Abstract
Acting through the Pelle and IRAK family of protein kinases, Toll receptors mediate innate immune responses in animals ranging from insects to humans. In flies, the Toll pathway also functions in patterning of the syncytial embryo and requires Tube, a Drosophila-specific adaptor protein lacking a catalytic domain. Here we provide evidence that the Tube, Pelle, and IRAK proteins originated from a common ancestral gene. Following gene duplication, IRAK-4, Tube-like kinases, and Tube diverged from IRAK-1, Pelle, and related kinases. Remarkably, the function of Tube and Pelle in Drosophila embryos can be reconstituted in a chimera modeled on the predicted progenitor gene. In addition, a divergent property of downstream transcription factors was correlated with developmental function. Together, these studies reveal previously unrecognized parallels in Toll signaling in fly and human innate immunity and shed light on the evolution of pathway organization and function.
Keywords: Death domain, Drosophila, Evolution, Gene duplication, NF-κB, Signal transduction
Introduction
Across a broad range of animals, the Toll signaling pathway mediates recognition of invading microorganisms and initiates defensive responses [1–5]. Receptor recognition of pathogen-associated molecular patterns initiates a signal transduction cascade that activates transcription factors responsible for innate immune effector expression. Pathway structure has been largely conserved from insects to humans, although there are notable variations, with some components being found only in certain lineages and others differing in number and structure. To understand how such a signal transduction system evolves requires distinguishing changes that affect function significantly from those that are neutral mutational events.
Drosophila melanogaster provides the opportunity to investigate modification of Toll signaling in response to the selective pressures imposed by multiple biological contexts. In addition to its ancestral role in innate immune signaling, the Toll pathway in flies functions in several developmental contexts [6–9]. Of these, the best characterized is the maternal specification of the embryonic dorsoventral axis [10]. In embryos, Toll signaling occurs in a spatially graded manner across a syncytium of >103 nuclei undergoing rapid division. This environment for Toll signaling is thus quite distinct from that in cells mediating immune responses to infection. A dissection of Toll signaling from both a genomic and functional perspective therefore has the potential to reveal how pathway organization and structure have evolved so as to fulfill specialized developmental as well as immune functions [11].
The effectors for Toll signaling are members of the NF-κB family of transcription factors. In vertebrates, these include p50 and p65, which heterodimerize to form NF-κB [12]. In flies, Toll activates the NF-κB family members Dorsal and Dif. In the absence of a stimulus, NF-κB proteins are held in the cytoplasm by an inhibitor: IκB in vertebrates and its counterpart, Cactus, in flies. Toll pathway activity triggers inhibitor degradation, releasing NF-κB factors for nuclear translocation and binding to cis-regulatory sites in DNA.
Most of the proteins that link Toll to NF-κB proteins in Drosophila immunity and development have known counterparts in mammalian immunity. In response to ligand-mediated activation, fruit fly Toll and many mammalian TLR proteins recruit the adaptor protein MyD88 [13, 14]. Binding to MyD88 occurs through an interaction mediated by the TIR domains present in the receptor and in MyD88. Similarly, MyD88 associates with protein kinases via a shared interaction motif, the death domain [15, 16]. In humans, recruitment of two protein kinases – IRAK-1 and IRAK-4 – promotes signaling; two additional kinases – IRAK-2 and IRAK-M (IRAK-3) – are thought to dampen or inhibit signaling [17, 18]. Drosophila has a single related kinase, Pelle, that is required for Toll signaling [19]. Although downstream events differ between flies and humans, signaling in both organisms relies on inhibitor phosphorylation and proteolysis [20].
Toll function in flies relies on Tube, a factor that has no identified counterpart in vertebrate genomes [21–23]. Tube has an amino-terminal death domain and a carboxyl-terminal region containing five copies of the Tube repeat, an 8-amino-acid motif [24]. Each half of Tube participates in one or more protein-protein interactions. The Tube death domain acts as a bridge between the death domains of MyD88 and Pelle [25, 26]. The repeat-containing domain mediates the stable association of Dorsal and Tube in embryos [27, 28].
Here we show that Drosophila Tube is derived from an ancestral gene that contained both a death domain and a kinase domain. We demonstrate that chimeras of Tube and Pelle are functional and can be used to delineate domain function in vivo. We also establish that the closely related transcription factors Dorsal and Dif differ in their ability to interact with a repeat-containing domain of Tube. Combining comparative genomics with functional assays, these studies provide a unique perspective on signal transduction pathway evolution and specialization.
Materials and Methods
Reagents
The Anopheles gambiae cDNA library was a gift from Carolina Barillas-Mury. The Apis mellifera RACE cDNA library was a gift from Michael Levine and was generated in his laboratory with the Clontech Marathon cDNA kit. S2 cell expression plasmids are all based on the vectors of the Drosophila expression system (Invitrogen). Tube, Pelle, Tube-Pelle chimeras, and Relish mutants were generated by PCR sewing [29]. Anti-V5 and anti-myc (9E10) antibodies were purchased from Invitrogen and Santa Cruz Biotechnology, respectively. Anti-Tube antiserum was described previously [24].
Gene Predictions and Computer Algorithms
D. melanogaster Tube and Pelle protein sequences were used to identify orthologs of these genes in the other insects investigated using the tBLASTN program (http://www.ncbi.nlm.nih.gov/BLAST/). In cases where full-length genes could not be reliably predicted from whole-genome BLAST data, we cloned overlapping cDNA fragments to verify our predictions. The structure of the A. gambiae TTLK (Tube and Tube-Like Kinase) was validated with cloned cDNA sequences that bridge the Tube-like and Pelle kinase-like portion of the gene. The absence of a kinase domain in the A. mellifera Tube gene was verified by 3′RACE. The TTLK from Tribolium castaneum, for which no cDNA was isolated, is completely contained within a 3-kb genomic contig and exhibits similarity throughout its length to TTLK genes from other insects.
We generated a gene structure prediction for the silkworm Bombyx mori TTLK, previously described as a Tube protein [25], and confirmed it with overlapping sequences from the silkworm EST project. However, other relevant sequence data for B. mori were of poor quality and coverage; B. mori was therefore excluded from further analyses.
Death domain sequences were aligned using the program Clustal W [30], and the phylogenetic tree from this alignment was generated with the neighbor-joining method using Phylip Drawtree [31]. The program iMol (http://www.pirx.com/) was used to highlight residues in the structure of the Drosophila Tube death domain that interact with MyD88 or Pelle, based on coordinates in the Protein Data Bank data set 1D2Z [25].
RNA Synthesis, Embryo Injection, and Cuticle Preparation
DNA templates of the Tube-PllC and TubeN-PllC chimeras were PCR amplified directly from pMT/V5-His constructs, using primers that incorporated an SP6 promoter at the 5′ end. Each construct thus includes a V5 tag and a 6XHis tag at the carboxyl terminus. RNA synthesis, sample preparation, and generation of single mutant (tubenull or pellenull) embryos were as described previously [25]. The recipient tubnull pllnull embryos were obtained from maternal flies of genotype h tubR5.6 e pll25/Df (3R)XM3 ri pp Df (3R)IR16. Embryos were collected, microinjected, prepared, and scored according to published methods [32–34].
S2 Cell Assays
S2 cells were maintained, transfected, and lysed as described previously [26]. Cell lysates were prepared with S2 cell lysis buffer (50 mm Tris, pH 7.5/150 mm NaCl/1% Nonidet P-40) supplemented with protease inhibitor mixture (Roche) on ice. Three hundred microliters of cleared lysate were prepared from each 35-mm well of transfected cells. Cell lysates were first incubated for 1 h at 4 °C with 2 μl of α-Tube antiserum and then with 10 μl (bed volume) of protein A/G agarose beads (Santa Cruz). After 3 h, the agarose beads were washed with S2 cell lysis buffer and resuspended in 40 μl of SDS-PAGE sample buffer. Immunoblot analysis was done as described [26].
Results
The Tube Death Domain Is Evolutionarily Conserved
Given the remarkable conservation of innate immune mechanisms between flies and humans, why has no mammalian counterpart been identified for Drosophila Tube? To trace the evolutionary origin of the Drosophila Tube protein, we set out to identify Tube orthologs in other species, beginning with a search for Tube-like death domains. Because death domains exhibit extensive sequence variation, a simple BLAST search was insufficient. Instead, we established criteria for identifying death domains orthologous to Tube based on the fact that the essential role of Tube is to interact, via its death domain, with the death domains of MyD88 and Pelle. Through biophysical and molecular genetic studies [25, 35], we have previously identified specific surfaces and residues that are required for these two interactions (fig. 1a). We could thus identify Tube-like death domains in other species by virtue of sequence conservation at the known interaction surfaces.
Fig. 1.
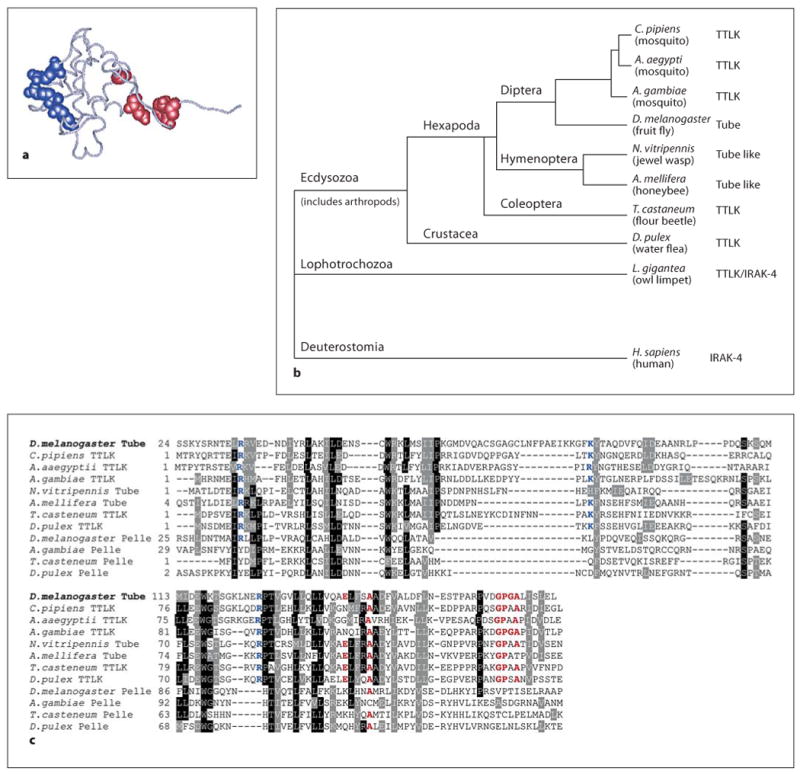
TTLK proteins, Pelle and IRAK are members of a kinase superfamily. a Diagram of the carbon backbone of the D. melanogaster Tube death domain; side chains required for binding to MyD88 (blue) and Pelle (red) are represented in space-filling form. b Location within the bilaterian animal phylogenetic tree of species encoding identified Tube orthologs. c Clustal W alignment of the death domains of TTLK and Pelle proteins; numbers indicate position in the polypeptide sequence. The TTLK death domains exhibit complete conservation of residues required for binding to MyD88 (blue), as well as strong conservation of residues required for binding to Pelle (red).
Using the tBlastN program [36] to probe whole genome sequence data, we identified Tube-like death domains all along the branches of the arthropod phylogeny leading to Drosophila (fig. 1b). These death domains exhibited very strong conservation of the three residues that in Drosophila Tube mediate binding to MyD88 and of the six residues that provide the surface for binding to Pelle (fig. 1c). Sequences neighboring these binding residues also showed a high degree of conservation among Tube orthologs. Consistent with conservation in Tube of surfaces for binding other Toll pathway death domain proteins, we identified an ortholog of MyD88 and of Pelle in each species studied. MyD88, Tube, and Pelle thus have a phylogenetic profile consistent with a common functional linkage [37] and a conserved role in innate immune signaling.
Many Tube Orthologs Are Protein Kinases
Having found that a number of arthropod genomes encode Tube-like death domains, we set about annotating the corresponding full-length proteins. A number of the genes had been studied previously, although in some cases an error in the annotation had resulted in the cataloguing of an incomplete or misassembled reading frame. For these examples, as well as uncharacterized loci, we pieced together coding sequence predictions, taking advantage of available cDNA sequences and generating new sequence data as needed (see Materials and Methods). These data are presented in supplemental figure 1 (see www.karger.com/doi/10.1159/000200773).
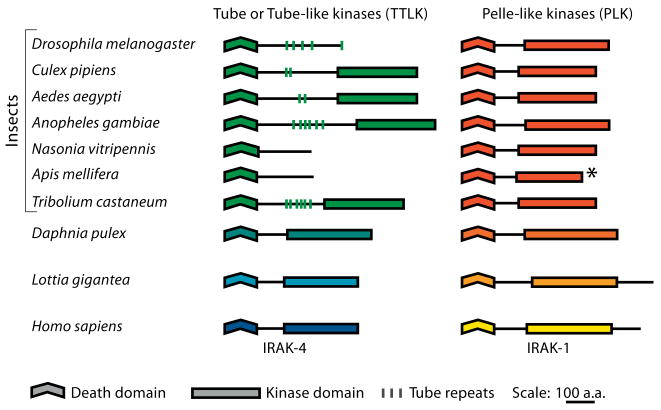
The structure of many of the insect Tube-like proteins was quite unexpected (fig. 2). Unlike Drosophila Tube, orthologs of Tube in five of the seven other species studied have a protein kinase domain. In each case, this protein kinase domain is C-terminal to the death domain. A kinase domain is absent from Nasonia and Apis Tube proteins.
Fig. 2.
Scale diagram of the structural elements comprising TTLK and Pelle proteins. The asterisk indicates that the A. mellifera Pelle sequence is incomplete.
BlastP analysis of kinase domains revealed that the Tube-like kinases are members of the RLK (receptor-like kinase)/Pelle family defined by the plant RLKs and the animal Pelle and IRAK proteins [38]. The catalytic domains of the Tube-like kinases are thus closely related to those of Pelle proteins. Furthermore, based on conservation of key sequence elements, all of the Tube-like kinases are predicted to be catalytically active (unlike a subset of IRAK kinases). We will hereafter refer to Tube and the set of arthropod Tube orthologs as the TTLK family.
Since each species examined encodes one MyD88, one Pelle and one TTLK, many arthropods have two kinases in the RLK/Pelle family. To explore the relationship between these pairs of kinases, we aligned catalytic domain sequences from arthropod Pelle and TTLK proteins (fig. 3). The alignment revealed a marked difference with regard to kinase type. Protein kinases can be classified as RD or non-RD based on a single position in sequence subdomain VI [39–42]. In RD kinase sequences, an arginine (R) residue immediately precedes the invariant aspartate (D) residue that is essential for catalytic activity. Upon phosphorylation of the kinase activation loop, interaction between this arginine and the phosphate residue converts the RD kinase from an inactive to an active state. Conversely, catalytic activity of the non-RD kinases does not appear to require phosphorylation of residues in the activation loop. All seven of the arthropod Pelle proteins have a glycine in place of the arginine and are thus non-RD kinases. In contrast, the five TTLK proteins with a catalytic domain are all RD kinases.
Fig. 3.
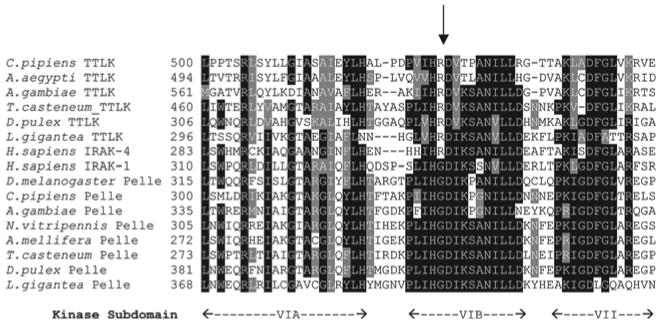
Tube-like kinases, but not Pelle proteins, are RD kinases. Alignment of sequence subdomains VI and VII from TTLK and Pelle proteins. Arrow indicates position of RD dipeptide that is hallmark of kinases regulated by activation loop phosphorylation.
Tube Is Closely Related to IRAK-4
Further comparative studies revealed that the presence of a pair of death domain-containing kinases, one similar to Pelle and one similar to TTLK proteins, is not limited to arthropods. We could, for example, find a pair of such proteins in a mollusk, Lottia gigantea (fig. 2). One kinase was a non-RD kinase (fig. 3), with a death domain that was most similar to Pelle death domains. The other kinase was an RD kinase and had a death domain that was most similar to the death domains of TTLK proteins.
We next analyzed human IRAK-1 and IRAK-4, both of which have previously been described as orthologs of Pelle [43–45], assessing the death and kinase domains independently. For the death domains, we aligned IRAK-1 and IRAK-4 with the eight arthropod TTLK proteins across the length of the six α-helices defining the core death domain structure [25]. For each of the 49 positions that were identical among four or more of the arthropod TTLKs, we scored whether the identical residue was present in IRAK-1 or IRAK-4. Overall, IRAK-4 had 47% identity with the conserved residues, whereas IRAK-1 had only a 22% match. For the kinase domains, when we carried our BLAST searches with any TTLK kinase domain against the human proteome, the top-scoring sequence was in every case IRAK-4. The statistical significance of the similarity to IRAK-4, as measured by E (expect) value, was greater by a factor of at least 108 in every case than the corresponding value for IRAK-1 (table 1). Furthermore, sequence alignment (fig. 3) revealed that IRAK-1, like Pelle, is a non-RD kinase, whereas IRAK-4 is an RD kinase. Thus, our results demonstrate that IRAK-4 is an ortholog of Drosophila Tube, while IRAK-1 is an ortholog of Pelle (fig. 2).
Table 1.
Sequence identity of TTLK kinase domains with human proteins
| TTLK kinase domain: a.a. | BLASTP vs. Homo sapiens | |
|---|---|---|
| top match | next match | |
| Culex: 285–678 | IRAK4 6e−37 | IRAK1 2e−23 |
| Aedes: 401–672 | IRAK4 3e−28 | MLK1 8e−19 |
| Anopheles: 471–743 | IRAK4 8e−34 | MLK2 5e−23 |
| Tribolium: 373–641 | IRAK4 7e−41 | IRAK1 1e−28 |
| Daphnia: 203–503 | IRAK4 3e−43 | IRAK1 2e−35 |
| Lottia: 275–568 | IRAK4 2e−41 | IRAK1 2e−33 |
Query sequences listed refer to the kinase domain of the TTLK proteins presented in figure 2. Each kinase domain begins with the conserved GXG motif and ends with the carboxyl terminus of the protein. The top two matches from BLASTP analysis against the NCBI database are listed with gene name and e (expect) value. a.a. = Kinase domain sequence boundaries. MLK = Mixed lineage kinase.
The Tube and Pelle Proteins Arose by Duplication and Divergence of a Common Ancestor
Although the Tube and Pelle death domain sequences have clear and significant differences, they are also recognizably similar (fig. 1c). The death domains of the TTLK proteins are in fact more similar to those of Pelle, the IRAKs, and PLK (Pelle-like kinase) proteins than to the death domains of any other proteins. Conservation between TTLK proteins and Pelle thus extends throughout both the kinase and death domains.
Given the structure and organization of TTLK and PLK proteins, the most parsimonious explanation is that they arose by duplication and divergence of an ancestral gene that included both a death domain and a protein kinase domain. Furthermore, the orthology of Tube to IRAK-4 and of Pelle to IRAK-1 indicates that the gene duplication predated the split between the invertebrate and vertebrate lineages. One gene copy gave rise to the PLK proteins, in which the death domain binds exclusively to a TTLK protein. The other lineage gave rise to the TTLK proteins, which contain a bivalent death domain, capable of binding both MyD88 and a PLK protein. By this model, the kinase domain of the TTLK protein was lost in certain lineages, including Apis, Nasonia, and Drosophila (fig. 2).
Tube and Pelle Functions Can Be Combined in a Single Protein
The clustering of the TTLK and Pelle proteins by death domain sequence indicates that the death domain structure diverged subsequent to duplication of the progenitor kinase gene. This divergence may have led to a fine-tuning of pathway function, e.g. enhancing regulation or matching output to other cellular systems. Alternatively, there may have been a partitioning of function that represents a fundamental alteration in the signaling mechanism. To examine this issue, we set out to determine whether a single death domain-containing kinase could effect Toll signaling.
Two lines of reasoning led us to conduct our experiment in Drosophila. First, the absence of either Tube or Pelle function abrogates Toll signaling. Thus, their functions are not redundant. Second, injection of RNA transcribed in vitro from a cDNA provides complete phenotypic rescue of Toll signaling in embryos, i.e., pelle RNA restores wild-type dorsoventral patterning to embryos null for pelle and tube RNA similarly rescues embryos null for tube. We could therefore assay for restoration of both Tube and Pelle function.
Since Tube is required for binding to MyD88 [26, 35, 46] and Pelle kinase activity is required for signal transduction [19, 47], we fused all of Tube (amino acids 1–462) to a carboxyl-terminal Pelle fragment (amino acids 130–501) containing the catalytic domain. The resulting chimeric gene, Tube-PllC, thus encodes the death domain of Tube, but not of Pelle (fig. 4a). To assess the activity of this chimera, we transcribed the fusion gene in vitro and microinjected the RNA into tubnull pllnull mutant embryos, which contain no Tube or Pelle protein.
Fig. 4.
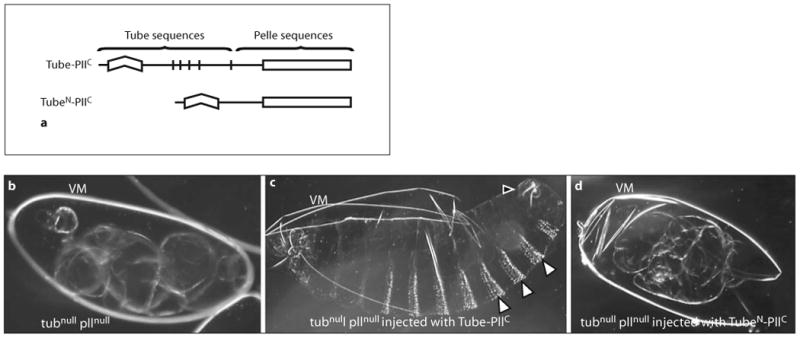
a Tube-Pelle chimera mediates robust, spatially regulated Toll signaling, a Diagram of the Tube-PllC and TubeN-PllC chimeras. The Tube-PllC construct encodes full-length Tube (amino acids 1–462) fused to amino acids 130–501 of Pelle. The TubeN-PllC construct encodes amino acids 1–185 of Tube fused to the same Pelle fragment. b–d Darkfield micrographs of cuticle preparations from an uninjected embryo (b) and embryos injected with 1 mg/ml RNA from an in vitro transcription reaction (c, d). VM = Intact or fragmented vitelline membrane that encases the developing embryo. b Uninjected tubnull pllnull embryo. c tubnull pllnul1 embryo injected with Tube-PllC RNA. White arrowheads point to the properly spaced bands of ventral denticles; a black arrowhead indicates the filzkörper apparatus, d tubnull pllnull embryo injected with TubeN-PllC RNA. No Toll-dependent patterning elements are observed.
In the absence of injected RNA, tubnull pllnull mutant embryos are dorsalized, forming a twisted tube of dorsal cuticle (fig. 4b). When injected with Tube-PllC RNA, however, these embryos produce cuticle that contains a full range of dorsal, lateral, and ventral markers (table 2, fig. 4c). Furthermore, cuticle structures such as the dorsolateral filzkörper apparatus and the ventral denticles have a wild-type arrangement across the embryo. Thus, the Tube-PllC construct not only restores robust signaling, but also mediates spatial regulation of signal strength along the dorsoventral axis.
Table 2.
Rescue of embryos with Tube-Pelle chimeras
| RNA injected | Scored n |
Cuticle phenotypes | |||
|---|---|---|---|---|---|
| D0 no rescue | D1 FK only | D2 FK and some VD twisted | D3 FK and VD in wild-type pattern | ||
| None | |||||
| tubnullpllnull | 50 | 50 | 0 | 0 | 0 |
| Tube-PllC | |||||
| tubnullpllnull | 100 | 47 | 10 | 37 | 6 |
| TubeN-PllC | |||||
| tubnullpllnull | 36 | 36 | 0 | 0 | 0 |
| None | |||||
| tubnull | 50 | 50 | 0 | 0 | 0 |
| TubeN-PllC | |||||
| tubnull | 74 | 9 | 23 | 42 | 0 |
| None | |||||
| pllnull | 50 | 50 | 0 | 0 | 0 |
| TubeN-PllC | |||||
| pllnull | 98 | 45 | 53 | 0 | 0 |
Columns represent increasing strength of signaling (D0–D3), with values indicating the number of injected embryos exhibiting the set of cuticle pattern elements diagnostic for that signal strength [32–34]. Filzkörper (FK) are dorsolateral patterning elements indicative of weak Toll signaling (D1). Ventral denticles (VD) are ventrolateral patterning elements that reflect a higher level of Toll signaling (D2 or D3). When VD and FK are both present and are arrayed in a wild-type pattern (D3), signal strength and regulation is nearly comparable to that in hatching embryos.
The restoration of a near wild-type cuticle pattern is a property of the chimera, not of the kinase domain. Even in the presence of full-length Tube, the kinase domain of Pelle (PllC) has no detectable activity [48], unlike the catalytic domains of many protein kinases [49, 50]. Furthermore, the activity of the chimera is not a trivial consequence of recruiting the Pelle kinase domain to the plasma membrane. For example, constructs in which the Pelle kinase domain is brought to the cell surface by fusion either to the Torso transmembrane receptor or to an Src peptide specifying myristylation activate nuclear translocation of Dorsal, but in a spatially unregulated manner [48, 51–53].
To determine whether signal transduction through the Tube-PllC chimera restored robust Toll signaling, we examined the tissues of injected embryos during development. Although none of these embryos hatched, a small fraction (6%) had readily apparent muscle contractions. Mesoderm induction leading to muscle formation requires the highest level of Toll signaling normally occurring during embryogenesis [54]. Given that the lack of either Tube or Pelle results in a fully dorsalized cuticle [55], and thus a loss of all lateral (neuroectoderm and filzkörper) and ventral (mesoderm) cell types, the Tube-PllC fusion largely reconstituted both Tube and Pelle activity in the injection experiments. The experimental observations thus fit well with the idea that a single protein containing both death and kinase domains mediated Toll pathway signaling in the common ancestor of the invertebrate and vertebrate lineages.
These findings strongly support the idea that the primary function of the Tube and Pelle death domains is to bring the Pelle kinase domain into indirect association with MyD88 and, hence, Toll. Indeed, since the Pelle death domain is absent from the fully functional chimera, the Pelle death domain appears to lack any requisite binding partner other than Tube. Moreover, we are led to the surprising conclusion that a covalent and hence irreversible interaction between the Tube death domain and the Pelle kinase domain functionally substitutes to a very large extent for their noncovalent wild-type association.
An Essential Function for the Tube Repeat Region in the Absence of the Pelle Death Domain
In addition to an amino-terminal death domain, Tube has a carboxyl-terminal region containing multiple copies of a conserved sequence motif, the Tube repeat [24]. The repeat-containing region has been shown to mediate the specific binding of Tube to Dorsal [27, 28]. Deletion experiments demonstrated that this region is largely dispensable for transducing signals from Toll during embryonic patterning [24, 28]. However, we have found that Dorsal also binds to Pelle, an interaction requiring both the death and kinase domains of Pelle [27, 28]. We therefore wondered whether the absence of a requirement for the Tube repeat region in signaling might reflect an overlap in function between Tube and Pelle. Specifically, we postulated that the binding of either Tube or Pelle to Dorsal promotes assembly of a functional signaling complex. Using Tube-Pelle chimeras, we set out to test this hypothesis.
The Tube-PllC construct described earlier lacks the Pelle death domain, but includes the repeat-containing region of Tube. To determine whether the repeat region has an essential function in Tube-Pelle chimeras, we generated a construct deleting this region. Specifically, the TubeN-PllC construct truncates Tube sequences at amino acid 185, eliminating the Tube repeats; there remains a 68-residue-long sequence linking the Tube death domain to the Pelle kinase domain (fig. 4a). The encoded protein thus lacks elements required for binding to Dorsal by Tube and by Pelle. We transcribed this construct in vitro and microinjected the synthetic RNA into embryos.
Unlike the chimera that contains the Tube C-terminal domain, TubeN-PllC failed to restore Toll signaling in tubnull pllnull embryos, resulting in no discernible effect on the cuticle pattern (table 2, fig. 4d). Next, we assessed the ability of either Tube or Pelle to rescue the activity of the TubeN-PllC chimera. To do so, we injected RNA transcribed from this construct into tubnull or pllnull single mutant embryos, which develop into completely dorsalized larvae in the absence of RNA injection. As shown in table 2, TubeN-PllC RNA was active in both genetic backgrounds, producing weak phenotypic rescue in pllnull embryos and stronger rescue in tubnull embryos. Thus, either Tube or Pelle can complement the chimera, consistent with our hypothesis that the Tube repeat region acts redundantly with Pelle in recruiting Dorsal.
Dorsal Has a Biochemical Activity Specific to Dorsoventral Patterning
To address whether binding of Tube to an NF-κB protein is specific to dorsoventral patterning, we carried out comparative binding studies with the three Drosophila NF-κB family members: Dorsal, Dif, and Relish. Dorsal is required for Toll pathway function in dorsoventral axis formation and in larval innate immunity [56, 57]. Dif functions in Toll signaling in larval and adult innate immunity, but is not required for embryonic development [58, 59]. Relish acts in an innate immune pathway distinct from Toll signaling [60–62].
We expressed Tube in Drosophila S2 cells together with individual Rel transcription factors or, as controls, MyD88 or Pelle. We then assayed for interaction in immunoprecipitation experiments with polyclonal α-Tube serum. All proteins were epitope-tagged for detection. As shown in figure 5, α-Tube efficiently immunoprecipitated Tube and associated MyD88, Pelle, or Dorsal protein (lanes 1–3). In contrast, there was no detectable interaction of Tube with either Dif or Relish (lanes 4–6). A Relish protein lacking the autoinhibitory carboxyl-terminal domain also failed to interact with Tube (lane 7).
Fig. 5.
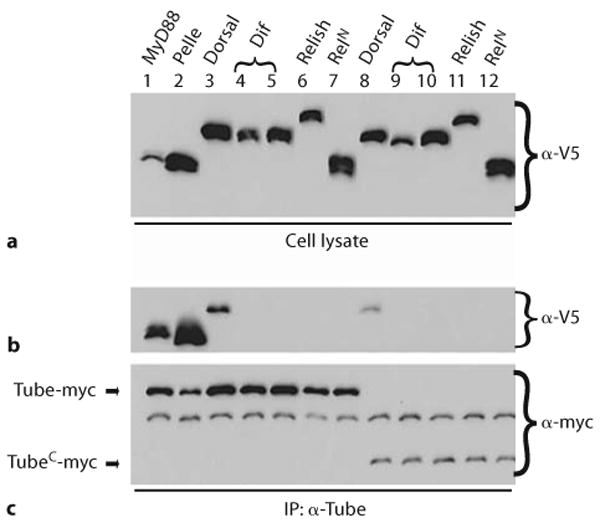
Tube interacts with dorsal, but not the related NF-κB transcription factors Dif and Relish. S2 cells were transfected with epitope-tagged Tube – either Tube-myc or TubeC-myc (amino acids 236–462 of Tube) – in combination with plasmids expressing V5-tagged forms of the proteins listed at the top of the figure. Immunoprecipitates were prepared with α-Tube antiserum. Tube proteins were detected with α-myc antibody (c); co-precipitating proteins were detected with α-V5 antibody (b). Samples of each cell lysate were probed with α-V5 antibody to assay expression of each V5-tagged protein (a). Dif was expressed under the control of two alternative 5′UTR sequences, which result in different levels of protein (top panel; lanes 4, 5 and 9, 10). The RelN construct (lanes 7 and 12) encodes amino acids 1–478 of Relish.
It has previously been demonstrated that Tube interacts with Dorsal via its carboxyl-terminal repeat-containing region [27, 28]. To rule out any potential inhibitory effect of the amino-terminal death domain on the interactions of the carboxyl terminal domain, we repeated the binding studies with a construct encoding only the carboxyl terminal half of the Tube protein (TubeC). Although TubeC was expressed at a lower level than the full-length construct, immunoprecipitation studies with this construct gave identical results. Specifically, TubeC associated with Dorsal, but neither with Dif nor with Relish (lanes 8–12).
These studies demonstrate that prior to signaling the Tube carboxyl-terminal domain can interact with Dorsal, the NF-κB family member required for embryo patterning, but not with either Dif or Relish, the NF-κB proteins that function solely in immune responses. It thus appears that, at least in Drosophila, the evolution of a developmental role for Toll has been accompanied by a specialization in NF-κB protein structure and function.
The identification of Tube repeats in the TTLK proteins of other insects provided several insights into the evolutionary basis for repeat generation. First, we note that the sequences of the third and fourth repeats in Anopheles TTLK are identical (QVPELSIF, suppl. fig. 2). Examining the context of these repeats, we found that each is contained within a 21-residue-long perfect repeat for which the encoding DNA is identical at 61 of 63 positions. In addition, a highly similar stretch of 21 amino acids spans the second Tube repeat in this TTLK protein. These results led us to speculate that the 8-amino-acid repeat originally defined in Tube is the most conserved remnant of a substantially longer region that underwent repeated duplication and modification.
We also found evidence that the repeat motif has undergone extensive change within the insect lineage. The repeat motif was originally defined in the D. melanogaster and D. virilis Tube proteins [22, 24] by the consensus NψPxψS/TxL, where x is any residue and ψ is a hydrophobic residue. The A. mellifera TTLK lacks sequences that closely match this consensus, but contains three copies of a motif that has an identical length and that begins with a sequence, ELP, that is very similar to the NψP that begins the previously defined Tube repeat (suppl. fig. 2). To what extent such changes in sequence reflect co-evolution of the motifs with the insect Dorsal proteins remains an open and stimulating question.
Discussion
Our analysis of insect Tube orthologs has led us to the conclusion that Tube arose from a gene for a protein kinase very similar in overall structure to Drosophila Pelle and the vertebrate IRAKs. That the ancestral gene for Tube would be a protein kinase was not evident from previous studies of Tube structure or function. A kinase domain is absent from the Tube protein in all eleven sequenced Drosophila genomes. In addition, the presence of a death domain in Tube did not suggest membership in a protein kinase family, since death domains are found in many proteins that lack a kinase domain or, indeed, any catalytic function.
Tube and IRAK-4 as Orthologous Components of Toll Pathways
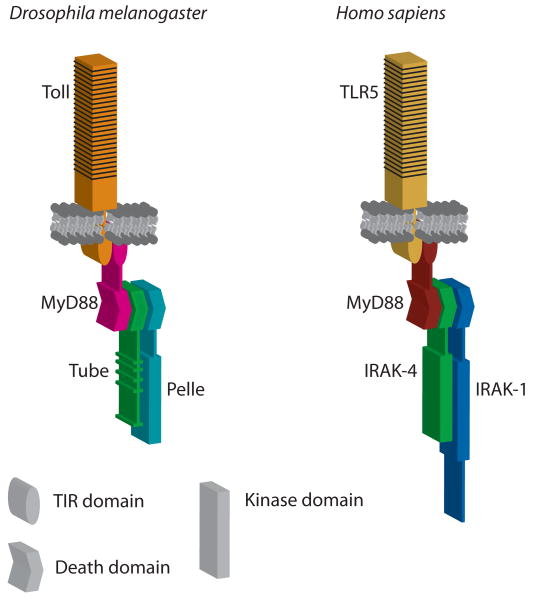
Our finding that Tube and Pelle represent an orthologous pair to IRAK-4 and IRAK-1 resolves a long-standing confusion regarding Toll signaling in flies and mammals. The absence of a clear counterpart to Tube had suggested a significant difference in the proximal Toll signaling complex of these species. Instead, we now discern a parallel arrangement of signaling components acting immediately downstream of the Toll receptors in humans and Drosophila. Thus, signaling in humans mediated by a TLR, MyD88, IRAK-4, and IRAK-1 corresponds to the pathway in flies formed by Toll, MyD88, Tube, and Pelle. As illustrated in figure 6, this suggests that a trimeric death domain complex, such as we have characterized in flies, functions in MyD88-dependent Toll responses in humans. Our newfound recognition of this conserved architecture immediately suggests that the mechanism for signal transduction likewise has greater conservation than previously suspected.
Fig. 6.
Conserved Toll-proximal signaling mechanism. Model for organization of signaling complexes immediately downstream of Toll receptors in flies and humans.
Although the Toll pathway functions in innate immunity in both humans and flies, the selective pressures on this pathway differ markedly between these organisms. In humans and other vertebrates, innate immunity has co-evolved with an adaptive response with which its function is intricately associated [4]. Furthermore, particular components of mammalian Toll pathways may have been co-opted for direct participation in adaptive responses [63]. In flies, the Toll pathway not only mediates immune responses, but also has multiple developmental functions, including a vital role in patterning of the early embryo [6–9]. It is not surprising, therefore, that the divergence between the Toll pathways in these two lineages has been extensive.
A Common Ancestor for TTLK, PLK, and IRAK Proteins
Although IRAK-4 and TTLKs differ in structure and function from IRAK-1 and PLKs, all of these proteins are apparently derived from a common ancestor. Furthermore, we have demonstrated that signaling can be reconstituted to a substantial degree by a chimera of Tube and Pelle. This suggests the earliest signaling cassette recognizable as a Toll pathway had a single protein kinase that contained a death domain. Prior to the split between the vertebrate and invertebrate lineages, the gene for this ancestral kinase duplicated and at least partially diverged. The death domain binding surfaces evolved to mediate distinct interactions and the kinase domains diverged into RD and non-RD types.
What advantage might duplication of the ancestral kinase gene have offered? The chimera injection experiments indicate that duplication did not radically alter the signaling mechanism. Rather, we believe that the cooperative activity of related but distinct kinase molecules provided the opportunity for regulatory fine-tuning and allowed for the incorporation into the pathway of additional factors specific for particular functional contexts.
Tube as a Model for IRAK-4 Death Domain Function
We have previously shown that the death domain of Tube is bivalent, mediating specific and essential interactions with MyD88 at one binding surface and with Pelle at the other. The conservation among TTLKs of residues required for these interactions indicates that assembly of a trimeric complex of MyD88, TTLK, and PLK proteins occurs similarly across the broad range of organisms we have surveyed. The finding that Tube and Pelle represent an orthologous pair to mammalian IRAK-4 and IRAK-1 extends the implications of this study to signal transduction in mammals. In particular, these findings support our proposal [26] that IRAK-4-mediated activation of IRAK-1 is equivalent to Tube-mediated activation of Pelle, with both Tube and IRAK-4 acting as an adaptor, linking Pelle or IRAK-1 to a Toll-associated MyD88 molecule (fig. 6).
We expect that IRAK-4 and IRAK-1, like Tube and Pelle, have a clear partitioning of death domain structure and function. We were not able, however, to identify in IRAK-4 the signature motifs indicative of the binding surfaces defined for Drosophila Tube. There are two likely explanations. First, conservation of primary sequence among death domains is remarkably low, with sequence identity across the entire domain sometimes being on the order of only 10% [25]. Second, mammalian TLR signaling involves multiple, partially overlapping combinations of receptors and adaptors [1]. It may be, therefore, that a conserved death domain complex architecture is masked by sequence variation that directs or accommodates alternative death domain pairings. Now that a structure for the IRAK-4 death domain is available [45], it may be possible to address this question experimentally.
Variable Requirement for Kinase Activity
The presence of a kinase domain in IRAK-4, but not Tube – its fly counterpart – has an intriguing precedent. Whereas the Drosophila Toll pathway requires Tube function, the other major innate immune pathway in flies requires Imd, another death domain protein. RIP, the mammalian counterpart to Imd, has a kinase domain; Imd does not. Thus, for each of two death domain kinases in mammals (IRAK-4 and RIP), the corresponding innate immune factor in flies lacks a kinase domain.
Why might a kinase domain be dispensable for signal transduction? The likely answer lies in pathway architecture distinct from that in a conventional kinase cascade [64]. If the essential functions of IRAK-4 and TTLK proteins are to act as scaffolds for the assembly of signaling complexes, the role of the kinase domain may be to modulate complex assembly rather than to directly activate a downstream kinase. It has been shown, for example, that IRAK proteins catalyze cross-phosphorylation and that Tube can act as a substrate for Pelle in vitro [44, 47, 51, 65, 66]. Thus, the involvement of both phosphoregulation and phosphorelay activities may underlie the presence of pairs of protein kinases, one RD and one non-RD, in many animals.
TTLK Adaptation for a Developmental Role
We have found that the repeat-containing region of Tube exhibits NF-κB factor binding specificity, interacting in cells with the morphogen Dorsal but not the immunity effector Dif. Abrogation of this Tube-Dorsal interaction likely contributes to the failure to achieve wild-type embryonic patterning in experiments in which either Tube is truncated carboxyl terminal to the death domain or Dif is substituted for Dorsal [24, 67].
Does the presence of Tube repeats in a TTLK protein indicate that Toll signaling in that species functions in dorsoventral patterning? The data are incomplete, but suggestive. Toll has a role in dorsoventral patterning in Drosophila and Tribolium [68, 69], and Tube repeats are present in the TTLK proteins of both these species. Dorsoventral patterning does not rely on TLR signaling in mammals and mammalian IRAK-4 proteins lack Tube repeats. Thus, it is possible that the co-option of Toll signaling for development occurred concomitantly with elaboration of an additional mechanism for morphogen binding. If so, the fact that the TTLK protein of Daphnia lacks Tube repeats suggests that Toll signaling is unlikely to be required for dorsoventral patterning in crustaceans.
Supplementary Material
Acknowledgments
We thank Russ Doolittle for assistance with the phylogenetic analyses; Carolina Barillas-Mury and Mike Levine for reagents; Peter Ung for technical support, and Bill McGinnis and Anthea Letsou for helpful comments on a draft manuscript. This work was supported by NIH grant 5R01-GM50545 to S.A.W.
References
- 1.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 2.Imler JL, Ferrandon D, Royet J, Reichhart JM, Hetru C, Hoffmann JA. Toll-dependent and Toll-independent immune responses in Drosophila. J Endotoxin Res. 2004;10:241–246. doi: 10.1179/096805104225005887. [DOI] [PubMed] [Google Scholar]
- 3.Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 4.Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Adv Exp Med Biol. 2005;560:11–18. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 5.Kim T, Kim YJ. Overview of innate immunity in Drosophila. J Biochem Mol Biol. 2005;38:121–127. doi: 10.5483/bmbrep.2005.38.2.121. [DOI] [PubMed] [Google Scholar]
- 6.Halfon MS, Keshishian H. The Toll pathway is required in the epidermis for muscle development in the Drosophila embryo. Dev Biol. 1998;199:164–174. doi: 10.1006/dbio.1998.8915. [DOI] [PubMed] [Google Scholar]
- 7.Anderson KV, Bokla L, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: the induction of polarity by the Toll gene product. Cell. 1985;42:791–798. doi: 10.1016/0092-8674(85)90275-2. [DOI] [PubMed] [Google Scholar]
- 8.Seppo A, Matani P, Sharrow M, Tiemeyer M. Induction of neuron-specific glycosylation by Tollo/Toll-8, a Drosophila Toll-like receptor expressed in non-neural cells. Development. 2003;130:1439–1448. doi: 10.1242/dev.00347. [DOI] [PubMed] [Google Scholar]
- 9.Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 10.Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 11.Govind S, Nehm RH. Innate immunity in fruit flies: a textbook example of genomic recycling. PLoS Biol. 2004;2:e276–e232. doi: 10.1371/journal.pbio.0020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baeuerle PA, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 13.Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem Sci. 2002;27:474–482. doi: 10.1016/s0968-0004(02)02145-x. [DOI] [PubMed] [Google Scholar]
- 14.Chen LY, Wang JC, Hyvert Y, Lin HP, Perrimon N, Imler JL, Hsu JC. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr Biol. 2006;16:1183–1193. doi: 10.1016/j.cub.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 15.Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 16.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. The death domain: a module shared by proteins with diverse cellular functions. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 17.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 18.Janssens S, Beyaert R. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell. 2003;11:293–302. doi: 10.1016/s1097-2765(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 19.Shelton CA, Wasserman SA. pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell. 1993;72:515–525. doi: 10.1016/0092-8674(93)90071-w. [DOI] [PubMed] [Google Scholar]
- 20.Silverman N, Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 21.Anderson KV, Nüsslein-Volhard C. Information for the dorsal-ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311:223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- 22.Letsou A, Alexander S, Orth K, Wasserman SA. Genetic and molecular characterization of tube, a Drosophila gene maternally required for embryonic dorsoventral polarity. Proc Natl Acad Sci USA. 1991;88:810–814. doi: 10.1073/pnas.88.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 24.Letsou A, Alexander S, Wasserman SA. Domain mapping of tube, a protein essential for dorsoventral patterning of the Drosophila embryo. EMBO J. 1993;12:3449–3458. doi: 10.1002/j.1460-2075.1993.tb06019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci USA. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards DN, Towb P, Wasserman SA. An activity-dependent network of interactions links the rel protein Dorsal with its cytoplasmic regulators. Development. 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Steward R. A multimeric complex and the nuclear targeting of the Drosophila Rel protein Dorsal. Proc Natl Acad Sci USA. 1997;94:14524–14529. doi: 10.1073/pnas.94.26.14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. PHYLIP – Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:16, 4–166. [Google Scholar]
- 32.Anderson KV, Jurgens G, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 33.Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila: A Practical Approach. Oxford: IRL Press; 1986. pp. 199–227. [Google Scholar]
- 34.Roth S, Hiromi Y, Godt D, Nüsslein-Volhard C. Cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development. 1991;112:371–388. doi: 10.1242/dev.112.2.371. [DOI] [PubMed] [Google Scholar]
- 35.Sun H, Towb P, Chiem DN, Foster BA, Wasserman SA. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. EMBO J. 2004;23:100–110. doi: 10.1038/sj.emboj.7600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 38.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 40.Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi H, Hendrickson WA. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 42.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 43.Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasker MV, Gajjar MM, Nair SK. Cutting edge: molecular structure of the IL-1R-associated kinase-4 death domain and its implications for TLR signaling. J Immunol. 2005;175:4175–4179. doi: 10.4049/jimmunol.175.7.4175. [DOI] [PubMed] [Google Scholar]
- 46.Charatsi I, Luschnig S, Bartoszewski S, Nüsslein-Volhard C, Moussian B. Krapfen/dMyd88 is required for the establishment of dorsoventral pattern in the Drosophila embryo. Mech Dev. 2003;120:219–226. doi: 10.1016/s0925-4773(02)00410-0. [DOI] [PubMed] [Google Scholar]
- 47.Towb P, Bergmann A, Wasserman SA. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development. 2001;128:4729–4736. doi: 10.1242/dev.128.23.4729. [DOI] [PubMed] [Google Scholar]
- 48.Galindo RL, Edwards DN, Gillespie SK, Wasserman SA. Interaction of the pelle kinase with the membrane-associated protein tube is required for transduction of the dorsoventral signal in Drosophila embryos. Development. 1995;121:2209–2218. doi: 10.1242/dev.121.7.2209. [DOI] [PubMed] [Google Scholar]
- 49.Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19:5614. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- 50.Seidel-Dugan C, Meyer BE, Thomas SM, Brugge JS. Effects of SH2 and SH3 deletions on the functional activities of wild-type and transforming variants of c-Src. Mol Cell Biol. 1992;12:1835–1845. doi: 10.1128/mcb.12.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosshans J, Bergmann A, Haffter P, Nüsslein-Volhard C. Activation of the kinase Pelle by Tube in the dorsoventral signal transduction pathway of Drosophila embryo. Nature. 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- 52.Towb P, Galindo RL, Wasserman SA. Recruitment of Tube and Pelle to signaling sites at the surface of the Drosophila embryo. Development. 1998;125:2443–2450. doi: 10.1242/dev.125.13.2443. [DOI] [PubMed] [Google Scholar]
- 53.Grosshans J, Schnorrer F, Nüsslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 54.Chasan R, Anderson KV. Maternal control of dorsal-ventral polarity and pattern in the embryo. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 387–432. [Google Scholar]
- 55.Hecht PM, Anderson KV. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics. 1993;135:405–417. doi: 10.1093/genetics/135.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 1999;18:3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, Tatei K, Levine M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 59.Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 60.Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki N, Suzuki S, Millar DG, Unno M, Hara H, Calzascia T, Yamasaki S, Yokosuka T, Chen NJ, Elford AR, Suzuki J, Takeuchi A, Mirtsos C, Bouchard D, Ohashi PS, Yeh WC, Saito T. A critical role for the innate immune signaling molecule IRAK-4 in T cell activation. Science. 2006;311:1927–1932. doi: 10.1126/science.1124256. [DOI] [PubMed] [Google Scholar]
- 64.Dardick C, Ronald P. Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2006;2:e2. doi: 10.1371/journal.ppat.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen B, Manley JL. Phosphorylation modulates direct interactions between the Toll receptor, Pelle kinase and Tube. Development. 1998;125:4719–4728. doi: 10.1242/dev.125.23.4719. [DOI] [PubMed] [Google Scholar]
- 66.Shen B, Manley JL. Pelle kinase is activated by autophosphorylation during Toll signaling in Drosophila. Development. 2002;129:1925–1933. doi: 10.1242/dev.129.8.1925. [DOI] [PubMed] [Google Scholar]
- 67.Stein D, Goltz JS, Jurcsak J, Stevens L. The Dorsal-related immunity factor (Dif) can define the dorsal-ventral axis of polarity in the Drosophila embryo. Development. 1998;125:2159–2169. doi: 10.1242/dev.125.11.2159. [DOI] [PubMed] [Google Scholar]
- 68.Chen G, Handel K, Roth S. The maternal NF-κB/dorsal gradient of Tribolium castaneum: dynamics of early dorsoventral patterning in a short-germ beetle. Development. 2000;127:5145–5156. doi: 10.1242/dev.127.23.5145. [DOI] [PubMed] [Google Scholar]
- 69.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo – shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.