Abstract
Bacterial microcompartments (BMCs) are specialized organelles that use proteinaceous membranes to confine chemical reaction spaces. The ethanolamine ammonialyase microcompartment of Escherichia coli represents such a class of cytosolic organelles that enables bacteria to survive on small organic molecules such as ethanolamine as the sole source for carbon and nitrogen. We present here the crystal structure of the shell protein EutL at 2.2-Å resolution. With 219 residues, it is the largest representative of this BMC's shell proteins. In the crystal, EutL forms a trimer that exhibits a hexagonally shaped tile structure. The tiles arrange into a tightly packed 2D array that is likely to resemble the proteinaceous membrane of the intact BMC. In contrast to other BMC shell proteins, which have only 1 pore per tile, EutL exhibits 3 pores per tile, thereby significantly increasing the overall porosity of this protein membrane. Each of the individual pores is lined with negatively charged residues and aromatic residues that are proposed to facilitate passive transport of specific solutes. The characteristic shape of the hexagonal tile, which is also found in the microcompartments of carbon-fixating bacteria, may present an inherent and fundamental building unit that may provide a general explanation for the formation of differently sized microcompartments.
Keywords: metabolosome, carboxysome, bacterial organelle
Most cellular and subcellular compartments are enclosed by semipermeable membranes that maintain specialized environments for many biochemical reactions. The lipid bilayer structure of the membranes serves as a barrier to prevent solutes from freely diffusing in or out of the organelle's lumen, despite its elastic and liquid dynamic properties. In most cases, organelle-specific proteins facilitate the controlled uptake or expulsion of solutes. In contrast to lipidic membrane-enclosed compartments, some bacteria have evolved compartments that are entirely enclosed by proteins. In the literature, these organelles are commonly referred to as metabolosomes, microcompartments, carboxysomes, or polyhedral bodies. We will here refer to them as bacterial microcompartments (BMCs).
BMCs encapsulate specific enzymes that are used for the fixation of carbon dioxide or the catabolism of small organic compounds. For example, Escherichia coli and Salmonella typhimurium have evolved these organelles for the efficient degradation of ethanolamine (EA) or propanediol (PD), respectively (1–4). In both cases, the function of the BMCs is to recycle these compounds and feed intermediate products such as acetyl-CoA back into the citric acid cycle. Consequently, BMCs are thought to enable bacteria to survive in specialized ecological niches because of their ability to live off PD or EA as the sole source for carbon, nitrogen, and energy (1).
One of the most fascinating aspects of these supramolecular assemblies is the formation of a selectively permeable shell structure that separates the lumen of the compartment from the cytosol. Recent studies suggest that specific shell proteins assemble into a semipermeable membrane (1, 2). Thus far, no lipids were found to be associated with the shell. Genetic analyses of the EA using operon (eut-operon) have revealed 17 genes that are directly or indirectly involved in the catabolism of EA (3). Among the gene products are EutA, EutB, EutC, EutD, EutE, and EutR, which are required for its degradation and the proteins EutK, EutL, EutM, EutN, and EutS, which are thought to be involved in the structural assembly of the microcompartment. Only the proteins EutK, EutM, and EutN exhibit some homology to other carboxysomal proteins and have therefore been suggested to be involved in the formation of the BMC shell structure. We report here the crystal structure of the protein EutL of the E. coli EA ammonialyase microcompartment. With 219 aa, EutL is the largest of the Eut BMC shell proteins. In contrast to EutK, EutM, and EutN, this protein exhibits weak homology only to the shell proteins EutS and PduB of the PD BMCs. No significant homologies were detected to other proteins of the Brookhaven Protein Databank (www.rcsb.org/pdb/home/home.do).
Results
Overall Structure.
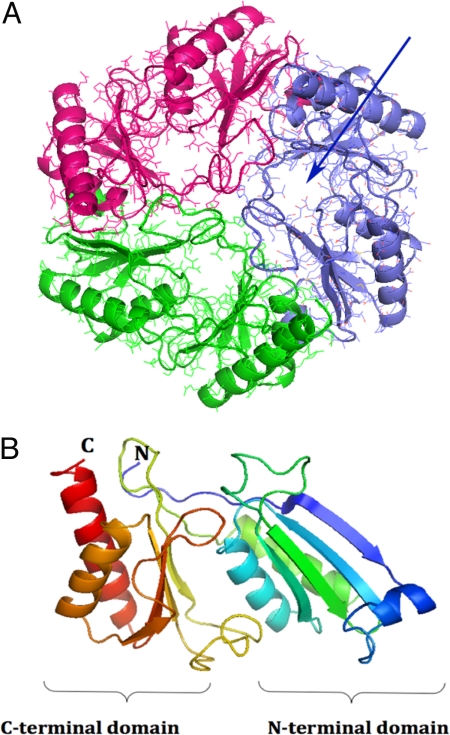
The structure of the EutL protein revealed a trimer that formed a hexagonally shaped tile structure of convex shape (Fig. 1). Analysis of the single EutL protein revealed 2 similarly folded domains. (Fig. S1). The core of each domain contains a 4-stranded β-sheet and 2 tightly packed antiparallel α-helices that are positioned on top of the sheet (Fig. 1). A third short α-helix was observed in the loop structure that connects strands 1 and 2 of each domain. Both of the domains superimpose with an overall rmsd of ≈2.7 Å composed of 102 common residues that were included in the superposition. Whereas the helices superimpose most accurately, the β-sheets appeared translated by >2 Å relative to the helices (Fig. S1). Interestingly, the sequence identity between both domains, i.e., between residues 1–100 and 101–208, was <16%, demonstrating significant divergence between the 2 sequences. Both domains are covalently connected by an extended 13-residue peptide that widens the β-sheet structure of the C-terminal domain by 1 additional strand (Fig. S2).
Fig. 1.
Overall structure of EutL. (A) Top view of the EutL trimer. Shown is a ribbon diagram superimposed onto an all-atom representation. The structure of each monomer is colored in blue, red, and green. The pseudo 6-fold symmetry is readily apparent as are the 3 pores in each of the monomers (blue arrow). (B) Ribbon diagram of the EutL monomer. The structure is rainbow-colored starting with the N terminus in blue and ending with the C terminus in red. The similar folds of the N-terminal and the C-terminal domains are apparent. The structure consists of residues 2–216, which could be built reliably into the electron density. The N-terminal methionine, the last 2 residues, and the His tag could not be modeled. Figs. 1–5 were generated with the program PyMol (http://pymol.sourceforge.net).
Structural Comparisons.
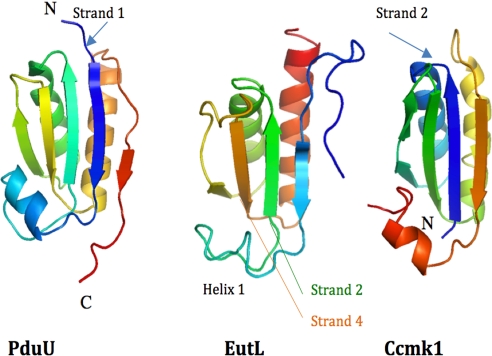
The structure of an individual domain of EutL closely resembles the architecture of other carboxysomal shell proteins. Structural searches with the program DALI (4) revealed similarity to the PD utilization protein PduU [Protein Data Bank (PDB) ID code 3CGI] (Fig. 2), which superimposes best onto the N-terminal domain of EutL with an rmsd of 2.2 Å (including peptide backbone atoms only) or the carboxysomal shell protein Ccmk1 (rmsd = 1.6 Å; PDB ID code 3BN4). The sequence identities, however, accumulate to only 18% or 16% for each of the proteins, respectively. Most of the structural differences are observed in the loop regions. Despite the low homology, a superposition of 2 adjacent PduU proteins of the hexameric assembly also superimposed accurately onto both domains of EutL, suggesting that the relative positioning of both domains has also remained the same (Fig. S2). Consequently, the individual domains of the EutL trimer closely resembled the hexameric arrangement of the PduU proteins.
Fig. 2.
Ribbon diagram of the structures of PduU [PDB ID code 3CGI (only residues 16–122 are shown for clarity)], EutL (C-terminal domain between residues 110 and 216), and Ccmk1 (PDB ID code 3BN4). The structures are displayed in rainbow coloring starting from the N terminus in blue in each case. Helix 1 of EutL is less regularly folded and consists of only 1 helix turn. Even though the overall fold of all proteins is very similar, the order of secondary structure elements is different. Whereas the structures EutL and PduU begin with and N-terminal β-strand (arrows), in the structure of Ccmk1 this strand and former helix 1 are now positioned at the C terminus.
Structure of the Pore.
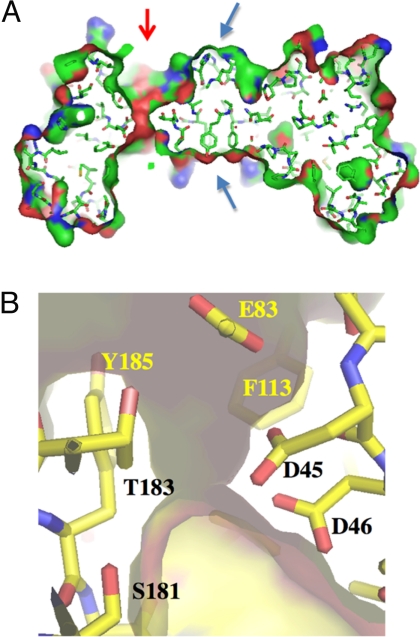
Calculation of a Connolly-type solvent-accessible surface area of the protein trimer revealed that, in contrast to other carboxysomal shell proteins, EutL showed a different arrangement of solvent channels. In contrast to the carboxysome proteins, which contain a single channel in the center of a tile structure, EutL contains 3 channels per tile (Fig. 3A and see Fig. 5A). Each pore is formed, in part, by 2 extensive loop structures between strands 3 and 4 of each domain, Helix B of the N-terminal domain and strands 3 and 4 of the C-terminal domain (Figs. 1B and 2) within a EutL monomer. Inside the channel Asp-45, Asp-46, Asp-53, and Glu-83 provide an acidic patch (Fig. 3B).
Fig. 3.
Structural features of the channel. (A) Vertical section through the center of a EutL trimer. Shown is a stick model of the structure and its solvent accessible surface area (atom color coding: N, blue; C, green; O, red). The only opening through the structure is asymmetrically positioned (red arrow), whereas the center of the tile is tightly packed by a ring of 3 tyrosine and 3 histidine residues (blue arrows). (B) Close-up view into the pore structure. The semitransparent surface of the pore is superimposed onto the stick model. The channel is lined with 3 closely spaced carboxyl residues (D45, D46, and E83) (residue D53, which is also part of the patch, is not shown) and 4 aromatic residues [F113, Y185, and Y48, F189 (both of which are not shown in this image)]. At the narrowest point, the diameter is of the pore is ≈2.2 Å wide.
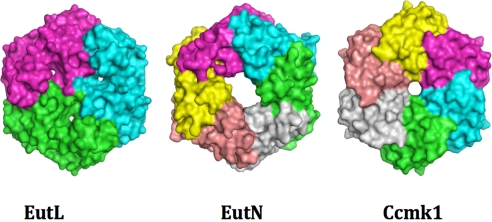
Fig. 5.
Representation of the solvent-accessible surface areas of EutL (Left), EutN (Center), and Ccmk1 (Right). In each structure, the monomers are colored differently. Even though EutN is also a hexameric structure, it displays an asymmetric central pore in contrast to Ccmk1. Despite the different functions and origin, all of the structures exhibit a hexagonal tile structure of very similar dimensions and shape.
The center of each tile, i.e., the location of the crystallographic 3-fold axis, is occupied by a ring of 3 Tyr-70 and His-75 residues (Fig. 3A). Because the packing of these residues is relatively tight at this position, with the Cβ-atoms of the tyrosine residues approaching 3.5 Å, a formation of a pore in the absence of temporary structural rearrangements is unlikely.
Sequence Similarity of EutS and PduB.
A comparison of EutL against other sequences of the protein sequence databank Swissprot (http://www.expasy.ch/sprot/) revealed some homology to the shell protein EutS. The sequence identity is most pronounced in the C-terminal domain of EutL (19% identity between residues 120 and 219). Mapping of the most-conserved residues onto the structure of EutL showed that most of the conserved residues are located within the domain, i.e., within helices and strands (Fig. S3). Some of these residues, however, are also located on the outer edge of the tile, which may indicate conservation of lateral tile packing interactions. No conservation of residues is observed in the pore structure, suggesting that it may be specific for other functions.
The only other protein that exhibits homology to EutL is the shell protein PduB of the PD utilization microcompartment from Salmonella enterica. Its sequence is similarly long (233 residues) and shares 24% sequence identity. Based on this relationship, we propose that PduB is also composed of 2 tandemly repeated domains. Modeling of all conserved residues onto the structure of EutL confirmed that the strongest homology resides within the core of each domain. Some sequence conservation was also observed in the pore structure of the modeled protein. At this point, however, it is not clear whether this structure remains the same because of significant insertions and deletions in both sequences.
As most carboxysomal shell proteins appear to assemble into a hexagonal tile structure, we propose that PduB may also do so by forming a hexagonally shaped trimer that resembles the pseudohexagonal domain structure of EutL.
Discussion
Electron microscopy of carboxysomes revealed highly ordered supramolecular assemblies of proteins exhibiting 5-, 3-, and 2-fold symmetries (5). Similar to the structure of icosahedral virus capsids, the assembly of these polyhedral organelles is likely to be constrained by the geometry of the proteinaceous membrane structure and thus its elementary units. In contrast to viruses, however, the carboxysomal polyhedra have been shown to exhibit different sizes, a property that may also be common to other microcompartments.
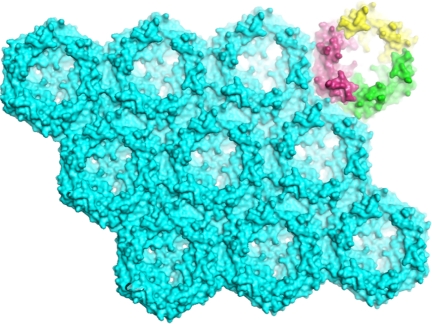
Because of its trimeric assembly and the significant structural similarity between the domains, the crystallographic trimer of EutL assumes a pseudo 6-fold symmetry in the crystal (Fig. 3B). A deliberate rotation of the trimer about its 3-fold axis by only 60° did not only result in a near-perfect superpositioning of the N-terminal domain onto the C-terminal, but it also revealed that the dimensions of the sides of the hexagon are nearly identical. In other words, despite the low sequence conservation, the equal-sided hexagonal geometry of the assembly was still maintained. In the crystal, the hexagonally shaped trimer structure assembles into a highly ordered 2D array of ≈37-Å thickness with no openings between the tiles (as calculated with a Connolly-type surface with a 1.4-Å probe radius) (Fig. 4). The tight packing is achieved by contacting side chains and by backbone–backbone interactions.
Fig. 4.
Structure of the EutL trimer sheet within the crystal. Symmetry-related molecules are displayed with their solvent-accessible surface areas. To enhance the 3D appearance, the structure has been tilted slightly. On the upper left, the individual monomers of 1 tile are colored in yellow, purple, and green. Within the sheet, all molecules display the same orientation. Clearly visible are the 3 pores of each hexagonal tile with its own funnel-shaped entrance. All tiles are tightly packed and no void spaces are observed in between.
Despite the recent structure determinations of proteins that are involved in the formation of the shell structure of bacterial metabolosomes, it is not understood how the shell proteins interact to form these polyhedral assemblies. In particular, the mechanisms that give rise to differently sized compartments remain to be discovered. With the crystal structure of EutL, another example of a hexagon-assembled protein membrane from a different metabolosome has become available. The similar congruent tiling of its hexagons may point to a common assembly principle that may reflect the physiological importance of this arrangement (6).The assembly of these tile units may not only facilitate the assembly into differently sized membrane facets, but may also explain the formation of differently sized compartments. Similar to the triangulation number of viral capsids (7), the number of hexagons within a facet may determine the overall size of the particle. To ensure a tightly sealed compartment, the joints between the facets and the vertices are also required to be sealed. Because the facets of the polyhedra are relatively large compared with those of some viruses, long seam structures between adjacent facets may be required. A first model of an icosahedral micrompartment was presented recently (8). At this point, however, it is not understood how the contacts between 2 adjacent facets are formed and sealed. Even though physiologically relevant interactions between individual shell proteins have not yet been established, the hexagonal assembly pattern raises the question of whether the lattice spacing between the tiles of 1 shell protein dictates the interactions and assembly of other proteins. For example, proteins that connect 2 adjacent facets will only have to match the size and the periodicity of the edges. Theoretically, longer facets can therefore easily be sealed with the recruitment of additional matching “seal proteins.” The hexagonally shaped tile structures may thus reflect a fundamental design principle that may explain the assembly of differently sized micrompartments.
To date, the only other known structure of a Eut shell protein is EutN [PDB ID code 2HD3; Northeast Structural Genomics Consortium (9)]. Its structure is closely related to the carboxysomal proteins OrfA and CcmL (8). In contrast to these pentamer-forming proteins, however, EutN forms a hexameric assembly with a somewhat asymmetric large pore in its center. Even though its structure also forms a hexagonal tile (Fig. 5), in the crystal, however, EutN hexamers do not assemble into a tight membrane-like sheet structure. Instead, they form a loosely packed sheet structure with large openings between the tiles. It therefore seems unlikely that this observed arrangement exhibits a structure of a physiologically relevant membrane. These observations thus raise the question of whether under physiological conditions EutN forms a differently structured protein membrane on its own, or if it assembles into a hexameric (or pentameric) structure while interacting with other shell components, such as EutL. As seen in Fig. 5, the overall dimensions and geometries of both hexamers are very similar. The edge length of a EutL hexagon also spans ≈67.5 Å, which is very similar to the tile dimensions of EutL and other carboxysomal tiles. In light of the matching tile dimensions, it seems plausible that EutN tiles pack orthogonally (below or above a EutL hexagon, for example) or laterally onto the membrane's edge without compromising the lattice order of EutL.
Stability of the Protein Membrane.
In an earlier study (10), the extraordinary stability of the EutL crystals with respect to varying solvent conditions was noted. Even though no covalent cross-links between proteins were observed in the crystal, as judged by SDS/PAGE and mass spectrometry of the crystals, the large and continuous interacting surfaces between adjacent tiles may strongly contribute to the stability. Because this lateral interaction pattern is observed within a single protein sheet, it may also significantly contribute to the stability of the microcompartment.
Consistent with the stability of the crystals, EutL was also observed to crystallize rapidly under many different crystallization conditions (10). In all cases, the proteins form similarly shaped triangular or hexagonal crystal plates that suggest that the packing arrangement has remained the same (in a number of cases this was also confirmed by preliminary unit cell determinations). Inspired by the rapid crystallization process, we have monitored the molecular mass of the protein with respect to time using size exclusion chromatography. Whereas freshly prepared protein eluted from the column as a trimer at pH 8.0, the protein's apparent molecular mass appeared to increase steadily at near-physiological pH values over time (Fig. S4). Thus, the rapid crystallization behavior of this protein and its tendency to oligomerize in physiologically buffered solution may present further evidence for its autonomous assembly into a physiologically relevant membrane facet. Because EutL crystallizes quickly under many different conditions, it may also do so in vivo. Such early assemblies could possibly serve as templates for the assembly of other Eut proteins.
Pore Structure.
Even though the structural composition of a functional BMC shell has not yet been resolved experimentally, the structure of the EutL membrane exhibits 3 distinct openings within each tile that are likely to function as solute channels. This unprecedented arrangement of the channels is likely to increase the overall porosity of the protein membrane considerably compared with its carboxysomal counterparts. Because each channel is lined by aromatic residues and negatively charged residues (Fig. 3B) favorable interactions of the positively charged EA with the channel are proposed that may facilitate its selection for passive transport. As described above, each pore is formed by 2 large loop structures that display increased structural flexibility as judged by the higher-than-average B values. Thus, temporary openings of the channel cannot be ruled out and may further increase the porosity of the membrane.
Tandem Duplication and Circular Permutation.
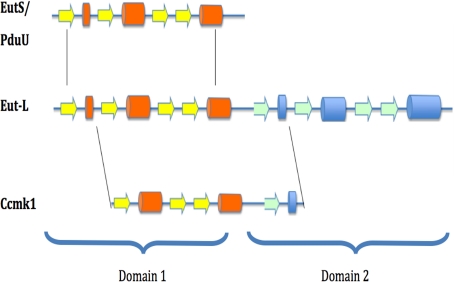
The structure of EutL is an example of a tandemly duplicated shell protein. Even though the protein fails to exhibit a direct repeat within its sequence, both domains reveal significant structural similarities that may have originated from a single gene duplication event.
As described above, a comparison of the EutL domain structure with other shell proteins revealed similarity to PduU of the PD-using microcompartment. Its structure also begins with an aminoterminal β-strand I (blue strand in Fig. 2). In contrast to these structures, however, other shell proteins of the carboxysomes display a different order of secondary structures. For example, in the shell protein Ccmk1, this β-strand is located at the C terminus, suggesting a circularly permuted version of the protein (Fig. 6). Circular permutations are rarely observed in natural proteins but have been shown to have little effect on a protein's structure (11). The observation of a tandemly repeated structure, however, may provide a plausible explanation for the evolutionary origins of this fold. The mechanism involves the duplication of a target gene into a tandem repeat and a subsequent partial degeneration of the 5′ and 3′ coding regions via the creation of new start and stop codons. Because the N and C termini in the structures of some shell proteins are found in close proximity, i.e., EutL and Ccmk1 for example, only small linker sequences may be required to covalently link the two for the creation of new termini at other places in the sequence. Similar relationships of homologous proteins have been characterized in other protein families and the feasibility of engineering such variants has also been demonstrated (11–13).
Fig. 6.
Possible relationship between tandem duplications and circular permutations among the shell proteins. Helices are shown as cylinders and β-strands are shown as arrows. The structure of EutL is composed of 2 very similar domains that are likely to have been the result of a direct duplication event of a gene like EutS. With the introduction of a new start codon and a stop codon, a permutated domain structure can be generated.
In addition to the structures of EutL and EutN, only a few other structures of the EA ammonialyase metabolosome from different organisms are known. The structures of EutB, EutD, and EutQ have been determined by structural genomics efforts (PDB ID codes 2QEZ, 1VMI, and 2PYT; Joint Center for Structural Genomics), whereas the structure of the large subunit of the EA ammonia lyase enzyme has been inferred from homology based model building studies (14). Because of the pronounced sequence similarity of EutL to EutS, we show here that its 3D structure may also be deduced. Based on a previously published alignment (3, 14) and a mapping of the conserved residues onto the structure of EutL, we propose that EutS folds similarly to the C-terminal domain of EutL. Because its sequence comprises only 1 domain, it appears plausible that its structure may also oligmerize into a hexamer.
Conclusion
The crystal structure of EutL contributes further to an understanding on the architecture, assembly, and evolution of BMCs. Because this structure also assembles into a proteinaceous membrane that is formed by hexagonally shaped tiles, the structural similarities among shell proteins may suggest that the hexagon tile may be a fundamental building unit that is common to all carboxysome-, PD-, and EA-using microcompartments. In contrast to viruses, the hexagonally shaped tile structures may allow for the formation of differently sized microcompartments while maintaining the selectively permeable qualities of the structure. Because these structures have evolved to optimize chemical reaction spaces, the crystal structure of EutL provides further insights into the architectural and physical properties of the proteinaceous membranes. Despite the lack of strong sequence homology of EutL to shell proteins of other BMCs, the significant structural similarities may present further evidence that points to a common evolutionary origin of BMCs.
Materials and Methods
Size Exclusion Chromatography.
Molecular mass and oligomerization state were determined by size-exclusion chromatography using a Sephadex 200 column operated by an Äkta Purifyer FPLC workstation (GE Healthcare). Molecular weight standards were obtained from GE Healthcare.
Crystallization.
A C-terminally tagged version of the protein (EutL_CHIS) was cloned, purified, and crystallized as described (10). Briefly, isolated EutL protein at a concentration of 1 mg/mL was dialyzed into 50 mM actetate buffer at pH 6.5 before crystallizations. Best crystals were obtained with 2 M NaCl, 100 mM phosphate, 100 mM Mes buffer (pH 6.5), and 5% PEG 400 by vapor diffusion.
Structure Determination.
Data collection and integration procedures have been described (10). The structure was determined by single isomorphous replacement using a single mercury derivative. For this purpose crystals were incubated for 2 or 3 h in mother liquor containing 1 μM HgCl2. Data were collected at the Stanford Synchrotron Radiation Laboratory (Menlo Park, CA) (Table S1). Heavy atoms positions were identified visually in difference Patterson maps and with the program SOLVE (15). Phasing and initial rounds of solvent flattening to 3.6-Å resolution were performed with the programs SOLVE and SHARP (16) (Fig. S5). Even though datasets from selenomethionine-derivatized crystals could not be used for phasing because of the pronounced radiation sensitivity of the crystals, the mercury-derived phases were of sufficiently good quality to identify the positions of the selenium atoms via anomalous Fourier calculations. Knowledge of the positions aided the threading of the amino acid sequence into the density. Through iterative cycles of phase combination and model building, the structure of the protein was built and refined against a 2.2-Å resolution dataset. Even though the entire model between residues 2 and 216 could be modeled into the density, the Rwork and Rfree values remained relatively high at 30% or 34%, respectively. Twinning analysis of the data using the UCLA twin server (17) and Britton plots (18) revealed a 42% partial twinning factor of the crystals with the twinning operator (k, h, −l). The degree of twinning was observed to vary among datasets from different crystals. These findings are also in agreement with the fact that refinement of the model in the alternative space group P321, which also scaled reasonably well, also resulted in unreasonably high R values. By using either CNS or REFMAC5 (19) with detwinned data, a significant drop in R values was observed. Further model building and refinement with this data resulted in a final Rwork = 22.7% and Rfree = 27% to 2.2-Å resolution. Model building and refinement were performed with the program COOT and REFMAC (19) and the program suite of CNS version 1.2 (20). Data and refinement statistics are given in Table S1. Identification of structural homologues was carried out with the program DALI (4). Alignment of the EutL and EutS amino acid sequences was performed with the program CLUSTAL (21, 22). Homology model building of EutS was performed interactively by using computer graphics and the sequence alignments.
Supplementary Material
Acknowledgments.
We thank Dr. R. R. Chapleau for comments on the manuscript and the staff at the Stanford Synchrotron Radiation Laboratory for beam time and kind help. This work was in part supported by National Science Foundation Grant DMR-0520415. A.O. gratefully acknowledges support by the International Training Program from Tokyo University of Agriculture and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3GFH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902324106/DCSupplemental.
References
- 1.Penrod JT, Roth JR. Conserving a volatile metabolite: A role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dou Z, et al. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem. 2008;283:10377–10384. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- 3.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid MF, et al. Structure of Halothiobacillus neapolitanus carboxysomes by cryoelectron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerfeld CA, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 7.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 9.Forouhar F, et al. Functional insights from structural genomics. J Struct Funct Genomics. 2007;8:37–44. doi: 10.1007/s10969-007-9018-3. [DOI] [PubMed] [Google Scholar]
- 10.Nikolakakis KOA, Newton K, Chworos A, Sagermann M. Preliminary structural investigations of the Eut-L shell protein of the ethanolamine ammonia-lyase metabolosome of E. coli. Acta Crystallogr F. 2009;65:128–132. doi: 10.1107/S1744309108042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luger K, Hommel U, Herold M, Hofsteenge J, Kirschner K. Correct folding of circularly permuted variants of a βα barrel enzyme in vivo. Science. 1989;243:206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: Circularly permuted amino acid sequences. Proc Natl Acad Sci USA. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Warncke K. Comparative model of EutB from coenzyme B12-dependent ethanolamine ammonia-lyase reveals a β8α8, TIM-barrel fold and radical catalytic site structural features. Proteins. 2006;64:308–319. doi: 10.1002/prot.20997. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 17.Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
- 18.Fisher RG, Sweet RM. Treatment of diffraction data from crystals twinned by merohedry. Acta Crystallogr A. 1980;36:755–760. [Google Scholar]
- 19.Collaborative Computational Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Aiyar A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol. 2000;132:221–241. doi: 10.1385/1-59259-192-2:221. [DOI] [PubMed] [Google Scholar]
- 22.Higgins DG. CLUSTAL V: Multiple alignment of DNA and protein sequences. Methods Mol Biol. 1994;25:307–318. doi: 10.1385/0-89603-276-0:307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.