Abstract
The predatory ecology of Varanus komodoensis (Komodo Dragon) has been a subject of long-standing interest and considerable conjecture. Here, we investigate the roles and potential interplay between cranial mechanics, toxic bacteria, and venom. Our analyses point to the presence of a sophisticated combined-arsenal killing apparatus. We find that the lightweight skull is relatively poorly adapted to generate high bite forces but better adapted to resist high pulling loads. We reject the popular notion regarding toxic bacteria utilization. Instead, we demonstrate that the effects of deep wounds inflicted are potentiated through venom with toxic activities including anticoagulation and shock induction. Anatomical comparisons of V. komodoensis with V. (Megalania) priscus fossils suggest that the closely related extinct giant was the largest venomous animal to have ever lived.
Keywords: evolution, phylogeny, squamate, protein, toxin
Predation by Varanus komodoensis, the world's largest extant lizard, has been an area of great controversy (cf. ref. 1). Three-dimensional finite element (FE) modeling has suggested that the skull and bite force of V. komodoensis are weak (2). However, the relevance of bite force and cranial mechanics to interpretations of feeding behavior cannot be fully evaluated in the absence of comparative data. Moreover, this previous analysis did not account for gape angle, which can significantly influence results (3). Irrespective of evidence for or against a powerful bite, V. komodoensis is clearly capable of opening wounds that can lead to death through blood loss (4). Controversially, the proposition that utilization of pathogenic bacteria facilitates the prey capture (4, 5) has been widely accepted despite a conspicuous lack of supporting evidence for a role in predation. In contrast, recent evidence has revealed that venom is a basal characteristic of the Toxicofera reptile clade (6), which includes the varanid lizards (7), suggesting a potential role of venom in prey capture by V. komodoensis that has remained unexplored. This is consistent with prey animals reported as being unusually quiet after being bitten and rapidly going into shock (4) and the anecdotal reports of persistent bleeding in human victims after bites (including B.G.F.'s personal observations). Shock-inducing and prolonged bleeding pathophysiological effects are also characteristic of helodermatid lizard envenomations (cf. ref. 8), consistent with the similarity between helodermatid and varanid venoms (6).
Here, we examine the feeding ecology of V. komodoensis in detail. We compare the skull architecture and dentition with the related extinct giant V. priscus (Megalania). In this 3D finite element modeling of reptilian cranial mechanics that applies a comparative approach, we also compare the bite force and skull stress performance with that of Crocodylus porosus (Australian Saltwater Crocodile), including the identification of optimal gape angle (an aspect not considered in previous nonreptilian comparative FE analyses). We also consider the relative roles of pathogenic bacteria vs. envenomation.
Results
Gape angles were adjusted to find the optima, which were 20° for V. komodoensis and 0° for C. porosus. Our adjustment for the optimal gape angle resulted in a V. komodoensis maximum posterior bite force (maximal predicted jaw muscle forces applied at optimal gape) of 39 N, considerably higher than in a previous analysis (2) yet still 6.5 times less than the 252 N produced by the bite of a C. porosus (Australian saltwater crocodile) with comparable skull size. In both simulations where only the influence of jaw adductors was considered (anterior and posterior bites), the mean stress in tetrahedral elements composing the V. komodoensis cranium was less than half that in C. porosus. When maximal jaw muscle forces predicted for the C. porosus cranium were applied to the V. komodoensis cranium, mean tetrahedral stress was 4.8 times greater than when forces predicted for V. komodoensis were applied; similar relative mean stresses in V. komodoensis mandible were 7.7 times greater (see Table S1). When maximal jaw muscle forces predicted for a similar-sized C. porosus were applied to V. komodoensis, mean tetrahedral stress was 4.8 times greater in the cranium and 7.7 times greater in the mandible (and see Table S1). This suggests that regions of the V. komodoensis skull would be likely to experience mechanical failure if subjected to forces exerted by the jaw muscles of a similar-sized C. porosus. In the 4 simulations applying varanid forces generated by postcranial musculature (lateral shake, pulling, head depression, and axial twisting), the mean tetrahedral stress in the skull of V. komodoensis was least during prey pulling and highest under axial twisting. These simulations were run at optimal gape angle and did not incorporate jaw adductor-generated forces. In all 4 simulations the comparative stress on the V. komodoensis skull was significantly greater than that seen in the C. porosus skull (Fig. 1 and Table S1).
Fig. 1.
Finite element models of (A) Varanus komodoensis and (B) Crocodylus porosus, assembled from computed tomography (CT) data and solved (C--H) to show stress distributions (Von Mises) under a range of loading cases and to determine maximal bite forces. (C and D), anterior bite; E and F, prey pull; G and H, axial twist.
Magnetic resonance imaging (MRI) of a preserved V. komodoensis head revealed a compound mandibular venom gland with a major posterior compartment (C1 in Fig. 2A) and 5 smaller anterior compartments (C2–C6 in Fig. 2A) (Movies S1, S2, and S3). Separate ducts were shown to lead from each compartment, opening between successive serrated pleurodont teeth (Fig. 2B), making this the most structurally complex reptile venom gland described to date (9–11). Consistent with the positioning of the ducts, the teeth lack the grooves commonly associated with venom delivery in helodermatid lizards (Fig. 3) or non-front-fanged snakes (9). The protein secreting venom glands are readily differentiated from the infralabial mucus glands (Fig. 2 A–C and E). The venom glands are encapsulated by a sheath of connective tissue and possess large distinct lumina (Fig. 2C). As in snakes, the central lumina are fed by an extensive network of tubular lumina and internal ducts (Fig. 2D), providing in a 1.6-m specimen a combined 1-mL liquid storage volume.
Fig. 2.
Anatomical investigation of the Varanus komodoensis venom system. (A) Magnetic resonance imaging of the V. komodoensis head showing the protein-secreting mandibular venom gland, with the 6 compartments colored in alternating red and pink (C1–C6), and the mucus-secreting infralabial gland in yellow (L). (B) Longitudinal MRI section showing the large duct emerging separately from each compartment of the mandibular venom gland and threading between the mucus lobes of the infralabial gland to terminate between successive teeth (black oval areas). (C) Transverse MRI section showing the large central lumen of the mandibular venom gland and individual lobes of the labial gland. (D) Transverse histology of Masson's Trichrome-stained section showing the intratubular lumina of the mandibular venom gland that feed into the large central lumen. (E) Transverse histology of Masson's Trichrome-stained section of a mucus infralabial gland showing numerous tightly packed internal lobules (note that the ∼6 large dark folds are histology artifacts).
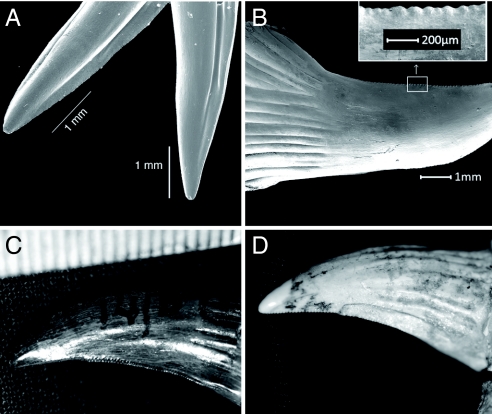
Fig. 3.
Scanning electron microscope views. (A) Both medial and anterior grooves (Left) and a sharp cutting edge at the bottom of a grooved Heloderma suspectum tooth (Right). (B) The structure and serrations of a Varanus komodoensis tooth. The Inset shows a magnified image of the serrations along the posterior (cutting) edge of the tooth. The length of the tooth does not show the presence of a discrete groove often associated with venom delivery systems. The maxillary teeth of the extinct Varanus (Megalania) priscus [C (QMF14/871) and D (QMF12370)] show clear similarity to those of V. komodoensis in overall shape and type of serration. V. (Megalania) priscus differs from V. komodoensis by possessing labial and lingual grooves that run from the base of the tooth (dorsal of the plicidentine) toward the tooth tip.
To further elucidate the potential role of envenomation in the predatory ecology of V. komodoensis we investigated the biochemical composition and toxinological properties of the venom. Mass spectrometry revealed a mixture of proteins (Fig. S1) as complex as that seen for snakes in similar analyses (12). Analysis of the mandibular venom gland cDNA library revealed a molecularly diverse transcriptome (Table 1) with 35% of the 2000 transcripts encoding known toxin types from other Toxicofera venoms (6, 9, 13). Molecular complexity and expression levels were comparable to those documented for venomous snakes (cf. ref. 14). Toxin types encoded were congruent with protein molecular weights revealed by mass spectrometry (Fig. S1). The toxin classes identified were AVIT, cysteine-rich secretory proteins (CRISP), kallikrein, natriuretic peptide, and type III phospholipase A2 protein scaffolds (GenBank accession nos. EU195455–EU195461). Isoforms isolated from V. komodoensis conserve the biochemistry of these coagulopathic, hypotensive, hemorrhagic, and shock-inducing toxins (Table 1; Figs. S2–S7). Crude venom cardiovascular studies revealed potent hypotensive effects mediated by an endothelium-independent vasodilator effect on vascular smooth muscle (Fig. 4). Pure V. komodoensis natriuretic toxin was demonstrated to have the same potent endothelium-independent hypotensive effect as the crude venom (Fig. 4) isoforms from V. varius (6) and Oxyuranus microlepidotus (15) venoms.
Table 1.
Molecular biodiversity of toxin types detected in V. komodoensis venom
| Toxin type | Previously characterized bioactivities (refs. 6, 9, and 13) |
|---|---|
| AVIT | Potent constriction of intestinal smooth muscle, resulting in painful cramping, and induction of hyperalgesia. |
| CRISP | Basal toxic activity of paralysis of peripheral smooth muscle and induction of hypothermia via blockage of L-type Ca2+- and BKCa K+-channels. Derived activities include blockage of cyclic nucleotide gated calcium channels. |
| Kallikrein | Basal toxic activity of increasing vascular permeability and production of hypotension in addition to stimulation of inflammation. Derivations affect the blood through the cleavage of fibrinogen. |
| Natriuretic | Basal activity potent induction of hypotension leading to loss of consciousness. Derived activities include cardiovascular effects independent of the GC-A receptor and antiplatelet activities evolved for emergent domains upstream of the natriuretic peptide domain. |
| PLA2 (T-III) | Anticoagulation via platelet inhibition. |
Fig. 4.
The depressor effect of Varanus komodoensis crude venom (A and B) or natriuretic toxin C on the blood pressure of anesthetized rats. The relaxant effect of V. komodoensis venom (D and E) or natriuretic toxin F on rat precontracted aorta is shown. The effects of the natriuretic toxin from V. varius are shown in C and F for comparison.
Discussion
Bite force has been established as a predictor of prey size in extant mammalian carnivores (16, 17). Moreover, in placental and marsupial carnivores a tendency to take relatively large prey is typically reflected in the skull's ability to withstand the high forces generated during prey subjugation (18). Previous work has suggested weak bite forces in V. komodoensis (2), but whether similar relationships exist among reptiles and their prey has remained unclear. Consideration of gape angle shows that maximal bite forces previously predicted for V. komodoensis by using FE techniques (2) were underestimates (and suggests that future studies should consider this factor). Nonetheless, bite force remains much lower than in a C. porosus of comparable size, suggesting that bite force may not be a reliable indicator of prey size in extant varanid lizards. Our findings suggest that, relative to C. porosus, the skull of V. komodoensis is poorly adapted to resist the erratic forces generated in a sustained bite and hold attack on large prey. Further, it is least well adapted to resist torsion. In contrast, the V. komodoensis skull is best adapted to resist forces generated in pulling on a prey item (or a prey item pulling back). These results are consistent with observational data showing that V. komodoensis opens wounds by biting and simultaneously pulling on prey by using postcranial musculature (4), thereby supplementing relatively weak jaw adductors by recruiting postcranial musculature. Our findings are also in accord with the view that the killing technique of V. komodoensis is broadly similar to that of some sharks and Smilodon fatalis (saber cat). Despite obvious anatomical differences, these unrelated predators kill or are thought to have killed (respectively) large prey by using relatively weak bite forces amplified by sharp teeth and postcranial input (19, 20).
It has been argued that as an alternative or adjunct to direct physical trauma V. komodoensis possesses pathogenic bacteria in its saliva (4, 5) capable of delivering lethal toxic effects through induction of sepsis and bacteremia in its prey (4). Supposedly V. komodoensis tracks the infected prey item or, alternatively, another V. komodoensis specimen benefits from an opportunistic feed. Neither of these scenarios, however, has actually been documented. Regardless, septicemia is popularly accepted as an integral part of the predatory ecology. The feeding behavior of V. komodoensis has also been interpreted within this framework, such as being an altruistic behavior with a group level benefit. Further, it has been speculated that bacterial growth and delivery are facilitated by the production of copious quantities of bloody saliva (4). Although wild-caught individuals have been shown to harbor a variety of oral bacteria, no single pathogen was found to be present in all V. komodoensis studied (5). Moreover, the bacterial species identified were unremarkable in being similar to those identified in the oral cavities of other reptiles or being typical gut contents of the mammalian species on which they prey (21–23).
We also note that the laboratory mouse study (5) that attributed lethal effects from V. komodoensis saliva to the pathogen Pasteurella multocida could not confirm the presence of the pathogen in the majority of donor animals. P. multocida is not typical reptile flora but is prevalent in mammals, especially individuals already under stress from disease or old age (21–23). Because these individuals are often the ones selected as prey items by V. komodoensis, it seems likely that P. multocida, and other bacterial species, are transiently acquired flora of the prey animals and other environmental sources. This would explain the observed variability in bacterial load within and among individuals (5). Such interindividual variability makes it exceedingly unlikely that toxic bacteria could reliably induce sepsis in prey animals to the extent that this would become an evolutionarily successful mechanism on which V. komodoensis could rely on for prey capture. We conclude that there is no compelling evidence for the hypothesized role of pathogenic bacteria in the predatory ecology of V. komodoensis.
Although we dismiss pathogenic bacteria as integral to the feeding ecology of V. komodoensis, we consider that envenomation may play an important role given the presence of venom delivery systems in other varanids (6). Absence of a modified dental architecture such as the delicate, grooved venom-delivering helodermatid teeth is likely one of the reasons that the venomous nature of V. komodoensis has been overlooked. Consistent with the skull performing best in response to pulling forces, V. komodoensis instead uses its robust serrated teeth to cut compliant tissue in an expanded use of the “grip-and-rip” mechanism (24), resulting in 2 parallel extremely deep wounds in prey items (4), which would allow ready entry of the venom.
However, our analyses point to a further feature that distinguishes the feeding ecology of V. komodoensis from other sharp-toothed predators such as sharks or saber cats: venom. Adult specimens (in the range of 1.4–1.6 m and 5–8 kg) of the closely related and known venomous lizard V. varius (6), with their proportionally smaller heads relative to V. komodoensis, yield up 10 mg dry weight material obtained by gentle squeezing of the glands to obtain the major lumen liquid contents or up to 50 mg through the utilization of pilocarpine stimulation (thus obtaining full lumen liquid contents intracellular stored material). Our results show that a 1.6-m V. komodoensis has an internal gland volume of 1 mL and, utilizing the V. varius results as a foundation, we estimate that the total protein (liquid plus storage contents) would be 150 mg, with 30 mg of this in the form of readily deliverable major lumen liquid contents. Because gland size and venom yield increase proportionally with head size in reptiles (25), a full-sized (3 m) adult V. komodoensis would thus potentially yield up to1.2 mL and 200 mg in major lumen liquid contents or 6 mL of liquid and 1 g of dry material (full contents including storage).
We have shown that in the species that have developed secondary forms of prey capture (e.g., constricting) or have switched to feeding on eggs, the reptile venom system undergoes rapid degeneration characterized by significant atrophying of the glands, reduction in fang length, and accumulated deleterious mutations in the genes encoding for the venom proteins (9, 26, 27). This is a consequence of selection pressure against the bioenergetic cost of protein production (28). The robust glands and high venom yield in V. komodoensis thus argue for continued active use of the venom system in V. komodoensis.
Our data thus suggest that V. komodoensis venom potentiates the deep laceration-induced bleeding and hypotension through anticoagulative changes in blood chemistry (PLA2 toxins) and shock-inducing lowering of blood pressure (CRISP, kallikrein, and natriuretic toxin types), with the prey item further immobilized by the hyperalgesic cramping AVIT toxins (Table 1). Our in vivo studies show that an i.v. dose of 0.1 mg/kg produces profound hypotension whereas 0.4 mg/kg is enough to induce hypotensive collapse (Fig. 4). Thus, a typical adult V. komodoensis prey item such as a 40-kg Sunda Deer would require 16 mg of protein to enter blood circulation to induce complete hypotensive collapse but only 4 mg to induce immobilizing hypotension. This is a realistic amount to deliver as even the weak delivery system of rear-fanged snakes, which may require a degree of mastication for venom delivery, can deliver >50% of the venom available in their glands (29). Such a fall in blood pressure would be debilitating in conjunction with blood loss and would render the envenomed prey unable to escape. These results are congruent with the observed unusual quietness and apparent rapid shock of prey items (4).
The predatory ecology of the V. komodoensis extinct gigantic close relative V. (Megalania) priscus is also unresolved. In particular, whether or not it was primarily a predator or a scavenger has remained an open question (20). Our recent finding of a common origin of the venom system in lizards and snakes (6) and the close evolutionary relationship between V. priscus and the clade of the predatory extant venomous lizards V. komodoensis, V. salvadori, and V. varius (30, 31) lends weight to the hypothesis that V. priscus was a combined-arsenal predator rather than a simple scavenger. Like the other members of this unique varanid lizard clade, the jawbones of V. priscus are also relatively gracile compared with the robust skull and the proportionally larger teeth similarly serrated (Fig. 3). Application of the “extant phylogenetic bracket” comparative approach (32) indicates that V. priscus used the same combined arsenal of large serrated teeth with anticoagulant and hypotension-inducing venom. Maximal body masses exceeding 2,000 kg and 7 m in length have been proposed for V. priscus (although such numbers rely on extrapolation well beyond available data ranges for extant lizards). However, even conservatively assuming geometric similitude (20) with large V. komodoensis suggests that its Pleistocene relative would have achieved at least 575 kg body weight and lengths exceeding 5.5 m. Scaling upward from V. komodoensis, we estimate that a varanid of this size range would produce a total stored venom protein yield (lumen liquid plus storage granules) reaching 6 g, with 1.2 g as readily deliverable major lumen liquid contents.
Our multidisciplinary analyses paint a portrait of a complex and sophisticated tooth/venom combined-arsenal killing apparatus in V. komodoensis and its extinct close relative V. priscus. Thus, despite a relatively weak skull and low bite force, we suggest that the combination of highly and very specifically optimized cranial and dental architecture, together with a capacity to deliver a range of powerful toxins, minimizes prey contact time and allows this versatile predator to access a wide range of prey including large taxa. These results indicate that V. priscus was the largest venomous animal to have ever lived.
Materials and Methods
Magnetic Resonance Imaging.
A Philips Achieva, 3T Tesla clinical MRI scanner (Philips Medical Systems) with an 8-channel knee receiver coil was used to scan the preserved head of V. komodoensis ZMB47873 from the Berlin Museum. A 3D fast field echo sequence was performed, comprising 400 slices with field of view, 160 mm; acquisition voxel size, 0.27 × 0.27 × 0.80 mm3; repetition time/echo time/flip angle, 12 ms/5.8 ms/20°; and scan time, 14:44 min. These 3T images then served as a guide for the acquisition of high-resolution images acquired on a Philips Intera, 1.5T clinical MRI scanner (Philips Medical Systems), by using a surface coil with a diameter of 23 mm. Subsequently several scout images were performed and the coil was repositioned to obtain a maximum signal-to-noise ratio in the anatomical area to be imaged. A 3D T2-weighted Turbo Spin Echo sequence was acquired with an echo train length of 11; repetition time/echo time/flip angle, 1000 ms/60 ms/90°; field of view, 100 mm (220 slices); acquisition voxel size, 0.2 × 0.2 × 0.2 mm3; number of signal averages, 2; and scan time, 49:32 min. Images with different angulations were reconstructed afterward on a Vitrea workstation (Vital Images). Image segmentation of the glands was performed manually in Amira 4.1 (Mercury Computer Systems) and 3D surface renderings were generated.
Tooth Structure Investigations.
V. komodoensis teeth were mounted on holders, sputter-coated with gold (20 nm thick), and imaged in an FEI XL30-FEG scanning electron microscope. Images were taken at 10 kV and a working distance of 7.1 mm and were postprocessed by using Adobe Photoshop (CS2). Heloderma suspectum teeth were air dried and specimens of tooth were mounted on duraluminium stubs using carbon adhesive paste (Agar Scientific) and coated with a 10- to 20-Å layer of gold palladium (3 min) by plasma discharge in an E300 diode sputter coater (Polaron) before being imaged in a JSM 840 scanning electron microscope. The specimen was examined at low power (50×) for orientation and at magnifications between 650× and 7,000× for observations of the grooves and sharp cutting edge. V. priscus teeth were from the Queensland Museum collection.
Surgical Excision of the Mandibular Venom Gland.
V. komodoensis venom glands for histological and cDNA analysis were obtained under anesthesia from “Nora,” a terminally ill animal at the Singapore Zoo. The animal was anesthetized with a combination of zolazepam and tiletamine (Zoletil, Virbac) at 3 mg/kg administered i.v. in the ventral tail vein. It was then intubated and maintained with Isoflurane (Attane, Minrad) at 1–3%. Respiration was assisted at a frequency of 2–3 breaths per minute. The animal was positioned in dorsal recumbency. A 5-cm incision was made between the second and the third row of mental (intermandibular) scales parallel to the lower jaw, thus exposing the capsule common to the 2 infralabial glands. Careful dissection was carried out to separate the mandibular venom gland from the mucus gland. The fibrous sheath between the 2 glands is very thin and does not separate readily in V. komodoensis. There are multiple ducts and blood vessels interlacing with one another. The posterior four-fifths of the left mandibular gland was separated and the affluent vessels were severed only seconds before it could be placed into a container with liquid nitrogen. The samples for histopathology were taken from the remaining anterior portion and fixed immediately in 10% formalin. On the right side the histology sections were taken midportion and the remaining gland was preserved in liquid nitrogen. The animal was killed by i.v. administration of 5 g of pentobarbital (Dorminal 20%, Alfasan).
Histology.
Formalin-fixed samples of the venom gland were dehydrated through a series of ascending ethanol concentrations and then transferred into isopropanol before being embedded via xylene in paraffin (Paraplast, Sherwood). After hardening, paraffin sections were cut at a thickness of 5 μm, by using a manual rotation microtome (Jung). For deparaffinization, the slides were transferred into Histoclear (Shandon), washed several times in 100% ethanol, and rehydrated via a series of descending ethanol concentrations. The slides were then stained by using the trichrome staining method of Masson-Goldner (applying light green as connective tissue stain). cDNA library construction, molecular modeling, and phylogenetic analyses were as described (6, 9).
Peptide Synthesis.
The natriuretic peptides IQPEGSCFGQLIDRIGHVSGMGCNKFDPNKESSSTG-NH2 (V. komodoensis) and LQPEGSCFGQKMDRIGHVSGMGCNKFDPNKGSSSTGKK-NH2 (V. varius) were synthesized on a CEM Liberty Peptide Synthesizer, by using Fmoc solid-phase peptide chemistry. The peptide was cleaved from the solid-phase resin with tfa/H2O/triisopropylsilane/3,6-dioxa-1,8-octane-dithiol (90:2.5:2.5:5) for 2 h. The crude peptide was isolated by ether precipitation, dissolved in 30% acetonitrile/water, and lyophilized. The crude linear peptide was reversed-phase HPLC purified (Agilent 1200 HPLC System) before forming the single disulfide bond by treatment with dipyridyldithiol (1 equivalent) in 100 mM ammonium acetate (peptide concentration 1 mg/mL, 30 min). The pure cyclic peptide was isolated by directly applying the ammonium acetate solution to a reversed-phase HPLC column, isolating pure peptide fractions, and lyophilization of the product. The identities of the pure cyclic peptides were confirmed by high-resolution mass spectrometry on an Agilent QTOF 6510 LC/MS mass spectrometer. Bioactivity studies using anesthetized rats and isolated blood vessels were as described in refs. 6 and 15.
SELDI-TOF MS.
Samples were analyzed by using the following arrays and wash buffers: Q10 (100 mM Tris·HCl, pH 9) and CM10 (20 mM sodium acetate, pH 5) (Bio-Rad). Arrays were initially assembled in a humidity chamber and preequilibrated with the appropriate wash buffer. Each spot was loaded with 5 μL of wash buffer, followed by an incubation step for 5 min on a shaking table. The buffer was wicked off by using a Kimwipe and the equilibration step was repeated. The samples (5 μL, 0.5 mg/mL diluted 1:2 into wash buffer) were applied to each spot and incubated for 1 h. Chips were washed with the appropriate wash buffer 3 times for 5 min, followed by two 1-min washes with 1 mM Hepes, pH 7.2. The chips were air dried and l μL of 50% saturated sinapinic acid (Bio-Rad) in 50% (vol/vol) acetonitrile, 0.5% trifluoroacetic acid was applied onto each spot twice, and arrays were air dried between each application. Chips were analyzed by SELDI-TOF MS by using a PBSIIc (Bio-Rad) and resulting spectra were examined by using ProteinChip software. Data were collected in both the low (<20 kDa) and high mass ranges (>20 kDa), and laser and sensitivity settings were optimized for each condition.
Supplementary Material
Acknowledgments.
We thank the Singapore Zoological Gardens for making V. komodoensis tissues available for the study and Dr. Rainer Guenther and Dr. Mark-Oliver Roedel (Museum of Natural History, Humboldt University) for generously loaning us a preserved specimen for the magnetic resonance imaging studies. H.S. is grateful to GlaxoSmithKline for an exclusive version of Swiss Protein Databank Viewer. We are particularly grateful to Lim Kok Peng Kelvin and the staff of Naturalis Museum for all of their kind help and to Nicolas Vidal for constructive criticisms. This work was funded by grants (to B.G.F.) from the Australian Academy of Science, the Australian French Association for Science and Technology, the Australia and Pacific Science Foundation, the Australian Research Council (DP0665971 and DP0772814 also to W.C.H. and J.A.N.), the CASS Foundation, the Ian Potter Foundation, the International Human Frontiers Science Program Organisation, the Netherlands Organisation for Scientific Research, the University of Melbourne (Faculty of Medicine and Department of Biochemistry and Molecular Biology), and a Department of Innovation, Industry and Regional Development Victoria Fellowship. This work was also funded by an Australian Government Department of Education, Science and Training International Science Linkages grant (to B.G.F. and J.A.N.) and funding from the Bio21 Molecular Science and Biotechnology Institute (to B.G.F., J.K., and D.S.) for peptide synthesis. Further funding came from Australian Research Council and University of New South Wales Internal Strategic Initiatives grants (to S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810883106/DCSupplemental.
References
- 1.Diamond J. Did Komodo dragons evolve to eat pygmy elephants? Nature. 1987;326:832. [Google Scholar]
- 2.Moreno K, et al. Cranial performance in the Komodo dragon (Varanus komodoensis) as revealed by high-resolution 3-D finite element analysis. J Anat. 2008;212:736–746. doi: 10.1111/j.1469-7580.2008.00899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourke J, Wroe S, Moreno K, McHenry C, Clausen P. Effects of gape and tooth position on bite force and skull stress in the Dingo (Canis lupus dingo) using a 3-dimensional finite element approach. PLoS ONE. 2008;3:e2200. doi: 10.1371/journal.pone.0002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auffenberg W. Behavioral Ecology of the Komodo Monitor. Gainesville, FL: Univ Press Florida; 1981. [Google Scholar]
- 5.Montgomery JM, Gillespie D, Sastrawan P, Fredeking TM, Stewart GL. Aerobic salivary bacteria in wild and captive Komodo dragons. J Wildl Dis. 2002;38:545–551. doi: 10.7589/0090-3558-38.3.545. [DOI] [PubMed] [Google Scholar]
- 6.Fry BG, et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- 7.Vidal N, Hedges SB. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C R Biol. 2005;328:1000–1008. doi: 10.1016/j.crvi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Tu AT, Murdock DS. Protein nature and some enzymatic properties of lizard Heloderma suspectum suspectum (gila monster) venom. Comp Biochem Physiol. 1967;22:389–396. doi: 10.1016/0010-406x(67)90602-0. [DOI] [PubMed] [Google Scholar]
- 9.Fry BG, et al. Evolution of an arsenal. Mol Cell Proteomics. 2008;7:215–246. doi: 10.1074/mcp.M700094-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Gabe M, Saint Girons H. Données histologiques sur les glandes salivaires des lépidosauriens. Mém Mus Natl Hist Nat Paris. 1969;58:1–118. [Google Scholar]
- 11.Kochva E. Biology of the Reptilia. London: Academic; 1978. [Google Scholar]
- 12.Fry BG, et al. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun Mass Spectrom. 2003;17:2047–2062. doi: 10.1002/rcm.1148. [DOI] [PubMed] [Google Scholar]
- 13.Fry BG. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagstaff SC, et al. Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem Biophys Res Commun. 2008;365:650–656. doi: 10.1016/j.bbrc.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Fry BG, et al. Novel natriuretic peptides from the venom of the Inland Taipan (Oxyuranus microlepidotus): Isolation, chemical and biological characterisation. Biochem Biophys Res Commun. 2005;327:1011–1015. doi: 10.1016/j.bbrc.2004.11.171. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen P, Wroe S. Bite forces and evolutionary adaptations to feeding ecology in carnivores. Ecology. 2007;88:347–358. doi: 10.1890/0012-9658(2007)88[347:bfaeat]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Wroe S, McHenry C, Thomason J. Bite club: Comparative bite force in big biting mammals and the prediction of predatory behaviour in fossil taxa. Proc R Soc B Biol Sci. 2005;272:619–625. doi: 10.1098/rspb.2004.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wroe S, Clausen P, McHenry C, Moreno K, Cunningham E. Computer simulation of feeding behaviour in the Thylacine and Dingo as a novel test for convergence and niche overlap. Proc R Soc B Biol Sci. 2007;274:2819–2828. doi: 10.1098/rspb.2007.0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McHenry CR, Wroe S, Clausen PD, Moreno K, Cunningham E. Supermodeled sabercat, predatory behavior in Smilodon fatalis revealed by high-resolution 3D computer simulation. Proc Natl Acad Sci USA. 2007;104:16010–16015. doi: 10.1073/pnas.0706086104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wroe S. A review of terrestrial mammalian and reptilian carnivore ecology in Australian fossil faunas, and factors influencing their diversity: The myth of reptilian domination and its broader ramifications. Aust J Zool. 2002;50:1–24. [Google Scholar]
- 21.Georghiou PR, Mollee TF, Tilse MH. Pasteurella multocida infection after a tasmanian devil bite. Clin Infect Dis. 1992;14:1266–1267. doi: 10.1093/clinids/14.6.1266. [DOI] [PubMed] [Google Scholar]
- 22.Gerardo SH, Goldstein EJC. Antimicrobial therapy & vaccines. In: Yu V, Merrigan T, editors. Pasteurella multocida and Other Species. Baltimore, MD: The Williams & Wilkins Co.; 1998. pp. 326–335. [Google Scholar]
- 23.Goldstein EJC, Agyare EO, Vagvolgyi AE, Halpern M. Aerobic bacterial oral flora of garter snakes—-development of normal flora and pathogenic potential for snakes and humans. J Clin Microbiol. 1981;13:954–956. doi: 10.1128/jcm.13.5.954-956.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abler WL. The serrated teeth of tyrannosaurid dinosaurs, and biting structures in other animals. Paleobiology. 1992;18:161–183. [Google Scholar]
- 25.Mirtschin PJ, et al. Influences on venom yield in Australian tigersnakes (Notechis scutatus) and brownsnakes (Pseudonaja textilis: Elapidae, Serpentes) Toxicon. 2002;40:1581–1592. doi: 10.1016/s0041-0101(02)00175-7. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Fry BG, Kini RM. Putting the brakes on snake venom evolution: The unique molecular evolutionary patterns of Aipysuras eydouxii (Marbled sea snake) phospholipase A(2) toxins. Mol Biol Evol. 2005;22:934–941. doi: 10.1093/molbev/msi077. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Fry BG, Kini RM. Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii) J Mol Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. [DOI] [PubMed] [Google Scholar]
- 28.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes WK, Lavinmurcio P, Kardong KV. Delivery of duvernoy secretion into prey by the Brown Tree Snake, Boiga irregularis (Serpentes, Colubridae) Toxicon. 1993;31:881–887. doi: 10.1016/0041-0101(93)90223-6. [DOI] [PubMed] [Google Scholar]
- 30.Ast JC. Mitochondrial DNA evidence and evolution in Varanoidea (Squamata) Cladistics. 2001;17:211–226. doi: 10.1111/j.1096-0031.2001.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 31.Head JJ, Barrett FLS PM, Rayfield EJ. Neurocranial osteology and systematic relationships of Varanus (Megalania) prisca Owen, 1859 (Squamata: Varanidae) Zool J Linn Soc. 2009;155:455–457. [Google Scholar]
- 32.Witmer L. The Extant Phylogenetic Bracket and the Importance of Reconstructing Soft Tissues in Fossils. Cambridge, UK: Cambridge Univ Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.