Abstract
Conserved chromosomal HP1 proteins capable of binding to histone H3 methylated at lysine 9 are believed to provide a dynamic platform for the recruitment and/or spreading of various regulatory proteins involved in diverse chromosomal processes. The fission yeast Schizosaccharomyces pombe HP1 family members Chp2 and Swi6 are important for heterochromatin assembly and transcriptional silencing, but their precise roles are not fully understood. Here, we show that Swi6 and Chp2 associate with histone deacetylase (HDAC) protein complexes containing class I HDAC Clr6 and class II HDAC Clr3 (a component of Snf2/HDAC repressor complex), which are critical for transcriptional silencing of centromeric repeats targeted by the heterochromatin machinery. Mapping of RNA polymerase (Pol) II distribution in single and double mutant backgrounds revealed that Swi6 and Chp2 proteins and their associated HDAC complexes have overlapping functions in limiting Pol II occupancy across pericentromeric heterochromatin domains. The purified Swi6 fraction also contains factors involved in various chromosomal processes such as chromatin remodeling and DNA replication. Also, Swi6 copurifies with Mis4 protein, a cohesin loading factor essential for sister chromatid cohesion, and with centromere-specific histone H3 variant CENP-A, which is incorporated into chromatin in a heterochromatin-dependent manner. These analyses suggest that among other functions, HP1 proteins associate with chromatin-modifying factors that in turn cooperate to assemble repressive chromatin; thus, precluding accessibility of underlying DNA sequences to transcriptional machinery.
Keywords: chromosome segregation, histone deacetylase, silencing, centromere, RNAi
Higher eukaryotic genomes contain large amounts of repetitive DNA elements, including satellite repeats, transposable elements, and their truncated derivatives. These sequences are major targets for the assembly of heterochromatin structures (1–4). The assembly of heterochromatin involves a complex array of histone modifications. In particular, heterochromatic regions are characterized by hypoacetylation of histones, and methylation of histone H3 at lysine 9 (H3K9me) (1, 5, 6). H3K9me has an important role in targeting HP1 family proteins that bind to methylated H3 tail via their amino-terminal chromodomain (7–10). HP1 proteins also contain a carboxy-terminal chromoshadow domain that is critical for their dimerization and interactions with diverse factors (11–13). Indeed, HP1 proteins contribute to diverse chromosomal processes, including silencing of target loci, proper segregation of chromosomes, and developmentally controlled long-range chromatin interactions (1). Also, heterochromatin prohibits illegitimate recombination at the repetitive DNA elements; thus, promoting genome stability (1, 14, 15).
In addition to their roles in the formation of repressive chromatin, HP1 proteins have also been found to associate with a subset of transcribed genes (16, 17). H3K9me and HP1γ are enriched at coding regions of transcribed genes in mammalian cells (18), and in Drosophila, HP1 localizes to actively transcribed heat shock puffs in polytene chromosomes (19). The significance of HP1 at transcribed loci remains largely unknown. It is possible that HP1 serves to recruit/stabilize factors that promote either polymerase (Pol) II transcription and/or processing of RNAs (1). In this regard, it should be noted that most eukaryotes contain multiple HP1 proteins that are believed to interact with a distinct set of proteins involved in different aspects of chromatin biology (1, 2).
In Schizosaccharomyces pombe, H3K9me and HP1 proteins localize to several sites, including loci within euchromatic domains, but these factors are highly enriched at pericentromeric regions, subtelomeres, and the silent mating-type locus (14). Each of these loci contains dg and dh repeats, which are transcribed by Pol II in a bidirectional manner, albeit at low levels, and serve as RNAi-dependent heterochromatin nucleation centers (1). Clr4/Suv39h methyltransferase responsible for H3K9 methylation (9) is essential for localization of chromodomain proteins Chp1, Chp2, and Swi6 (9, 14, 20, 21), which in turn are believed to mediate targeting of factors involved in different aspects of heterochromatin assembly and functions (1). Chp1, a component of the Argonaute (Ago1)-containing RNA-induced transcriptional silencing (RITS) complex (21) tethers RNAi machinery to heterochromatic loci, facilitating posttranscriptional silencing of repeats in cis (1). Similarly, Chp2 and Swi6 are required for localization of transcriptional silencing complex SHREC [Snf2/histone deacetylase (HDAC) repressor complex], which contains class II HDAC Clr3 and Snf2 ATPase Mit1 among other factors (22, 23). Swi6 also recruits a JmjC domain-containing antisilencing protein Epe1, cohesin, and factors involved in cell-type switching (1), and stabilizes chromatin association of RNAi factors such as RNA-dependent RNA polymerase (24).
Despite significant advances in our understanding of heterochromatin assembly and functions, the full extent of factors associated with HP1 proteins, in particular effectors that collaborate with these proteins to assemble repressive chromatin, remains to be fully explored. Also, it has been argued that heterochromatic silencing occurs largely because of posttranscriptional processing of transcripts, and that heterochromatin has little or no effect on Pol II occupancy at the target loci (25, 26). In this article, we use a combination of biochemical and genomics approaches to identify silencing complexes associated with Chp2 and Swi6, and explore their comparative roles in the silencing of heterochromatic repeat elements. Results presented also suggest potential mechanisms for Swi6 role in cohesin recruitment to chromatin, via its interaction with Mis4 cohesin loading factor, and for heterochromatin involvement in establishment of CENP-A chromatin.
Results
Effects of Different Heterochromatin Factors on Centromeric Silencing.
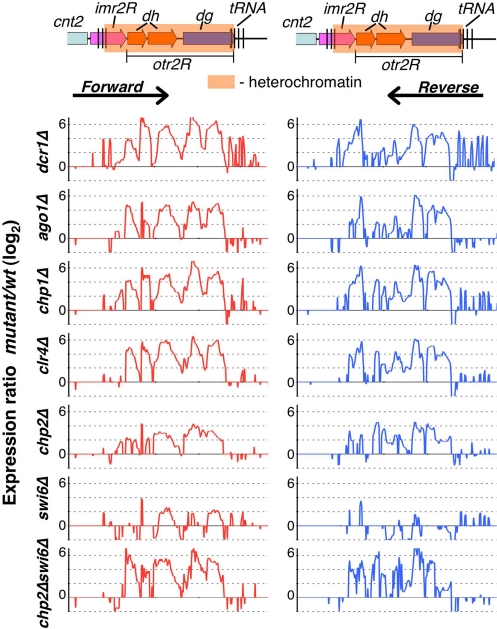
Heterochromatic silencing in S. pombe involves both transcriptional gene silencing (TGS) and posttranscriptional processing of transcripts in cis (cis-PTGS) by the RNAi machinery (1, 22). To explore the contributions of different factors to heterochromatic silencing, we used high-density custom tiling arrays containing probes corresponding alternatively to plus and minus strands to perform strand-specific expression profiling at centromere 2. Deletions of RNAi components Dcr1, Ago1, and Chp1 (a RITS subunit) resulted in widespread up-regulation of transcripts, including dg and dh repeat transcripts, at levels comparable to clr4Δ cells (Fig. 1), in which loss of H3K9me results in disruption of heterochromatin (9). Because RNAi machinery is required for targeting H3K9me and its associated factors involved in transcriptional repression and processing repeat transcripts (1), the observed up-regulation of transcripts in RNAi mutants likely represents cumulative defects in both cis-PTGS and TGS.
Fig. 1.
Chp2 and Swi6 contribute cooperatively to heterochromatic silencing. Schematic representation of S. pombe right pericentromeric repeat region at cen2 (Upper) is shown, including part of the central core region (cnt2), the innermost repeat (imr2R), and the outermost repeat (otr2R), which consist of dh and dg repeat regions. The shaded box represents the heterochromatin region covered by H3K9me and heterochromatin proteins. Expression profiles in indicated strain backgrounds (Lower); cDNAs isolated from wt and mutant strains were labeled and hybridized to high density microarrays containing alternating forward and reverse strand 60-mer probes at every 50 nt. Expression ratios (mutant/wt) for forward strand (Left) and reverse strand (Right) probes were plotted on a log2 scale. Note that, because of the repetitive nature of centromeric probes, the observed expression changes represent average values of centromeric repeat regions.
We next investigated the roles of Chp2 and Swi6 in heterochromatic silencing. These factors largely act downstream of H3K9me at centromeres (9, 20, 23). Compared with clr4Δ, deletion of chp2 or swi6 resulted in a partial loss of centromeric repeat silencing (Fig. 1). Levels of transcripts were generally higher in chp2Δ than in swi6Δ cells (Fig. 1). Because both Chp2 and Swi6 are believed to mediate localization of repressor complexes (22, 23), we tested whether the chp2Δswi6Δ double mutant would be more defective in silencing of centromeric repeat loci. Indeed, the double mutant showed a cumulative increase in the levels of repeat transcripts (Fig. 1).
Besides pericentromeric repeat transcripts, levels of transcripts corresponding to tRNAs surrounding centromeres were also affected, particularly in dcr1Δ, chp1Δ, and swi6Δ mutants (Fig. 1). Therefore, centromeric tRNA genes are also likely subject to RNAi and heterochromatic silencing.
Swi6 Associates with Mis4 Cohesin Loading Factor, CENP-A, and HDAC Repressors.
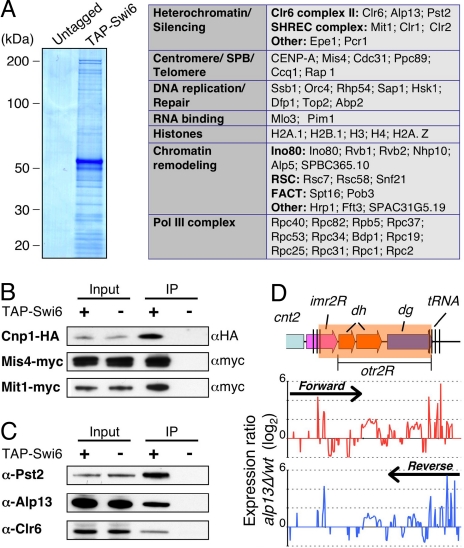
As mentioned above, Swi6 has been implicated in diverse chromosomal functions (1). To gain further insight into Swi6 functions, we performed tandem affinity purification (TAP) using extracts from strain expressing amino-terminal TAP-tagged Swi6 (TAP-Swi6). The fusion protein was expressed under the control of its native promoter at levels comparable with untagged Swi6, and was functional in silencing assays (Fig. S1). Also, ChIP-chip analysis showed that TAP-Swi6 localizes to heterochromatic loci at levels comparable to untagged Swi6 (Fig. S1). MS analysis of purified samples identified several factors specifically present in the TAP-Swi6 purified fraction (Fig. 2A). These factors included proteins involved in heterochromatin silencing, chromatin remodeling, RNA polymerase III transcription, DNA replication, and RNA binding (Fig. 2A and Fig. S2). Many peptides corresponded to FACT subunits Spt16 and Pob3 (Fig. 2A and Fig. S2) (27). Also, subunits of the INO80 (28) and RSC (29) chromatin remodeling complexes, Snf2-related proteins (such as Snf21 and Fft3), and a chromodomain helicase DNA-binding (CHD) family remodeling factor Hrp1 (30) copurified with Swi6 (Fig. 2A). Other notable proteins included Sap1, which has a role in DNA replication and mating-type switching (31, 32), telomere associated proteins (such as Ccq1 and Rap1) implicated in heterochromatin assembly (22, 33), and components of the microtubule organizing center known as the spindle pole body (SPB) (Fig. 2A and Fig. S2).
Fig. 2.
Swi6 associates with the Clr6 HDAC, and factors involved in chromosome segregation. (A) Coomassie blue staining of TAP Swi6 from cells expressing amino-terminally TAP-tagged Swi6 and untagged Swi6 are shown. The purified proteins were subjected to tandem MS (LC-MS/MS) analyses. The identified nuclear proteins were sorted into functional groups, as indicated in the table. (B and C) Fractions immunoprecipitated from indicated strains were subjected to Western blot analyses by using α-HA antibody (12CA5) to detect Cnp1-HA, α-myc antibody (9E10) to detect Mis4-myc or Mit1-myc (B) or α-Alp13, and α-PstII and α-Clr6 antibodies (C) to detect Clr6 complex II. (D) Expression profiling at the right pericentromeric repeat region of cen2 was performed for the wild-type and alp13Δ cells. Expression ratios (mutant/wt) for forward strand (Upper, red) and reverse strand (Lower, blue) probes were plotted on a log2 scale.
Several peptides derived from core histones, as well as variant histones, H2A.Z, and CENP-A (Cnp1) involved in chromosome segregation, were also identified (Fig. 2A). To explore whether indeed Cnp1 copurifies with Swi6, we constructed a strain expressing TAP-Swi6 and HA-tagged Cnp1 (Cnp1-HA). Western blot analysis of purified fractions confirmed the presence of Cnp1 specifically in the TAP-Swi6 purified fraction (Fig. 2B). Another factor critical for chromosome segregation that copurified with Swi6 is Mis4, implicated in the loading of cohesin complex onto chromosomes (34, 35). This result has important implications for understanding the mechanism linking Swi6 to preferential loading of cohesin across heterochromatic domains (Fig. S3A) (36, 37). To establish further interaction between Swi6 and Mis4, we tagged Mis4 at its caboxy-terminus with myc epitope (Mis4-myc), and performed coimmunoprecipitation analysis. Mis4-myc was detected in the affinity-purified Swi6 fraction, whereas no band was visible in the control fraction (Fig. 2B). Moreover, ChlP analyses showed that Mis4 was preferentially enriched across pericentromeric heterochromatic domains in wild-type cells but its localization was severely affected in swi6Δ mutant (Fig. S3A), which was also defective in cohesin (Rad21) recruitment to heterochromatic loci (Fig. S3A) (36, 37).
We also identified peptides, albeit few, corresponding to previously described Swi6-associated proteins, such as JmjC protein Epe1 (38, 39), Hsk1-Dfp1 (40, 41), and Pcr1 involved in heterochromatic silencing (42). Also detected were peptides representing different subunits of SHREC (Fig. 2A). The scarcity of peptides derived from these proteins may be because of their limited interactions with Swi6 at heterochromatic loci, or because these interactions occur in a specific chromatin context. Indeed, their interactions were significant as Swi6 was required for Epe1 localization throughout a pericentromeric domain (Fig. S3B). Also, Swi6 contributes to localization of SHREC at heterochromatic loci (22, 23). Supporting the SHREC association with Swi6, the Mit1 subunit of SHREC readily coimmunoprecipitated with both TAP-Swi6 (Fig. 2B) and untagged Swi6 (Fig. S4).
Interestingly, we found that a previously characterized Clr6 HDAC complex, which was shown to have silencing activity (43), also copurified with Swi6 (Fig. 2A). Western blot analyses with antibodies against Alp13, Pst2, and Clr6, 3 different subunits of a Clr6-containing HDAC complex, confirmed their interactions with Swi6 (Fig. 2C). These interactions are functionally important, because loss of Alp13 causes up-regulation of transcripts across the pericentromeric domain in a pattern similar to the swi6Δ mutant (Fig. 2D). Based on these data, Swi6 associates with various factors involved in diverse chromosomal functions, including SHREC and Clr6 HDAC complexes implicated in repression of heterochromatic repeats and euchromatic loci.
Chp2 Interacts with Clr3 HDAC-Containing SHREC.
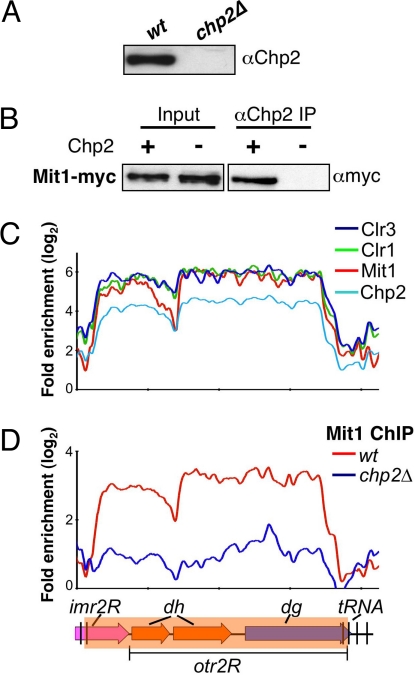
Chp2 and Swi6 contribute to the recruitment of SHREC to mediate transcriptional silencing at heterochromatic loci (22, 23). Swi6 interacts with the SHREC components (this study and ref. 23), but the exact in vivo function of Chp2 in this process is not known. To explore Chp2 function, we attempted to purify carboxyl terminal TAP-tagged Chp2 (Chp2-TAP). However, we noticed that tagged Chp2 was only partially functional in silencing assays (Fig. S5). MS analysis of a limited number of peptides recovered from Chp2-TAP purified fraction identified peptides matching the Mit1 subunit of SHREC. To determine whether Mit1 indeed interacts with Chp2, we generated rabbit polyclonal antibody against full-length recombinant Chp2 (Fig. 3A) and used affinity-purified antibody to perform immunoprecipitation. Our results show that Mit1 coimmunoprecipitates with Chp2 (Fig. 3B). This result, together with previous data (23), suggests that Chp2 physically associates with SHREC to mediate localization of this repressor complex to heterochromatic loci. Consistent with this view, ChIP-chip showed identical distribution patterns for Chp2 and SHREC components across the pericentromeric heterochromatic domain (Fig. 3C), and loss of Chp2 affects heterochromatic localization of Mit1 (Fig. 3D).
Fig. 3.
Chp2 interacts with SHREC. (A) The antibody specifically recognizes Chp2 protein. Extracts prepared from wt and chp2Δ strains were analyzed by Western blot analysis using α-Chp2 antibody. (B) Mit1, a component of the SHREC interacts with Chp2. Extracts prepared from wt and chp2Δ cells expressing Myc-tagged Mit1 were immunoprecipitated using the α-Chp2 antibody. Immunoprecipitated (IP) fractions were analyzed by Western blot analysis using α-myc antibody. (C) Chp2 colocalizes with SHREC subunits. Distribution profiles of indicated factors were determined by ChIP-chip. (D) Chp2 dependent localization of Mit1 at centromeric repeats. Mit1-myc distributions in wt or in chp2Δ backgrounds are shown. The probes are aligned to correspond with the schematic representation below.
We next probed the extent to which SHREC contributes to heterochromatic silencing as compared with Chp2. Mutation in clr3, which encodes a catalytic subunit of SHREC, showed up-regulation of both forward and reverse transcripts across the pericentromeric domain, as we also observed in chp2Δ cells (Figs. 1 and 4C). However, transcript levels were, in general, higher in the clr3 mutant compared with chp2 mutant cells. In particular, transcripts derived from a region corresponding to the inner most (imr) repeat and its adjacent dh fragment showed greater up-regulation in the clr3 mutant (Fig. 4C). Therefore, SHREC has a Chp2-independent role in heterochromatic silencing that may involve Swi6 and/or other factors capable of recruiting this repressor to repeat loci. Consistent with Swi6 involvement, we note that the chp2Δswi6Δ double mutant showed enhanced accumulation of forward and reverse transcripts corresponding to the imr heterochromatic region (Fig. 1).
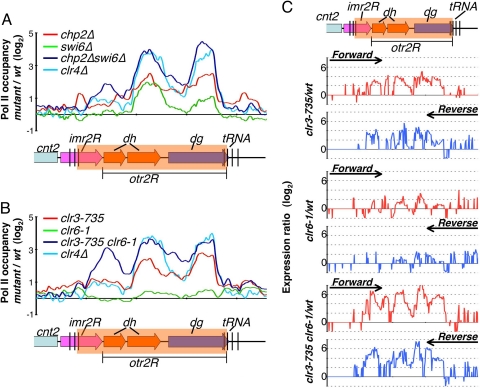
Fig. 4.
Chp2 and Swi6 proteins and their associated HDACs collaborate to limit Pol II occupancy at pericentromeric heterochromatic repeats. (A) Chp2 and Swi6 cooperatively contribute in transcriptional silencing of the pericentromeric heterochromatin regions. Pol II distributions were determined by ChIP-chip analysis in the indicated mutants and wt strains. Changes in Pol II occupancy in the mutants compared with wt (mutant/wt) were plotted on a log2 scale. (B) Clr3 and Clr6 cooperatively contribute to transcriptional silencing of the pericentromeric heterochromatin regions. Differential Pol II occupancy in indicated mutant strains compared with wt strains (mutant/wt) were plotted on a log2 scale. (C) Expression profiles for the indicated strains, at the right pericentromeric repeat region of cen2, are shown. Expression ratios (mutant/wt) for forward strand (Upper, red) and reverse strand (Lower, blue) probes were plotted on a log2 scale.
Chp2 and Swi6 Cooperate to Limit Pol II Occupancy at Centromeric Repeats.
Based on the results presented above, both Chp2 and Swi6 associate with HDAC silencing activities. Also, double deletion of chp2 and swi6 causes cumulative increase in the levels of centromeric repeat transcripts (Fig. 1). To determine the contribution of Chp2 and Swi6 to heterochromatic transcriptional repression, we performed ChIP-chip to investigate the effects of their deletions on Pol II occupancy across a pericentromeric heterochromatin domain. As compared with cells carrying deletion of clr4, which creates H3K9me binding sites for HP1 proteins (9, 20) and strongly inhibits Pol II occupancy at centromeric repeats (44), chp2Δ and swi6Δ mutants were only partially defective in limiting Pol II occupancy at heterochromatic repeats (Fig. 4A). Both chp2Δ and swi6Δ mutant strains showed similar levels of increased Pol II occupancy as compared with wild-type cells, with the exception that chp2Δ cells displayed relatively higher Pol II occupancy specifically at dg elements (Fig. 4A). We also examined the chp2Δswi6Δ double mutant for Pol II levels across the pericentromeric heterochromatin domain. Interestingly, we observed a cumulative increase in Pol II occupancy at centromeric repeats, to levels comparable with those observed in the clr4Δ mutant (Fig. 4A). We conclude from these data that Chp2 and Swi6, which largely act downstream of H3K9 methylated by Clr4 at centromeres, cooperate to suppress Pol II occupancy at heterochromatic repeats.
Clr3 and Clr6 HDACs Collaborate to Restrict Pol II Access to Heterochromatin.
The dramatic increase in Pol II levels observed in the chp2Δswi6Δ double mutant background led us to consider whether their associated HDACs collaborate to inhibit Pol II accessibility to repeat loci. Mutations in the catalytic sites of Clr3 (clr3-735) and Clr6 (clr6-1) (45) resulted in increased Pol II occupancy at pericentromeric repeats (Fig. 4B). Relatively higher Pol II levels were detected in the mutant clr3-735 compared with the clr6-1 mutant. We noticed that, compared with clr4Δ, both clr3 and clr6 single mutants were only partially defective in restricting Pol II access to heterochromatic repeats. However, the clr3clr6 double mutant combination resulted in an additive increase in Pol II levels at pericentromeric loci (Fig. 4B). The levels of Pol II observed in the clr3clr6 double mutant were similar to the levels displayed by the chp2Δswi6Δ double mutant. Together with the results presented above, these data suggest that Chp2 and Swi6, and their interacting HDACs, cooperate to restrict Pol II accessibility across pericentromeric regions.
To test whether loss of Clr3 and Clr6 HDACs results in heterochromatic derepression to levels equivalent to chp2Δswi6Δ double mutants, we performed expression profiling in single and double HDAC mutants. Compared with single clr3 or clr6 mutant cells, clr3clr6 double mutant cells displayed a strong cumulative derepression of heterochromatic repeats across the entire pericentromeric region (Fig. 4C). Also, transcript levels in the clr3clr6 mutant were similar to those observed in both chp2Δswi6Δ and clr4Δ mutant cells (Fig. 4C). We conclude from these data that Chp2 and Swi6 proteins, which are bound to H3K9 methylated by Clr4, collaborate with their associated SHREC and Clr6 HDAC complexes to mediate TGS at heterochromatic loci.
Discussion
HP1 proteins bound to methylated H3K9 have important roles in mediating various heterochromatin functions, including TGS and chromosome segregation (1). In S. pombe, HP1 family proteins Chp2 and Swi6 are critical for heterochromatic silencing (46, 47), and both factors facilitate localization of SHREC, a complex involved in TGS (13, 22, 23). However, the exact contributions of these proteins, as well as their associations with other factors involved in the assembly of repressive chromatin, remain poorly understood. Here, we show that Swi6 associates with a Clr6-containing HDAC complex that, along with SHREC bound to Chp2, acts to preclude Pol II accessibility to heterochromatin. Together with results showing that Swi6 and Chp2 act downstream of H3K9me at centromeres (9, 20), these analyses suggest that the distribution of HP1 proteins across heterochromatin domains (this study and refs. 13, 14) facilitates the localization of repressive chromatin-modifying activities.
Clr6 has been shown to exist in at least 2 physically and functionally distinct Sin3-HDAC complexes, referred to as complex-I (Clr6 CI) and complex-II (Clr6 CII) (43). We found that Swi6 interacts specifically with Clr6 CII, but not with Clr6 CI (Fig. S4B). Among other factors, Clr6 CII contains Alp13, which shares homology to mammalian MRG15, and a Sin3-related protein PstII (Fig. 2C). This complex, which has been implicated in the global protective function of chromatin, including DNA damage protection and suppression of antisense transcription (43), could be recruited to chromatin via histone modifications established by Pol II-interacting proteins (48). However, association of Clr6 CII with Swi6 might allow it to stably associate with chromatin and/or act broadly across extended domains, to protect against the detrimental effects of unwanted transcription of repetitive DNA elements. Indeed, loss of either Alp13 or Clr6 results in derepression of the repeat elements present within pericentromeric heterochromatin domains (Figs. 2D and 4C) (43, 49).
Both SHREC and Clr6 CII are required to efficiently inhibit Pol II access to heterochromatin (Fig. 4). How do these activities affect transcriptional silencing? Evidence suggests that silencing requires HDAC and ATPase activities associated with the Clr3 and Mit1 subunits of SHREC (22). Also, Clr6 HDAC activity contributes to heterochromatic silencing (43). Whereas Clr3 preferentially targets histone H3 lysine 14, Clr6 displays broad substrate specificity for lysine residues on both histone H3 and H4 tails (50). Deacetylation of histones by these activities may prevent binding of factors such as SWI/SNF that remodel chromatin to provide access to the transcriptional machinery (51). Alternatively, HP1-associated Clr6 and SHREC complexes might act together to remove acetyl groups from histones, and to facilitate the proper positioning of nucleosomes required for higher-order chromatin assembly (22, 23, 52). Considering that hypoacetylated histones are a hallmark of heterochromatin in most eukaryotes (5), and that HP1 interacts with HDAC and chromatin remodeling factors in other organisms (53, 54), similar mechanisms may exist in other species.
Apart from heterochromatic silencing, HP1 proteins have an important role in proper segregation of chromosomes (1, 46). In S. pombe, Swi6 is critical for preferential loading of cohesin to heterochromatic loci (36, 37). Although Swi6 is believed to directly recruit cohesin (37), the exact mechanism has remained elusive. We show that Swi6 associates with Mis4 protein (Fig. 2), essential for loading of cohesin complex onto chromosomes (34, 35). This interaction is functionally important, because Swi6 is required for Mis4 localization across pericentromeric heterochromatic domains (Fig. S3), and swi6Δ exacerbates cohesion defects caused by the partial loss of function alleles of Mis4 cohesin-loading complex (34). Similar interaction between HP1 proteins and cohesin-loading factors may help explain heterochromatin requirement for chromosome segregation in higher eukaryotes. Our work also suggests a connection between Swi6 and centromere-specific histone H3 variant CENP-A. Considering that the S. pombe centromeres are organized into 3D structures reminiscent of mammalian centromeres (55), Swi6 localized across pericentromeric regions may provide the base to directly anchor CENP-A at the centromere core. Additional efforts are needed to explore whether Swi6 directly interact with CENP-A or cooperates with other CENP-A-associated factors. Indeed, facilitates chromatin transcription (FACT) complex, known to associate with CENP-A in mammals (56), copurifies with Swi6. In either case, CENP-A association with Swi6 may have implications for the recently described role of heterochromatin in CENP-A loading at centromeres (57, 58).
Swi6 purified fraction also contained factors implicated in a wide range of chromosomal processes. Additional work is required to confirm their interaction with Swi6. Chromatin remodelling factors Ino80 and RSC might not be required for silencing at heterochromatic loci, but it is possible that Swi6 partners with these factors to exert control over other chromosomal processes (41). While this work was under review, another independent study described purifications of S. pombe HP1 proteins (59). The authors also demonstrated Chp2 associating with SHREC to exert repressive influence on heterochromatic repeats, and identified diverse set of proteins copurifying with Swi6. However, certain Swi6-associated factors important for TGS and genome stability were not identified. This disparity could be because of limited interaction of factors such as Epe1, Mis4, Hsk1-Dbf1, and HDACs with Swi6 in the context of constitutive heterochromatin representing a small fraction of total chromatin. Another difference is that Motamedi et al. (59) purified Swi6 tagged at its carboxy-terminus, which contains the chromoshadow domain implicated in dimerization and interactions of HP1 proteins with other factors (11, 12). In this regard, we note that amino-terminal TAP-tagged Swi6 purified in our study is functional, and interacts with TGS and chromosome segregation factors linked to heterochromatin biology. Also, our analyses suggest that chromatin-modifying factors associated with both Chp2 and Swi6 cooperate to assemble repressive chromatin; thus, limiting Pol II accessibility to the target loci.
Materials and Methods
Yeast Strains.
A strain expressing amino-terminal tagged TAP-Swi6, under the control of its native promoter, was generated by inserting ura4+ reporter at the promoter region of the corresponding gene and then replacing ura4+ with the TAP-tagged swi6+. The chp2+-TAP, mit1+-myc, mis4+-myc, rad21+-HA, epe1+-TAP, and deletions used were constructed by using the standard PCR based module method (see also 22, 43). The clr6-1 and clr3-735 mutant alleles have been described previously (45). Strain expressing Cnp1-HA was a gift from M. Yanagida (Kyoto University, Kyoto).
Protein Purification.
For purification of TAP-Swi6 or Chp2-TAP associated proteins, cells harvested from 4-L cultures (OD595 2.5) were suspended in LB buffer [50 mM Tris·HCl, pH 7.5/150 mM NaCl/1.5 mM MgCl2/0.15% (vol/vol) Nonidet P-40/0.5 mM DTT] containing complete protease inhibitors (Roche) and PMSF (1 mM). All subsequent steps were performed at 4 °C. Cells were ground with 25 mL of glass beads by using a Pulverisette 6 system (Fritsch Gmbh, Germany). Beads/lysate was transferred into a 50-mL syringe and pressed into Falcon tubes. Cell debris was removed by centrifugation at 3,000 × g for 10 min, and lysate was cleared by centrifugation at 27,000 × g for 1 h. The cleared lysate was incubated with 400 μL IgG Sepharose (Amersham) for 2 h at 4 °C. Beads were washed by gravity flow-through with 20 mL LB buffer, 10 mL LB + 5 mM EGTA buffer, followed by a final wash of 10 mL LB buffer. Bound proteins were eluted from the beads by rocking overnight in 500 μL LB buffer containing 4 μg TEV protease (Invitrogen). The TEV eluate was incubated with 300 μL calmodulin Sepharose (Amersham) in LB + 2 mM CaCl2 buffer for 2 h at 4 °C. The calmodulin beads were washed with 20 mL LB + 2 mM CaCl2 buffer. The bound proteins were eluted by 600 μL LB + 5 mM EGTA buffer and precipitated by TCA. Purified samples were either resolved by SDS/PAGE and visualized by Coomassie Blue staining or subjected to tandem MS (LC-MS/MS) analyses. Western blot analyses of purified samples were performed by using previously described antibodies against Clr6, Alp13, and PstII (43). Anti-Chp2 rabbit antibody was raised against purified full-length recombinant Chp2 protein expressed in E. coli. The crude rabbit antisera were affinity-purified by using NHS-activated HiTrap system (Amersham).
ChIP-chip.
ChIP-chip using antibody against the largest subunit of Pol II (8WG16; Covance), Swi6 (9), Mit1-myc (9E10; Santa Cruz), Epe1-TAP (IgG; Amersham) or Chp2 (this study) were performed as described previously (14), except microarray containing probes corresponding alternately to plus and minus strands of cen2 at 50-bp resolution were used. Enrichment values were corrected for nonspecific signals from control samples with the same antibodies used to perform ChIP.
Expression Profiling.
Total RNA was processed by using Invitrogen RiboMinus Yeast kit to partly deplete ribosomal RNA before reverse transcription step by using random hexamer and anchored oligodT primer mix; 3 μg of RNA was labeled with SuperScript Indirect cDNA labeling kit (Invitrogen). Labeled samples from mutant (Cy3) and wild type (Cy5) were mixed and hybridized to the microarray described above. Data were extracted with Agilent Feature Extraction Software. Expression ratios were calculated as Cy5processed signal/Cy3processed signal. Background signal was estimated as a median processed signal of 152 oligonucleotides with no homology to the S. pombe genome.
Supplementary Material
Acknowledgments.
We thank S. Jia, T. Yamada, and T. Sugiyama for strain constructions; S. Jia for generating anti-Chp2 antibody; K. Takahashi and M. Yanagida (Kyoto University, Kyoto, Japan) for providing a strain expressing Cnp1-HA; and I. Hall and H. Cam for comments on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813063106/DCSupplemental.
References
- 1.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 2.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 3.Selker EU. Repeat-induced gene silencing in fungi. Adv Genet. 2002;46:439–450. doi: 10.1016/s0065-2660(02)46016-6. [DOI] [PubMed] [Google Scholar]
- 4.Birchler JA, Bhadra MP, Bhadra U. Making noise about silence: Repression of repeated genes in animals. Curr Opin Genet Dev. 2000;10:211–216. doi: 10.1016/s0959-437x(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 7.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 8.Lachner M, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama J, et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs SA, et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasher SV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 13.Sadaie M, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cam H, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 15.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci USA. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cryderman DE, et al. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 18.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Piacentini L, et al. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Yamada T, et al. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama T, et al. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 27.Lejeune E, et al. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 29.Cairns BR, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 30.Walfridsson J, et al. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucl Acid Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcangioli B, Copeland TD, Klar AJ. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:2058–2065. doi: 10.1128/mcb.14.3.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi C, Noguchi E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007;175:553–566. doi: 10.1534/genetics.106.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanoh J, Ishikawa F. Composition and conservation of the telomeric complex. Cell Mol Life Sci. 2003;60:2295–2302. doi: 10.1007/s00018-003-3245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard P, et al. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 2001;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 37.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 38.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Isaac S, et al. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics. 2007;175:1549–1560. doi: 10.1534/genetics.106.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailis JM, et al. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat Cell Biol. 2003;5:1111–1116. doi: 10.1038/ncb1069. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi MT, et al. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- 42.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 43.Nicolas E, et al. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 44.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 45.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allshire RC, et al. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 47.Thon G, Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Hansen KR, et al. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjerling P, et al. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 52.Shogren-Knaak M, et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 53.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: Potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen AL, et al. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J. 2002;21:5797–5806. doi: 10.1093/emboj/cdf560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kniola B, et al. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 57.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 59.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.