Abstract
The process of human erythrocyte invasion by Plasmodium falciparum parasites involves a calcium-dependent serine protease with properties consistent with a subtilisin-like activity. This enzyme achieves the last crucial maturation step of merozoite surface protein 1 (MSP1) necessary for parasite entry into the host erythrocyte. In eukaryotic cells, such processing steps are performed by subtilisin-like maturases, known as proprotein convertases. In an attempt to characterize the MSP1 maturase, we have identified a gene that encodes a P. falciparum subtilisin-like protease (PfSUB2) whose deduced active site sequence resembles more bacterial subtilisins. Therefore, we propose that PfSUB2 belongs to a subclass of eukaryotic subtilisins different from proprotein convertases. Pfsub2 is expressed during merozoite differentiation and encodes an integral membrane protein localized in the merozoite dense granules, a secretory organelle whose contents are believed to participate in a late step of the erythrocyte invasion. PfSUB2’s subcellular localization, together with its predicted enzymatic properties, leads us to propose that PfSUB2 could be responsible for the late MSP1 maturation step and thus is an attractive target for the development of new antimalarial drugs.
Malaria remains one of the major human parasitic diseases, principally in subtropical regions. Most of the fatal cases are caused by Plasmodium falciparum, one of the four Plasmodia species that infect humans. The propagation of multidrug-resistant parasites and insecticide-resistant mosquitoes has led to major difficulties in treatment and control of malaria. Therefore, the identification of novel therapeutic targets essential for parasite development is an important first step in the development of new antimalarial drugs.
Erythrocyte invasion initiates the intra-erythrocytic asexual cycle that is responsible for all known malaria symptoms. Electronmicroscopy studies have shown that red blood cell (RBC) invasion by merozoites is a rapid multistep process, starting with the adhesion of the merozoite to the cell surface, followed by its reorientation and its entry into the host cell (1). This last step is concomitant with content release of three different merozoite organelles (rhoptries, micronemes, and dense granules) and can be blocked by antibodies to merozoite proteins as well as serine protease inhibitors (2–5). One of the best documented steps affected by serine protease inhibitors is the last processing step of the major merozoite surface protein 1 (MSP1). MSP1 is synthesized as a 200-kDa precursor that is proteolyzed in two successive steps (6). The second concerns the C-terminal glycosylphosphatidylinositol-anchored polypeptide (MSP1–42), where cleavage produces the MSP1–33 and MSP1–19 polypeptides (7). This processing is achieved by a parasite membrane-bound calcium-dependent serine protease, and its inhibition by serine protease inhibitors, or by antibodies, interrupts RBC entry of merozoites (6, 8).
In eukaryotic cells the maturation of polypeptide precursors is a conserved process usually achieved by calcium-dependent serine proteases of the subtilase family, the proprotein convertases (PCs) (9, 10). Among other roles, PCs are implicated in the maturation of bacterial toxins and retroviral surface glycoproteins (11, 12). Considering that the second MSP1 maturation step is performed by a calcium-dependent serine protease, we hypothesized that P. falciparum encodes a subtilisin-like protease involved in MSP1–42 proteolysis.
In this work, we report the identification of a P. falciparum gene (Pfsub2, for P. falciparum-subtilisin-like protease 2), which codes for a serine protease similar to the recently described PfSUB1 (13). Interestingly, even though P. falciparum is an eukaryotic organism, the active site of PfSUB2 is highly similar to prokaryote subtilisins. Indeed, active site phylogenetic analysis of PfSUB2 compared with subtilases of eubacteriae, plants, yeast, or higher eukaryotic organisms suggests that PfSUB2 belongs to a subclass of prokaryotic-like eukaryotic enzymes, which includes PfSUB1, the Dictyostelium discoideum serine protease TAGB, and the recently described mammalian S1P/SKI-1 maturase (14–16). The Pfsub2 gene is transcribed during the merozoite differentiation, producing an integral membrane precursor protein with the fully processed form of PfSUB2 being secreted into dense granules. The characterization of PfSUB2 leads us to propose that this enzyme might achieve the maturation step of MSP1–42 and thus be a crucial enzyme for parasite entry into the erythrocyte.
MATERIALS AND METHODS
Isolation of Pfsub2 Full-Length Nucleotide Sequence.
The original clone (GenBank accession no. N97791) was a 2,043-bp cDNA obtained from the University of Florida P. falciparum expressed sequence tag collection (17) and was sequenced on an ABI DNA sequencer (Genome Express, Grenoble, France). The complete ORF was obtained by initiating PCR using the oligonucleotide K50 5′-AACGTCAAAATGACTAGAGC-3′ in combination with M13 universal primer on the original cDNA library (17). The amplified fragment, corresponding to nucleotides 1554–2645, was cloned by using the TopoTA cloning kit (Invitrogen) and sequenced by using the Sequenase reaction kit (United States Biochemical-Amersham Pharmacia). Then, the oligonucleotide K51 5′-CGAATGAAGATCTAATTCTCC-3′ was used to prime an asymmetric PCR on a DraI Palo Alto genomic DNA vectorette library (18), which yielded nucleotides 648 and 1755. Finally, PCR performed on the original cDNA library by using the oligonucleotide K53 5′-TTTTGTTACTAGCCAAATTC-3′ and M13 universal primer amplified a 750-bp fragment corresponding to the 5′ end of Pfsub2 cDNA sequence. Each amplified fragment was sequenced in triplicate to eliminate potential PCR-introduced errors.
Pulsed-Field Gel Electrophoresis.
Pulsed-field gel electrophoresis of Palo Alto chromosomal blot was a kind gift from A. Scherf (Institut Pasteur, Paris) and was performed as described (19). The Pfsub2 cDNA probe was produced by reverse transcriptase (RT)–PCR using the oligonucleotides Khis, 5′-TCCTACAGATGCTTAGGTC-3′, and SUB1B, 5′-ATCGAATTCATACAAATTATATAAGGC-3′. The 1,027-bp cDNA fragment was α-32P-labeled by random priming (Megaprime, Amersham Pharmacia), hybridized overnight in 6× SSC, 0.1% SDS, and 2.5% nonfat milk at 65°C, and washed at the same temperature for 30 min in 0.5× SSC (16.5 mM NaCl/15 mM sodium citrate, pH 7.0), 0.1% SDS.
Computer Analysis of PfSUB2 and Construction of the Phylogenetic Tree.
The prediction of PfSUB2 signal peptide and its cleavage site was accomplished following Nielsen et al. (20), while the PfSUB2 proposed transmembrane domain was identified by using Rost et al.’s program (21). The phylogenetic analysis of the 32 subtilisin-like proteases active sites was performed as described (22), except that the multiple sequence alignment of the 32 subtilisin-like proteases was done by using clustalw 1.7 program with low gap penalty (23). The 259 residues corresponding to the full active site region PfSUB2 were aligned to the corresponding region of 31 other sequences referred to in ref. 22 together with PfSUB1 (13). The multiple alignment was analyzed by using the phylip software to produce an unrooted tree (24).
RT-PCR and Intron Identification.
Total RNA was prepared by using Trizol (GIBCO/BRL), following the manufacturer’s conditions. RT-PCR has been performed by using RNase-free/DNase1 (Boehringer Mannheim)-treated RNA corresponding to approximately 106 parasites. cDNA synthesis was initiated by the SUB1B primer in the presence of Moloney murine leukemia virus RT (GIBCO/BRL). An 880-bp fragment was further amplified by using SUB1B and KB 5′-GCATCCGGAAATAAAAGTAAC-3′ primers. For Pfrab6 RT-PCR analysis, oligonucleotides were used to amplify a 203-bp cDNA bridging exons 1 and 2 (25). To map the precise intron-exon junction, the Pfsub2-specific genomic DNA fragment was cloned and sequenced.
Western Blot and Immunoprecipitation Analysis of PfSUB2.
A gluthatione S-transferase (GST)-PfSUB2 fusion protein was generated by using oligonucleotides SUB2A 5′-TATATGGATCCGCGTAAACATAGTGATAAG-3′ and SUB2B 5′-GTAGAATTCTTTGGTCTCTTTCTTTC-3′ to amplify a 351-bp fragment that was ligated into pGEX-A (Amersham Pharmacia). The GST-PfSUB2 fusion protein was prepared as described (18) except that the purified protein was eluted by using 0.2% SDS in PBS (10 mM potassium phosphate, 145 mM NaCl, pH 7.4). CD1 mice were immunized s.c. with 20 μg of GST-PfSUB2 in the presence of complete Freund’s adjuvant, followed by five additional injections performed with incomplete Freund’s adjuvant. Protein extracts were produced from synchronous knob+ Uganda Palo Alto-infected RBCs cultivated in vitro; segmented schizonts were enriched by Percoll-Sorbitol gradient centrifugation, and merozoites were prepared as described (26). Parasitized RBCs and the equivalent amount of noninfected erythrocytes were pelleted and lysed in 1 vol of water by freeze-thaw treatment. The insoluble pelleted proteins were further extracted by 100 mM alkaline Na2CO3 treatment (30 min, 4°C), which is known to discriminate between peripheral membrane proteins that are solubilized and integral membrane proteins that are insoluble and pelleted by a 30-min centrifugation at 100,000 g at 4°C (27). Protein extracts corresponding to approximately 5.106 parasitized RBCs were separated on 7.5% SDS/PAGE. After transfer to nitrocellulose, Western blot assays were performed as described except that antiserum were used at 1:100 dilution (18). Immunoprecipitations were performed as described (18), except that highly synchronous parasites were cultivated in the presence of 175 μCi/ml of 35S-labeled methionine/cysteine (ICN) for the last 7 hr of schizogony.
Immunolocalization of PfSUB2 by Indirect Fluorescence Assay and Immunoelectron Microscopy (IEM).
Mouse anti-GST-PfSUB2 or mouse anti-GST (diluted 1:50) sera were incubated 30 min at 37°C with air-dried parasitized RBCs. Slides then were washed in PBS and incubated with goat anti-mouse IgG and IgM conjugated with fluorescein (diluted 1:100, Immunotech) in the presence of 25 μg/ml of propidium iodide. Slides were washed in PBS and sealed in the presence of 0.1% p-phenylenediamine (in 0.5% glycerol) and analyzed with a Leica microscope at 1,500-fold magnification under UV light.
For IEM, P. falciparum (Palo Alto) parasites were fixed for 30 min at 4°C with 1% formaldehyde, 0.1% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. Fixed samples were washed, dehydrated, and embedded in LR White resin (Polysciences) as described (1). Thin sections were blocked in PBS containing 5% (vol/vol) nonfat dry milk and 0.01% (vol/vol) Tween 20. Grids were incubated with primary antibodies (mouse anti-GST-PfSUB2 or mouse anti-GST) diluted 1:50 to 1:200 in PBS containing 1% (wt/vol) BSA and 0.01% (vol/vol) Tween 20 (PBT) for 2 hr at room temperature. Negative controls included normal mouse serum and PBT applied as the primary antibody. After washing, grids were incubated for 1 hr in 15 nm gold-conjugated goat anti-mouse IgG (Amersham Pharmacia) diluted 1:20 in PBT, rinsed with PBT, and fixed with glutaraldehyde to stabilize the gold particles. Samples were stained with uranyl acetate and lead citrate and examined in a Zeiss CEM902 electron microscope.
RESULTS
The Pfsub2 Gene Encodes a Serine Protease Belonging to a Subclass of Eukaryotic Subtilases Similar to the Bacterial Subtilisins.
We have identified a P. falciparum expressed sequence tag in the University of Florida collection having an ORF displaying strong similarities to the bacterial subtilisin active site. This clone starts at nucleotide 2669 of the complete Pfsub2 (GenBank accession no. AJ132006) and includes the 3′ untranslated end (Fig. 1). A combination of different asymmetric PCR-based techniques allowed us to obtain the full-length Pfsub2 4.3-kb ORF, a size confirmed by Mung bean nuclease digestion (data not shown, ref. 28).
Figure 1.
Predicted protein sequence of PfSUB2 (GenBank accession no. AJ132006) and the alignment of its predicted catalytic domain with B. amyloliquefaciens BPN subtilisin (GenBank accession no. X00165). The translation initiation and stop codons are in bold and underlined. PfSUB2 protein sequence begins with a signal peptide shown in bold. The four residues that compose the subtilase active site are heavily overlined, and the C-terminal transmembrane segment is double underlined. The interruption of the ORF by an intron is shown by ↕. Brackets define the 259 residues of the catalytic domain used to perform the phylogenetic analysis, and the sequence corresponding to the GST-fusion protein is presented in italics and dashed underlined. The two proposed activation sites are underlined.
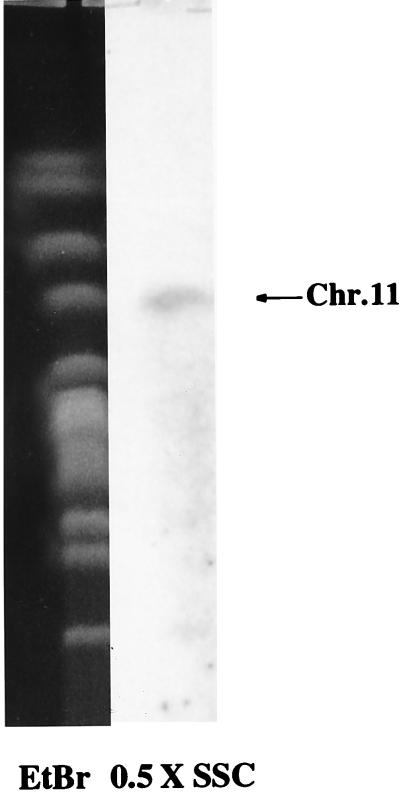
The deduced 1,337-residue PfSUB2 protein sequence corresponds to a theoretical 147-kDa polypeptide (Fig. 1). It starts with a 21-residue signal peptide and possesses a 19-aa putative hydrophobic transmembrane domain close to the C terminus. No glycosylphophatidylinositol addition motif was detected. Taken together, these observations suggest that PfSUB2 is a secreted type 1 integral membrane protein. Importantly, PfSUB2 contains the four consensus sequences known to form the active site of subtilisin-like serine proteases of the superfamily S8 (22, 29). The universally conserved catalytic triade is composed of Asp750, His793, and Ser956, whereas the oxyanion-hole implicated in the enzyme-substrate stabilization involves Asn878 (Fig. 1). This region of PfSUB2’s active site exhibits 33% identity and 44% similarity with Bacillus amyloliquefaciens bacterial proteinase novo (BPN) subtilisin (Fig. 1). Interestingly, Asn878 and Ser956 of PfSUB2’s active site are encoded by different exons separated by a 143-bp intron (Fig. 1). Finally, a 1,027-bp probe corresponding to the active site hybridizes specifically to chromosome 11 (Fig. 2) and under the same conditions produced a genomic Southern blot pattern consistent with a unique Pfsub2 gene (data not shown).
Figure 2.
Pfsub2 gene specifically hybridizes to chromosome 11. Chromosome blocks from P. falciparum Palo Alto strain were probed with 1,027 bp of cDNA corresponding to the catalytic domain of PfSUB2.
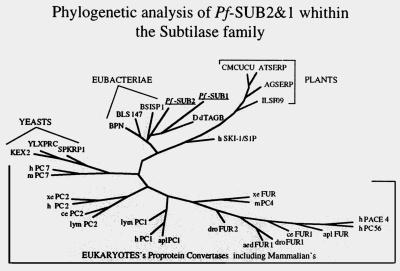
Phylogenetic analysis of PfSUB2’s 259-residue active site region compared with 31 subtilisin-like active sites expressed by various organisms is presented in Fig. 3 and shows that PfSUB2 clusters with bacterial subtilases. It displays 44% similarity to the active site of Bacillus subtilis subtilisin and only 29% similarity with kex2 from Saccharomyces cerevisiae, 34% with Arabidopsis thaliana serine protease from A. thaliana, and 36% with the human PC7, the mammalian PC most related to the lower eukaryote subtilisin-like proteases (22). Among eukaryotic subtilases, only TAGB (36%) from D. discoideum and the recently described mammalian S1P/SKI-1s (35%) (14, 15) are closely related to PfSUB2. Our phylogenetic analysis suggests that PfSUB2 and PfSUB1 belong to the pyrolysin subfamily of subtilases, as previously shown for DdTAGB and S1P/SKI-1 (22).
Figure 3.
Phylogenetic tree analysis based on sequence alignment of catalytic domains of subtilases from various organisms. PfSUB2 and PfSUB1 appear to diverge from known eukaryotic subtilases and to be more similar to the bacterial enzymes. DdTAGB and human S1P/SKI-1 are the most related eukaryotic subtilases and with PfSUB2 and PfSUB1 define a subclass of eukaryotic subtilases.
Two characteristics of the PfSUB2 active site are worthy of mention. The serine consensus site (GTS956 MA) contains a methionine that is conserved in all bacterial, fungal, and plant subtilisins, but is absent in PCs (22). This methionine is present in DdTAGB’s active site (16), whereas S1P/SKI-1’s active site has a valine (22). PfSUB2’s active site (D750SG) is identical to bacterial subtilisin’s (22), whereas PCs have DDG. In the case of DdTAGB and S1P/SKI-1’s active sites, the plant-related motif DTG is found (22). Taken together, these observations strongly suggest that PfSUB2 with PfSUB1, DdTAGB, and S1P/SKI-1, define a subclass of eukaryotic pyrolysin-like subtilases similar to the bacterial enzymes and differing from PCs.
Pfsub2 Is Transcribed During P. falciparum Schizogony, Allowing the Synthesis of an Integral Membrane Protein That Is Transported into Merozoite Dense Granules.
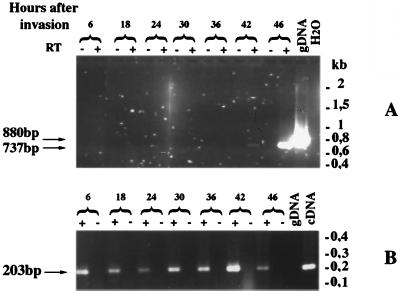
RT-PCR was performed on total RNA prepared from P. falciparum rings, trophozoites, and schizonts. Pfsub2-specific oligonucleotides amplified the expected 737-bp spliced cDNA fragment from RNA prepared 42 and 46 hr postinvasion (Fig. 4A). Therefore, Pfsub2 transcription is stage-specifically regulated to occur during P. falciparum schizogony, differing (Fig. 4B) from the constitutive expression of Pfrab6 (30).
Figure 4.
Analysis of Pfsub2 mRNA expression through the P. falciparum asexual cycle. RT-PCR was performed on total RNA isolated between 6 and 46 hr after invasion of erythrocytes by merozoites of P. falciparum Palo Alto strain cultivated in vitro. (A) Pfsub2 oligonucleotides have been chosen to flank the Pfsub2 intron. The expected sizes of the amplified fragments are 880 bp and 737 bp on genomic DNA and cDNA, respectively. (B) RT-PCR was performed by using two Pfrab6 oligonucleotides, where one oligonucleotide crossed the junction of the two first exons of Pfrab6 and therefore only cDNA was amplified. As negative controls RT-PCR on water were performed.
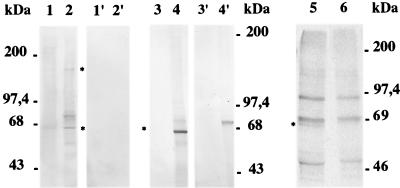
To identify the Pfsub2 gene product, Western blot and immunoprecipitation assays were performed. Anti-GST-PfSUB2 antibodies identify a 160-kDa polypeptide (Fig. 5), a size predicted by the ORF (Fig. 1). This polypeptide is detected only in Na2CO3-insoluble protein extracts consistent with PfSUB2 being an integral membrane protein (27). Several smaller polypeptides also are identified by Western blot analysis, which probably correspond to processing or degradation products, because only the integral membrane 65-kDa polypeptide is detected by immunoprecipitation of 35S-labeled segmented schizont protein extracts (Fig. 5, lane 5). Furthermore, this 65-kDa polypeptide is the only PfSUB2 form that persists in merozoites (Fig. 5, lane, 4), being probably the final processed form produced from 160-kDa precursor (Fig. 5, lane 2). By analogy with bacterial subtilisins, the 160-kDa polypeptide would correspond to the inactive pro-enzyme and the 65-kDa form to the active enzyme.
Figure 5.
Western blot analysis on protein extracts prepared from P. falciparum segmented schizonts and merozoites. Lanes 1 and 2 correspond to Na2CO3-soluble and Na2CO3-insoluble protein extracts prepared from segmented schizonts. Lanes 1′ and 2′ contain the same proportion of Na2CO3-soluble and –insoluble protein extracts prepared from noninfected RBCs. Lanes 3 and 3′ and lanes 4 and 4′ show Western blot analysis performed on Na2CO3-soluble and -insoluble merozoite protein extracts. Lanes 1 and 2, lanes 1′ and 2′, and lanes 3 and 4 have been incubated with anti-GST-PfSUB2 antibodies, and lanes 3′ and 4′ have been incubated with antibodies to GST alone. Lanes 5 and 6 correspond to the immunoprecipitation of segmented schizont S35-labeled proteins performed with mouse anti-GST-PfSUB2 and anti-GST.
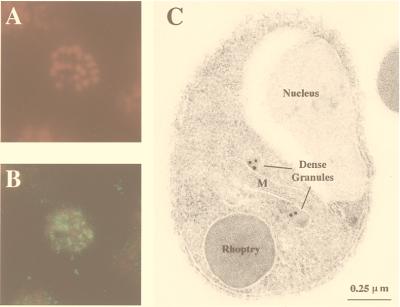
Indirect fluorescence assay with anti-GST-PfSUB2 antibodies produces a punctuated pattern on schizonts (Fig. 6B) distant from the nucleus (Fig. 6A). IEM studies established that PfSUB2 is transported to the dense granules of both intra-erythrocytic (data not shown) and free merozoites (Fig. 6C). Because only the 65-kDa form of PfSUB2, proposed to be the active enzyme, can be detected in merozoite protein extracts (Fig. 5, lane 4), and considering that the content of dense granules is secreted during the late steps of erythrocyte invasion by merozoites (5), PfSUB2 is likely to play a role in this crucial process.
Figure 6.
Localization of PfSUB2 protein by immunofluorescence on segmented schizonts and by IEM on fixed merozoites. (A and B) An air-dried fixed segmented schizont incubated with 25 μg/ml of propidium iodide to label the parasite nuclei (A) and mouse anti-GST-PfSUB2 antibodies (B). Bound antibodies then were detected with FITC-coupled anti-mouse antibodies (magnification: ×1,500). (C) The IEM analysis of thin sections of resin-embedded merozoites probed with mouse anti-GST-PfSUB2 antibodies and revealed with gold-labeled anti-mouse IgG antibody. M, mitochondrion.
DISCUSSION
PfSUB2 is a serine protease synthesized during schizogony and transported to merozoite dense granules (Fig. 6). PfSUB2 is detectable only in Na2CO3-insoluble protein extracts, and because it contains a C-terminus transmembrane domain (Fig. 1) it can be classified as a type 1 integral membrane protein (27). However, a solubilized form could escape detection as antibodies were raised to a polypeptide C terminal to the transmembrane domain. Specific antibodies to a region N terminal to this domain should allow us to address this question.
Interestingly, the active site of PfSUB2 is 48% similar to the equivalent region of the recently described PfSUB1 protease (13), suggesting that the two genes may be derived from a common ancestor. However, outside their active site regions the proteins show no significant similarity. Moreover, only Pfsub2 has an intron that places the active site Ser956 on a different exon to the three other catalytic residues (Fig. 1). This finding contrasts with higher eukaryotic PCs, where each catalytic residue is encoded by a separate exon (9).
Phylogenetic analysis of PfSUB2 and PfSUB1 active sites showed that the closest eukaryotic sequences are DdTAGB and the recently described mammalian S1P/SKI-1 (14, 16). Although these two proteases previously have been shown to belong to the pyrolysin subfamily of subtilases, they were not assigned to any particular subclass (22). We propose that DdTAGB and S1P/SKI-1 belong to a subclass of bacterial-like eukaryotic pyrolysin subtilases that includes both PfSUB2 and PfSUB1. The natural substrate specificity of S1P/SKI-1 has been determined recently. It maturates the transcription factor SREBP at a Leu↕Ser dipeptide bond and participates in the regulation of the animal cell lipid biosynthesis (14). Therefore, this subclass displays different substrate specificities to the eukaryotic PCs (9, 10), being able to cleave after hydrophobic residues. We also performed phylogenetic analysis using the parsimony procedure rooting the tree on the B. subtilis BPN sequence (data not shown), which suggests that the DdTAGB diverged from the bacterial enzymes after PfSUB2 and PfSUB1 in accordance with Sogin and Silberman (31), who propose that protists diverged before plants and animals in the eukaryotic kingdom.
The crucial role of parasite serine proteases in the invasion of erythrocytes by merozoites is well established (3). Two major roles have been attributed for these enzymes: (i) the proteolysis of the erythrocyte plasma membrane anion transporter band 3 by the chymotrypsin-like Pfgp76 participating in the formation of the parasitophorous vacuole (32), and (ii) the maturation of MSP1–42 by a merozoite membrane-bound and calcium-dependent serine protease (6, 33). In addition to their difference of size and maturation pattern, because PfSUB2 has an active site similar to prokaryotic subtilisins and contains in its C terminus a transmembrane domain without any sequence typical of a glycosylphosphatidylinositol addition, it is unlikely that PfSUB2 corresponds to Pfgp76, which is a 76-kDa glycosylphosphatidylinositol-anchored merozoite serine protease with specificity for substrates with Arg at the P1 position and hydrophobic residues at the P3 and P4 positions (32, 34).
PfSUB2’s similarity to calcium-dependent subtilisins raises the question of its role in MSP1–42 maturation and a number of observations support this hypothesis. The location of PfSUB2 in merozoite dense granules suggests a role for this enzyme during the invasion process, as the contents of these organelles are known to be secreted after junction formation between merozoites and erythrocytes (5) and are believed to participate in parasitophorous vacuole formation (2). However, IEM studies have shown that a part of the dense granule contents also can be seen around the surface of merozoites during their entry (5). This event is both the time and place where MSP1–42 maturation is thought to occur (6, 33). Hence, by analogy with other Apicomplexa parasites, it has been proposed that P. falciparum merozoites may have subpopulations of dense granules with different properties (13). Thus, PfSUB2, like PfSUB1, might be located in a subset of dense granules whose contents are released during invasion, allowing PfSUB2 to access and cleave MSP1–42, producing the MSP1–19 fragment. Finally, PfSUB2 apparently does not persist through to ring stages, because indirect fluorescence assay studies performed on young rings by using the anti-GST-PfSUB2 antibodies were negative (data not shown).
B. amyloliquefaciens BPN subtilisin, S. cerevisiae kex2, and most of the known subtilases remove their pro-regions by an auto-catalytic process, and importantly, the sequence of the auto-activation site mimics the cleavage site of their specific substrates (35). For example, BPN subtilisin auto-activates after a tyrosine, whereas the active enzyme hydrolyses a peptide bond of broad specificity. PfSUB2’s auto-activation site has to be located between residues 680 and 720 to produce the 65-kDa polypeptide observed in merozoite extracts (Fig. 5, lane 4), which would place the auto-activation site about 50 residues before Asp750 (Fig. 1), a distance typical for subtilases (22). The alignment of the PfSUB2 region located 30–50 residues before its catalytic aspartate with the Leu↕Asn dipeptide MSP1–42 cleavage site region shows that this motif is tandemly conserved (Figs. 1 and 7). Moreover, these dipeptides are located in regions of conserved charge or group residues in both MSP1–42 and PfSUB2 sequences (Fig. 7), suggesting that not only the cleavage site sequence is conserved, but also its environment, which is important in terms of substrate recognition. Similarly, S1P/SKI-1’s substrate cleavage site (ArgXXLeu↕Ser) is found as expected 50 aa before its catalytic Asp218, which is consistent with the observed S1P/SKI-1 processing product (14, 15). In contrast, analysis of PfSUB1 did not lead to the identification of a putative Leu↕Asn auto-activation site in the appropriate region, suggesting that PfSUB2 and PfSUB1 could have different substrate specificities.
Figure 7.
Alignment of the 22 residues surrounding the Leu↕Asn MSP1–42 maturation site with the 38 residues corresponding to the predicted PfSUB2 auto-activation site region. Identities are shown by vertical bars, and residues of the same group are shown by double dots.
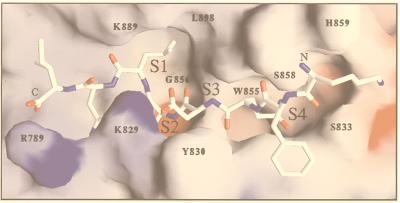
To investigate the properties of the PfSUB2 active site, we carried out homology modeling of the PfSUB2 catalytic domain based on the available crystal coordinates of the most similar subtilisin sequences (Fig. 8). The overall predicted architecture for the PfSUB2 active site is similar to that of BPN, but there are interesting differences. The major determinant of substrate specificity in subtilase proteases is the distinct S1 pocket, which is involved in the interaction with the P1 residue of the substrate. In the case of BPN, the essential residue involved in these interactions is Gly166, which contributes to the creation of a large S1 pocket. The equivalent residue in PfSUB2 is Leu898, which reinforces the general hydrophobic character of the S1 pocket and is consistent with the presence of a Leu residue at the P1 position of the proposed substrate (MSP1–42). Another distinct subsite characteristic of subtilisins, subsite S4, also is predicted to be conserved in PfSUB2. At the bottom of this pocket, a tryptophan residue (Trp855) is found in PfSUB2, rather than Leu126 in BPN sequence. The polar group of the glutamine side chain at substrate position P4 could interact with the NH group of the Trp855 side chain and with amino and carbonyl main chain groups in this region. More significant differences are predicted for the PfSUB2 S2 and S3 binding subsites. The presence of Lys829-Tyr830 in PfSUB2 (instead of Gly100-Ser101 for BPN) should render less accessible the S2-S3 pocket, leading to a more stringent substrate specificity. It is interesting to note that the decrease of solvent accessibility for the S2 subsite (because of the aliphatic moiety of the Lys829 side chain) would be consistent with a hydrophobic side chain at position P2 (methionine) of MSP1–42. Furthermore, the amino group of Lys829 could interact with the aspartic acid carboxylate at position P3 (pointing outward). In summary, the three-dimensional model of the PfSUB2 active site appears to be compatible with the proposed substrate specificity, leading additional support to the hypothesis that this enzyme could be a MSP1–42 maturase.
Figure 8.
Homology model of PfSUB2 predicted catalytic domain in complex with MSP1–42 cleavage site (Nterm-K-F-Q-D-M-L-N-I-Cterm). Residues involved in the PfSUB2 predicted S1, S2, S3, and S4 subsites are numbered. Modeling was carried out with computer programs quanta and charmm by using the atomic coordinates of B. amyloliquefaciens BPN subtilisin in complex with the eglin inhibitor (PDB code 2SNI) and B. licheniformis Carlsberg’s subtilisin in complex with eglin C (PDB code 2SEC) as templates. Framework residues differing between proteins were mutated, and all required insertions (Fig. 1) were modeled in hypothetical conformations taken from the loop library implemented in quanta. These insertions occur in surface-exposed loops located far from the substrate-binding region, with the possible exception of the segment preceding the catalytic His793 residue (including Arg789), which may influence the local structure of the S1′-S2′ subsites. The model was manually adjusted to remove unreasonable contacts and subjected to energy minimization by using the program charmm.
Further work obviously is needed to confirm the proposed function and specificity of PfSUB2. The characterization of active recombinant PfSUB2 enzyme or purification from the parasite of the active protease should enable us to confirm its role. Nonetheless, the identification of a P. falciparum merozoite-specific serine protease is an important step toward a better understanding of erythrocyte invasion and may lead to the definition of a new target for the development of antimalarial drugs.
Acknowledgments
The support of Luiz Pereira da Silva during the initiation of this work is gratefully recognized. We thank Nabil Seidah, Alain Boudreault, and Claude Lazure for fruitful discussion on subtilases, Geneviève Milon and Denise Mattei for helpful comments, and Solange Touron for parasite culture. We thank Debopam Chakrabarti for the gift of the original expressed sequence tag, and we acknowledge the financial support of the Pasteur Institute, the Centre National de la Recherche Scientifique, and North Atlantic Treaty Organization. J.-C.B. received awards from the Pasteur-Weizmann foundation and the Fondation pour la Recherche Médicale and is currently a fellow of the Association Française pour la Recherche Thérapeutique. M.A. and H.F. received support from the National Institute of Allergy and Infectious Diseases/National Institutes of Health (AI-35827), U.S. Agency for International Development (DPE-936–6001-29), and a grant-in-aid for scientific research on priority areas from the Ministry of Education, Science, Sports, and Culture of Japan.
ABBREVIATIONS
- RBC
red blood cell
- RT
reverse transcriptase
- IEM
immunoelectron microscopy
- MSP1
merozoite surface protein 1
- GST
gluthatione S-transferase
- PBT
PBS containing 1% (vol/vol) BSA and 0.01% (vol/vol) Tween 20
- PC
proprotein convertase
- BPN
bacterial proteinase novo
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ132006).
References
- 1.Aikawa M. In: Malaria: Principles and Practice of Malariology. Wernsdorfer W H, McGregor I, editors. Vol. 1. Edinburgh: Churchill Livingstone; 1988. pp. 97–129. [Google Scholar]
- 2.Bannister L H, Mitchell G H. J Protozool. 1989;36:362–367. doi: 10.1111/j.1550-7408.1989.tb05527.x. [DOI] [PubMed] [Google Scholar]
- 3.Braun-Breton C, Pereira da Silva L. Parasitol Today. 1993;9:92–96. doi: 10.1016/0169-4758(93)90212-x. [DOI] [PubMed] [Google Scholar]
- 4.Perrin L H, Ramirez E, Lambert P H, Miescher P. Nature (London) 1981;289:301–303. doi: 10.1038/289301a0. [DOI] [PubMed] [Google Scholar]
- 5.Torii M, Adams J H, Miller L H, Aikawa M. Infect Immun. 1989;57:3230–3233. doi: 10.1128/iai.57.10.3230-3233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackman M J, Holder A A. Mol Biochem Parasitol. 1992;50:307–316. doi: 10.1016/0166-6851(92)90228-c. [DOI] [PubMed] [Google Scholar]
- 7.Blackman M J, Ling I T, Nicholls S C, Holder A A. Mol Biochem Parasitol. 1991;49:39–34. doi: 10.1016/0166-6851(91)90127-r. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J A, Bujard H. Mol Biochem Parasitol. 1992;56:151–160. doi: 10.1016/0166-6851(92)90162-d. [DOI] [PubMed] [Google Scholar]
- 9.Seidah N G, Day R, Marcinkiewicz M, Chrétien M. Ann NY Acad Sci. 1998;839:9–24. doi: 10.1111/j.1749-6632.1998.tb10727.x. [DOI] [PubMed] [Google Scholar]
- 10.Steiner D. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 11.Gordon V M, Leppla S H. Infect Immun. 1994;62:333–340. doi: 10.1128/iai.62.2.333-340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk H-D, Garten W. Nature (London) 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 13.Blackman M J, Fujioka H, Stafford W H L, Sajid M, Cough B, Fleck S L, Aikawa M, Grainger M, Hackett F. J Biol Chem. 1998;273:23398–23409. doi: 10.1074/jbc.273.36.23398. [DOI] [PubMed] [Google Scholar]
- 14.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 15.Seidah N G, Mowla S J, Hamelin J, Mamarbachi A M, Benjannet S, Touré B B, Basak A, Munzer J S, Marcinkiewicz J, Zhong M, et al. Proc Natl Acad Sci USA. 1999;96:1321–1326. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaulsky G, Kuspa A, Loomis W F. Genes Dev. 1995;9:1111–1122. doi: 10.1101/gad.9.9.1111. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti D, Reddy G R, Dame J B, Almira E C, Laipis P J, Ferl R J, Yang T P, Rowe T C, Schuster S M. Mol Biochem Parasitol. 1994;66:97–104. doi: 10.1016/0166-6851(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 18.Barale J-C, Attal-Bonnefoy G, Brahimi K, Pereira da Silva L, Langsley G. Mol Biochem Parasitol. 1997;87:169–181. doi: 10.1016/s0166-6851(97)00065-0. [DOI] [PubMed] [Google Scholar]
- 19.Hinterberg K, Scherf A. Parasitol Today. 1994;10:225. doi: 10.1016/0169-4758(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Rost B, Fariselli P, Casaido R. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siezen R J, Leunissen J A M. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein F. phylip (Phylogeny Inference Package), version 3.5c. Seattle: Univ. of Washington; 1993. [Google Scholar]
- 25.Templeton T J, Kaslow D C. Mol Biochem Parasitol. 1998;94:149–153. doi: 10.1016/s0166-6851(98)00038-3. [DOI] [PubMed] [Google Scholar]
- 26.Pasvol G, Wilson R J M, Smalley M E, Brown J. Ann Trop Med Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- 27.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCutchan T F, Hansen J L, Dame J B, Mullins J A. Science. 1984;225:625–628. doi: 10.1126/science.6330899. [DOI] [PubMed] [Google Scholar]
- 29.Rawlings N D, Barrett A J. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alves de Castro F, Ward G E, Jambou R, Attal G, Mayau V, Jaurreguiberry G, Braun-Breton C, Chakrabarti D, Langsley G. Mol Biochem Parasitol. 1996;80:77–88. doi: 10.1016/0166-6851(96)02670-9. [DOI] [PubMed] [Google Scholar]
- 31.Sogin M L, Silberman J D. Int J Parasitol. 1998;28:11–20. doi: 10.1016/s0020-7519(97)00181-1. [DOI] [PubMed] [Google Scholar]
- 32.Roggwiller E, Morales-Betoulle M E, Blisnick T, Braun-Breton C. Mol Biochem Parasitol. 1996;82:13–24. doi: 10.1016/0166-6851(96)02714-4. [DOI] [PubMed] [Google Scholar]
- 33.Blackman M J, Chappel J A, Shai S, Holder A A. Mol Biochem Parasitol. 1993;62:103–114. doi: 10.1016/0166-6851(93)90182-w. [DOI] [PubMed] [Google Scholar]
- 34.Braun-Breton C, Pereira da Silva L. Biol Cell. 1988;64:223–231. doi: 10.1016/0248-4900(88)90081-0. [DOI] [PubMed] [Google Scholar]
- 35.Shinde U, Li Y, Inouye M. In: Intramolecular Chaperones and Protein Folding. Shinde U, Inouye M, editors. Austin, TX: Landes; 1995. pp. 11–34. [Google Scholar]