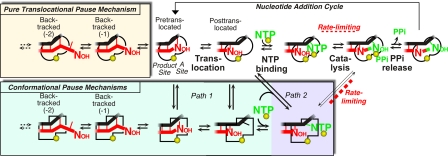
Transcriptional pausing by multisubunit RNA polymerases (RNAPs) plays key roles in gene regulation by coordinating RNAP movement with interactions of regulators and folding of the nascent RNA and, in metazoans, by helping program cycles of promoter-proximal transcription that poise RNAPII for gene expression (1, 2). However, mechanistic understanding of pausing is incomplete. Proposed mechanisms can be divided broadly into 2 classes (Fig. 1): backtrack pausing, in which reverse translocation of RNAP dislodges the transcript 3′ end from the active site and thereby prevents RNA synthesis; and (ii) nonbacktrack pausing, in which conformational rearrangements in the RNAP active site block the nucleotide addition cycle. Only a single example of nonbacktrack pausing, one for which the pause lifetime is increased by a nascent RNA hairpin, has been studied in biochemical detail (refs. 3 and 4 and references therein). Considerable disagreement exists over the contribution of nonbacktrack pauses to the ubiquitous pausing observed during both ensemble and single-molecule in vitro transcription experiments (5–9). An article in this issue of PNAS by Kireeva and Kashlev (10) now provides definitive evidence that, at least for bacterial RNAPs, significant nonbacktrack pausing occurs even without the contribution of pause RNA hairpins.
Fig. 1.
Nucleotide addition cycle and alternative pausing pathways. The active site is depicted as semicircles with red RNA, black DNA, green NTP substrate, and yellow Mg2+ ions. Pausing might occur either by direct, reverse translocation of RNAP into backtrack states (pale yellow box; e.g., ref. 5) or isomerization of the active site into an inhibited conformation (squares) that could also facilitate backtracking (pale green box; e.g., ref. 4). Path 1 (pale green) and path 2 (pale blue) are alternative paths of isomerization that could occur before or during NTP binding, respectively.
Hairpin-stabilized pausing was initially studied for its role in synchronizing transcription with translation in the attenuation control regions of amino acid biosynthetic operons in enteric bacteria (e.g., the his and trp operons). Like all pauses studied to date, these sites cause kinetic partitioning of RNAPs between active and paused states (as opposed to affecting all transcribing RNAPs uniformly). These pause signals were found to be multipartite and to include synergistic contributions of different parts of RNAP's nucleic-acid scaffold both to the pause efficiency (the probability of entering the pause state at the kinetic branch between pausing and active elongation) and to pause lifetime (although the pause hairpin contributes only to pause lifetime). Detailed study of the his pause signal revealed that (i) it halted RNAP largely in the pretranslocated register (Fig. 1; i.e., not backtracked), (ii) the active site was primarily inhibited for catalysis rather than for translocation or NTP binding, and (iii) inhibition appeared to arise by blocking formation of a 3-helix bundle composed of the RNAP bridge helix and trigger loop (4). This 3-helix bundle makes contacts to the NTP substrate in the A site required for rapid catalysis (11).
A few other pause signals also have been studied to a limited extent, including (i) those in the early-transcribed region of bacteriophage T7, some of which were found to be RNA hairpin-independent (12), (ii) the Escherichia coli ops pause at which RNAP can enter backtrack states (13), and (iii) the HIV-1 +62 pause (affecting human RNAPII), which appears to involve direct partitioning into a backtracked state (14). Single-molecule experiments with bacterial RNAP revealed that pauses of similar efficiency and strength to those characterized biochemically could be detected on average at least once per 100 bp within transcription units (9). Application of force to resist or assist transcription affected a small subset, as expected for backtrack pauses, but for most neither the probability nor the duration were affected by applied force. Thus, these so-called ubiquitous pauses were proposed to be nonbacktracked and arise from the effect of nucleic acid sequences on the conformation of the RNAP active site (7, 9). In contrast, experiments with yeast RNAPII found that the probability and duration of most pauses were strongly affected by applied force, suggesting they resulted from backtracking (6). A mathematical model based on this work is argued to explain most, if not all, pausing, including the apparently force-insensitive ubiquitous pauses observed for bacterial RNAP, purely by a random walk among backtrack registers (5, 8). However, fitting of the mathematical model does not exclude other mechanisms, and direct biochemical characterization of ubiquitous pausing has been lacking.
Kireeva and Kashlev (10) have now provided this detailed biochemical characterization for one of the hairpin-independent pause signals originally identified by Levin and Chamberlin (12) in bacteriophage T7 DNA (the T7 A1 D111 C37 pause, which halts RNAP after addition of C37). Using exonuclease III to map the DNA footprint of E. coli RNAP at the C37 pause, antisense oligonucleotides to block backtracking along the RNA, and nonhydrolyzable NTPs to detect substrate binding, Kireeva and Kashlev establish that the C37-paused RNAP is not backtracked, is not inhibited in translocation from pretranslocated to posttranslocated registers, and is not inhibited for substrate binding. As was concluded for the hairpin-stabilized his leader pause (4), the C37 pause appears to result from an active-site rearrangement that inhibits catalysis, rather than from effects on translocation. The C37 pause was strongly affected not only by the identity of the RNA 3′ nucleotide, but also by the identity of the incoming NTP. Changing the DNA template to alter the incoming NTP from ATP to UTP decreased both the fraction of RNAP entering the paused state (from 98% to 20%) and the lifetime of the pause (from 40 to 17 s at 10 μM substrate NTP). These results definitely establish incoming NTPs or their cognate nucleotides in the DNA as components of pause signals. Based on this result, it is suggested the paused state involves misalignment of the NTP substrate in the active site that might arise as the NTP binds (path 2; Fig. 1). Path 2 contrasts with the proposal that the his leader pause forms when RNAP arrives at pause site and before NTP binding (path 1; Fig. 1). Although this mechanistic detail is important (see below), the central finding from Kireeva's and Kashlev's elegant work is that ubiquitous pauses can result from an altered active-site configuration, rather than from backtracking.
Does this finding help explain the disparity in interpretation of single-molecule pausing by bacterial RNAP and yeast RNAPII (5–9)? Kireeva and Kashlev (10) show that yeast RNAPII does not recognize the C37 pause, although some other T7 DNA sites cause RNAPII to pause by backtracking. Their results clearly establish that the sequence determinants for bacterial RNAP and yeast RNAPII pausing differ. At a minimum, it is clear that ubiquitous pausing by bacterial RNAP must include, if not be dominated by, nonbacktrack pause sites. Whether nonbacktrack pauses also exist for RNAPII, as Kireeva and Kashlev hint, will require further study.
What can we conclude about the fundamental mechanism of pausing? First, confirmation of the existence of a nonbacktrack paused state is broadly consistent with the view that all pauses may arise through an initial or elemental isomerization of the active site that inhibits nucleotide addition. Subsequent events that prolong pausing, which may involve backtracking, RNA hairpin formation, or regulator binding, may be aided by this initial isomerization, either by allowing time for them to occur or by weakening contacts to the RNA 3′ end. Although misalignment of NTP substrates may well occur in the paused active site, at least 2 arguments favor formation of the pause state by path 1 rather than path 2 (Fig. 1). First, withholding NTP from RNAP halted at a pause site increases the fraction of RNAP that enters the paused state. Further, the rate of this NTP-independent isomerization is dramatically slowed by amino acid substitutions that affect the active-site conformation (4). In other words, the paused state forms by a time-dependent isomerization of the active site before NTP addition. Second, the transcription inhibitor streptolydigin, which binds the unfolded trigger loop, fails to inhibit nucleotide addition at the his leader pause site (4). This result is most easily explained if the paused active-site conformation is incompatible with streptolydigin binding (presumably because a “paused” conformation of the trigger loop is incompatible with streptolydigin binding), whereas simple misalignment of NTP seemingly would enhance streptolydigin action because such misalignment is observed in an RNAP–NTP–streptolydigin crystal structure (11).
Significant nonbacktrack pausing occurs even without the contribution of pause RNA hairpins.
Why should these mechanistic details of pausing matter? In both bacteria and eukaryotes, paused RNAPs are targets for regulators. The ability of GreB or TFIIS to rescue bacterial RNAP or eukaryotic RNAPII from backtracked states by transcript cleavage is well known (15, 16). However, where should we look for the mechanistic targets of bacterial regulators like NusG and NusA or eukaryotic regulators like DSIF/NELF, hepatitis δ antigen, Elongin, or ELL, all of which modulate pausing but do not stimulate transcript cleavage (16, 17)? If most or all pauses arise by a direct backtracking pathway (Fig. 1), then we should look principally for interactions that affect translocation. However, if pausing occurs by an active-site isomerization, then we should look for interactions that affect this isomerization. The results of Kireeva and Kashlev (10), and the arguments advanced here, favor the latter view.
Acknowledgments.
Work on transcriptional pausing in my laboratory is supported by National Institutes of Health Grants GM38660 and GM72795.
Footnotes
See companion article on page 8900.
The author declares no conflict of interest.
References
- 1.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 2.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 4.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 7.Herbert R, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejia YX, Mao H, Forde NR, Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J Mol Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuman K, Abbondanzieri E, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 10.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassylyev D, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 12.Levin JR, Chamberlin MJ. Mapping and characterization of transcriptional pause sites in early genetic region of bacteriophage T7. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- 13.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J Mol Biol. 2001;311:265–282. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- 15.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 17.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]