Abstract
The degradation of the RpoS (σS) subunit of RNA polymerase in Escherichia coli is a prime example of regulated proteolysis in prokaryotes. RpoS turnover depends on ClpXP protease, the response regulator RssB, and a hitherto uncharacterized “turnover element” within RpoS itself. Here we localize the turnover element to a small element (around the crucial amino acid lysine-173) directly downstream of the promoter-recognizing region 2.4 in RpoS. Its sequence as well as its location identify the turnover element as a unique proteolysis-promoting motif. This element is shown to be a site of interaction with RssB. Thus, RssB is functionally unique among response regulators as a direct recognition factor in ClpXP-dependent RpoS proteolysis. Binding of RssB to RpoS is stimulated by phosphorylation of the RssB receiver domain, suggesting that environmental stress affects RpoS proteolysis by modulating RssB affinity for RpoS. Initial evidence indicates that lysine-173 in RpoS, besides being essential of RpoS proteolysis, may play a role in promoter recognition. Thus the same region in RpoS is crucial for proteolysis as well as for activity as a transcription factor.
Keywords: Sigma-S, sigma factor, Clp protease, two-component system, stress
RpoS or σS is a sigma subunit of RNA polymerase that is present at very low levels in exponentially growing Escherichia coli cells. In response to various stress conditions, RpoS is strongly up-regulated and activates 50–100 genes, which results in multiple stress resistance and other physiological and morphological alterations (for recent reviews, see refs. 1 and 2). The control of the cellular RpoS content occurs at the levels of rpoS transcription and translation as well as RpoS proteolysis. In exponentially growing cells, RpoS is a very unstable protein (with a half-life of approximately 2 min), but RpoS is stabilized in response to carbon starvation or shift to high osmolarity, high temperature, or low pH (3–7).
Some trans-acting factors involved in the control of RpoS proteolysis have been described. The relevant protease is ClpXP (8), a complex ATP-dependent protease consisting of proteolytic (ClpP) and chaperone (ClpX) subunits that form a proteasome-like assembly (9, 10). In addition, a two-component-type response regulator, RssB (also termed SprE or MviA), is essential for RpoS degradation (3, 11, 12). The C-terminal output domain of RssB is unlike that of any other response regulator and also does not show similarity to other proteins of known function. So far, its molecular function has remained unknown. RpoS degradation in vivo is positively modulated by acetyl phosphate, which readily phosphorylates the D58 residue in the RssB receiver domain in vitro (13).
In addition to these trans-acting factors, a “turnover element” within RpoS is required for its proteolysis. The turnover element confers instability upon other proteins, e.g., RpoS-β-galactosidase hybrid proteins (5, 8). Thus, it may be functionally comparable to proteolysis-promoting elements in various eukaryotic proteins, such as the “destruction box” or D-box (14). The exact location of the turnover element in RpoS and its molecular function have not been demonstrated.
To understand the recognition of RpoS as a substrate for the proteolytic machinery, we have localized the turnover element by site-directed mutagenesis and have used the resulting mutants to characterize the molecular function of this element both in vivo and in vitro. From the data presented here, we conclude that the turnover element in RpoS (with K173 as an essential amino acid) is a binding site for the response regulator RssB. Moreover, binding of RssB to RpoS is modulated by phosphorylation of the RssB receiver domain.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions.
The strains used in this study are derivatives of MC4100 (15), into which various alleles of rpoS, rssB, and clpP were introduced by P1 transduction (16). Specifically, these alleles are rpoS359∷Tn10 (17), rssB∷Tn10 (11), rssB∷cat (kindly provided by F. Moreno, Hospital Ramón y Cajal, Madrid), and clpP1∷cat (18). Strains carrying reporter gene fusions to RpoS-dependent genes were the following: RO151(MC4100 carrying csi-5(osmY)∷lacZ(λplacMu55); ref. 19), RH95 (MC4100 carrying λMAV103∷ bolAp1∷lacZYA; ref. 20), DW12 (MC4100 carrying csi-12(csiD)∷lacZ(λplacMu15); ref. 19), LB83 (MC4100 carrying otsB∷lacZ(λplacMu55), with the otsB fusion derived from FF1112 (21), and their respective rpoS, rssB, and clpP derivatives obtained as mentioned above. These strains were used as recipients for plasmids derived from pBAD18, which express different variants of RpoS under the control of the pBAD promoter (see below).
Cultures were grown at 37°C under aeration in LB medium or in minimal medium M9 (16) supplemented with 0.4% glycerol. Ampicilline (100 μg ml−1) was used to grow plasmid-containing strains. For selecting transductants, various antibiotics were added as recommended (16). Growth was monitored by measuring the OD at 578 nm.
Introduction of Single Amino Acid Substitutions in RpoS by in Vitro Mutagenesis.
For the isolation of rpoS mutations by site-directed mutagenesis, an EcoRI–HindIII fragment carrying the rpoS structural gene was obtained from pRL40.1 (which contained rpoS under the control of the ptac promoter; ref. 4) and cloned into the multiple cloning site of pBAD18 directly downstream of the pBAD promoter, which yielded pRpoS18. For replacement of a rpoS-internal fragment containing the putative turnover element, unique XmaIII and Eco721 restriction sites were used (with the latter being directly downstream of the codons to be mutated). PCR fragments (376 bp) were isolated with an upstream primer containing the XmaIII site (5′-GATTGGTTATTCACCACTGTTAACGGCCG-3′) and several downstream primers, which all contain the Eco721 site as well as small alterations that replace single codons in rpoS for those present at the corresponding positions in rpoD (K173E: 5′-GGACAACTCACGTGCGGTTCGCAGGTAAACGTTCAGCTCCTCTACGATGTGAATCG-3′; E174T: 5′-GGACAACTCACGTGCGGTTCGCAGGTAAACGTTCAGGGTCTTTACGATGTGAATCG-3′; V177K: 5′-GGACAACTCA CGTGCGGTTCGCAGGTACTTGTTCAGCTCCTTTA CGATGTGAATCG-3′; Y178L: 5′-GGACAACTCACGTGCGGTTCGCAGGAGAACGTTCAGCTCCTTTACGAT GTGAATCG-3′; K173E/V177L: 5′-GGACAACTCACGTGCGGTTCGCAGGTACTTGTTCAGCTCCTCTACGAT GTGAATCG-3′; with relevant codons underlined, and altered nucleotides given in bold). PCR fragments were digested with XmaIII and Eco721 and were used to replace the corresponding fragment in equally digested pRpoS18. The PCR-derived parts in the resulting RpoS mutant plasmids were sequenced.
SDS/PAGE and Immunoblot Analysis.
Sample preparation for SDS/PAGE (22) and immunoblot analysis were performed as described (4). Polyclonal sera against RpoS and RssB or the penta-His antibody (Qiagen, Hilden, Germany), a goat anti-rabbit IgG alkaline-phosphatase conjugate (Sigma), and a chromogenic substrate (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium; Boehringer Mannheim) were used for visualization of RpoS and RssB bands.
Pulse Labeling of Cells and Immunoprecipitation.
Pulse labeling of cells with l-[35S]methionine and immunoprecipitation of RpoS has been described (4). Exponentially growing cells were harvested at an OD578 of 0.6 and were pulse-labeled for 2 min. Chase times varied between 0.4 and 30 min. For immunoprecipitation, a polyclonal serum against RpoS was used. Protein bands on autoradiographs were densitometrically quantitated. The intensity of bands representing RpoS was calculated relative to the intensity of bands representing stable proteins that weakly crossreacted with the antisera used.
Overexpression and Purification of RpoS and RssB Proteins.
rssB was cloned onto a S-tag-TRX-His6 vector (Novagen) as described (13). The rpoS coding region was cloned into the pQE30 vector (Qiagen): a BamHI/HindIII-digested PCR fragment obtained with pRL 40.1 (4) as the template and the primers 5′-GACGTGGATCCAGTCAGAATACGCTGAAAGTTC-3′ and 5′-ACACGTAAGCTTTCATTACTCGCGGTAACAGCGCTTCG-3′ was used (with underlined nucleotides indicating additions to or deviations from the wild-type sequence introduced to create BamHI and HindIII restriction sites respectively, as well as an additional stop codon in the second primer). The insert of the resulting plasmid (pRpoS30), which encodes His6-RpoS, was sequenced.
For the construction of the RpoSK173E and RpoSE174T overexpressing plasmids, internal fragments containing these mutations were obtained from the corresponding pRpoS18 derivatives by digestion with XmaIII and Eco721 and were cloned into pRpoS30 (equally XmaIII/Eco721 digested) to replace the rpoS wild-type fragment. The PCR-derived parts of the resulting rpoS mutant plasmids were sequenced.
S-tag-TRX-His6-RssB and His6-RpoS were overexpressed and purified as described (13). When necessary, the S-TRX-His6 tag was cleaved off from RssB by using enterokinase according to the directions given by the manufacturer (Novagen).
Purified proteins were dialyzed overnight against storage buffer (50 mM Tris⋅HCl, pH 7.5/50% glycerol/0.1 mM EDTA/0.1 mM DTT/250 mM NaCl) at 4°C for RpoS and at room temperature in the presence of 1 mM DTT for RssB. Protein concentrations were determined by using the Bio-Rad protein assay.
In Vitro Interaction of RpoS and RssB.
Fifty microliters of S-protein agarose (Novagen) was equilibrated in binding buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/5 mM MgCl2) and incubated with 400 μl of BSA (30 mg⋅ml−1) for 30 min at 4°C. The agarose was pelleted and washed with 1 ml of binding buffer. Equimolar amounts (0.5 nmol) of His6-RpoS and S-TRX-His6-RssB, with or without 50 mM acetyl phosphate (Sigma), were incubated for 1 hr at room temperature in 25 mM Tris⋅HCl, pH 7.5/25% glycerol/0.05 mM EDTA/0.05 mM DTT/125 mM NaCl (inverting the test tube every few minutes) and then added to the agarose. After incubation for 30 min at room temperature the agarose was pelleted and the supernatant was removed. After washing the S-protein agarose three times with 500 μl of binding buffer, 50 μl of 0.2 M sodium citrate (pH 2) was added, incubated for 5 min, and after centrifugation, 20 μl of the supernatant was run on a 10% SDS gel. Proteins were subjected to immunoblot analysis by using a Penta His antibody (Qiagen).
Analogous binding experiments also were performed with His6-RpoS and tag-free RssB with Ni-nitrilotriacetic acid agarose (Ni-NTA; Qiagen) in the buffer described above. A buffer containing 50 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 10 mM or 250 mM imidazole was used for washing and elution, respectively.
Computational Analyses.
Secondary structure prediction of proteins was performed with the PHD neural network systems (23–25) by using the http://www.embl-heidelberg.de/predictprotein website.
RESULTS
Isolation of Mutations in the Putative Turnover Element in RpoS.
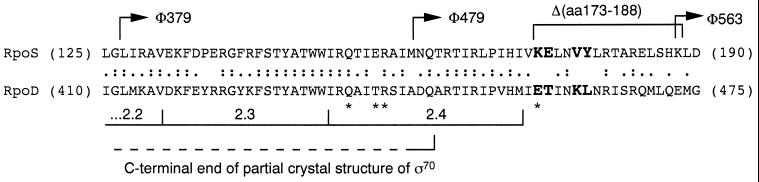
The analysis of rpoS∷lacZ fusions indicated a location of the turnover element somewhere in the central part of RpoS. A hybrid protein containing the first 126 aa of RpoS (encoded by rpoS379∷lacZ) was shown to be stable, whereas a hybrid with the fusion joint after E247 (rpoS742∷lacZ) exhibited regulated proteolysis just as RpoS itself (5). β-Galactosidase activities of hybrid proteins with the reporter inserted after M159 and H187 suggested stability of the former and degradation of the latter (8). In addition, the data obtained with a deletion between V172 and K188 suggested that the deletion eliminated turnover (8). These observations localized an essential part of the turnover element somewhere between M159 and H187 and also mean that the turnover element is close to or may even overlap with region 2.4, which is involved in recognition of −10 promoter regions (26, 27) (Fig. 1).
Figure 1.
Alignment of the turnover element-containing region of RpoS with the corresponding region of RpoD. Partial amino acid sequences of RpoS and RpoD between the positions indicated are given. Amino acids at positions that were mutated in the present study are given in larger and bold capital letters. Regions important for transcription initiation as well as RpoD residues that are included in a partial crystal structure (34) are indicated. Amino acids in RpoD with a direct role in promoter recognition are indicated by ∗. Arrows indicate fusion joints of translational rpoS∷lacZ fusions (5, 8). In addition, the position of an in-frame deletion that interferes with RpoS proteolysis is shown (8).
RpoS is the sigma factor most closely related to the vegetative RpoD (σ70) (28). The two sigmas recognize very similar promoter sequences, which is reflected in a high degree of homology in their 2.4 regions (Fig. 1). On the other hand, the two proteins differ completely with respect to stability, because RpoD is not subject to proteolysis, i.e., obviously it does not contain a turnover element. Together with the findings mentioned above, this indicated that the turnover element in RpoS probably is located in a region between M159 and H187 that is not conserved between RpoS and RpoD. Indeed, a short region of divergence is found directly downstream of region 2.4. Clearly different amino acids are K173 (corresponding to E458 in RpoD), E174 (T459 in RpoD), V177 (K462 in RpoD), and Y178 (L463 in RpoD) (Fig. 1). These amino acids in RpoS were substituted by the corresponding ones present in RpoD. For both wild-type proteins as well as in the mutant RpoS proteins with single amino acid exchanges (K173E, E174T, V177K, and Y178L), these regions are strongly predicted to be in α-helical conformation. Nevertheless, because E458 and K462 in RpoD would be predicted to be almost at the same side of an α-helix, which may be stabilized by the opposite charge of these two amino acids, we also isolated the K173E/V177K double mutation.
Mutagenesis was performed by using a derivative of the pBAD18 vector (29), into which the rpoS structural gene was cloned directly downstream of the arabinose-inducible pBAD promoter. Because of medium copy number and tight regulation of pBAD, this construct (pRpoS18) produces RpoS levels that correspond to approximately 30% of wild-type levels when cultures are grown in the absence of inducer (data not shown). RpoS expressed from pRpoS18 also exhibited physiological regulation with respect to RssB and ClpXP, i.e., proteolysis. During exponential growth, RpoS levels as well as the expression of the RpoS-dependent gene osmY were similarly enhanced in rssB and clpP mutants, when RpoS was expressed from the wild-type gene in the chromosome or from pRpoS18 under the control of the pBAD promoter. In addition, RpoS exhibited stationary phase induction when expressed from pRpoS18 (data not shown). Translational regulation of RpoS, however, is altered with the pRpoS18 construct, because the extended 5′ untranslated regions present in wild-type rpoS mRNA, which have been implicated in secondary structure formation and translational control (30–32), are not present in pRpoS18. Therefore, mutations within the rpoS part on pRpoS18 that affect RpoS levels most likely influence RpoS proteolysis.
Effects of Single Amino Acid Exchanges on RpoS Levels and Proteolysis: K173 Is Crucial for RpoS Turnover.
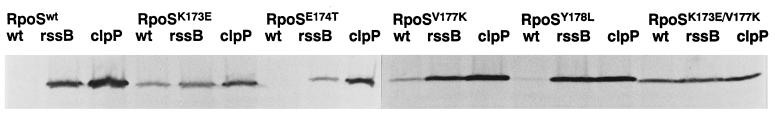
Cellular RpoS levels were compared in exponentially growing cultures of strains expressing either wild-type or mutant RpoS (heretofore termed RpoSwt, RpoSK173E, RpoSE174T, RpoSV177K, RpoSY178L, and RpoSK173E/V177K). Strains proficient in RpoS proteolysis or proteolysis-deficient rssB and clpP mutants were analyzed. Fig. 2 demonstrates that RpoSK173E, RpoSK173E/V177K, and to a lesser extent RpoSV177K accumulate in growing cells even when the proteolytic machinery is present, whereas levels of RpoSE174T and RpoSY178L appear as low as that of RpoSwt, i.e., at the limit of detection. For the K173E and the double-mutant forms of RpoS, hardly any further increase was observed in an rssB mutant, while the cellular level of RpoSV177K was further elevated (Fig. 2). In stationary phase, all variants of RpoS were present at similarly high levels (data not shown).
Figure 2.
Cellular levels of various mutant forms of RpoS in exponential phase. Cells expressing wild-type or mutant versions of RpoS from pBAD18 in wild-type, rssB∷cat, or clpP∷cat backgrounds were grown in minimal medium M9 with 0.4% glycerol. Immunoblots are shown demonstrating cellular RpoS levels during exponential phase (at an OD578 of 0.6).
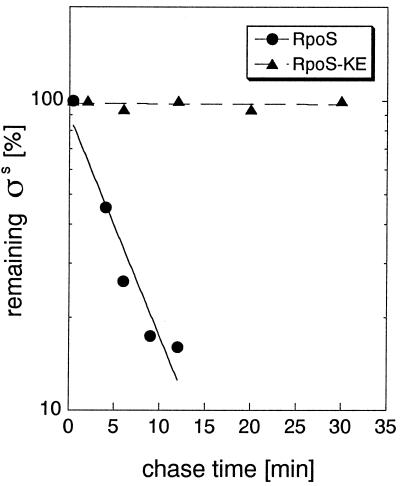
To see whether increased levels of these mutant forms of RpoS reflected defects in proteolysis, their rate of degradation was determined in pulse–chase experiments with exponentially growing cells (Fig. 3 and data not shown). RpoS half-lives were calculated from the pulse–chase data and are summarized in Table 1. The K173E as well as the K173E/V177K mutations were found to virtually eliminate RpoS turnover. The E174T substitution increased RpoS half-life approximately 2-fold, which seems not enough for RpoS levels to be clearly detectable by immunoblotting (Fig. 2). V177K increased RpoS half-life 3-fold, consistent with RpoSV177K being weakly detectable on the immunoblot (Fig. 2).
Figure 3.
RpoSK173E is not subject to proteolysis. Cells expressing RpoSwt (●) or RpoSK173E (▴) from pBAD18 were grown in minimal medium M9 with 0.4% glycerol. RpoS degradation was demonstrated by pulse–chase labeling and immunoprecipitation of exponential phase samples (harvested at an OD578 of 0.6) as detailed in Materials and Methods.
Table 1.
In vivo half-lives of mutant RpoS
| RpoS | Half-life, min |
|---|---|
| Wild type | 4.6 |
| K173E | — (stable) |
| E174T | 10 |
| V177K | 13 |
| Y178L | 4.5 |
| K173E/V177K | — (stable) |
We conclude that a single amino acid in RpoS, K173, is absolutely essential for RpoS proteolysis, and thus probably represents the core amino acid of the turnover element. E174 and V177 appear to play auxiliary roles, whereas Y178 is unimportant for RpoS degradation.
The Response Regulator RssB Directly Interacts with the Turnover Element in RpoS in a Phosphorylation-Dependent Manner.
With the hypothesis in mind that RssB could act as a recognition factor for RpoS proteolysis, we tested a putative direct interaction between RssB and RpoS in vitro and the effect of the K173E mutation thereon. RpoSwt, RpoSK173E, and RpoSE174T were purified as His6-tagged proteins. For RssB, a S-thioredoxin(TRX)-His6-tagged derivative was purified (thioredoxin improves solubility of RssB and if required, the complete tag can be cleaved off after purification by the use of an enterokinase cleavage site). Because the two proteins carry different tags, either one can be bound to the respective affinity materials, and retention and coelution of the other protein can be tested.
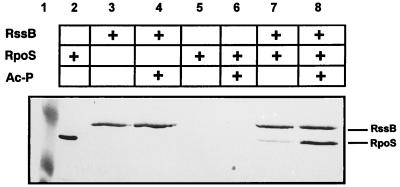
Fig. 4 demonstrates retention and coelution of RpoS on S-protein agarose to which S-TRX-His6-RssB was adsorbed. RpoS did not bind to S-protein agarose in the absence of S-TRX-His6-RssB. Interaction was found to be strongly stimulated by the addition of acetyl phosphate (Fig. 4). Because acetyl phosphate efficiently phosphorylates the D58 residue in the receiver domain of RssB (13), this finding indicates that phosphorylation of RssB improves binding of RpoS. Similarly, acetyl phosphate-stimulated interaction also was observed for His6-RpoS bound to Ni-chelating material (Ni-nitrilotriacetic acid) and RssB (from which the S-TRX-His6-tag had been cleaved off). Moreover, RssB did not interact with another His-tagged protein (His6-NtrC), indicating that retention and coelution of RssB and RpoS was the result of specific interaction (data not shown).
Figure 4.
Coelution of RssB and RpoS from S-protein agarose in the presence and absence of acetyl phosphate. Equimolar amounts (0.5 nmol) of purified S-TRX-His6-RssB and/or His6-RpoS were incubated for 1 hr at room temperature in the presence or absence of 50 mM acetyl-phosphate. Proteins adsorbed to S-protein agarose (added for another 30 min) were washed and eluted with sodium citrate (pH 2). Proteins were separated by SDS/PAGE and visualized by immunoblotting using Penta-His antibodies. Lane 2 shows purified His6-RpoS. Size standard proteins (49.5 and 32 kDa) are shown in lane 1.
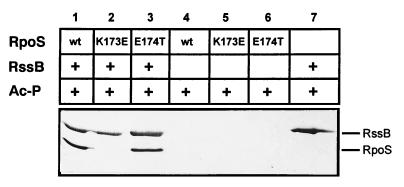
We then asked whether the K173E mutation in RpoS, which in vivo eliminates proteolysis, had any effect on the interaction between RssB and RpoS in vitro. When the ability to bind to S-TRX-His6-RssB on S-protein agarose was compared for His-tagged RpoSwt, RpoSK173E, and RpoSE174T, the K173E mutation resulted in a complete loss of interaction (Fig. 5). The E174T exchange, however, which hardly affects RpoS turnover in vivo, also did not affect interaction between RpoS and RssB. Similar results were obtained with the His-tagged RpoS variants bound to Ni-nitrilotriacetic acid and tag-free RssB (data not shown).
Figure 5.
RpoSK173E does not interact with RssB adsorbed to S-protein agarose. Equimolar amounts (0.5 nmol) of S-TRX-His6-RssB and His6-RpoSwt, His6-RpoSK173E, or His6-RpoSE174T (lanes 1–3) were incubated with 50 mM acetyl phosphate, adsorbed to S-protein agarose, washed, eluted, and visualized as described in the legend to Fig. 4. As controls, either S-TRX-His6-RssB (lane 7) or the different His6-RpoS proteins alone (lanes 4–6) were subject to the same treatment.
We conclude that K173 in RpoS is crucial for in vivo proteolysis as well as for in vitro binding of RpoS to RssB, and that therefore the molecular function of the turnover element in RpoS is that of a binding site for the response regulator RssB. Moreover, binding of RssB to RpoS is stimulated by phosphorylation of the RssB receiver domain. Thus, RssB serves as a phosphorylation-modulated recognition factor in the proteolysis of RpoS.
In Addition to Its Role in Proteolysis, K173 in RpoS Contributes to Transcription at RpoS-Dependent Promoters.
The turnover element in RpoS, and especially K173, is located immediately downstream of the −10 promoter-recognizing region 2.4 (Fig. 1). We therefore tested whether the mutations in the turnover element also affect RpoS activity as a transcription factor. Because under certain conditions the binding of RssB to RpoS also can affect RpoS activity (ref. 33, and G.B. and R.H.-A., unpublished work), and RssB binding is altered for some of the mutant variants of RpoS isolated in the present study, these experiments were done in RssB-deficient genetic backgrounds.
LacZ fusions to the RpoS-dependent genes osmY, bolA, csiD, and otsB were chosen as reporters to monitor in vivo activity of RpoS. In parallel, RpoS levels were determined. Because levels of the various RpoS mutants did not vary by more than approximately 10% (data not shown), the β-galactosidase activities obtained with RpoS-dependent reporter fusions directly reflect relative in vivo activities of the respective RpoS variants. Measurements were performed 2 hr after entry into stationary phase with strains that express RpoSwt, RpoSK173E, RpoSE174T, RpoSV177K, or RpoSY178L.
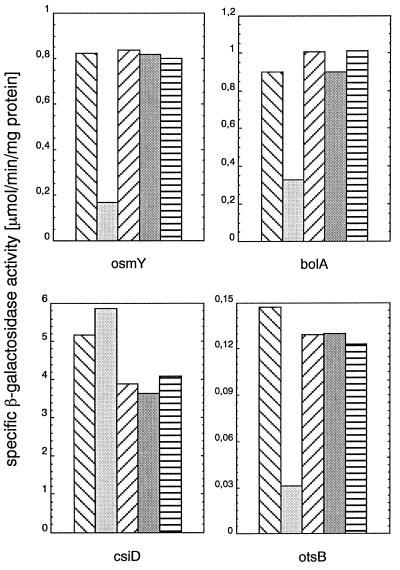
We observed that the K173E mutation differentially affected the expression of the genes tested. Although the expression of osmY, bolA, and otsB was clearly reduced, csiD expression was not affected (Fig. 6). The other mutations in RpoS did not significantly alter transcriptional activity. These findings can be taken as evidence that K173 in RpoS is also important for transcription, perhaps in promoter recognition, because the effects observed are promoter specific. Moreover, the observation that RpoSK173E has wild-type activity in the expression of csiD also eliminates the possibility that its resistance to proteolysis may be the result of general denaturation and/or aggregation.
Figure 6.
The K173E mutation in RpoS differentially affects the expression of various RpoS-dependent genes. Strains carrying rssB∷cat, rpoS∷Tn10, and lacZ fusions in the RpoS-controlled genes osmY, bolA, csiD, and otsB, which express different RpoS variants from pBAD18-derived plasmids, were grown in LB. Two hours after entry into stationary phase, specific β-galactosidase activities were determined. In each panel representing one of the reporter gene fusions, the bars represent strains carrying the following RpoS variants (from left to right): RpoSwt, RpoSK173E, RpoSE174T, RpoSV177K, and RpoSY178L. Values given are averages of two independent cultures, each of which was sampled in triplicate.
DISCUSSION
The cellular level of the global regulator RpoS (σS) is controlled by rapid alterations in its rate of proteolysis in response to changing environmental conditions (1). For this regulated proteolysis, at least three components are absolutely essential: ClpXP protease (8), the response regulator RssB (3, 11, 12), and a hitherto uncharacterized cis-acting turnover element located somewhere between M159 and H187 in RpoS (5, 8) (see Fig. 1 for relevant RpoS sequence).
In the present study, we have localized the turnover element by demonstrating that a single amino acid, lysine-173, is essential for RpoS proteolysis. The K173E mutation (E is the amino acid present at this position in the naturally stable RpoD homolog of RpoS) is sufficient to completely eliminate RpoS proteolysis (Fig. 3, Table 1). In addition, we have identified E174 and V177 as amino acids that optimize RpoS degradation, but are not absolutely essential (Table 1). We therefore propose that the [K173/E174-V177] element is the core of the turnover element in RpoS. Although these amino acids are crucial for RpoS degradation, it is not yet clear whether they are also sufficient. Experiments in which this small patch of amino acids will be engineered into surface-exposed positions of other originally stable proteins will answer this question.
Unfortunately, the partial crystal structure available for RpoD (34) ends shortly upstream of the region, which in RpoS contains the turnover element (Fig. 1). However, the region downstream of H170 is strongly predicted to be in α-helical conformation. If so, the [K173/E174–V177] element would be located at one face of this α-helix, which, considering its function, should be surface-exposed. The [K173/E174–V177] element also is located directly downstream of region 2.4 in RpoS, which in RNA polymerase-bound sigma factors is the site of interaction with the core −10 promoter region (26, 27, 35). Not only its sequence but also this location of the turnover element in RpoS is unlike that of any recognition site in other ClpXP substrates, which usually are positioned at the C termini (36). The RpoS turnover element is therefore a unique proteolysis-promoting element.
Here, we demonstrate that this element, with K173 in particular, serves as a binding site for the response regulator RssB (Figs. 4 and 5). As RssB is essential for RpoS proteolysis in vivo, this finding indicates a role of RssB as a direct recognition factor for RpoS. RssB may be RpoS specific, because it is not required for proteolysis of another ClpXP substrate, λO protein (33). It appears that so far only one other example of a specific direct recognition factor promoting proteolysis of an unstable protein has been described in prokaryotes. This factor is the MecA protein in Bacillus subtilis, which, in a complex with ClpC, binds the competence-inducing transcriptional regulator ComK and thereby initiates ComK degradation by the ClpCP protease (37, 38). The site of interaction with MecA in ComK, i.e., a cis-acting signal such as the turnover element in RpoS, has not been identified. Unlike RssB, MecA is not a response regulator, and the two factors do not exhibit any sequence similarity. Despite extensive analyses of regulated proteolysis in eukaryotic cells, comparable specific recognition factors (e.g., for the recognition of the destruction box) are still elusive (39). In the degradation of the heat shock sigma factor RpoH (σ32), the DnaK chaperone machine acts as recognition factor. Yet, DnaK is not RpoH specific but binds to a large number of different substrates, i.e., denatured proteins, a property that the heat shock system elegantly uses for signal transduction (40, 41).
Taken together, RpoS proteolysis is the result of a specific recognition factor, RssB, that binds to the turnover element in RpoS and presents RpoS, which by itself would not be a substrate for proteolysis, to the ClpXP machinery. Its function in RpoS proteolysis makes RssB unique among all known response regulators, most of which are DNA-binding regulatory proteins (42). The use of a response regulator as a recognition factor allows RpoS proteolysis to be controlled by environmental signals. Rapid phosphorylation of D58 in the RssB receiver domain previously has been shown in vitro with acetyl phosphate as a phosphodonor. The observation that acetyl phosphate-free mutants have increased RpoS half-life (13), as well as the finding that phosphorylation of RssB improves binding of RpoS (Fig. 4) indicate that phosphorylated RssB is the active form that promotes RpoS proteolysis. This finding suggests that environmental stresses that inhibit RpoS proteolysis trigger the dephosphorylation of RssB. As a consequence, RssB affinity for RpoS would be reduced and RpoS would be stabilized. In contrast to this effect on the activity of RssB in the RpoS system, proteolysis of RpoH is controlled by sequestration of the DnaK chaperone system (40, 41). In the control of ComK proteolysis mentioned above, recognition by MecA/ClpC is disrupted by yet another factor, the quorum-sensing peptide ComS (38). It is striking that in these three examples of regulated proteolysis in prokaryotes the nature of the recognition factors as well as the mechanisms of their regulation are completely different.
Finally, this study provides evidence that K173 in RpoS also may play a role in RpoS-mediated transcription initiation. RpoSK173E exhibits a promoter-specific defect in the expression of certain RpoS-dependent genes that suggests that K173 could be involved in promoter recognition (Fig. 6). In fact, the corresponding amino acid in RpoD (E458) has been implicated in the recognition of extended −10 promoter regions, and the part of RpoD containing E458 has been termed region 2.5 (43). A model for interaction between regions 2.4/2.5 and promoter sequences downstream of position −20 was presented recently (44). In the case of RpoS, genetic evidence indicates that interaction with the −10 region is especially important, such that for some promoters contacts in the −35 region are dispensible (45). Future studies will have to show whether K173 as the core of region 2.5 in RpoS can directly interact with certain nucleotides in an extended −10 region of RpoS-controlled promoters.
Acknowledgments
We thank Verena Weiss and Ralf Peist for advice in biochemical experiments and Felipe Moreno and Susan Gottesman for bacterial strains. Part of this study was carried out in the laboratories of Winfried Boos, whose support is gratefully acknowledged. Financial support was provided by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm “Regulatory Networks in Bacteria”), the State of Baden-Württemberg (Landesforschungspreis), and the Fonds der Chemischen Industrie.
References
- 1.Hengge-Aronis R. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1497–1512. [Google Scholar]
- 2.Loewen P C, Hengge-Aronis R. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 3.Bearson S M D, Benjamin W H, Jr, Swords W E, Foster J W. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange R, Hengge-Aronis R. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 5.Muffler A, Traulsen D D, Lange R, Hengge-Aronis R. J Bacteriol. 1996;178:1607–1613. doi: 10.1128/jb.178.6.1607-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muffler A, Barth M, Marschall C, Hengge-Aronis R. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takayanagi Y, Tanaka K, Takahashi H. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 8.Schweder T, Lee K-H, Lomovskaya O, Matin A. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 10.Grimaud R, Kessel M, Beuron B, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 11.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 12.Pratt L A, Silhavy T J. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouché S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 14.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:1132–1138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 15.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 17.Lange R, Hengge-Aronis R. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 18.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 19.Weichart D, Lange R, Henneberg N, Hengge-Aronis R. Mol Microbiol. 1993;10:407–420. [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giaever H M, Styrvold O B, Kaasen I, Strøm A R. J Bacteriol. 1988;170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 24.Rost B, Sander C, Schneider R. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 25.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 26.Siegele D A, Hu J C, Walter W A, Gross D A. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 27.Waldburger C, Gardella T, Wong R, Susskind M M. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 28.Gross C A, Chan C L, Lonetto M A. Philos Trans R Soc London B. 1996;351:475–482. doi: 10.1098/rstb.1996.0045. [DOI] [PubMed] [Google Scholar]
- 29.Guzman L M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 32.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou A N, Gottesman S. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malhotra A, Severinova E, Darst S A. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 35.Dombroski A J. J Biol Chem. 1997;272:3487–3494. [PubMed] [Google Scholar]
- 36.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 37.Turgay K, Hamoen L W, Venema G, Dubnau D. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- 38.Turgay K, Hahn J, Burghoorn J, Dubnau D. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamano H, Tsurumi C, Gannon J, Hunt T. EMBO J. 1998;17:5670–5678. doi: 10.1093/emboj/17.19.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bukau B. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 41.Craig E A, Gross C A. Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 42.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 43.Barne K A, Bown J A, Busby S J W, Minchin S D. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bown J A, Owens J T, Meares C F, Fujita N, Ishihama A, Busby S J W, Minchin S D. J Biol Chem. 1999;274:2263–2270. doi: 10.1074/jbc.274.4.2263. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka K, Kusano S, Fujita N, Ishihama A, Takahashi H. Nucleic Acids Res. 1995;23:827–834. doi: 10.1093/nar/23.5.827. [DOI] [PMC free article] [PubMed] [Google Scholar]