Abstract
Spider silks are renowned for their excellent mechanical properties and biomimetic and industrial potentials. They are formed from the natural refolding of water-soluble fibroins with α-helical and random coil structures in silk glands into insoluble fibers with mainly β-structures. The structures of the fibroins at atomic resolution and silk formation mechanism remain largely unknown. Here, we report the 3D structures of individual domains of a ≈366-kDa eggcase silk protein that consists of 20 identical type 1 repetitive domains, one type 2 repetitive domain, and conserved nonrepetitive N- and C-terminal domains. The structures of the individual domains in solution were determined by using NMR techniques. The domain interactions were investigated by NMR and dynamic light-scattering techniques. The formation of micelles and macroscopic fibers from the domains was examined by electron microscopy. We find that either of the terminal domains covalently linked with at least one repetitive domain spontaneously forms micelle-like structures and can be further transformed into fibers at ≥37 °C and a protein concentration of >0.1 wt%. Our biophysical and biochemical experiments indicate that the less hydrophilic terminal domains initiate the assembly of the proteins and form the outer layer of the micelles whereas the more hydrophilic repetitive domains are embedded inside to ensure the formation of the micelle-like structures that are the essential intermediates in silk formation. Our results establish the roles of individual silk protein domains in fiber formation and provide the basis for designing miniature fibroins for producing artificial silks.
Keywords: NMR, spider silk, structural transition, TuSp1
Spider silks are renowned for their excellent mechanical properties and biomimetic and industrial potentials. The orb-web spiders employ up to seven types of abdominal glands to produce silks for various purposes, ranging from prey capture to offspring protection in egg cases (1). Among seven different types of silks, tubuliform silk (eggcase silk) is unique because of its high serine and low glycine content (2, 3). Eggcase silk fibroins are synthesized only in female tubuliform (cylindrical) silk glands for the construction of protective egg cases, where eggs undergo development. A number of eggcase silk cDNA clones from orb-weaver superfamilies have recently been isolated (4–8). The length of eggcase silk cDNA varies from 10 to 13 kb in different species. The sequence of the inferred protein, tubuliform spidroin 1 (TuSp1) accounts for the amino acid (aa) composition of tubuliform silk very well, indicating that TuSp1 is the major component of eggcase silk (4–8). Both in situ hybridization and immunoblot analyses show that TuSp1 is expressed specifically in the tubuliform gland (4). Eggcase silk must be sufficiently robust to resist different threats, such as predator/parasitoid invasion or temperature fluctuations. Compared with the widely studied spider dragline silk, eggcase silk is slightly less tough but more resistant to aqueous environments (9). Furthermore, the good biocompatibility and slow biodegradability of the eggcase silk are an advantage for use in biomaterial application (10).
The mechanical properties of silks depend on the structures of the silk constituent proteins before and after silk fiber formation. The fibroins in the tubuliform and other glands contain both α-helical and disordered conformations as revealed by circular dichroism (CD) and infrared spectroscopy (11, 12). Yet the three-dimensional (3D) structures of silk proteins in aqueous solution and spider silk glands before fiber formation are completely unknown. In spider silks, it is well known that the proteins exist in mainly β-sheet structures (13), but the exact 3D protein structure in fibers remains unknown. Nevertheless, it is obvious that structural transition is a necessary step in the process of silk fiber formation. Despite numerous studies, the mechanism of the structural transition and fiber formation still remains mysterious.
Here, we report the solution structures of individual domains of a ≈366-kDa spider eggcase silk protein. We determined the minimal fibroin motif (fragment) that can form macroscopic fibers and demonstrated the roles of individual domains in fiber formation based on the solution structures and biochemical and biophysical experiments.
Results and Discussion
Molecular Architecture of TuSp1.
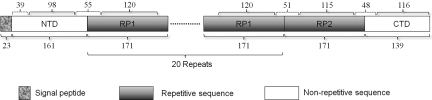
In an earlier study, we cloned the C terminus with two repetitive sequences of TuSp1 from the total silk gland cDNA library of the golden web spider, Nephila antipodiana (4). Here, we cloned the N terminus with partial repetitive sequences [supporting information (SI) Fig. S1] from both the genomic DNA and the cDNA library. Genomic sequences of TuSp1 obtained by PCR are not interrupted by introns, consistent with TuSp1 genes from other orb-weaving spiders and suggesting a single large exon (5). The aa sequence of TuSp1 shows a typical spider silk molecular architecture (Fig. 1) (5–8). TuSp1 contains a signal peptide at the N terminus, one nonrepetitive sequence just after the signal peptide (NTD) and one at the C terminus (CTD), 20 identical type 1 repetitive sequences (RP1) that were deduced from our identified ≈12.0-kb mRNA (4), and one type 2 repetitive sequence (RP2) linked to CTD. The signal peptide at the N terminus was predicted by signalP3.0 software with a probability of >99% (14). The most likely cleavage site is just after A23 (Fig. 1). The boundaries of RP1 were established simply based on the repetitive aa sequence. The boundary between RP2 and CTD was determined by assuming that RP2 and RP1 contain the same number of aa residues. The assumption is based on the sequence similarity of RP2 and RP1 and the 3D structures of RP1, RP2, and CTD described below. NTD and CTD share low sequence identity with the repetitive sequences, whereas RP2 is nearly identical to RP1 in the first 130 residues (Fig. S2).
Fig. 1.
TuSp1 architectures. NTD, RP1, RP2, and CTD domains are represented by bars. The aa number of each domain is indicated below the corresponding bar. The repeat number of RP1 domains is also indicated below the bars. The aa number of structural regions (solid brace) and linkers between the structural regions (dashed brace) are indicated above the bars.
NMR Structures of TuSp1 Domains.
We found that the N- and C-terminal regions of NTD were unstable and susceptible to degradation, and a short form within residues E40-A155 (sNTD) was stable (Fig. S2). Both NTD and sNTD form soluble aggregates in aqueous solution as indicated by gel filtration and dynamic light scattering (DLS) (Fig. S3A). In the presence of 100 mM dodecylphosphocholine (DPC), sNTD exists in monomers as demonstrated by its small overall tumbling time (6.8 ns) estimated from 15N NMR relaxation times. The CD spectra of sNTD do not change with DPC concentration ranging from 0 to 100 mM (Fig. S4A), showing that sNTD still retains its native conformation in the presence of DPC. Thus, we determined the structure of sNTD in DPC using NMR experiments (Table 1 and Fig. S5A). The residues E40–S137 in sNTD form a four-helix bundle through hydrophobic interactions among helices (Fig. 2A), and 18 residues in the C terminus of sNTD are unstructured. The ensemble of the 10 lowest-energy conformers is depicted in Fig. S6A, and the detected long-range nuclear Overhauser effects (NOEs) mapped to its structures are shown in Fig. S7A. Twenty-six of 39 (≈67%) hydrophobic residues in the structural region of NTD are exposed to solvent and cluster to form three large, contiguous, and protrusive hydrophobic patches on the surface (Fig. 2E). Thus, NTD tends to form oligomers by burying the solvent-exposed large hydrophobic patches in aqueous solution.
Table 1.
NMR and refinement statistics for TuSp1
| Parameters | NTD | RP1 | RP2 | CTD |
|---|---|---|---|---|
| NMR distance and dihedral constraints | ||||
| Distance constraints | ||||
| Total NOE | 1,187 | 2,125 | 2,011 | 1,733 |
| Intraresidue | 242 | 389 | 559 | 436 |
| Interresidue | ||||
| Sequential (|i–j| = 1) | 487 | 745 | 690 | 716 |
| Medium-range (|i–j| ≤4) | 424 | 732 | 518 | 538 |
| Long-range (|I–j| ≥5) | 34 | 259 | 244 | 43 |
| Total dihedral angle restraints* | 125 | 229 | 211 | 148 |
| Structure statistics | ||||
| Violations, mean ± SD | ||||
| Distance constraints, Å | 0.31 ± 0.05 | 0.30 ± 0.05 | 0.23 ± 0.04 | 0.32 ± 0.07 |
| Dihedral angle constraints, ° | 4.08 ± 0.54 | 4.08 ± 0.39 | 2.55 ± 0.87 | 3.47 ± 0.78 |
| Max. dihedral angle violation, ° | 5.48 | 4.67 | 4.02 | 5.10 |
| Max. distance constraint violation, Å | 0.40 | 0.40 | 0.33 | 0.43 |
| Ramachandran plot, allowed region | 98.9% | 99.6% | 99.6% | 96.6% |
| Average rms deviation, ņ | ||||
| Heavy atoms | 1.62 ± 0.19 | 0.98 ± 0.13 | 0.81 ± 0.08 | 1.95 ± 0.31 |
| Backbone atoms | 1.25 ± 0.19 | 0.55 ± 0.10 | 0.46 ± 0.07 | 1.59 ± 0.31 |
*Dihedral angle constraints were generated by TALOS based on Cα and Cβ chemical shifts.
†Average rms deviation to the mean structure in each structural region was calculated among 10 refined structures. The total AMBER energies for NTD, RP1, RP2, and CTD are −3,556 ± 34, −3,834 ± 41, −3,220 ± 29 and −4,231 ± 25 kcal/mol, respectively.
Fig. 2.
Solution structures of TuSp1 domains. Ribbon drawing of the lowest-energy conformers of NTD (A), RP1 (B), RP2 (C), and CTD (D). Hydrophobic and charged surface of NTD (E), RP1 (F), RP2 (G), and CTD (H). Unstructured regions are not shown. Color code is yellow for hydrophobic, blue for positive charges, red for negative charges, and white for neutral surface. Hydrophobic patches on the surfaces are circled in black. (Scale bars: 3 nm.)
RP1 and RP2 domains are natively monomers in aqueous solution at room temperature, as indicated by gel filtration and DLS (Fig. S3D). Although RP1 and RP2 have 78% sequence identity (Fig. S2), their heteronuclear single-quantum correlation (HSQC) spectra show great differences (Fig. S5B) (15). Thus, the NMR structures of RP1 and RP2 were determined independently in aqueous solution (Table 1). Thirty-one and 20 residues are disordered in the N and C termini of RP1, respectively, and the rest of the residues form an orthogonal bundle with six helices (Fig. 2B and Fig. S6B). Although ≈46% hydrophobic residues in the structural region (Y32–Q151) are solvent-exposed, unlike NTD there is no large protrusive hydrophobic patch on the surface. Some of the solvent-exposed hydrophobic residues form a long hydrophobic groove, and the rest are scattered on the surface (Fig. 2F). Thus, RP1 could not form oligomers even at high protein concentration (>1 mM) in aqueous solution. Similar to RP1, the N- and C-terminal regions of RP2 are unstructured, and the structural region (Y32–N146) forms an orthogonal bundle with six helices (Fig. 2C and Fig. S6C). The distribution of hydrophobic residues on the surface is nearly identical for both RP1 and RP2 (Fig. 2 F and G), consistent with their similar solubility in water. Pairwise structural comparison (16, 17) shows that RP1 and RP2 own highly similar structures (≈95% structural overlay; Table S1), indicating that RP1 and PR2 can be classified into the same sequence family level and that they could share similar or the same functions. Hence, we define RP2 as type 2 repetitive domain.
To determine the boundary between RP2 and CTD, an N-terminal extended CTD (eCTD), which consists of the C-terminal region of RP2 (22 residues) and the full-length CTD, was used for structural determination. eCTD and CTD form soluble oligomers (dimer, trimer, and tetramer) at very low protein concentration in aqueous solution at 20 °C (Fig. S3B). eCTD also exists as monomers in the presence of 100 mM DPC as evidenced by its small overall tumbling time (7.8 ns) estimated from 15N relaxation times. DPC titration shows that eCTD retains its native structure in the presence of DPC because no structural changes were detected during the titration. Therefore, we also solved the NMR structure of eCTD in DPC (Table 1 and Fig. S5C), which can represent its native structure in aqueous solution. Forty-five residues in the N terminus of eCTD adopt a random coil structure, whereas the rest of the residues fold into a five-helix bundle (Fig. 2D and Figs. S6D and S7B). According to the structures of RP2 and eCTD, the orthogonal helical bundle in RP2 is connected to the helical bundle in CTD by a long unstructured linker (48 residues), implying that an exact definition of the boundary of CTD is not crucial to domain structures. Because RP2 and RP1 are very similar in both aa sequences and structures, it is reasonable to assume that RP2 and RP1 contain the same number of residues. Taking this assumption, the boundary between RP2 and CTD is located nearly in the middle of the linker. The protein constructs of RP2 and CTD designed based on this boundary could be expressed well, indicating that the boundary is not incorrect. The helices in CTD are also held together through hydrophobic interactions. Thirty-eight of 58 (≈66%) hydrophobic residues in the structural region of CTD are solvent-exposed to form four large, contiguous, and protrusive hydrophobic patches on the surface (Fig. 2H). Obviously, there are significantly more solvent-exposed hydrophobic residues in CTD than NTD. Therefore, CTD is more susceptible to form oligomers than NTD in aqueous solution.
Domain–Domain Interaction.
To investigate potential domain–domain interactions, we applied DLS to detect the change in molecular size by mixing different domains (Fig. S3). At 20 °C, only NTD could interact with CTD to form larger soluble aggregates. NMR titration as monitored by HSQC experiments on 15N-labeled RP1 further confirms that RP1 does not interact with other domains at 20 °C. Similarly, RP2 does not interact with all other domains. These results are expected from the surfaces of individual domains. At 37 °C, however, the interactions between the terminal domains and repetitive domains become noticeable (Fig. S3 F–I). Moreover, RP1/RP2 can interact with themselves to form large oligomers although the population of the oligomers is quite small (≈2%) (Fig. S3E). The enhanced interdomain interactions should result from temperature-induced conformational changes of individual domains and increased hydrophobic interaction at higher temperature.
Roles of Domains in Silk Fiber Formation.
To understand the role of each domain in eggcase silk fiber formation, we first studied structural transitions of individual domains. All of the 4 domains show temperature-dependent irreversible structural transition from α to β conformations (Fig. S8 A–D). At 90 °C, RP1 and RP2 exhibit mainly a β-sheet conformation, whereas sNTD and CTD contain both β-sheet and β-turn structures. Upon cooling to room temperature, RP1, RP2, and sNTD still nearly retain the same conformations, similar to the CTD of major ampullate spidroin I (MaSp1) (18). Surprisingly, the structure of CTD at 90 °C is different from that after cooling to room temperature (Fig. S8D). Even when the concentrations of individual domains were >0.2 wt%, only water-soluble aggregates and insoluble precipitates were observed, but no macroscopic fibers could be formed within a temperature range of 20–90 °C.
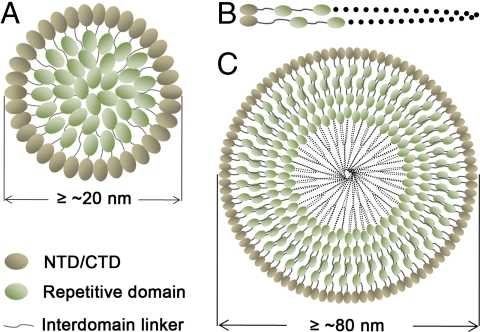
We second introduced one RP1 to the C terminus of NTD (NRP) and one RP2 to the N terminus of CTD (RPC). DLS indicates that NRP and RPC also form water-soluble oligomers at 20 °C. Transmission electron microscopy (TEM) shows that the oligomers adopt micelle-like structures (Fig. 3 A–F), but the oligomers formed by respective sNTD and CTD are amorphous and do not form micelle-like structures (Fig. S9). The size of the micelles observed varies from ≈20 to ≈400 nm. The smaller micelles can fuse to form larger micelles (Fig. 3 E and F), implying that the interactions among monomers are not very strong. Both NRP and RPC in the micelles still adopt an α-helical conformation as evidenced by CD (Fig. S8 E and F). The smooth and nonprotrusive surfaces of the NRP and RPC micelles (Fig. 3) suggest that the repetitive domains are embedded inside to form the inner core, whereas the N-/C-terminal domains form the outer surface. If instead the polar repetitive domains formed the surface, the micelle surfaces would be spiky because the repetitive domains do not interact with each other in water at 20 °C. Our conjecture is verified by dot blotting of His6-tagged NRP/RPC with anti-His antibodies (Fig. 3 H–J). The His6 tagged to the N terminus of NRP (His6-NRP) or the C terminus of RPC (RPC-His6) were immunodetected, whereas the His6 tagged to the C terminus of NRP (NRP-His6) or the N terminus of RPC (His6-RPC) failed to show positive signals. Moreover, thrombin cleavage could release His6 from His6-NRP micelle, but not from NRP-His6. Based on these results, we propose models of the micelle structures for NRP/RPC and the full-length TuSp1 (Fig. 4). In the models, terminal domains form the outer layer of the micelle whereas repetitive domains are packed inside. Native fibroins in spider silk glands indeed exist in micelle-like structures as observed by TEM (Fig. 3G), supporting our structural model. The micelle-like structure proposed here shares similarities to the proposed model of silkworm fibroin in terms of domain arrangement (19), i.e., repetitive and terminal domains are inside and outside the micelle, respectively. However, the underlying mechanisms of micelle formation are different. The repetitive blocks of silkworm fibroin were thought to be more hydrophobic than the terminal blocks and were presumed to direct the assembly of fibroin molecules into micelles, whereas the hydrophilic nonrepetitive terminal blocks could interact with each other to form the outer edges of the micelles. It is noteworthy that the hydrophobicities of the repetitive and terminal blocks of silkworm fibroin were predicted purely based on aa sequences instead of 3D structures. The structural region of TuSp1 NTD seems more hydrophilic than those of RP1 and RP2 based on aa sequences alone. In fact, sNTD CTD monomer is less water-soluble than RP1/RP2, and sNTD CTD is more hydrophobic in terms of protein surface. The driving force for the formation of the micelle-like structure of TuSp1 should be the hydrophobic interactions among NTD (CTD) domains that have solvent-exposed hydrophobic patches. The micelle structure could be further stabilized by the intermolecular hydrogen bonds among repetitive (RP) domains that are packed inside the micelle because serine/threonine residues are very abundant in RP domains (≈25%), and almost all of these residues are exposed on the surfaces of RP domains.
Fig. 3.
Micelle structures of NRP and RPC at 20 °C. TEM of stained NRP (A) and RPC (B) micelles, enlarged surface structures (C and D), nonstained NRP (E) and RPC (F) micelles, and stained TuSp1 micelles isolated from tubuliform gland (G). (E and F) Self-fusion of micelles is indicated by arrows. [Scale bars: (A), 50 nm; (B), 50 nm; (C), 20 nm; (D), 10 nm; (E), 50 nm; (F), 50 nm; (G), 200 nm.] (H) SDS/PAGE of His6-NRP (Lane 1, Upper), NRP-His6 (Lane 2, Upper), His6-RPC (Lane 1, Lower), and RPC-His6 (Lane 2, Lower). (I) Protein staining of His6-NRP (Left, Upper), NRP-His6 (Right, Upper), His6-RPC (Left, Lower) and RPC-His6 (Right, Lower). (J) Dot blotting of His6-NRP (Left, Upper), NRP-His6 (Right, Upper), His6-RPC (Left, Lower), and RPC-His6 (Right, Lower).
Fig. 4.
Models of TuSp1 micelles. (A) NRP/RPC micelle. The N-terminal/C-terminal domains form the outer layer of the micelle, whereas the repetitive domains with flexible long unstructured linkers are randomly packed into the inner core of the micelle. (B) Single full-length TuSp1 molecule with the N- and the C-terminal domains interacting with each other. (C) Full-length TuSp1 micelle. The sizes of micelles were estimated based on our DLS and TEM results and the structures of NTD, RP1, RP2, and CTD.
Similar to individual domains, NRP and RPC display temperature-dependent irreversible structural transition from α- to β-conformations (Fig. S8 E and F). We found that NRP had a lower thermal transition temperature than RP1 but higher than NTD. Similarly, the transition temperature for RPC was between those for RP2 and CTD. The results show that the two domains in NRP and RPC undergo structural changes cooperatively, implying that the two domains interact with each other at high temperature. At ≥37 °C, both NRP and RPC could self-assemble and polymerize into macroscopic fibers within a couple of hours without physical shear when the concentrations of NRP and RPC were only 0.1–0.2 wt%, which are much lower than 25–30 wt%, the fibroin concentration in spider glands (20). During the polymerization, structural transition from mainly α-conformation to β-conformation occurs because the protein molecule adopts a mainly β-conformation in silk fibers (13). Similar to NRP and RPC of TuSp1, the CTD of MaSp1 linked with its repetitive domain could also form fibers at room temperature without physical shear (21).
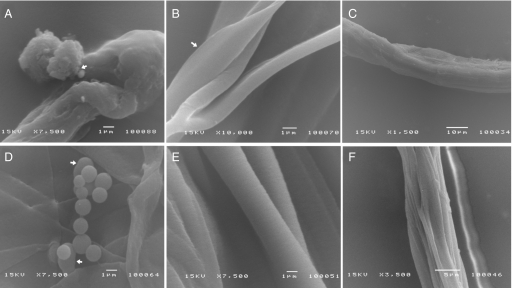
Different size and shape morphologies of fibers, including spherical and film-like structures, are observed in scanning electron microscopy (SEM) (Fig. 5). Although fibers are the major product in the formation of eggcase silk, some intermediates such as micelles and micrometer globules also exist (Fig. 5 A and D). These intermediate structures were proposed as the prerequisite for fiber formation (19, 22). They may result from the merger of smaller micelles that were observed in aqueous solution at 20 °C. The intermediates further assemble into small single fibers with diameters of 1–3 μm (Fig. 5 B and E), whose size is close to the native single eggcase silk (23). These small fibers can form larger assembly with diameters of 10–20 μm (Fig. 5 C and F).
Fig. 5.
SEMs of eggcase silk fibers formed from NRP and RPC. (A) Spherical NRP assemblies (arrow). (B) Small NRP fibers and intermediate film structures (arrow). (C) Large NRP fibers. (D) Spherical RPC assemblies (arrow). (E) Small RPC fibers. (F) Large RPC fibers. [Scale bars: (A), 1 μm; (B), 1 μm; (C), 10 μm; (D), 1 μm; (E), 1 μm; (F), 5 μm.]
Without NTD or CTD, single and multiple repetitive domains cannot form oligomers, micelle-like structures, or macroscopic fibers. Thus, the role of the terminal domains should be to initiate the assembly of protein molecules to form micelles through hydrophobic interactions among the terminal domains. The micelles are quite soluble in aqueous solution by sequestering most hydrophobic residues. This explains why fibroin proteins exist in a soluble form even at very high concentrations (≥ 25 wt%) in silk glands although the conserved nonrepetitive terminal domains contain many hydrophobic residues.
Without repetitive domains, NTD and CTD could not form micelle-like structures or macroscopic fibers. Our results show that at least one repetitive domain linked to one terminal domain is required for the formation of micelle-like structures and fibers. The sample of two repetitive domains linked to CTD displayed similar micelle/fiber morphology too. Thus, the role of the repetitive domains is to ensure the formation of the micelle-like structures. The micelles (protein oligomers) are one of the key intermediates in the fiber formation process as shown here and previously (19, 22). Interestingly, intermediate oligomers were also observed in the formation of fibrils resulting from protein (amyloid) misfolding (24, 25), indicating that the fiber formation mechanism may be similar to or the same for silk fiber proteins and amyloid proteins.
Because the terminal domains among all spider silk proteins are relatively conserved in aa sequence (4, 26) and conserved in secondary structure (12), the roles of the terminal and repetitive domains in fiber formation could be similar for all of the silk proteins, and our findings can be applied to other silk proteins. According to the results obtained here, the minimal silk protein fragment (miniature fibroin) for the formation of silk fibers must contain at least one terminal domain and one repetitive domain. This conclusion is supported by a previous study on MaSp1 (21). The structures determined here lay a foundation for further understanding the structural transition and fiber formation mechanism by experimental and computational techniques. The domain boundary identified here, which is the critical information for protein engineering, may also be applied to other spider silk proteins, especially for the conserved terminal domains. Therefore, our structural work will provide the basis for designing various miniature fibroins with combinations of domains and linkers from the same or different silk proteins as the starting materials of artificial silks.
Physical shear and elongation stress acting on the high-concentration fibroin solution in spider glands are proved to cause fibroin crystallization in solution. Furthermore, this process involves liquid crystal spinning (2). Shear-induced fibroin crystallization in vivo and in vitro promotes the formation of fibers with better morphology during a shorter time at ambient temperature, but it requires a high fibroin concentration. The heat-induced fiber formation shown here could become an alternative approach for producing artificial silks without using physical shear (2, 22, 27).
Methods
Cloning of the N-Terminal Sequence of TuSp1 from N. antipodiana.
N-terminal fragments (≈200 bp) of TuSp1 were obtained, respectively, from a genomic DNA and cDNA library by PCR using degenerated primer sets (5′-ATG GTT TGG RTN ACN XYN ATQ GC-3′ and 5′-RTC XTG XTG XTC XTG RAA NGG RAA-3′) designed on the basis of the conserved amino acid sequences of the N termini from known TuSp1s (1, 28). One of the two fragments obtained from genomic DNA was the same as the one from the cDNA library. The N- and C-terminal fragments with partial repetitive sequences were further amplified from both genomic DNA and cDNA library. The sequence alignment was performed in ClustalW2 (29).
Preparation of Individual Domain Constructs.
On the basis of secondary structure prediction, five cDNA constructs coding the full-length NTD and its various truncated forms were subcloned into pET32-derived expression vectors. Only the shortest one, sNTD, did not degrade in solution within a week and hence was selected for structural determination. The cDNAs coding RP1, RP2, CTD, NRP, and RPC domains were also subcloned into pET32-derived vectors, respectively. To determine the boundary between CTD and RP2, a construct consisting of the full-length CTD and the C-terminal region of RP2 (with 22 residues) was prepared. Its corresponding protein denoted as eCTD was used for structure determination. Protein expression was carried out in Escherichia coli strain BL21 (DE3). RP1 and RP2 were purified by using Ni-nitrilotriacetic acid affinity chromatography (Qiagen), FPLC gel filtration, and/or ionic exchange (GE Healthcare), as described in ref. 15. NTD, sNTD, CTD, and eCTD were expressed in inclusion body and purified under denatured condition. Then they were refolded into ice-cold buffer [50 mM Tris (pH 7.0)], respectively. For DPC titration, DPC was gradually added into the respective purified sNTD and eCTD domains with a final DPC:protein molar ratio of 10,000:1. Isotopic labeling was carried out by expressing the proteins in M9 minimal medium containing [15N]NH4Cl and/or 13C-labeled glucose as the sole source of nitrogen and carbon.
NMR Spectroscopy and Structure Calculations.
NMR experiments on RP1 and eCTD were performed on a Bruker 800-MHz NMR spectrometer with a cryogenic probe at 17 °C and 35 °C, respectively. The experiments on sNTD and RP2 were done on a 500-MHz NMR spectrometer with a cryogenic probe at 30 °C and 17 °C, respectively. To obtain more methyl–methyl NOEs, 3D double constant-time 13C,13C methyl NOE spectroscopy (NOESY) (30) with suppression of diagonal peaks (31) was recorded for sNTD and eCTD, respectively. All spectra were processed with NMRPipe (32) and analyzed with NMRView with an NOE assignment plug-in (33). Resonance assignments of backbone, aliphatic, and aromatic side chains were obtained using our recently developed methods (34–36). NOE restraints were obtained from 3D and/or 4D NOESY spectra. Ambiguous NOEs were assigned with iterated structure calculations by DYANA (37). Final structure calculation was started from 100 conformers. Energy minimization of the 10 conformers with the lowest final target function values was performed in AMBER force field (38). The mean structure was obtained from the 10 energy-minimized conformers for each domain. The helical borders at the N and C termini of the mean structure were used to present the structural borders of each domain. PROCHECK-NMR (39) was used to assess the quality of the structures. All of the structural figures were made by using MOLMOL (40).
Structure-Based Alignments and Structural Comparison.
Pairwise structure-based alignment and comparison were carried out by using SSAP server (www.cathdb.info/cgi-bin/cath/SsapServer.pl).
DLS Measurement.
The apparent hydrodynamic radii of TuSp1 domains and fragments were examined by DLS (DynaPro; Protein Solutions) at 20 °C or 37 °C. The data were analyzed by using dynamics 5.0 software.
Immunoblotting of His6-NRP, NRP-His6, His6-RPC, and RPC-His6.
A His6 tag was linked to the N terminus (His6-NRP) and C terminus of NRP (NRP-His6), respectively. The His6 tag was also linked to the N (His6-RPC) and C termini of RPC (RPC-His6), respectively. Recombinant proteins were overexpressed in E. coli BL21 (DE3) and then purified. All of the four proteins were diluted to the same concentration. A 1-μL sample of each protein was applied onto nitrocellulose membrane. Anti-His antibody (Qiagen) was used to detect the exposed His6 tag (QIAexpress Detection and Assay Handbook, 2002). Proteins were stained with Ponceau as positive controls.
Fiber Formation in Vitro.
The His6-tagged NRP or RPC fibroin solution with a concentration of 0.1–0.2 wt% and purity of ≈90% was incubated at different temperature (20 °C, 37 °C, and 50 °C) for hours to days.
TEM.
Fresh-prepared fibroin samples were stained with phosphotungstic acid for 1 minute. Stained or nonstained fibroins were applied to an electron microscope specimen grid covered with a thin carbon support. TEM micrographs were then recorded on a JEOL JEM 2010F HRTEM operating at a voltage of 200 kV and room temperature.
SEM.
Assembled silk fibers were applied on SEM stubs and air-dried overnight. The samples were vacuum-coated with a 20-nm layer of gold. Specimens were observed on a JEOL JSM T220A SEM and photographed at a voltage of 15 kV and room temperature.
Supplementary Material
Acknowledgments.
We thank G. L. Loy and P. L. Chong for help in TEM and SEM micrographs. We also thank Dr. Y. K. Mok for valuable discussions and Dr. D. Li for critical reading of the manuscript. This work was supported by National University of Singapore, Singapore Ministry of Education, and Singapore Millennium Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence of the N terminus of TuSp1 has been deposited in the GenBank database (accession no. EU730637). The atomic coordinates and structure factors of sNTD, RP1, RP2, and eCTD have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2K3Q, 2K3N, 2K3O, and 2K3P, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813255106/DCSupplemental.
References
- 1.Foelix RF. The Biology of Spiders. New York: Oxford Univ Press; 1996. [Google Scholar]
- 2.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 3.Guerette PA, Ginzinger DG, Weber BHF, Gosline JM. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science. 1996;272:112–115. doi: 10.1126/science.272.5258.112. [DOI] [PubMed] [Google Scholar]
- 4.Huang W, et al. Characterization and expression of a cDNA encoding a tubuliform silk protein of the golden web spider Nephila antipodiana. Biochimie. 2006;88:849–858. doi: 10.1016/j.biochi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Garb JE, Hayashi CY. Modular evolution of eggcase silk genes across orb-weaving spider superfamilies. Proc Natl Acad Sci USA. 2005;102:11379–11384. doi: 10.1073/pnas.0502473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, et al. Araneoid eggcase silk: A fibroin with novel ensemble repeat units from the black widow spider, Latrodectus hesperus. Biochemistry. 2005;44:10020–10027. doi: 10.1021/bi050494i. [DOI] [PubMed] [Google Scholar]
- 7.Tian M, Lewis RV. Molecular characterization and evolutionary study of spider tubuliform (eggcase) silk protein. Biochemistry. 2005;44:8006–8012. doi: 10.1021/bi050366u. [DOI] [PubMed] [Google Scholar]
- 8.Zhao A, et al. Unique molecular architecture of eggcase silk protein in a spider, Nephila clavata. J Biochem. 2005;138:593–604. doi: 10.1093/jb/mvi155. [DOI] [PubMed] [Google Scholar]
- 9.Viney C. From natural silks to new polymer fibers. J Tex Inst. 2000;91:2–23. [Google Scholar]
- 10.Gellynck K, et al. Biocompatibility and biodegradability of spider egg sac silk. J Mater Sci: Mater Med. 2008;19:2963–2970. doi: 10.1007/s10856-007-3330-0. [DOI] [PubMed] [Google Scholar]
- 11.Dicko C, Knight D, Kenney JM, Vollrath F. Secondary structures and conformational changes in flagelliform, cylindrical, major, and minor ampullate silk proteins. Temperature and concentration effects. Biomacromolecules. 2004;5:2105–2115. doi: 10.1021/bm034486y. [DOI] [PubMed] [Google Scholar]
- 12.Rising A, Hjälm G, Engström W, Johansson J. N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules. 2006;7:3120–3124. doi: 10.1021/bm060693x. [DOI] [PubMed] [Google Scholar]
- 13.Van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc Natl Acad Sci USA. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendtsen JD, Nielsen H, von Heijne HG, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Lin Z, Huang W, Yang D. Resonance assignments of a repeated domain of the eggcase silk from Nephila antipodiana. J Biomol NMR. 2006;36:17. doi: 10.1007/s10858-005-5584-5. [DOI] [PubMed] [Google Scholar]
- 16.Orengo CA, Taylor WR. SSAP: Sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 17.Orengo CA, Michie AD, Jones DT, Swindells MB, Thornton JM. CATH: A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 18.Hedhammar M, et al. Structural properties of recombinant nonrepetitive and repetitive parts of major ampullate spidroin 1 from Euprosthenops australis: Implications for fiber formation. Biochemistry. 2008;47:3407–3417. doi: 10.1021/bi702432y. [DOI] [PubMed] [Google Scholar]
- 19.Jin HJ, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature. 2003;424:1057–1061. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 20.Holland C, Terry AE, Porter D, Vollrath F. Comparing the rheology of native spider and silkworm spinning dope. Nat Mater. 2006;5:870–874. doi: 10.1038/nmat1762. [DOI] [PubMed] [Google Scholar]
- 21.Stark M, et al. Macroscopic fibers self-assembled from recombinant miniature spider silk proteins. Biomacromolecules. 2007;8:1695–1701. doi: 10.1021/bm070049y. [DOI] [PubMed] [Google Scholar]
- 22.Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc Natl Acad Sci USA. 2008;105:6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu X, et al. Eggcase protein-1: A new class of silk proteins with fibroin-like properties from the spider Latrodectus hesperus. J Biol Chem. 2005;280:21220–21230. doi: 10.1074/jbc.M412316200. [DOI] [PubMed] [Google Scholar]
- 24.Walsh DM, et al. Amyloid β-protein fibrillogenesis: Structure and biological activity of protofibrillar intermediates. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 25.Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi CY, Blackledge TA, Lewis RV. Molecular and mechanical characterization of aciniform silk: Uniformity of iterated sequence modules in a novel member of the spider silk fibroin gene family. Mol Biol Evol. 2004;21:1950–1959. doi: 10.1093/molbev/msh204. [DOI] [PubMed] [Google Scholar]
- 27.Lazaris A, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 28.Zhao A, et al. Novel molecular and mechanical properties of eggcase silk from wasp spider, Argiope bruennichi. Biochemistry. 2006;45:3348–3356. doi: 10.1021/bi052414g. [DOI] [PubMed] [Google Scholar]
- 29.Larkin MA, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Zwahlen C, et al. An NMR experiment for measuring methyl-methyl NOEs in 13C-labeled proteins with high resolution. J Am Chem Soc. 1998;120:7617–7625. [Google Scholar]
- 31.Wu J, Fan JS, Pascal SM, Yang D. General method for suppression of diagonal peaks in heteronuclear-edited NOESY spectroscopy. J Am Chem Soc. 2004;126:15018–15019. doi: 10.1021/ja045300l. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Lin Z, Xu Y, Yang S, Yang D. Sequence-specific assignment of aromatic resonances of uniformly 13C,15N-labeled proteins with use of 13C- and 15N-edited NOESY spectra. Angew Chem Int Ed. 2006;45:1960–1963. doi: 10.1002/anie.200503558. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Zheng Y, Fan JS, Yang D. A new strategy for structure determination of large proteins in solution without deuteration. Nat Methods. 2006;3:931–937. doi: 10.1038/nmeth938. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Long D, Yang D. Rapid data collection for protein structure determination by NMR spectroscopy. J Am Chem Soc. 2007;129:7722–7723. doi: 10.1021/ja071442e. [DOI] [PubMed] [Google Scholar]
- 37.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment by using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 38.Case DA, et al. AMBER 7. San Francisco: University California; 2002. [Google Scholar]
- 39.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 40.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.