Abstract
Tight control of cellular redox homeostasis is essential for protection against oxidative damage and for maintenance of normal metabolism as well as redox signaling events. Under oxidative stress conditions, the tripeptide glutathione can switch from its reduced form (GSH) to oxidized glutathione disulfide (GSSG), and thus, forms an important cellular redox buffer. GSSG is normally reduced to GSH by 2 glutathione reductase (GR) isoforms encoded in the Arabidopsis genome, cytosolic GR1 and GR2 dual-targeted to chloroplasts and mitochondria. Measurements of total GR activity in leaf extracts of wild-type and 2 gr1 deletion mutants revealed that ≈65% of the total GR activity is attributed to GR1, whereas ≈35% is contributed by GR2. Despite the lack of a large share in total GR activity, gr1 mutants do not show any informative phenotype, even under stress conditions, and thus, the physiological impact of GR1 remains obscure. To elucidate its role in plants, glutathione-specific redox-sensitive GFP was used to dynamically measure the glutathione redox potential (EGSH) in the cytosol. Using this tool, it is shown that EGSH in gr1 mutants is significantly shifted toward more oxidizing conditions. Surprisingly, dynamic reduction of GSSG formed during induced oxidative stress in gr1 mutants is still possible, although significantly delayed compared with wild-type plants. We infer that there is functional redundancy in this critical pathway. Integrated biochemical and genetic assays identify the NADPH-dependent thioredoxin system as a backup system for GR1. Deletion of both, NADPH-dependent thioredoxin reductase A and GR1, prevents survival due to a pollen lethal phenotype.
Keywords: redox homeostasis, redox imaging, redox-sensitive GFP, thioredoxin reductase
Thiol-redox biochemistry is a common feature of life and is involved in a broad range of physiological and pathological processes. Thus, tight control of cellular redox homeostasis is essential for maintenance of normal metabolism and redox-dependent signaling. In general, several metabolic reactions and, to a larger extent, stress-induced processes lead to the formation of reactive oxygen species (ROS). The concomitant oxidation is buffered through a tight network of antioxidant enzymes and low-molecular weight antioxidants (1). The most prevalent nonprotein thiol-based redox buffer is the tripeptide glutathione, which is present in low millimolar concentrations in most eukaryotic cells. Under nonstress conditions, cytosolic glutathione is present mainly in the reduced form (GSH) with only nanomolar concentrations of the oxidized form, glutathione disulfide (GSSG) (2, 3). In plants, ROS are at least partially detoxified through the glutathione-ascorbate-cycle at the expense of electrons from the GSH pool (4), causing a transient oxidation of the GSH pool. This transient change in glutathione redox potential (EGSH) has been suggested to be part of signaling cascades leading to changes in gene expression during stress responses and developmental processes (5, 6). The transmission of such signals is likely to be facilitated by glutaredoxins (GRXs) (6), which equilibrate EGSH with the redox potential of target protein thiols. Indeed, GRXs have been shown to be involved in disease resistance and developmental processes, suggesting that downstream events in these processes are modified by EGSH (7, 8).
NADPH-dependent glutathione reductase (GR), a member of the FAD-binding disulfide reductase superfamily, is the major enzyme responsible for reduction of GSSG to GSH in most organisms, with only few GR-deficient exceptions (9, 10). The Arabidopsis genome contains 2 GR genes. One gene codes for an organellar isoform (GR2), which is dual-targeted to chloroplasts and mitochondria (11). GR2 is essential for plant development, which is evident from lethality of deletion mutants in early embryo development (12). The second gene codes for the cytosolic GR1. Expression analysis by scrutiny of the Genevestigator microarray database (13) shows GR1 to be responsive to several stress factors. Although GR activity is one of the most frequently monitored enzyme activities after stress application, there is still very little information on the biochemical properties of Arabidopsis GR1. Because gr1 deletion mutants have not been reported in plants, we questioned whether other oxidoreductases may provide a functional backup system for GR1 and, thus, conceal function on gene disruption (14).
Like GRs, NADPH-dependent thioredoxin reductases (NTRs) also belong to the FAD-binding disulfide reductase superfamily (15). In Arabidopsis, NADPH-dependent thioredoxin reductase A (NTRA) and NADPH-dependent thioredoxin reductase B (NTRB) result from recent gene duplication and constitute the major route to reduce mitochondrial and cytosolic thioredoxins (TRXs) (16, 17). Nevertheless, the double knockout ntra ntrb was found to be viable, although it was hypersensitive to depletion of the GSH pool (18). A different, although less efficient, reduction system consisting of GRX and TRX was apparently able to complement the NTR function (18). To be maintained in the reduced form, GRX requires GSH, GR, and NADPH as the primary electron donor. Partial redundancies between the NTR/TRX system and the GSH redox system have also been observed in Escherichia coli and Saccharomyces cerevisiae (19, 20). However, in fission yeast, GR is indispensable for growth under aerobic conditions (21).
We previously showed that the cytosolic EGSH is ≈−320 mV under nonstress conditions (2), indicating a very high reduction capacity for GSSG. Therefore, we focused on the homeostatic function of GR1, and assessed it at phenotypic, genetic, and biochemical level. The impact of GR1 on cytosolic redox homeostasis was investigated with state-of-the-art redox-sensitive (ro)GFP tools that allow dynamic measurements of EGSH in vivo (3). The results demonstrate TRXh3 in conjunction with NTRA as a GR-independent mechanism of GSSG reduction capable of partially complementing gr1 mutants. The fundamental function of the NTR/TRX system for GSSG reduction is emphasized by pollen lethality when both reduction systems are lost.
Results
Expression, Purification, and Characterization of Arabidopsis GR1.
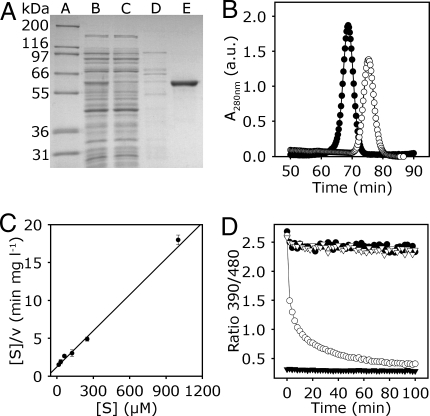
Based on homology, the gene function of locus At3g24170 has been assigned as a cytosolic GR1. The predicted protein shows 61.4% similarity and 23.1% identity with GRs from E. coli and S. cerevisiae, respectively (Fig. S1). However, the Arabidopsis protein contains motifs not found in other GRs including 11 aa before the active centre and a C-terminal extension of 18 aa. To verify that GR1 is a genuine GR, the cDNA was cloned into an expression vector and purified using an introduced N-terminal His-tag. SDS/PAGE analysis of purified GR1 indicated a monomer of ≈60 kDa, which corresponds to the predicated molecular mass of 57.1 kDa calculated for GR1 including the N-terminal His-tag (Fig. 1A). Native GR1 is composed of 499 aa and has a molecular mass of 53.8 kDa. Size exclusion chromatography indicated that purified native GR1 eluted significantly earlier than BSA (66 kDa) as a control, suggesting native GR1 was present as a dimer (Fig. 1B). The optimal pH for GR1 activity was found to be in the range of 7.2 to 7.6 (Fig. S2A); thus, all subsequent experiments were performed at pH 7.4, unless indicated otherwise. Analysis of the catalyzed reaction revealed Michaelis-Menten kinetics with a Km for the substrate GSSG of 77 μM and a Vmax of 63 μmol min−1 mg−1 (Fig. 1C). The Km for the cosubstrate NADPH was 33 μM (Fig. S2B). Whereas animal and bacterial GRs are inhibited by 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) (22, 23), significant inhibition of recombinant GR1 was observed only at high BCNU concentration >1 mM (Fig. S2C), making this compound not suitable for in vivo application.
Fig. 1.
Biochemical characterization of cytosolic GR1. (A) Expression and purification of GR1. SDS/PAGE Gel stained with Coomassie Blue. Lane A, molecular mass standard. Lanes B–E, purification of GR1 expressed in E. coli HMS 174 cells, as follows: 8 μg of crude extract (lane B); 8 μg of flow through after loading the crude extract on Ni2+-NTA column (lane C); wash step with 80 mM imidazole (lane D); 2 μg of purified GR1 eluted with 150 mM imidazole (lane E). (B) Size exclusion chromatography of GR1 and BSA. Purified GR1 (peak A, ●) or BSA (peak B, ○) was chromatographed in 50 mM Tris, pH 8, 250 mM NaCl. Peak A corresponds to ≈120 kDa and peak B to ≈70 kDa. a.u., arbitrary units. (C) Kinetic mechanism of GR1. Hanes plot for kinetic analysis of GR1 regarding the substrate GSSG is shown; experimental data are represented by symbols and the global fit of all data by a line (R2 = 0.92). The enzyme assay was performed with 200 μM NADPH and 15–1,000 μM GSSG. Mean values ± SD are shown (n = 3). (D) GR assay using GRX1-roGFP2 as a sensor for the actual EGSH generated from the mixture of substrate (GSSG) and product (GSH). Fully oxidized GRX1-roGFP2 was mixed with NADPH (100 μM) and with (○) or without (▿) 0.1 μM AtGR1 at pH 7.0. 2 min after start of the measurement freshly prepared GSH solution was added to a final concentration of 2 mM. For control, full reduction of the probe was achieved with 10 mM DTT (▼), and full oxidation with 10 mM H2O2 (●).
To further elucidate the efficiency of GR1 for reduction of GSSG, we used recombinant GRX1-roGFP2 as a highly sensitive sensor for the actual EGSH (2, 3). Addition of 2 mM freshly prepared GSH to oxidized GRX1-roGFP2 did not cause a reduction of the sensor unless GR1 and NADPH were present (Fig. 1D). In the presence of GR1, roGFP2 was almost completely reduced within 100 min, which was detected as a decrease in the 390/480-nm excitation ratio (Fig. 1D). Based on its midpoint potential of −280 mV (24), roGFP2 approaches full reduction only when the degree of GSH oxidation (OxDGSH) drops <0.001%. Thus, GR1 is capable of reducing virtually all GSSG leaving only nanomolar concentrations of GSSG at steady state.
Isolation and Biochemical Characterization of gr1 Mutants.
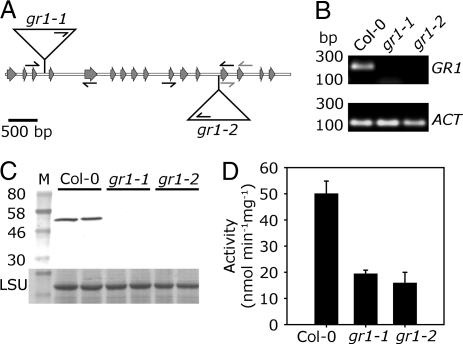
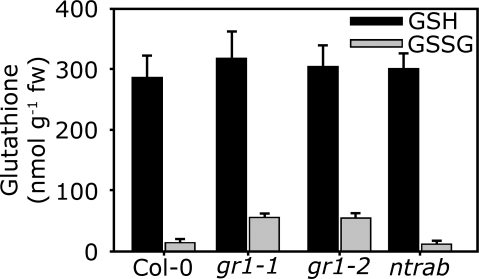
To further elucidate the function of GR1, 2 different gr1 T-DNA insertion alleles were obtained from the Salk collection. The homozygous mutants, designated gr1-1 and gr1-2, contain T-DNA insertions in the 3rd and the 12th intron, respectively (Fig. 2A). For both mutants, the sequence of genomic DNA flanking the left T-DNA border was sequenced to verify the insertion site (Fig. 2A). self-fertilization of heterozygous gr1 plants resulted in ≈25% homozygous mutants for both alleles (Table S1). Semiquantitative RT-PCR with primers binding to the 13th and the 14th exon (Fig. 2A) revealed that both deletion mutants were completely devoid of GR1 mRNA (Fig. 2B). Protein gel blot analysis with antiserum against GR1 showed the absence of GR1 protein in both mutant lines (Fig. 2C). In total protein extracts from both mutant lines GR activity was reduced by 65% (Fig. 2D). The residual GR activity of <20 nmol min−1 mg−1 is most likely due to the organellar isoform GR2, which is not affected in its abundance in the gr1 mutant (Fig. S3). To a lower extend, this residual activity can also be attributed to the activity of the NTS system (see below). Loss of GR1 did not affect the amount of GSH. However, the whole cell amount of GSSG determined by HPLC increased ≈4-fold from 13 ± 2 nmol g−1 FW in wild-type leaves to 54 ± 7 nmol g−1 FW in gr1 mutants (Fig. 3). Thus, both gr1 mutants are complete knockouts of GR1.
Fig. 2.
Isolation and characterization of gr1 deletion mutants. (A) Physical map of the GR1 gene (At3g24170) and T-DNA insertion sites for alleles gr1-1 and gr1-2. Gray arrows indicate exons. Triangles represent the inserted T-DNAs, small black arrows indicate primers used for genotyping and gray arrows primers used for RT-PCR. (B) Saturated RT-PCR with primers specific for GR1 cDNA. PCR was performed using cDNA from Col-0, gr1-1, and gr1-2, respectively, as template. As reference, primers specific for actin 7 were used. (C) Protein gel blot analysis with antiserum raised against GR1; 15 μg of total protein of Col-0, gr1-1, and gr1-2 were separated on a 10% SDS/PAGE and electro blotted to nitrocellulose in duplicates. M, prestained molecular mass standard. Equal loading in all lanes is confirmed by staining of the large subunit (LSU) of 1,5-bisphosphate carboxylase/oxygenase (Rubisco). (D) GR activity of total leaf protein. Means ± SD (n = 3).
Fig. 3.
Glutathione content of gr1 and ntra ntrb deletion mutants. Reduced GSH and GSSG was measured after extraction of 6-week old rosette leaves and derivatisation of thiols with monobromobimane. Means ± SD (n = 8).
The gr1 Plants Have Decreased Buffer Capacity Against ROS.
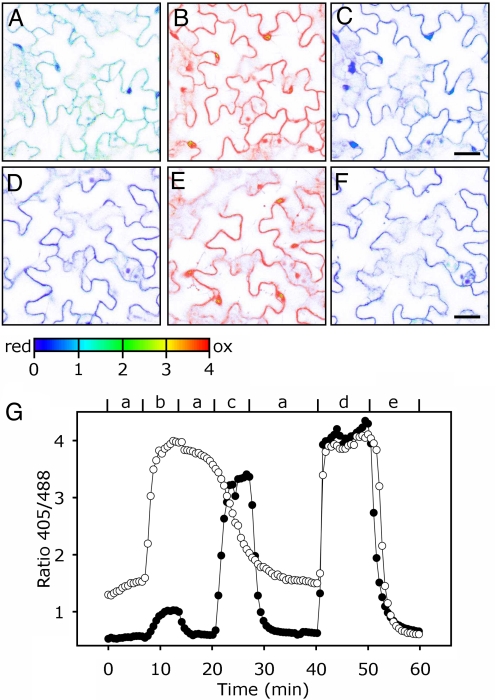
To further elucidate whether the increased GSSG affects EGSH, we expressed GRX1-roGFP2 (3) in the cytosol of gr1-1 mutants and wild-type plants. Under nonstress conditions, the fluorescence ratio was markedly higher in gr1 plants (1.5 compared with 0.5 in wild-type plants) (Fig. 4 A and D). Calibration against 100 mM H2O2 to fully oxidize (Fig. 4 B and E) and 10 mM DTT to fully reduce the sensor (Fig. 4 C and F) led to similar fluorescence ratio values in both mutant and wild-type. These calibration values and an assumed cytosolic pH of 7.4 gave a value of EGSH = −270 ± 9 mV in gr1 and ≈−315 ± 9 mV in wild-type leaves. The less negative EGSH in the cytosol of gr1 plants indicated an increased GSSG concentration in the cytosol, which is in agreement with the HPLC data (Fig. 3). EGSH in plastids and mitochondria was not affected in gr1 mutants (Fig. S4), indicating that the prime physiological effect of gr1 deletion was confined to the cytosol.
Fig. 4.
Oxidation and reduction of glutathione in the cytosol of Col-0 and gr1-1 plants. Images of wild-type and mutant leaves expressing GRX1-roGFP2 in the cytosol were taken by CLSM with excitation at 405 and 488 nm, respectively. Single images were used for the calculation of the ratio images. (A–F) Steady state ratio images of gr1 (A–C) and Col-0 (D–F). (A) The gr1 leaf, control, (B) gr1 oxidized with 100 mM H2O2, (C) gr1 treated with 10 mM DTT, (D) Col-0, untreated control, (E) Col-0 oxidized with 100 mM H2O2, and (F) Col-0 reduced with 10 mM DTT. (Scale bars, 10 μm.) (G) Typical time course showing the dynamic response of the cytosolic EGSH in Col-0 (●) and gr1 (○) to transient oxidation induced by H2O2. Small letters indicate perfusion steps: a, ½ MS medium; b, 1 mM H2O2; c, 5 mM H2O2; d, 100 mM H2O2; e, 10 mM DTT. Because treatment of gr1 leaves with 1 mM H2O2 already led to full oxidation of the probe, perfusion step c in this case was done with ½ MS instead of 5 mM H2O2.
To test the cytosolic buffer capacity and the ability to counteract an induced oxidation, seedlings were perfused with oxidizing and reducing solutions (Fig. 4G). In wild-type plants, GRX1-roGFP2 showed a very small increase in the 405/488-nm ratio from 0.5 to ≈1 after treatment with 1 mM H2O2. In contrast, the same treatment of gr1 seedlings resulted in an increase of the fluorescence ratio from ≈1.4 to ≈4, corresponding to the ratio for fully oxidized GRX1-roGFP2. In contrast, in wild-type seedlings GRX1-roGFP2 was not yet fully oxidized after treatment with 5 mM H2O2, showing that the capacity for maintaining GSH in reduced form after H2O2 treatment is clearly limited in gr1 plants. Also, in gr1 mutants, the GSSG was only reduced very slowly after washout of H2O2 (Fig. 4G; 13–19.5 min).
Attenuated Cytosolic GSSG Reduction Does Not Confer a Strong Phenotype.
The absence of functional GR1 did not result in any obvious phenotype under physiological conditions (Fig. S5). This lack of phenotype is surprising in regard to the high GSSG level measured in the gr1 mutant, and suggests that accumulation of GSSG is not toxic for the plant or that GSSG is exported from the cytosol. Also, the gr1 mutant is not hypersensitive to severe stress conditions like exposure to oxidative stress generating agents (Cu2+ ions, H2O2) or the heavy metal Cd2+ that inactivates metalloproteins and is detoxified by the GSH polymers phytochelatins (Fig. S6). The low level of reduction in absence of a distinct phenotype suggests an alternative cytosolic system for reduction of GSSG.
The Cytosolic TRX System Can Reduce GSSG In Vitro.
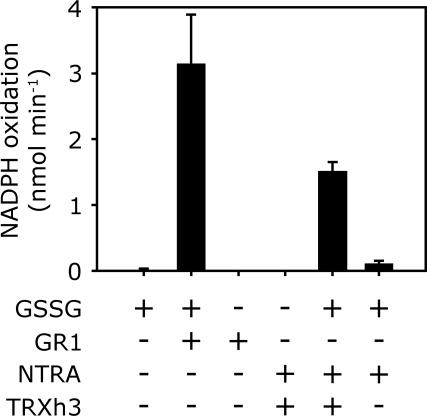
Therefore, we examined whether the cytosolic NADPH-dependent TRX system can reduce GSSG in vitro. For this purpose, recombinant NTRA and TRXh3 were expressed in E. coli and purified. Based on the same amount of protein, the rate of GSSG reduction by NTRA/TRXh3 was ≈200-fold lower than by GR1. In the absence of TRXh3, NTRA on its own did not significantly reduce GSSG (Fig. 5).
Fig. 5.
Biochemical activity of GSSG reduction by NTRA and TRXh3. TRXh3 is first reduced for 30 min with NTRA and NADPH. Activity is monitored as NADPH oxidation. Note that the amount of disulfide reductase protein varied between the assays. Enzymes and substrates were used at the following concentrations: NADPH, 200 μM; GSSG, 400 μM; GR1, 0.01 μM; NTRA, 1 μM; TRXh3, 10 μM. Means ± SD (n = 3).
Triple Knockouts Are Pollen Lethal.
The ntra ntrb double mutant had wild-type GSH levels and GSH/GSSG ratios (Fig. 3), but showed smaller rosettes (Fig. S5). To test for functional redundancy, we examined whether the TRX system is substituting the lack of GR1 in the gr1 mutants, we generated triple mutants lacking NTRA, NTRB, and GR1. Both mutant lines, gr1-1 and gr1-2, were crossed with an ntra ntrb double knockout. From the F2 generation, mutants homozygous for ntra and ntrb, but heterozygous for gr1 were selected and selfed. These mutants segregated a ratio of 1 (GR1/gr1):1 (gr1/gr1) with no homozygous triple mutant for either gr1 allele (Table S2), suggesting that triple mutants are lethal. Lack of aborted seeds (Fig. S7) indicated a pollen lethal effect while female transmission of gr1 was not impaired.
To further test whether the triple mutation affects male fertility, pollen of a GR1/gr1-1 ntra ntrb was transferred to wild-type stigma. Progeny testing failed to identify plants heterozygous for gr1-1 (Table 1), suggesting that no gr1-1 ntra ntrb pollen had successfully fertilized the wild-type ovules. Alexander staining (25) revealed no cytoplasmic or pollen wall abnormalities. The reciprocal cross did result in 50% GR1/gr1 plants, which again confirmed that female transmission of the defective gr1 allele in the ntra ntrb background was normal.
Table 1.
Reciprocal cross between GR1/gr1-1 ntra/ntra ntrb/ntrb and Col-0
| A: | Female parent | × | Male parent | Progeny genotype | |
|---|---|---|---|---|---|
| GR1/gr1-1 | GR1/gr1-1 | GR1/GR1 | |||
| ntra/ntra | × | Col-0 | NTRA/ntra | NTRA/ntra | |
| ntrb/ntrb | NTRB/ntrb | NTRB/ntrb | |||
| Frequency* | 13 | 16 | |||
| B: | Female parent | X | Male parent | Progeny genotype | |
|---|---|---|---|---|---|
| GR1/gr1-1 | GR1/gr1-1 | GR1/GR1 | |||
| Col-0 | × | ntra/ntra | NTRA/ntra | NTRA/ntra | |
| ntrb/ntrb | NTRB/ntrb | NTRB/ntrb | |||
| Frequency | 0 | 38 | |||
*Twenty-nine progeny were genotyped by PCR. χ 2 = 0.416 for 1:1 segregation, P = 0.52.
†Thirty-eight progeny were genotyped by PCR. χ 2 = 29 for 1:1 segregation, P = 0.
The genetic evidence suggests that at least one of the cytosolic disulfide Rs, GR1, NTRA, or NTRB, is required for development of the male gametophyte. To test this hypothesis, the presence of the 3 transcripts was measured in mature pollen. Semiquantitative RT-PCR indicated that GR1 and NTRA were both expressed in pollen, whereas no signal could be detected for NTRB Fig. S8. In flowers, GR1 and both NTR isoforms were detected. Selfing of a mutant homozygous for ntra and heterozygous for gr1 resulted in a 1:1 segregation of the progeny (15:15, χ2 = 0, P = 1) expected if the double knockout is lethal. In combination with the ntrb knockout, gr1 segregated with a 1:2:1 ratio (7:17:6; χ2 = 0.6, P > 0.5).
Discussion
Catalytic Activity of Arabidopsis GR1.
Arabidopsis GR1 is present as a FAD-bound homodimer (Fig. 1) similar to homologous flavoprotein disulfide reductases from other organisms (15). The Km value of 77 μM for the substrate GSSG is similar to Km values reported for GRs from E. coli (61 μM), yeast (55 μM), and human (72 μM) (26–28). GRX1-roGFP2 reports the actual EGSH (3), and due to its midpoint potential of −280 mV (24), allows the determination of residual amounts of GSSG in the presence of GR and the second substrate NADPH. Both, in vitro (Fig. 2) and in vivo (Fig. 4) GR1 maintain an EGSH of ≈−310 mV, which is consistent with earlier in vivo measurements in Arabidopsis and HeLa cells (2, 3). Due to irreversibility of the reaction, GR1 can, thus, maintain nanomolar GSSG concentrations in the cytosol.
GR1 Is Not Essential for Plant Development.
In mammals and bacteria, the inhibitor BCNU has been used (22, 23). However, it does not inhibit plant GR1. Thus, to confirm the importance of GR1, T-DNA knockout mutants were selected and characterized. In the absence of detectable GR1 transcript and GR1 protein in both isolated mutants and no elevation of organellar GR2 protein, the remaining GR activity of total protein extract of ≈35% in gr1 mutants compared with wild-type plants can be attributed to GR2 activity (29) and possibly other cytosolic reduction systems. The 65% share of the overall GR activity in leaf extracts for cytosolic GR1 is much higher than 20% activity associated with the cytosol in pea leaves (30).
ATP-binding cassette (ABC) transporters with micromolar Km values have been proposed as backup systems for insufficient cytosolic GR activity (31). Although roGFP at this stage is not capable of detecting increased accumulation of GSSG in the ER or the vacuole, the absence of continuous accumulation of GSSG and the less reducing cytosolic EGSH of gr1 mutants rather suggest an alternative cytosolic reduction system with lower efficiency than GR1.
TRX and NTR Can Replace GR In Vitro.
An EGSH of −270 mV in the cytosol is still sufficiently negative to maintain metabolic functions under normal conditions, which is consistent with reports of GR deletion mutants in S. cerevisiae (32). The use of a ratiometric probe in this work allowed dynamic in vivo measurements of EGSH under induced oxidative stress. These measurements, showed (i) that the buffer capacity of gr1 against ROS is significantly diminished, and (ii) that another less efficient reduction system is in place (Fig. 4G). In mammalian cells, GR and NTR share many common features, including similar primary and tertiary structures and high similarities of active site residues (33). However, lack of electrostatic attraction in the potential GSSG binding pocket prevents efficient GSSG reduction by human NTR (34). However, this observation cannot be directly transferred to plants, because GR1 and NTRA in this case are not related to each other (35). Also, our results for Arabidopsis clearly disproved the hypothesis of direct GSSG reduction for NTRA (Fig. 5). Only together with TRX and NADPH as electron donor, NTRA is capable of reducing GSSG, suggesting that the TRX system as a whole may constitute a backup system for GR1. When measured in vitro, the activity of NTRA/TRXh3 toward GSSG was 200-fold lower than the activity of GR1 (Fig. 5). This large difference in activity highlights the difficulties of direct extrapolation from in vitro data to the in vivo situation where the gr1 mutant still has a considerable reduction capacity for GSSG (Fig. 4). Within the pool of 11 cytosolic type h TRX (36), some isoforms may be more efficient than the tested TRXh3. Also, the in vitro assay does not consider the relative abundance of the respective oxidoreductases. The abundance of documented ESTs indicates that NTR (60 ESTs) and TRXh (>100 ESTs) may be far more abundant than GR1 (30 ESTs), at least at mRNA level. The situation in the Arabidopsis gr1 mutant to some extent resembles the naturally occurring situation in Drosophila melanogaster, where the TRX system substitutes for the lacking GR (9). It can also not be excluded that some other alternative GSSG reduction systems may act in Arabidopsis and contribute to maintenance of GSH redox homeostasis in gr1 mutants. A multifunctional TRX GR has been found in Schistosoma mansoni, which has an N-terminal GRX-like domain for TRX-independent GSSG reduction (37). In Synechocystis, which also lacks a genuine GR, NTR-reducible GRXs may act as intermediate reducers of GSSG (38). Due to the high number of isoforms, further dissection of individual contributions to GSSG is a difficult challenge. However, the genetic evidence provided here (see below) clearly shows the need of either GR1 or NTR in plants. The NTRs are highly similar across the plant kingdom, and wheat NTR is a highly efficient reducer of Arabidopsis TRXh. Thus, it can be assumed that the backup of cytosolic GR1 by the NTR/TRX system is universal across plant species. In contrast to S. cerevisiae, where mutual functional backup between NTR/TRX and GR has been shown (20), redundancy between GR and the NTR/TRX system in plants seems to be restricted to the cytosol, because mutants lacking organellar GR2 are embryo lethal (12). Deletion of GR1 may be tolerable, because oxidative stress in the cytosol is relatively low compared with plastids and mitochondria (39). A drop in cytosolic EGSH is likely to gradually affect downstream signaling and developmental processes. Antisense lines for Arabidopsis GSH1 indicated that 5% of wild-type GSH, which would result in an EGSH of ≈−240 mV, already limits the growth rate and renders plants more sensitive to environmental stress (40). Progressive growth inhibition and increasing stress sensitivity is also apparent for the even more severely affected rml1 mutant, which has <5% of wild-type GSH, and embryo lethal gsh1 deletion mutants (41, 42). Deletion of GR and a concomitant 21-fold increase in GSSG have been reported to cause oxidation of GRXs in S. cerevisiae (20). Even though this oxidation does not have an immediate effect on plant development, it can be assumed that excessive oxidation of GRXs can affect downstream processes under conditions of severe stress.
Either GR1 or NTRA Is Required for Pollen Viability.
Genetic evidence for the functional overlap between GR1 and the TRX system was gained from the combination of the gr1 mutation with a ntra ntrb double knockout. In Arabidopsis, the ntra and ntrb deletion mutants on their own do not show any phenotype (16). A growth phenotype of the ntra ntrb double knockout indicated functional redundancy between the 2 NTRs in the diploid phase (18). Interestingly, our present results also show functional overlap between cytosolic GR1 and NTRA during the haploid phase. Cross-pollination experiments showed that the lethal phenotype can be attributed to the male gametophyte, whereas oocytes with the triple mutation in GR1, NTRA, and NTRB genes are viable, as shown by the segregations after pollination with the wild-type (Table 1, Fig. S7). The difference between male and female gametophytes remains elusive, but it may be that pollen depends on a more robust redox buffer system. Pollen tube growth is associated with increased ROS production (43), which may be deleterious in the triple mutant. Oocytes, in contrast, are resting and also much better protected by maternal tissues. Transmission of defective gr1 alleles in pollen can only occur in the presence of functional NTRA, suggesting that GR1 and NTRA proteins have a major role in pollen fertility. Indeed, detection of GR1 and NTRA transcripts in pollen are also supported by proteomic analysis (44). However, NTRB mRNAs were hardly detected in pollen, and this protein has never been found in the pollen proteome. The fact that no NTRB transcript was found in pollen and that gr1 in a homozygous ntrb background segregated 1:2:1 unambiguously shows that NTRB is not important for gr1 transmission. However, this does not exclude the possibility that NTRB might contribute to cytosolic GSH redox homeostasis in mature plants, especially under conditions of severe oxidative stress. Because gr1 ntra pollen are not viable, homozygous mutants required to test for the function of GR1 and NTRs in embryonic tissue and mature plants can only be generated from conditional knockouts activated after fertilization. RNAi and inducible expression constructs will be helpful to investigate this question in more detail.
In summary, our study shows that NTRA together with TRXh3 exhibit functional redundancy with cytosolic GR1. Biochemical and genetic evidence shows that GR1 constitutes the main GR activity in Arabidopsis. Despite the presence of an efficient backup system, deletion of GR1 significantly lowers the buffering capacity of the cytosolic GSH pool against ROS. The delayed reduction of GSSG after an exogenously triggered oxidation in the gr1 mutant indicates that gr mutants expressing GRX1-roGFP2 may constitute a suitable system for detection of stress-induced ROS signals.
Materials and Methods
Plant Material and Growth Conditions and Treatments.
Plants were soil grown in 8.5:15.5-h day/light cycle at a temperature between 18 and 22 °C, 100 μE m−2 s−1 and 50% humidity. For Confocal laser scanning fluorescence microscopy (CLSM) analysis, transgenic seeds were surface sterilized and grown on 0.5× Murashige and Skoog (MS) medium (pH 5.7, 1% sucrose, 0.7% agar). Rosette leaves of 19- to 22-d old T2-seedlings were used for imaging.
For stress treatments of in vitro seedlings, seeds were surface sterilized and plated on 0.5× MS medium including Gamborg B5 vitamins [pH 5.7, 1% (wt/vol) sucrose, 0.8% (wt/vol) plant agar] (Duchefa). To supplement the growth medium with H2O2, CdCl2, and CuSO4 at various concentrations, appropriate amounts of the respective filter-sterilized stock solutions were added to 0.5× MS medium before gelling.
Isolation of Total RNA and Semiquantitative RT-PCR.
Total RNA was extracted from Arabidopsis leaves, pollen, or inflorescences, and reverse transcribed in cDNA; cDNA was subsequently used for semiquantitative RT-PCR. For details, see SI Materials and Methods.
Molecular Cloning and Plasmid Constructs.
Cloning and expression of proteins is described in SI Materials and Methods.
Protein Purification.
Recombinant roGFP protein was cloned, expressed, and purified as previously described (2). For all other proteins, details are described in SI Materials and Methods.
Identification of T-DNA Insertion Lines.
Two T-DNA insertion lines, gr1-1 and gr1-2, were obtained form the SALK collection, and characterized as described in SI Materials and Methods. Alleles of ntra and ntrb were identified as described (18).
Mutant Crosses and Pollen Competition.
For generation of ntra ntrb gr1 triple mutant, the ntra ntrb double mutant was crossed with either gr1-1 or gr1-2. The F1 plants were selfed, and plants with the desired genotype were selected in the F2 generation. Pollen competition experiments were performed as described (18).
Protein Extraction from Arabidopsis Leaves.
Protein was extracted from 250-mg ground leaf material using extraction buffer (50 mM Hepes, pH 7.4/10 mM KCl/1 mM EDTA/1 mM EGTA/10% vol/vol glycerol) supplemented with 1 mM PMSF. The extract were desalted with NAP5 columns (GE Healthcare), and resuspended in 1 mM K2HPO4/KH2PO4, pH 7.4/1 mM EDTA. Protein concentrations were determined by Bradford.
GR Activity.
Twenty micrograms of proteins were assayed in 100 mM K2HPO4/KH2PO4, pH 7.4/1 mM EDTA/750 μM DTNB/200 μM NADPH/400 μM GSSG. Absorbance was measured at 412 nm. NADPH and GSSG were always freshly prepared. For inhibition experiments, BCNU was dissolved in ethanol and added to the concentrations indicated.
Antibody Production and Protein Gel Blot Analysis.
Experimental details are provided in SI Materials and Methods.
HPLC Analysis of Glutathione.
GSH and GSSG were analyzed from 7-week-old, soil grown Arabidopsis plants as described (2). The amount of GSH measured after extraction with NEM refers to the amount of GSSG present in the sample. After subtraction form the total GSH pool measured after extraction with DTT, the difference corresponds to the amount of GSH present in the sample.
Redox-Sensitive GFP Imaging and GR Assay Using GRX1-roGFP.
Ratiometric imaging of roGFP, perfusion experiments, and image analysis were done as described previously (45).
Supplementary Material
Acknowledgments.
This work was supported by Centre National de la Recherche Scientifique Genoplante Project GNP0508, the Agence Nationale de la Recherche (ANR), ANR-Blanc Grant 06-0047 (to W.S.), and in part by grants to R.H.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900206106/DCSupplemental.
References
- 1.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plants Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Meyer AJ, et al. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- 3.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 4.Noctor G, Foyer CH. Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 5.Ball L, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16:2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer AJ. The integration of glutathione homeostasis and redox signaling. J Plant Physiol. 2008;165:1390–1403. doi: 10.1016/j.jplph.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Xing S, Rosso MG, Zachgo S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development. 2005;132:1555–1565. doi: 10.1242/dev.01725. [DOI] [PubMed] [Google Scholar]
- 8.Ndamukong I, et al. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 9.Kanzok SM, et al. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 10.Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. BBA-Gen Subjects. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Chew O, Whelan J, Millar AH. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem. 2003;278:46869–46877. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- 12.Tzafrir I, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouché N, Bouchez D. Arabidopsis gene knockout: Phenotypes wanted. Curr Opin Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 15.Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: Advances in chemistry and function. In: Moldave K, editor. Progress in Nucleic Acid Research and Molecular Biology. Vol 78. San Diego: Elsevier Academic; 2004. pp. 89–142. [DOI] [PubMed] [Google Scholar]
- 16.Reichheld JP, Meyer E, Khafif M, Bonnard G, Meyer Y. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 2005;579:337–342. doi: 10.1016/j.febslet.2004.11.094. [DOI] [PubMed] [Google Scholar]
- 17.Laloi C, et al. Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA. 2001;98:14144–14149. doi: 10.1073/pnas.241340898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichheld J-P, et al. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell. 2007;19:1851–1865. doi: 10.1105/tpc.107.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae response to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 20.Trotter EW, Grant CM. Non-reciprocal regulation of the redox state of the glutathione-glutaredoxin and thioredoxin systems. EMBO Rep. 2003;4:184–188. doi: 10.1038/sj.embor.embor729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song J-Y, Cha J, Lee J, Roe J-H. Glutathione reductase and a mitochondrial thioredoxin play overlapping roles in maintaining iron-sulfur enzymes in fission yeast. Eukaryot Cell. 2006;5:1857–1865. doi: 10.1128/EC.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 23.Starke PE, Farber JL. Endogenous defenses against the cytotoxicity of hydrogen peroxide in cultured rat hepatocytes. J Biol Chem. 1985;260:86–92. [PubMed] [Google Scholar]
- 24.Dooley CT, et al. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 25.Alexander M. Differential staining of aborted and nonaborted pollen. Stain Technol. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- 26.Savvides SN, Karplus PA. Kinetics and crystallographic analysis of human glutathione reductase in complex with a xanthene inhibitor. J Biol Chem. 1996;271:8101–8107. doi: 10.1074/jbc.271.14.8101. [DOI] [PubMed] [Google Scholar]
- 27.Henderson G, et al. Engineering the substrate specificity of glutathione reductase toward that of trypanothione reduction. Proc Natl Acad Sci USA. 1991;88:8769–8773. doi: 10.1073/pnas.88.19.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massey V, Williams CH., Jr On the reaction mechanism of yeast glutathione reductase. J Biol Chem. 1965;240:4470–4480. [PubMed] [Google Scholar]
- 29.Chew O, Rudhe C, Glaser E, Whelan J. Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol Biol. 2003;53:341–356. doi: 10.1023/b:plan.0000006939.87660.4f. [DOI] [PubMed] [Google Scholar]
- 30.Edwards EA, Rawsthorne S, Mullineaux P. Subcellular distribution of multiple forms of glutathione reductase in leaves of pea (Pisum sativum L. ) Planta. 1990;180:278–284. doi: 10.1007/BF00194008. [DOI] [PubMed] [Google Scholar]
- 31.Foyer CH, Theodoulou FL, Delrot S. The functions of inter- and intracellular glutathione transport systems in plants. Trends Plants Sci. 2001;6:486–492. doi: 10.1016/s1360-1385(01)02086-6. [DOI] [PubMed] [Google Scholar]
- 32.López-Mirabal HR, Winther JR. Redox characteristics of the eukaryotic cytosol. BBA-Mol Cell Res. 2008;1783:629–640. doi: 10.1016/j.bbamcr.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 33.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: Implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci USA. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urig S, Lieske J, Fritz-Wolf K, Irmler A, Becker K. Truncated mutants of human thioredoxin reductase 1 do not exhibit glutathione reductase activity. FEBS Lett. 2006;580:3595–3600. doi: 10.1016/j.febslet.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 35.Meyer Y, et al. Evolution of redoxin genes in the green lineage. Photosynth Res. 2006;89:179–192. doi: 10.1007/s11120-006-9095-3. [DOI] [PubMed] [Google Scholar]
- 36.Meyer Y, et al. Glutaredoxins and thioredoxins in plants. BBA-Mol Cell Res. 2008;1783:589–600. doi: 10.1016/j.bbamcr.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Alger HM, Williams DL. The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase. Mol Biochem Parasitol. 2002;121:129–139. doi: 10.1016/s0166-6851(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 38.Marteyn B, Domain F, Legrain P, Chauvat F, Cassier-Chauvat C. The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803. Mol Microbiol. 2009;71:520–532. doi: 10.1111/j.1365-2958.2008.06550.x. [DOI] [PubMed] [Google Scholar]
- 39.Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. [Google Scholar]
- 40.Xiang C, Werner BL, Christensen EM, Oliver DJ. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 2001;126:564–574. doi: 10.1104/pp.126.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vernoux T, et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell. 2000;12:97–110. doi: 10.1105/tpc.12.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006;141:446–455. doi: 10.1104/pp.106.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardenas L, McKenna ST, Kunkel JG, Hepler PK. NAD(P)H Oscillates in Pollen Tubes and Is Correlated with Tip Growth. Plant Physiol. 2006;142:1460–1468. doi: 10.1104/pp.106.087882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noir S, Brautigam A, Colby T, Schmidt J, Panstruga R. A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun. 2005;337:1257–1266. doi: 10.1016/j.bbrc.2005.09.185. [DOI] [PubMed] [Google Scholar]
- 45.Schwarzländer M, et al. Confocal imaging of glutathione redox potential in living plant cells. J Microsc. 2008;231:299–316. doi: 10.1111/j.1365-2818.2008.02030.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.