cell wall formation during the bacterial cell cycle awaits future investigation.
Orientation of PBP1b in the Membrane.
The presence of the TM helix in our structure allowed us to postulate the orientation of the E. coli PBP1b molecule in lipid bilayers. It is commonly accepted that tryptophan and tyrosine residues have a higher frequency to be found at the lipid–water interface in membrane proteins (21). We examined all plausible tryptophan and tyrosine residues in the TG domain and TM helix and found a plane consisting of tryptophan and tyrosine residues that might be associated with lipids (Fig. 1A). As a result, the established membrane orientation made the bottom of the TG domain partially embedded in lipid bilayers. Also, based on this model, the C terminus of the TM helix (residues 88–96; ≈2 helical turns) is not embedded in the membrane.
To further test the validity of this membrane orientation model, we performed molecular dynamics (MD) simulations (22). In the MD simulations, the proposed orientation of E. coli PBP1b in lipid bilayers was observed to be energetically stable (Movie S1 and Movie S2). The stable orientation recurred in different MD simulations, strengthened our proposed model, and also suggested that the contact between TM and TG is not an artifact caused by crystal packing.
The E. coli TP domain closely resembles the corresponding region in the SaPBP2 structure; however, the relative orientation between the TG and TP domains are dissimilar between our structure and SaPBP2 (Fig. 4A) (6, 23). Despite the discrepancy, we considered all different orientations plausible because of the possibly inherent flexibility of a hinge region (23). Different crystal structures can simply represent different structural states of the bifunctional transglycosylases. The changes in the relative orientation of SaPBP2 had been proposed to be correlated to the regulation of TG activity (23).
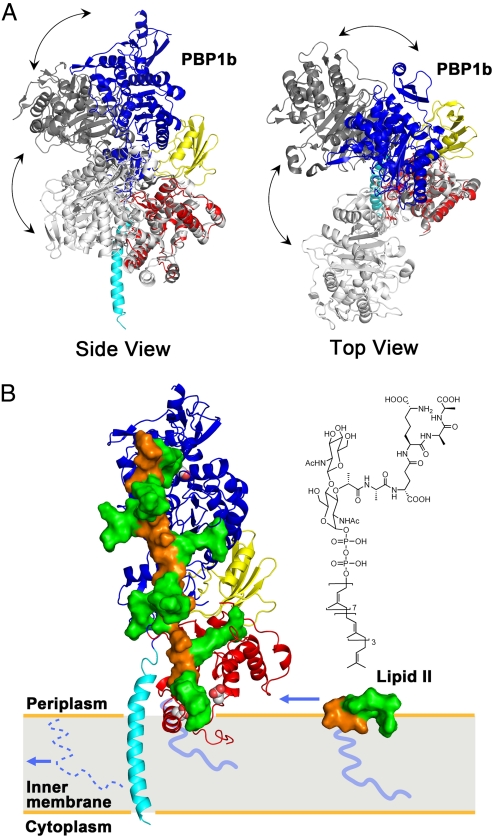
Fig. 4.
Interdomain flexibility and a model of peptidoglycan synthesis. (A) E. coli PBP1b and 2 SaPBP2 conformers (6, 23) were indicated by colored, light gray (PDB ID code 3DWK), and dark gray (PDB ID code 2OLV), respectively. The TG domain of SaPBP2 was superimposed onto the TG of E. coli PBP1b. Side view (Left) and top view (Right) of the comparison reveals possible flexibility of a hinge region between TG and TP domains. (B) Active sites of TG and TP were shown by van der Waals spheres. The disaccharide, pentapeptide, and lipid tail of lipid II and peptidoglycan were shown in orange surface, green surface, and blue line, respectively. A single strand of the proposed peptidoglycan model (25) was docked onto the structure of E. coli PBP1b, with lipid IV portion replacing moenomycin. The incoming lipid II, of which the chemical structure is shown on top, diffuses in the plane of the membrane. After the TG reaction, the lipid moiety of this lipid II is kept as the membrane anchor, whereas the original lipid tail (shown as dotted line) is recycled. The polymerized peptidoglycan grows perpendicularly to the membrane and toward the TP domain, where the cross-linking reaction of the pentapeptides takes place (see also Movie S3).
We have previously observed that the binding affinity of moenomycin to E. coli PBP1b is TM domain dependent (9). However, no direct interaction between moenomycin and the TM helix was observed in our crystal structure. Furthermore, removal of the TM helix does not affect the structure of TG domain in the binding site, when comparing our structure and SaPBP2 in their moenomycin binding pockets (Fig. 2A). We therefore suggest that the TM helix simply stabilizes the protein–membrane interaction, and the resulting orientation limits the interaction between PBP1b and moenomycin or lipid II in the membrane in a 2D lateral diffusion fashion. Removal of TM may destabilize the protein–membrane interaction, thus affecting moenomycin or lipid II binding to TG. Indeed, stable protein–membrane interaction has been reported recently to be crucial for the normal function of some membrane proteins, and hence it has been suggested to be a target for drug discovery (24).
A Model for Peptidoglycan Synthesis.
Lovering et al. (6, 23) have elegantly proposed that the moenomycin molecule in the binding site of transglycosylase structurally mimics lipid IV, the dimerized peptidoglycan from 2 molecules of lipid II, and suggested a mechanism of peptidoglycan elongation where the growing glycan chain acts as an acceptor for the nucleophilic attack with lipid II as a donor. Recently, the architecture of peptidoglycan has been modeled based on the NMR structure of a lipid IV derivative (25). Using the proposed peptidoglycan model, we docked a single strand of the peptidoglycan onto the structure of E. coli PBP1b, with the lipid IV portion replacing moenomycin (Fig. 4B). We noted that the distance (65.8 Å) between the active-site residues of the TG and TP domains in our structure corresponds well to the distance (67.1 Å) between the reaction sites on the peptidoglycan. In this model, the surface of PBP1b in contact with peptidoglycan is largely composed of loops, which are possibly flexible and capable of accommodating the polymerizing peptidoglycan (Movie S3). Therefore, the membrane orientation of PBP1b established by the transmembrane helix implies that its product, peptidoglycan, can be synthesized perpendicularly to the membrane surface. In contrast to the conventional views that cell wall consists of layers of cross-linked peptidoglycans with their glycan backbones lying parallel to the membrane surface, our structure and model suggest the possibility of vertical orientation of peptidoglycans at the membrane surface, at least when they are initially synthesized. We, however, acknowledge the intrinsic flexibility of both the bifunctional transglycosylases and their product peptidoglycan strands. As the peptidoglycans grow longer, the complete polymerized and cross-linked cell wall may have different appearance where the peptidoglycan strands lie parallel to the membrane surface or even form a coiled-coil cable as demonstrated by recent electron microscopic studies (26, 27).
Materials and Methods
Cloning, Expression, and Purification.
Purified full-length PBP1b degraded readily into a slightly smaller protein. After N-terminal sequencing accompanying with molecular weight determination by MALDI-TOF Mass spectrometry, we identified the stable region containing amino acids 58–804. PBP1b (residues 58–804) was amplified from E. coli genomic DNA and was cloned into the expression vector pET15b (EMD Biosciences) at the NdeI and BamHI restriction enzyme sites. BL21(DE3) E. coli host cells transformed with expression vectors were grown at 37 °C until OD600 reached 0.6, and protein expression was induced with 1 mM IPTG for 3 h. Cell pellets were resuspended in 20 mM Tris (pH 8.0) and 300 mM NaCl and broken by Microfluidizer (Microfluidics). Recombinant protein containing an N-terminal (His)6 tag was solubilized with 20 mM N-dodecyl-β-d-maltopyranoside (DDM; Anatrace) and purified by nickel chelation chromatography in the presence of 1 mM DDM. The N-terminal (His)6 tag was removed by thrombin cleavage (Sigma–Aldrich) at room temperature overnight. Tag-free PBP1b was further purified by using a Superdex 200 size-exclusion column (GE LifeSciences) in 20 mM Tris (pH 8.0), 300 mM NaCl, and 4.5 mM N-decyl-β-d-maltopyranoside (Anatrace). Peak fractions were concentrated, and the detergent was exchanged to 0.28 mM N-dodecyl-N,N-dimethylamine-N-oxide (Anatrace) by using Amicon Ultra filter units (Millipore). The purified stable PBP1b construct (residues 58–804) showed similar TG enzymatic activity to that of full-length PBP1b. (When the TG activity of stable PBP1b construct was normalized to 100%, the TG activity of full-length PBP1b was 84%). The selenomethionine (SeMet; Anatrace) -labeled protein was expressed by using minimal medium supplemented with selenomethionine and purified as described above.
Crystallization, Data Collection, and Structure Determination.
PBP1b–moenomycin complex was cocrystallized by using sitting-drop vapor diffusion at 16 °C. Crystals were obtained by mixing 12 mg/mL protein containing additional 1.4 mM moenomycin with the same volume of reservoir solution containing 1.2 M sodium formate. For cryoprotection, crystals were gradually transferred into 3 M sodium formate and flash-frozen in liquid nitrogen.
The native dataset was collected at beamline BL44XU of the Japan Synchrotron Radiation Research Institute (Hyogo, Japan) and SeMet derivative datasets were collected at beamline BL13B1 at National Synchrotron Radiation Research Center (Hsinchu, Taiwan). All data were indexed, integrated, and scaled with HKL2000 (28). MAD method was used to collect anomalous datasets from SeMet derivatized crystals. The selenium locations (22 of 24) and structure phase were obtained by SOLVE (29). Density modification by solvent flattening was carried out by RESOLVE (29) to generate an interpretable 3.4-Å electron density map. The model was manually built by using COOT (30). This model was subsequently refined to 2.16-Å resolution by using the native dataset. Phenix (31) and Refmac (32), using the same Rfree set (5% of reflections), were used at the refinement process, with the group atomic displacement parameter, and TLS options turned on. TLS groups were determined by the TLSMD (33) server.
Morphological Aberration and DNA Segregation.
The morphological differences and DNA Segregation between JE5702/pPBP1bWT and JE5702/pPBP1bΔUB2H were examined by microscope. The 30 °C overnight-cultured transformants were diluted 1:100 with the fresh LBC liquid media (LB containing 0.5% NaCl and 12.5 μg/mL chloramphenicol), and then cultured with 0.5 mM IPTG at 42 °C. Bacterial cells were used at stationary growth phase (OD600 = 1–1.1) and spotted on the glass slides. The photograph of bacteria morphology was taken at a magnification of 10 × 100 by phase contrast microscope (BX15; Olympus). The bacterial cells (50 μl, OD600 = 1–1.4) for DAPI staining were incubated by 1 μl DAPI solution (0.25 mg/ml). After being fixed with 80% methanol and then washed by PBS, the cells were spotted on glass slides. The differential interference contrast and fluorescence images were taken at a magnification of 10 × 100 using an upright microscope (DM 6000B, Leica).
Pull-Down Assay.
Purified PBP1b variants (including full-length PBP1b, PBP1bΔUB2H, and UB2H only) and UvrB were coupled with CNBr-activated Sepharose (GE LifeSciences). A control Sepharose without proteins was treated in the same procedure. His tagged MltA, PBP3, FtsN, and UvrA were overexpressed in BL21(DE3) cells at 37 °C for 3 h. Overexpressed cells were extracted in lysis buffer [10 mM Tris (pH 6.8), 10 mM sodium maleate (Sigma–Aldrich), 10 mM MgCl2, and 2% Triton X-100] at 4 °C overnight and then centrifuged at 20,000 × g for 30 min at 4 °C. The resulting supernatant, the detergent-solubilized membrane fractions, was incubated with PBP1b variants-coupled Sepharose beads at 4 °C overnight. Beads were washed with lysis buffer 30 times column volume (CV) and further washed with 5 CV lysis buffer supplemented with 150 mM NaCl. The interacting protein was eluted with lysis buffer containing 1 M NaCl. The eluate was analyzed by Western blotting with anti-His antibody.
Supplementary Material
Acknowledgments.
We sincerely thank T.-H. Chang, S.-D. Tsen, C.-C. Chou, E. Yamashita, A.-S. Yang, and T.-J. Cheng for very insightful discussions; S. Mobashery for providing the coordinates of peptidoglycan model; National BioResource Project, National Institute of Genetics, Japan, for E. coli strain JE5702. X-ray diffraction data were collected at BL13B1 of the National Synchrotron Radiation Research Center, Taiwan, and BL44XU of the SPring-8, Japan. This work was supported by Academia Sinica and National Health Research Institute, Taiwan, Grant NHRI-EX95-9515SC (to C.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3FWL and 3FWM).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904030106/DCSupplemental.
References
- 1.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 2.Payne DJ. Microbiology. Desperately seeking new antibiotics. Science. 2008;321:1644–1645. doi: 10.1126/science.1164586. [DOI] [PubMed] [Google Scholar]
- 3.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halliday J, McKeveney D, Muldoon C, Rajaratnam P, Meutermans W. Targeting the forgotten transglycosylases. Biochem Pharmacol. 2006;71:957–967. doi: 10.1016/j.bcp.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 6.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, et al. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc Natl Acad Sci USA. 2007;104:5348–5353. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan Y, et al. Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem Biol. 2008;3:429–436. doi: 10.1021/cb800078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng TJ, et al. Domain requirement of moenomycin binding to bifunctional transglycosylases and development of high-throughput discovery of antibiotics. Proc Natl Acad Sci USA. 2008;105:431–436. doi: 10.1073/pnas.0710868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz B, Markwalder JA, Seitz SP, Wang Y, Stein RL. A kinetic characterization of the glycosyltransferase activity of Eschericia coli PBP1b and development of a continuous fluorescence assay. Biochemistry. 2002;41:12552–12561. doi: 10.1021/bi026205x. [DOI] [PubMed] [Google Scholar]
- 11.Berezovsky IN, Shakhnovich EI. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci USA. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terrak M, et al. Importance of the conserved residues in the peptidoglycan glycosyltransferase module of the class A penicillin-binding protein 1b of Escherichia coli. J Biol Chem. 2008;283:28464–28470. doi: 10.1074/jbc.M803223200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovering AL, Gretes M, Strynadka NC. Structural details of the glycosyltransferase step of peptidoglycan assembly. Curr Opin Struct Biol. 2008;18:534–543. doi: 10.1016/j.sbi.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truglio JJ, et al. Interactions between UvrA and UvrB: The role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 17.Vollmer W, von Rechenberg M, Holtje JV. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- 18.Bertsche U, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 19.Muller P, et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 20.Lommatzsch J, Templin MF, Kraft AR, Vollmer W, Holtje JV. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl E, Sansom MS. Membrane proteins: Molecular dynamics simulations. Curr Opin Struct Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Lovering AL, De Castro L, Strynadka NC. Identification of dynamic structural motifs involved in peptidoglycan glycosyltransfer. J Mol Biol. 2008;383:167–177. doi: 10.1016/j.jmb.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Segers K, et al. Design of protein membrane interaction inhibitors by virtual ligand screening, proof of concept with the C2 domain of factor V. Proc Natl Acad Sci USA. 2007;104:12697–12702. doi: 10.1073/pnas.0701051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meroueh SO, et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan L, Chen SY, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci USA. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie AG, et al. Automation of the collection and processing of X-ray diffraction data—A generic approach. Acta Crystallogr D. 2002;58:1924–1928. doi: 10.1107/s0907444902016864. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.