Abstract
Perinuclear aggresome formation is a key mechanism to dispose of misfolded proteins that exceed the degradative capacity of ubiquitin–proteasome and autophagy–lysosome systems. Functional blockade of either degradative system leads to an enhanced aggresome formation. The tuberous sclerosis complex–Ras homologue enriched in brain–mammalian target of rapamycin (TSC–Rheb–mTOR) pathway is known to play a central role in modulating protein synthesis and autophagy. However, in spite of the constitutive activation of mTOR and the abrogated autophagy activity in TSC1- or TSC2-deficient cells, the TSC mutant cells are defective in aggresome formation and undergo apoptosis upon misfolded protein accumulation both in vitro and in vivo. High Rheb activity in TSC mutant cells inhibits aggresome formation and sensitizes cell death in response to misfolded proteins. Surprisingly, this previously unrecognized function of Rheb is independent of TOR complex 1. Active Rheb disrupts the interaction between dynein and misfolded protein cargos, and therefore blocks aggresome formation by inhibiting dynein-dependent transportation of misfolded proteins. This study reveals a function of Rheb in controlling misfolded protein metabolism by modulating aggresome formation.
Keywords: mammalian target of rapamycin, tuberous sclerosis complex
Cells mainly deploy 3 mechanisms to counteract misfolded proteins: up-regulating chaperones to assist protein refolding (1), proteolytic degradation of the misfolded/damaged proteins involving ubiquitin–proteasome and autophagy–lysosome systems (2), and formation of detergent-insoluble aggresome by transporting the misfolded proteins to the juxtanuclear region in a microtubule-dependent manner (3, 4). In addition, inhibition of translation is also associated with misfolded protein response as a means to relieve overloading of the folding and degradation machineries (1). Dysregulation of these mechanisms has been implicated in the pathogenesis of a plethora of clinical diseases exemplified by cystic fibrosis and Parkinson's disease (2).
The mammalian target of rapamycin (mTOR) integrates diverse signals to regulate fundamental cellular processes, such as translation, cell growth, autophagy, and stress response (5, 6). Recent studies not only have positioned mTOR downstream of the tuberous sclerosis complex 1 (TSC1) and TSC2 tumor suppressors (7), but also revealed the existence of 2 different mTOR complexes (8) (named mTORC1 and mTORC2), which differ in molecular composition, cellular function, upstream regulation, and sensitivity to rapamycin. TSC1 and TSC2 form a dimer and function as a GTPase-activating protein (GAP) to inactivate the small GTPase Ras homologue enriched in brain (Rheb) (9–13), which directly binds to and activates mTORC1 kinase activity (14). Both genetic and biochemical studies have established a linear TSC1/2–Rheb–mTORC1 pathway (5–7, 9–13), in which Rheb is the key downstream effector of TCS1/2 in controlling mTORC1 activity. However, whether Rheb has mTORC1-independent function is currently unclear. Here, we show that high Rheb activity inhibits aggresome formation and sensitizes cell death in response to misfolded proteins. Surprisingly, this function of Rheb is independent of mTORC1 activity. We further show that Rheb blocks aggresome formation by inhibiting dynein-dependent transportation of misfolded proteins. This study reveals a previously uncharacterized function of Rheb in controlling misfolded protein metabolism by modulating aggresome formation.
Results
Autophagy Defect Leads to Enhanced Aggresome Formation.
Autophagy is an intracellular degradation system that delivers cytoplasmic constituents to lysosomes (15, 16). Recent studies have not only identified a large number of autophagy genes (ATGs) but also expanded the autophagic cargos from intracellular pathogens to misfolded proteins (15–18). During autophagy, light chain 3-I (LC3-I) is converted to LC3-II by phosphatidylethanolamine conjugation, and this LC3 conversion is frequently used as an autophagy marker (19). As expected, no LC3 conversion was observed in the autophagy-defective ATG5−/− cells (Fig. S1A). Upon accumulation of misfolded protein triggered by proteasome inhibition with MG132, the ATG5−/− cells underwent dramatic aggresome formation that was reflected by the enhanced deposition of ubiquitinated proteins into detergent-insoluble fraction (Fig. S1A). The ATG5−/− cells also showed a significant increase in aggresomes as determined by immunostaining for ubiquitin–protein conjugates, an aggresome marker (4) (Fig. S1B). Staining for vimentin and histone deacetylase 6 (HDAC6), 2 known aggresome markers (4), confirmed that the ubiquitin conjugate stains for aggresome (Fig. S1C). The enhanced aggresome formation in ATG5−/− cells supports the notion that autophagy plays a critical role in clearing aggresome (17, 18).
TSC Null Cells Are Aggresome-Defective Despite Abrogated Autophagy.
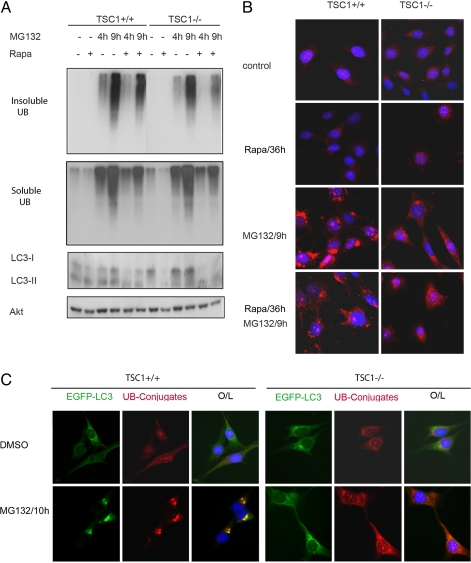
Consistent with the inhibitory role of mTORC1 in autophagy regulation (15, 16), the TSC1−/− mouse embryonic fibroblast (MEF), which has high mTORC1 activity (20), displayed a lower basal conversion of the autophagy marker LC3-I to LC3-II than the TSC1+/+ MEF (Fig. 1A). One would predict that the TSC1−/− MEFs should accumulate more aggresomes when proteasome degradation is inhibited. Surprisingly, the abrogated autophagy activity in TSC1−/− MEFs was not accompanied by an enhanced aggresome formation upon MG132 treatment, as indicated by the immunoblot for detergent-insoluble ubiquitinated proteins (Fig. 1A). In fact, the TSC1−/− cells displayed less ubiquitinated protein in the insoluble fraction. The distribution of polyubiquitin staining was dispersed in TSC1−/− MEFs as opposed to the perinuclear staining of aggresome observed in TSC1+/+ MEFs (Fig. 1B). A total of 78% ± 5.2% of TSC1+/+ MEFs underwent aggresome formation, whereas only 19.7% ± 3.8% of TSC1−/− cells formed aggresome with 9 h of MG132 treatment (P < 0.01). Consistent with the expected role of autophagy in counteracting aggresome, EGFP-LC3 fluorescent signals encroached the aggresome perinuclearly in TSC1+/+ MEFs and HeLa cells (Fig. 1C and Fig. S1D). In contrast, EGFP-LC3 remained as a diffusive pattern in TSC1−/− MEFs, further supporting an autophagy defect in TSC1−/− MEFs.
Fig. 1.
TSC1−/− MEF is defective in aggresome formation. (A) TSC1−/− cells have less insoluble ubiquitinated protein, even in the presence of rapamycin (Rapa). TSC1−/− MEF was a spontaneously immortalized MEF cell line derived from TSC1−/− embryos. TSC1+/+ and TSC1−/− MEFs with or without 20 nM rapamycin pretreatment for 36 h were treated with DMSO or 5 μM MG132 for 4 or 9 h before harvest. RIPA-insoluble cell lysates were normalized and blotted for ubiquitin (UB). RIPA-soluble lysates were blotted for UB, LC3, and Akt (for loading control). (B) Rapamycin does not restore aggresome formation in TSC1−/− cells. TSC1+/+ and TSC1−/− MEFs with or without 20 nM rapamycin pretreatment for 36 h were treated with DMSO or 5 μM MG132 for 9 h before fixation. Cells were immunostained for ubiquitin–protein conjugates (red), and nuclei were counterstained with DAPI (blue). (C) TSC1−/− cells are defective in autophagy and aggresome formation. TSC1+/+, TSC1−/− MEFs expressing EGFP-LC3 (green) treated with 5 μM MG132 or DMSO for 10 h were immunostained for ubiquitin–protein conjugates. Nuclei were stained with DAPI. The data show that LC3 signals encroach aggresome in TSC1+/+ but not in TSC1−/− cells. O/L denotes overlay.
TSC1 and TSC2 function as a complex in vivo. We wanted to test whether this aggresome deficiency also exists in TSC2 null cells. Rat LExF2 cells, which have TSC2 mutation and fail to express detectable TSC2 protein (21), were indeed unable to form aggresome, and this defect was rescued by TSC2 reconstitution (TSC2+/+; 78% ± 2.6% of TSC2+/+ vs. 16% ± 2.9% of TSC2−/− cells formed aggresomes; P < 0.01; Fig. S2). These results show that loss of TSC2 also leads to aggresome deficiency.
Defective Aggresome Formation in TSC Null Cells Is mTORC1-Independent.
The lack of aggresome formation despite an abrogated autophagy in TSC-deficient cells was totally unexpected. If constitutive activation of mTORC1 were the underlying mechanism causing this defect in TSC cells, then rapamycin pretreatment would restore aggresome formation in these cells. However, rapamycin treatment did not restore the detergent-insoluble ubiquitinated proteins in TSC−/− cells (Fig. 1A and Fig. S2A). Consistently, polyubiquitin immunostaining also revealed that rapamycin failed to reconstitute the aggresome formation in TSC-deficient cells (Fig. 1B and Fig. S2B). In fact, rapamycin reduced ubiquitinated proteins in the detergent-insoluble fractions in both TSC+/+ and TSC−/− cells, possibly because of the fact that rapamycin inhibits translation and induces autophagy, thus counteracting misfolded protein accumulation. Therefore, the high mTORC1 activity in TSC mutant cells is not responsible for the defects in aggresome formation in these cells.
Rheb Regulates Aggresome Formation.
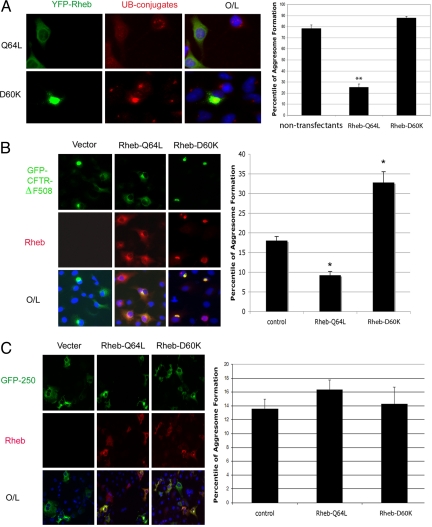
The defective aggresome formation in TSC1−/− and TSC2−/− cells cannot be explained by mTORC1 activation but suggests a possible involvement of Rheb in this process. We tested this possibility and found that expression of constitutively active Rheb-Q64L inhibited aggresome formation (Fig. 2A). Moreover, Rheb-Q64L also inhibited aggresome formation by cystic fibrosis transmembrane conductance regulator (CFTR)-ΔF508, which is a normally ubiquitinated and aggresome-prone mutant associated with cystic fibrosis (3), whereas Rheb-D60K enhanced CFTR-ΔF508 aggresome formation (Fig. 2B). Interestingly, Rheb-D60K was colocalized with aggresomes (Fig. 2 A and B). Taken together, our data show that active Rheb inhibits aggresome formation, whereas dominant-negative Rheb promotes this process. In contrast, Rheb had no significant effects on aggresome formation of GFP-250 (Fig. 2C), which forms aggresomes without being ubiquitinated (4), indicating that the aggresome-regulatory activity of Rheb applies only to ubiquitinated proteins.
Fig. 2.
Rheb regulates ubiquitinated aggresome formation. (A) Rheb regulates endogenous aggresome formation. YFP-Rheb-Q64L (constitutively active) or YFP-Rheb-D60K (dominant-negative) constructs were transfected into A549 cells, followed by 5 μM MG132 treatment for 12 h. (Left) Cells were immunostained for ubiquitin (UB)–protein conjugates, and nuclei were stained with DAPI. (Right) Percentage of aggresome-harboring cells was counted among the transfectants and nontransfectants. Values represent means ± SD of 3 independent experiments. **, P < 0.01 (Student's t test). (B) Rheb regulates aggregation of CFTR-ΔF508. At 18 h after cotransfection of EGFP-CFTRΔF508 with myc-Rheb-Q64L, myc-Rheb-D60K, or control vector, COS-7 cells were immunostained for MYC and nuclei (DAPI). (Left) Representative images are shown from 3 independent experiments. (Right) Percentage of aggresome-harboring cells was scored among the cotransfectants. Values represent means ± SD of 3 independent experiments. *, P < 0.05. (C) Rheb does not regulate nonubiquitinated aggresome formation. (Left) At 18 h after cotransfection of GFP-250 plasmids with myc-Rheb-Q64L, myc-Rheb-D60K, or vector constructs, COS-7 cells were immunostained for MYC and nuclei (DAPI). (Right) Percentage of aggresome-harboring cells was scored among the cotransfectants as shown. Values represent means ± SD of 3 independent experiments. O/L denotes overlay.
TSC Deficiency or Rheb Overexpression Sensitizes Cells to Apoptosis in Response to Misfolded Proteins.
TSC mutant cells are defective to cope with various stresses (6). For example, energy starvation induces apoptosis in TSC mutant cells (20). We found that TSC1−/− but not TSC1+/+ cells displayed massive apoptosis upon MG132 treatment (Fig. 3 A and B). Given the fact that the TSC1−/− cells are defective in aggresome formation (Fig. 1 A and B), our data support a beneficial role for aggresome formation in cell survival in misfolded protein response (4, 22, 23). Similar results were observed in the TSC2−/− cells (Fig. S3 A and B). We then tested whether high Rheb activity sensitizes cells to misfolded protein-induced apoptosis. As expected, expression of Rheb-Q64L indeed sensitized cells to apoptosis upon MG132 treatment (Fig. 3C and Fig. S3C), recapitulating the TSC−/− phenotype. The TSC-deficient cells were also sensitive to PS341 (bortezomib), a Food and Drug Administration-approved proteasome inhibitor for chemotherapy (Fig. S4). Although rapamycin pretreatment did produce some protection for cell viability, presumably because of its effect on autophagy induction, a substantial population of cells still underwent apoptosis in response to MG132 (Fig. 3 A and B). This observation is in dramatic contrast to the energy starvation response, in which rapamycin efficiently blocked the cell death induced by glucose starvation in TSC1−/− MEFs (20) (Fig. S3D). Therefore, misfolded protein-induced apoptosis is largely mTORC1-independent but correlates with high Rheb activity and defects in aggresome formation.
Fig. 3.
TSC1 deletion and Rheb activation sensitize misfolded protein-induced apoptosis. (A) TSC1−/− cells are sensitive to MG132. TSC1+/+ and TSC1−/− MEFs with or without 20 nM rapamycin (Rapa) pretreatment for 1 h were challenged with 5 μM MG132 or DMSO for 12 h. Phase-contrast images were taken. (B) MG132 induces apoptosis in TSC1−/− cells. TSC1+/+ and TSC1−/− MEFs with or without 20 nM rapamycin pretreatment for 1 h were challenged with 5 μM MG132 or DMSO for 4 or 9 h before harvest. Cell lysates were blotted for cleaved caspase-3, cleaved PARP, and Akt (for loading control). (C) Rheb-Q64L sensitizes cell death to MG132. (Left) MCF-7 stable cell clones expressing YFP or YFP-Rheb-Q64L treated with 5 μM MG132 for 24 h were immunostained with cleaved caspase-3 antibody, and nuclei were stained with DAPI. (Right) Percentage of cleaved caspase-3 positively stained cells for different clones is shown in diagram.
TSC1 Deletion Causes Aggresome Formation Deficiency in Vivo.
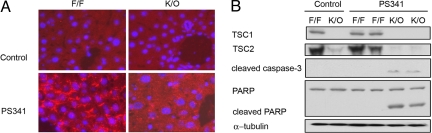
Here, we have demonstrated that Rheb inhibits aggresome formation accompanied by increased apoptosis upon proteasome inhibition in vitro. We wanted to test whether this phenomenon also exists in vivo. To this end, we generated liver-specific TSC1 knockout mice by using TSC1 flox and albumin Cre, which specifically expressed the Cre recombinase in liver. As shown in Fig. 4A, upon PS341 injection, ubiquitinated proteins aggregated in the wild-type hepatocytes, whereas they appeared as dispersed foci throughout cytoplasm in TSC1 knockout hepatocytes. Therefore, the TSC1−/− hepatocytes are defective in aggresome formation. Furthermore, the TSC1 knockout liver tissues also showed apoptosis upon PS341 injection, as indicated by the presence of cleaved caspase 3 and poly(ADP-ribose) polymerase (PARP; Fig. 4B).
Fig. 4.
TSC1 knockout causes defective aggresome formation and sensitizes misfolded protein-induced apoptosis. (A) TSC1 knockout hepatocytes are compromised in aggresome formation. Shown are representative images of ubiquitin–protein conjugate immunostaining and nuclei staining (DAPI) of liver frozen sections from TSC1flox/flox (F/F) and TSC1flox/flox, albumin-Cre (K/O) littermates with or without (control) PS341 treatment. (B) PS341 induces apoptosis in TSC1−/− liver. PS341 preferentially induces apoptosis in TSC1−/− hepatocytes. Liver homogenates from TSC1flox/flox (F/F) and TSC1flox/flox, albumin-Cre (K/O) littermates with or without (control) PS341 treatment were immunoblotted with TSC1, TSC2, cleaved caspase-3, PARP, and α-tubulin.
Rheb Regulates Microtubule/Dynein-Mediated Transportation of Misfolded Proteins.
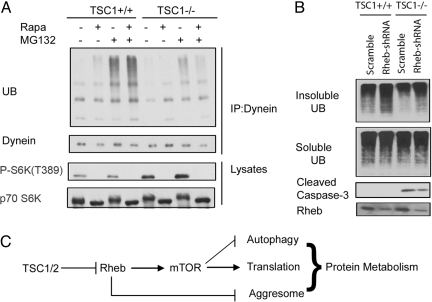
Aggresome formation necessitates retrograde transportation along the microtubule network, involving the minus end-directed motor protein dynein (4, 22). We found that far fewer ubiquitinated proteins were coimmunoprecipitated with dynein in TSC1−/− (Fig. 5A) or TSC2−/− (Fig. S5) cells, indicating a defect in the transportation of ubiquitinated protein to aggresome. However, rapamycin treatment not only had no effect on the interaction between dynein and ubiquitinated protein in TSC1+/+ cells but also failed to restore this interaction in TSC1−/− cells, indicating an mTORC1-independent function of Rheb in regulating microtubule–dynein-mediated misfolded protein transportation (Fig. 5A). We further investigated the role of Rheb in regulating the association between dynein and ubiquitinated protein by resorting to RNA interference. Short hairpin RNA (shRNA) against Rheb was lentivirally introduced into TSC1 MEFs to achieve Rheb knockdown. Consistent with an inhibitory role for Rheb in aggresome formation, Rheb knockdown not only enhanced ubiquitinated proteins in the detergent-insoluble fraction in both TSC1+/+ and TSC1−/− MEFs but also increased cell viability in TSC1−/− MEFs, as indicated by a reduced caspase-3 activation in response to MG132 (Fig. 5B). These results strongly support the notion that active Rheb inhibits aggresome formation by disrupting dynein-mediated transportation of ubiquitinated protein cargos, providing a biochemical basis for the defect of aggresome formation in the TSC mutant cells.
Fig. 5.
High Rheb activity inhibits the interaction between the dynein motor and ubiquitinated protein cargos. (A) The association between dynein and ubiquitinated proteins is disrupted by TSC1 deletion. TSC1+/+, TSC1−/− MEFs with or without 20 nM rapamycin (Rapa) pretreatment for 1 h were treated with 5 μM MG132 or DMSO for 4 h before harvest. Lysates were normalized and immunoprecipitated (IP) with dynein antibody. Immunoprecipitates were immunoblotted with antibodies for ubiquitin (UB) and dynein. Whole-cell lysates were blotted for P-S6K (T389) and p70 S6K. (B) Rheb knockdown increases cell viability in response to MG132. TSC1 MEFs lentivirally introduced with Rheb shRNA or scramble shRNA were challenged with 5 μM MG132 at 9 h before harvest. RIPA-insoluble cell lysates were normalized and blotted for UB. Soluble lysates were blotted for UB, Rheb, and cleaved caspase-3. (C) A proposed model for TSC1/2–Rheb–mTOR in the regulation of misfolded protein metabolism.
Discussion
The observations described in this report suggest a model in which the TSC–Rheb–mTOR pathway plays a major role in cellular protein metabolism by regulating translation, autophagy, and aggresome (Fig. 5C). The regulation of translation and autophagy is mediated by mTORC1, whereas the regulation of aggresome formation is Rheb-dependent but mTORC1-independent. mTORC1 activation by Rheb promotes general protein synthesis and inhibits aggresome-mediated, nonselective degradation. In addition, active Rheb also inhibits agrresome formation. Our observations not only establish an mTORC1-independent function of Rheb but also suggest a molecular basis for the coordination between aggresome and autophagy mediated by Rheb.
Rheb inhibits the association between the ubiqitinated protein cargos and the dynein motor, therefore inhibiting perinuclear aggresome formation. Because motor protein dynein itself does not bind to cargos directly (24), and a direct interaction between Rheb and dynein motor was not observed (Fig. S6), we reason that additional components acting between Rheb and dynein may exist. Interestingly, a substantial portion of Rheb-Q64L is colocalized with Rab7 (25), which plays a critical role in endocytosis and autophagy processes also involving the microtubule–dynein transportation system (26, 27), indicating the possibility that there might be some interplay between Rheb and the retrograde microtubule–dynein transportation system.
The observations described in this study have several implications. First, the segregation between cell death and aggresome formation under Rheb activation conditions supports a beneficial role for aggresome in coping with misfolded proteins (4, 23, 28). Second, mTORC1 inhibition has been shown to have a beneficial effect on aggresome-related disease models (29, 30). Strategies aimed at modulating Rheb activity may be more advantageous, because Rheb affects both autophagy via mTORC1 and aggresome independent of mTORC1 to counteract the toxicity of misfolded protein. Therefore, Rheb offers an attractive therapeutic target for diseases associated with misfolded protein. Moreover, our data show that high Rheb activity sensitizes cells to proteasome inhibitors. This observation suggests a potential for using proteasome-inhibiting drugs, such as bortezomib, to selectively kill tumor cells that have high Rheb activity.
Materials and Methods
Preparation of Detergent-Insoluble Fraction of Cell Lysates.
Cells were lysed in RIPA lysis buffer. After centrifugation for 15 min at 16,800 × g, the insoluble pellets were washed 2 times with PBS, and then they were suspended with RIPA buffer and underwent sonication for protein solubilization.
Immunostaining and Microscopy.
Cells were either fixed with 4% paraformaldehyde/PBS followed by permeabilization with 0.1% Triton X-100/PBS (CFTR, GFP-250, MYC, cleaved caspase-3), or fixed with methanol at −20 °C (ubiquitin–protein conjugates, HA, vimentin, MYC), then blocked with 20% FBS/PBS. Cells were then incubated with primary antibodies (1:100 dilution in blocking solution) at 4 °C overnight, then labeled with fluorescent-conjugated secondary antibody and mounted with Vectashield H-1200 (with or without DAPI; Vector Laboratories). Fluorescence signals were visualized with an Ultraview LCI Confocal Imaging system (Perkin–Elmer).
Generation of Liver-Specific TSC1-Deficient Mice.
Liver-specific TSC1 knockout mice were generated by breeding TSC1flox/flox mice (31) with albumin-Cre mice (Jackson Laboratory). Mice were maintained on the mixed genetic background (C57BL/6 × 129Sv × BALB/c). All animal experiments were conducted following protocols approved by the University Committee on the Use and Care of Animals at the University of Michigan. For genotyping, genomic DNA from mice tails was isolated and amplified by PCR. Primers F4536 (5′-AGGAGGCCTCTTCTGCTACC-3′) and R4830 (5′-CAGCTCCGACCATGAAGTG-3′) were used to detect the TSC1 conditional allele. PCR products were 295 bp for the wild-type allele and 486 bp for the conditional allele. The presence of the albumin-Cre allele was assessed by amplification of the Cre recombinase sequence using universal Cre primers.
Peritoneal Injection and Sample Processing.
PS341 was injected peritoneally into 6-week-old transgenic mice twice a week for 2 weeks at a dosage of 2 mg/kg body weight, followed by 2 injections every other day at a dosage of 8 mg/kg body weight. Liver resections were either homogenized in RIPA buffer or embedded in OCT compound (Sakura Finetek) for frozen section processing. Liver frozen sections were fixed with 1% paraformaldehyde/PBS for 10 min at room temperature before undergoing immunostaining.
Western Blotting and Immunoprecipitation.
For Western blot analysis, cells were lysed in RIPA lysis buffer with protease inhibitor tablet (Roche), and protein assays were routinely carried out to normalize the proteins by using a BCA protein assay kit (Pierce). For immunoprecipitation, cells were lysed in 1% Nonidet P-40 lysis buffer with protease inhibitor tablet as described previously (20). The same amount of proteins was incubated with 3 μg/mL dynein antibody and protein M-agarose beads (Sigma) and incubated for 3 h at 4 °C. Immunoprecipitates were washed 4 times in the lysis buffer before Western blot analysis.
Quantification of EGFP-CFTR-ΔF508 and GFP-250 Aggresome Formation and Cell Death.
EGFP-CFTR-ΔF508 or GFP-250 constructs were cotransfected with myc-tagged Rheb Q64L, D60K, or control vector (pRK5) in a 1:3 ratio into COS-7. At 18 h after transfection, cultures were stopped and underwent immunostaining. For aggresome quantification, random fields were selected, and the percentage of cells containing aggresomes was counted from 300–450 costained cells. Three independent experiments and Student's t test were carried out for statistical comparison. For cell death quantification, MCF-7 stable cell clones expressing YFP or YFP-Rheb-Q64L treated with 5 M MG132 for 24 h were immunostained for cleaved caspase-3. Random fields were selected for photographing.
Quantification of Endogenous Aggresome Formation.
Aggresome formation was detected by immunofluorescence staining with an anti-ubiquitin conjugate antibody as described previously (4). The presence of perinuclear inclusions was considered indicative of aggresome formation. For quantification, 10 fields from each sample were randomly selected. The percentage of cells (among ≈200 cells) containing aggresomes was counted. Student's t test was used for statistical comparison.
Other.
Supplementary Material
Acknowledgments.
We thank Drs. N. Mizushima (Tokyo University, Tokyo) for providing ATG5 MEF, D. Kwiatkowski (Harvard University, Boston) for TSC1 F/F mice, R. R. Kopito (Stanford University, Stanford, CA) for pEGFP/CFTR-ΔF508 plasmid, E. S. Sztul (University of Alabama, Birmingham) for GFP-250 construct, and R. S. Yeung (University of Washington, Seattle) for rat LExF2 cells. This work is supported by grants from the National Institutes of Health and the Department of Defense (to K.-L.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903621106/DCSupplemental.
References
- 1.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 3.Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 5.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 7.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 8.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 9.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 10.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 11.Stocker H, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 12.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 14.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Shintani T, Klionsky DJ. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 17.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 19.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 21.Jin F, et al. Suppression of tumorigenicity by the wild-type tuberous sclerosis 2 (Tsc2) gene and its C-terminal region. Proc Natl Acad Sci USA. 1996;93:9154–9159. doi: 10.1073/pnas.93.17.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- 23.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 24.Ross JL, Ali MY, Warshaw DM. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 28.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 29.Berger Z, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 30.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 31.Kwiatkowski DJ, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.