Abstract
Biogenesis of the flagellum, a motive organelle of many bacterial species, is best understood for members of the Enterobacteriaceae. The flagellum is a heterooligomeric structure that protrudes from the surface of the cell. Its assembly initially involves the synthesis of a dedicated protein export apparatus that subsequently transports other flagellar proteins by a type III mechanism from the cytoplasm to the outer surface of the cell, where oligomerization occurs. In this study, the flagellum export apparatus was shown to function also as a secretion system for the transport of several extracellular proteins in the pathogenic bacterium Yersinia enterocolitica. One of the proteins exported by the flagellar secretion system was the virulence-associated phospholipase, YplA. These results suggest type III protein secretion by the flagellar system may be a general mechanism for the transport of proteins that influence bacterial–host interactions.
Gram-negative bacteria secrete a variety of proteins into the extracellular milieu, which requires secretion systems that are designed specifically for the transport of macromolecules across two biological membranes. At least four different mechanisms for extracellular protein secretion have been described and are termed types I-IV (1). Recent examinations of proteins that are secreted by a type III mechanism have shown that they often influence bacterial–host interactions for pathogens of animals and plants (2, 3). These proteins, which have been referred to as effectors or toxins, have a wide range of functions, including cytotoxicity, hemolysis, proteolysis, protein phosphorylation, and protein dephosphorylation. They are transported by a dedicated machinery (the type III secretion apparatus), which is composed of subunits that are homologous among distantly related bacteria. This apparatus often functions only when bacteria are intimately associated with a host to specifically transport targeted proteins, without any apparent modification, to the extracellular environment of the host or directly into host cells. Because of the observed functional requirement for host association these type III secretion systems have been categorized as contact-dependent.
Assembly of a flagellum, the motive organelle produced by many bacteria, requires the export of protein subunits from the cytoplasm to the outer surface of the cell by a mechanism that resembles type III secretion (4, 5). The structure of a flagellum generally is described as consisting of three main components: (i) the basal body, (ii) the hook, and (iii) the filament. Flagellum biogenesis is highly ordered such that the basal body is assembled initially and includes subunits that share a high degree of homology with subunits that form the secretion machinery of the contact-dependent type III secretion apparatus (2, 4). The basal body traverses from the cytoplasm to the outside of the cell and is structurally similar to the type III secretion apparatus (6). Further maturation of the flagellum requires the export of other flagellum nascent subunits without modification by export machinery of the basal body. This sequential export of flagellum subunits results in the assembly of the hook and the filament onto the basal body on the outer surface of the cell.
Among bacteria belonging to the Enterobacteriaceae, regulation of the genes that are involved in flagellum biogenesis and chemotaxis occurs in coordination with flagellar assembly and in response to environmental signals (4). Expression of these genes is organized into a regulatory hierarchy of three major classes: I, II, and III. Class I genes, consisting of flhD and flhC, form a single operon that is expressed at the top of the hierarchy and is required for the expression of all other flagellar genes. The FlhD and FlhC proteins form a heteromultimeric transcriptional activator that is required for the expression of class II genes. Class II genes encode structural and accessory proteins required for assembly of the basal body and hook components of the flagellum as well as two genes that encode the FlgM and σ28 regulatory proteins. Class III genes are transcribed from σ28-dependent promoters and encode proteins required for maturation of the flagellum and chemosensory system. Some of these proteins, such as flagellin, are exported to the outer surface of the cell by the flagellar type III secretion machinery. FlgM functions to limit σ28 activity until flagellar basal body assembly is complete and competent for the export of flagellin.
The homology between subunits that form the protein transport apparatus of the flagellum and the contact-dependent type III secretion apparatus combined with evidence that substrates of these apparatus are transported without modification suggest that the two systems are functionally related. This similarity has led to the hypothesis that these two types of apparatus share a common evolutionary origin (3). Although the flagellum transport machinery previously was thought to have a dedicated role in organelle biogenesis, the results presented in this study demonstrate that it is also required for the transport of proteins to the extracellular environment in the pathogenic bacterium Yersinia enterocolitica. One of these proteins was shown to be a previously characterized phospholipase that is required for Y. enterocolitica pathogenesis (7). These results indicate the flagellar regulon can influence bacterial–host interactions in a manner independent of motility per se.
MATERIALS AND METHODS
Bacterial Strains, Culture Conditions, and Plasmid Constructs. All Y. enterocolitica strains used in this study were derived from JB580v [ΔyenR (R−M+)] (8). The flgE, flgF, flgMN, flhA, flhB, flhDC, fliA, and yplA mutants were described previously (7, 9, 10): JO1v (flgM∷nptI), YCK10 (flgE∷cat), YCK11 (flgF∷cat), YMS12 (flhA∷cat), YMS13(flhB∷cat), GY460 (ΔflhDC), VK1 (fliA∷aadA), and YEDS10 (yplA∷pEP185.2). Escherichia coli S17–1λ(pir) was used to conjugally transfer plasmids to Y. enterocolitica (11). Bacteria were grown in LB at 26°C for procedures requiring general growth (12). When strains were examined for extracellular proteins or motility, growth was in 1% wt/vol tryptone (T medium) at 26°C (9). For the detection of phospholipase activity, bacteria were grown on phospholipase indicator plates consisting of T media or MacConkey agar base (Difco) supplemented with 1% Tween 80 and 1 mM CaCl2. The following concentrations of antibiotics were used when appropriate: 12.5 μg/ml of chloramphenicol, 50 μg/ml of gentamicin, 50 μg/ml of kanamycin, 20 μg/ml of naladixic acid, 50 μg/ml of streptomycin, and 7.5 μg/ml of tetracycline.
The mobilizable plasmids encoding ptac-flhDC (pGY20) and ptac-yplAB (pDHS32) transcriptional fusions are derivatives of pVLT33 (13). Construction of pGY20 was described (9). To construct pDHS32, a 1.8-kb EcoRI fragment was subcloned from plasmid pDHS42 into pVLT33. Plasmid pMalE-YplA was constructed by cloning a 1.7-kb AvaI-NheI fragment from pDHS20 (7) into pMAL-p2 (New England Biolabs). Other plasmids used in this study were described (7, 9): pACYC4GM (flgEF+), pGY15 (flhDC+), pSWIM1 (flgMN+), pMSflh (flhBAE+), pVM200 (yplAB+), and pVM201 (yplAB+).
Preparation of Extracellular Proteins and SDS/PAGE.
Extracellular proteins were prepared as described with minor modifications (9). Liquid cultures were grown in T medium, and cells were collected by centrifugation. The supernatant was passaged through a nonpyrogenic, 0.22-μm filter (Gelman) and then extracted with 0.3 vol of chloroform/methanol (2:1). Extracellular proteins were concentrated by precipitation with 10% (wt/vol) ice-cold trichloroacetic acid and washed with ice-cold acetone. All samples were resuspended in sample buffer containing 2-mercaptoethanol and normalized according to the OD600 of the culture. Samples were heated to 95°C for 10 min, and proteins were separated on SDS containing polyacrylamide gels by SDS/PAGE (14). Proteins were visualized by staining with silver (15).
Phospholipase Assays.
Crude preparations of YplA were prepared from liquid cultures grown as indicated. Cells were collected by centrifugation at 8,000 × g for 3 min and then resuspended in phospholipase buffer (PLA buffer), which consisted of 0.1 M Tris, pH 8/0.5% sodium chloride/0.2% SDS/100 μg/ml of streptomycin. Cells were lysed by sonication, with 3 × 15-sec pulse at maximum setting. Proteins in the supernatant fraction were precipitated with 4 vol of ice-cold acetone and collected by centrifugation at 8,000 × g for 15 min. The protein pellet was washed with ice-cold acetone and resuspended in PLA buffer. Samples were normalized based on the OD600 of the cultures used for the preparations. Phospholipase activity in a sample was determined by a modified radial-diffusion assay (16). Thin 1% agarose gels containing PLA buffer, 1% Tween 80, and 1 mM CaCl2 were poured onto glass plates. Samples were loaded into wells made in the gel and incubated in a humidified chamber at 26°C for 48 or 72 hr. Phospholipase activity in a sample was detected as circular zone of precipitation emanating from the well.
Preparation of Antibodies and Immunoblot Analysis.
The fusion protein MalE-YplA was affinity-purified from crude cell extracts of E. coli/pMalE-YplA by passage over an amylose resin column as instructed by the manufacturer (New England Biolabs). Polyclonal antibodies against MalE-YplA were raised in New Zealand White rabbits by using standard procedures (Cocalico Biologicals, Reamstown, PA). Nonspecific antibodies were removed by adsorption of the sera with an acetone powders prepared from a cultures of Y. enterocolitica YEDS10 and E. coli containing pMAL-p2 (17). Polyclonal sera directed against chloramphenicol acetyl transferase and invasin were described (18, 19). Polyclonal sera directed against β-lactamase was a gift (K. Postle, Washington State University). Immunoblot analysis was done as described (17). Reactive antigen was detected by using a chemiluminescent substrate (ECL, Amersham).
RESULTS
Identification of Fops, Extracellular Proteins That Require the Flagellar Regulon for Production.
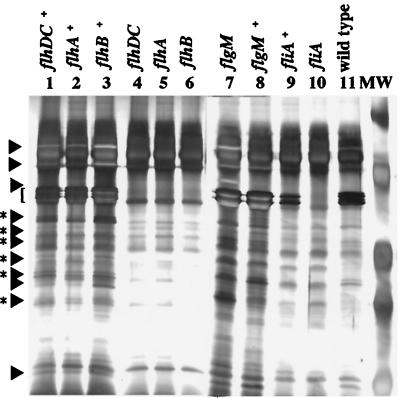
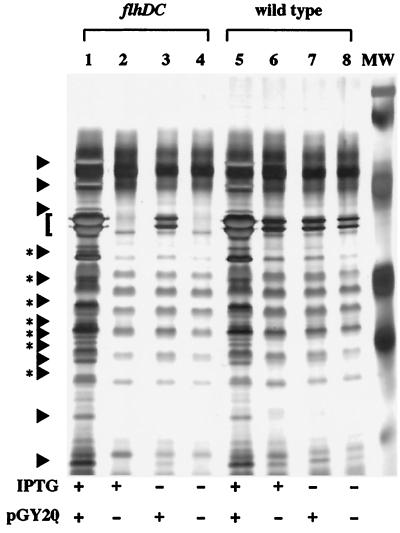
In Y. enterocolitica, motility is controlled by the master regulatory operon, flhDC (9). Examination of culture supernatants from a flhDC mutant to confirm the loss of flagellin production revealed that several secreted proteins produced at low levels were missing compared with the wild-type strain (Fig. 1, lanes 4 and 11). Complementation of the flhDC mutation with a plasmid-encoded copy of flhDC restored the production of flagellin and the secreted proteins (Fig. 1, lane 1). Similar results were seen when flhDC was placed under control of an exogenous ptac promoter such that expression became dependent on the presence of the inducer isopropyl β-d-thiogalactopyranoside (IPTG). When the ptac-flhDC construct (pGY20) was expressed in the flhDC mutant, the amount of extracellular proteins detected in culture supernatants corresponded to levels of flhDC expression (Fig. 2). In the wild-type strain, expression of the ptac-flhDC construct resulted in overexpression of the secreted proteins (Fig. 2). These proteins were designated Fops (flagellar outer proteins) because expression of the flagellar regulon was required for their appearance in the culture supernatant.
Figure 1.
Extracellular protein secretion by Y. enterocolitica requires the flagellar export apparatus. Extracellular proteins were concentrated from culture supernatants, separated by SDS/PAGE in 12.5% gels, and stained with silver. Lanes: 1, GY460/pGY15; 2, YMS12/pMSflh; 3, YMS13/pMSflh; 4, GY460 (flhDC); 5, YMS12 (flhA); 6, YMS13 (flhB); 7, JO1v (flgM); 8, JO1v/pSWIM1; 9, VK1/pJB222; 10, VK1 (fliA); and 11, JB580v (wild type). Each lane contained the equivalent of 3 ml supernatant from a culture at an OD600 of 1.0. Molecular mass standards (MW) from top to bottom are 81, 46.9, 34.1, 28.5, and 20 kDa. The location of flagellin is indicated by the bracket at the left. The locations of Fops are indicated by arrows (regions with a single species) and arrows with asterisks to denote regions where multiple species comigrate.
Figure 2.
Fop production is affected by expression of the flagellar regulon. Extracellular proteins were examined from concentrated culture supernatants of strains grown under conditions that modulate the expression of motility. Expression of motility was modulated by expressing flhDC from a ptac promoter. Transcription from the ptac promoter was positively controlled by the inclusion of IPTG in the growth medium. Proteins were separated by SDS/PAGE and stained as described in Fig. 1. Lanes: 1, GY460/pGY20 grown in T medium + 50 μM IPTG; 2, GY460/pVLT33 grown in T medium + 50 μM IPTG; 3, GY460/pGY20 grown in T medium; 4, GY460/pVLT33 grown in T medium; 5, JB580v/pGY20 grown in T medium + 50 μM IPTG; 6, JB580v/pVLT33 grown in T medium + 50 μM IPTG; 7, JB580v/pGY20 grown in T medium; and 8, JB580v/pVLT33 grown in T medium. Each lane contains the equivalent of 1.5 ml supernatant from a culture at an OD600 of 1.0. The locations of Fops are indicated by arrows (regions with a single species) and arrows with asterisks to denote regions where multiple species comigrate.
A survey of known secreted proteins showed that the Fops were a novel set of extracellular proteins. The Fops are not flagellin because they did not cross-react with a flagellin-specific mAb (data not shown). In addition, they are not related to the virulence plasmid-encoded proteins (Yops) known to be transported by a different type III secretion apparatus because the Fops were produced by plasmid-cured strains (data not shown). To ensure the Fops were secreted by Y. enterocolitica and did not accumulate in culture supernatants as a result of general cell lysis, culture supernatants were examined for contamination by proteins that are localized and retained in specific bacterial cell compartments by immunoblot analysis. Examination of culture supernatants prepared from Y. enterocolitica strains revealed no contamination by the outer membrane protein invasin (data not shown). There also was no detectable contamination for strains engineered to express the periplasmic protein β-lactamase or the cytoplasmic protein chloramphenicol acetyl transferase (data not shown).
To determine whether regulators of the flagellar regulon other than flhDC would affect the production of Fops, culture supernatants from different regulatory mutants were analyzed. Fop production first was examined in a strain carrying a mutation in fliA that encodes σ28 and is necessary for flagellar class III gene transcription (10). The fliA mutant did not produce flagellin, as was shown previously (10), and it also did not secrete the Fops (Fig. 1, lane 10). Complementation of the fliA mutation restored Fop and flagellin production (Fig. 1, lane 9). Examination of a strain with a mutation in flgM, which encodes a negative regulator of flagellar genes, revealed that this regulatory defect caused overproduction of flagellin and increased secretion of the Fops (Fig. 1, lane 7). Complementation of the flgM mutation reduced extracellular protein production to wild-type levels (Fig. 1, lane 8). This indicated that expression of motility and the appearance of Fops in the supernatant are coupled.
Fops levels also were affected by genes encoding flagellum structural subunits. Strains carrying mutations in flhA or flhB, which encode components of the basal body, did not produce Fops (Fig. 1, lanes 5 and 6). The appearance of Fops in the supernatant was restored by complementation of the flhA and flhB mutations (Fig. 1, lanes 2 and 3). These results suggest that secretion of the Fops requires the flagellar type III export system. Furthermore, because Fops (>14 proteins) outnumber proteins known to compose extracellular components of the flagellum organelle (6–9 proteins), some Fops should have functions not specifically related to motility. These observations led us to hypothesize that the flagellar type III export machinery has two functions: (i) to export flagellum subunits for organelle assembly and (ii) to transport Fops for release into the extracellular space.
yplA Is a Class III Gene of the Flagellar Regulon.
Regulation of genes that encode components of the type III secretion apparatus and their substrates often is coordinated (3). If Fop secretion requires the flagellar type III secretion apparatus, then genes encoding Fops probably belong to the flagellar transcriptional regulon. One such candidate gene fitting these criteria was yplA. This gene was shown previously to encode a phospholipase that is required for virulence and is believed to be transcribed from a σ28-dependent promoter (7), suggesting it is a class III flagellar gene. In addition, yplA is similar to phlA of Serratia liquefaciens, which has been shown previously to require flhDC for expression and encodes an extracellular protein (16, 20). To eliminate the possibility that yplA may have a general role in flagellum biogenesis, a Y. enterocolitica yplA mutant was compared with wild type for changes in levels of flagellin production and motility (data not shown). The two strains were indistinguishable, suggesting the phospholipase encoded by yplA is not a functional component of the flagellum.
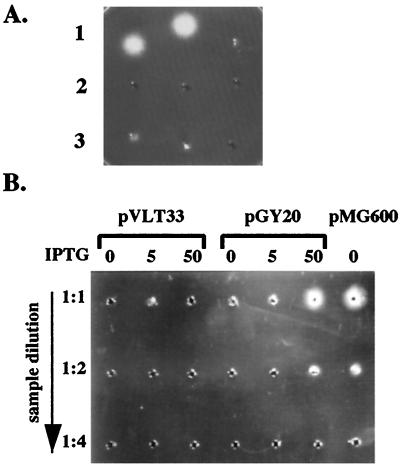
As an initial test to determine whether YplA was secreted by Y. enterocolitica, cultures grown under conditions that induce motility were fractionated and phospholipase activity for whole-cell extracts and for culture supernatant fractions was determined (Fig. 3A). The results showed that phospholipase activity was detected only in the culture supernatant fractions, indicating that YplA is exported out of the cell. Further examination of YplA activity revealed it was tolerant to heat treatment at 90°C for 10 min (Fig. 3A); a biochemical property that may be expected for an extracellular enzyme that has to survive extreme environmental conditions.
Figure 3.
YplA is an extracellular protein whose production requires expression of the flagellar regulon. (A) Concentrated whole-cell lysates and culture supernatants of strain JB580v grown in LB or T medium were examined for phospholipase activity by a radial diffusion assay. A positive reaction resulted in the formation of a precipitate emanating from the sample well in gels containing the artificial substrate Tween 80. Each well contained an equivalent of cells or supernatants from 1.5 ml culture at an OD600 of 3.0. Row 1, culture supernatant after growth in T medium (Left), culture supernatant after growth in T medium and preheated to 90°C for 10 min (Center), and cell lysate after growth in T medium (Right). Row 2, culture supernatant after growth in LB (Left), culture supernatant after growth in LB and preheated to 90°C for 10 min (Center), and cell lysate after growth in T medium and preheated to 90°C for 10 min (Right). Row 3, cell lysates after growth in LB (Left), cell lysate after growth in LB and preheated to 90°C for 10 min (Center), and buffer control (Right). (B) Phospholipase activity of secreted YplA was measured for strain GY460 containing plasmid pVLT33 (vector control), pGY20 (ptac-flhDC from Y. enterocolitica), and pMG600 (ptac-flhDC from S. liquefaciens). Conditions of the assay were as described in A. Each well contained concentrated culture supernatant for each strain grown in T medium, with the inducer IPTG at a concentration of 0, 5, or 50 μM (indicated at the top). From top to bottom each well was loaded with an increased dilution of each sample.
To demonstrate that expression of the flagellar regulon is required for YplA secretion, levels of YplA activity were determined under conditions known to affect expression of motility. YplA activity was measured for culture supernatants of Y. enterocolitica grown in LB, conditions known to repress the expression of motility (9). Under these growth conditions, no YplA activity was detected (Fig. 3A). YplA activity also was measured for supernatants from cultures of the Y. enterocolitica flhDC mutant strain in which motility (9) and Fop production was controlled ectopically by expressing flhDC from the ptac promoter (Fig. 2). Results from this analysis demonstrated levels of phospholipase activity were completely dependent on the level of flhDC expression (Fig. 3B). No phospholipase activity was detected when the same flhDC mutant contained the cloning vector alone (Fig. 3B). As a positive control, the Y. enterocolitica flhDC mutant was transformed with a plasmid containing the Serratia liquefaciens flhDC locus expressed from the ptac promoter, a condition previously shown to result in constitutive expression of motility (9). This also resulted in constitutive levels of phospholipase (Fig. 3B). Consistent with these results YplA activity was abolished in a fliA mutant but not a flgM mutant (Table 1). Flagellin and Fop production is eliminated only in the former strain, which does not express flagellar class III genes.
Table 1.
YplA activity requires genes involved in flagellum biogenesis
| Strain | Mutation | Plasmid | YplA activity* |
|---|---|---|---|
| JB580v | + | ||
| YCK10 | flgE | None | − |
| pACYC4GM | + | ||
| YCK11 | flgF | None | − |
| pACYC4GM | + | ||
| JO1v | flgM | None | + |
| pSWIM1 | + | ||
| YMS12 | flhA | None | − |
| pMSflh | + | ||
| YMS13 | flhB | None | − |
| pMSflh | + | ||
| GY460 | flhDC | None | − |
| pGY15 | + | ||
| VK1 | fliA | None | − |
| pJB222 | + |
Yp1A activity was determined for strains on phospholipase-indicator plates.
A screen of other motility mutants for YplA activity also implicated yplA as belonging to the flagellar regulon (Table 1). Mutations in genes such as flgE, flgF, flhA, and flhB prevent assembly of a complete flagellum basal body and the expression of class III flagellar genes (4). The assembly and transcription defects of the mutants eliminated extracellular flagellin and Fop production (Fig. 1, data not shown). They also eliminated YplA activity, which was restored in each strain by complementation of the mutation (Table 1). In agreement with this analysis a yplA-lacZ transcriptional fusion was found recently to require both flhDC and fliA for expression (unpublished results).
YplA Is One of the Fops.
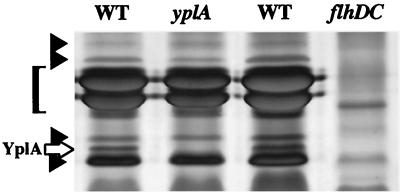
Examination of motility mutants combined with regulatory analysis suggested yplA encodes a protein exported by the flagellar type III secretion apparatus. To identify YplA among the numerous other extracellular proteins, culture supernatants were examined after fractionation by SDS/PAGE (Fig. 4). These results showed that wild-type Y. enterocolitica, grown under conditions that induced motility, produced an extracellular protein with an apparent mass of 35.5 kDa that was not produced by the yplA mutant (Fig. 4). The apparent mass of the missing protein was similar to the predicted molecular mass of YplA if it is secreted without modification by a type III mechanism. This analysis also showed that the same 35.5-kDa protein was one of the Fops because it was not secreted by a flhDC mutant (Fig. 4). YplA was confirmed to be the 35.5-kDa Fop because its production was restored by complementation of the yplA mutation with a plasmid-encoded copy of yplA, and this protein specifically reacted with an anti-YplA polyclonal antibody when examined by immunoblot (Fig. 5A, lanes 1–3) and immunoprecipitation analyses (data not shown).
Figure 4.
Identification of YplA as a 35.5-kDa secreted protein. Extracellular proteins were concentrated from culture supernatants of strains YEDS10/pGY20, GY460/pGY20, and GY460/pVLT33 grown in T medium with 50 μM IPTG to induced transcription from the ptac promoter. Proteins were separated on 12.5% SDS-polyacrylamide gel and stained with silver. Lanes: 1, GY460/pGY20; 2, YEDS10/pGY20; 3, GY460/pGY20; and 4, GY460/pVLT33. Each lane contains the equivalent of 1.5 ml culture at an OD600 of 1.0. The position of YplA is marked with an open arrow, and other Fops are marked with solid arrowheads. The location of flagellin is indicated with a bracket.
Figure 5.
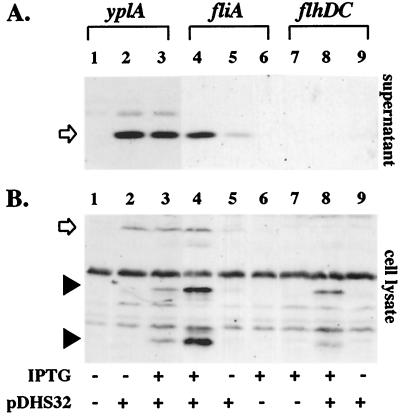
Detection of YplA in culture supernatants by immunoblot analysis. Concentrated protein from whole-cell lysates or culture supernatants from strains grown in T medium with 0 and 100 μM IPTG were separated on 12.5% SDS-polyacrylamide gel, transferred to nitrocellulose membranes, and probed with anti-YplA antibody. (A) Levels of YplA in culture supernatants. Lanes: 1, YEDS10, 0 μM IPTG; 2, YEDS10/pDHS32, 0 μM IPTG; 3, YEDS10/pDHS32, 100 μM IPTG; 4, VK1/pDHS32, 100 μM IPTG; 5, VK1/pDHS32, 0 μM IPTG; 6, VK1/pVLT33, 100 μM IPTG; 7, GY460/pVLT33, 100 μM IPTG; 8, GY460/pDHS32, 100 μM IPTG; and 9, GY460, 0 μM IPTG. (B) Levels of YplA detected in whole cells. Lanes are the same as those listed in A. Full-length YplA is indicated by the open arrow. YplA degradation products detected in whole cells are indicated by the solid arrowheads.
Secretion of YplA Requires Only the Flagellar Basal Body.
To more clearly determine whether the flagellar regulon is required for YplA secretion we examined export of YplA expressed ectopically in various motility mutants. For this analysis yplA expression was placed under the control of a ptac promoter. This allowed transcription of yplA to be controlled in response to the presence of IPTG and to be independent from mechanisms that control flagellar gene expression, thus allowing the direct examination of how specific mutations affect secretion. Secretion of YplA was evaluated by using two assays: (i) YplA activity was determined for strains on phospholipase indicator media, and (ii) YplA export was determined by localization of the protein to whole-cell or supernatant fractions of cultures by immunoblot analysis. These two assays showed for the yplA mutant, which produces a complete flagellum, that expression of ptac-yplA restored YplA activity (Table 2) and it was exported out of the cell (Fig. 5A). YplA activity and export were also restored when the ptac-yplA construct was expressed in the fliA mutant (Table 2 and Fig. 5A). The fliA mutant expresses flagellar class II but not class III genes (including the chromosomal copy of yplA). Thus, fliA mutants that produce the flagellum basal body (including the flagellar type III secretion machinery) but not the filament are competent for YplA secretion.
Table 2.
Activity of YplA when produced ectopically in different flagellar mutants
| Strain | Mutation | YplA activity* |
|---|---|---|
| YCK10 | flgE | − |
| YXK11 | flgF | − |
| YMS12 | flhA | − |
| YMS13 | flhB | − |
| GY460 | flhDC | − |
| VK1 | fliA | + |
| YEDS10 | yplA | + |
YplA activity for strains on phospholipase-indicator plates. YplA was expressed from pDHS32 in each strain examined; expression was induced by the addition of 50 μM IPTG to the medium.
In contrast, no YplA activity or export was detected when the ptac-yplA construct was introduced into the flhDC mutant, which does not express class II or III genes (Table 2 and Fig. 5A). In this case the mutant does not produce the flagellar type III secretion machinery or other flagellum components. When YplA was produced in this strain it was detected in the whole-cell fractions but it appeared to be degraded rapidly (Fig. 5B). Similarly, no YplA activity or export was detected when ptac-yplA was expressed in strains carrying mutations in class II genes such as flgE, flgF, flhA, or flhB (Table 2 and data not shown). These class II genes encode subunits of the flagellum basal body including the type III secretion machinery (4, 21). For each of these secretion-defective strains, expression of ptac-yplA resulted in YplA production, but it was only detected in whole-cell fractions and appeared to be rapidly degraded. Collectively, examination of YplA secretion in the different flagellar mutants demonstrated that expression of yplA is sufficient for phospholipase activity as long as the cell is able to produce a competent flagellar type III secretion apparatus. These results make it unlikely that YplA is secreted by another secretion system that is regulated by flhDC because expression of such a hypothetical secretion system should not be affected by flagellar class II mutations. These results also show that mislocalized YplA was retained by cells and degraded (Fig. 5B), a phenomenon that was accentuated in secretion-defective strains (Fig. 5B).
DISCUSSION
Examination of Y. enterocolitica motility mutants for the loss of flagellin production revealed the surprising result that mutations that abolished flagellum biosynthesis also eliminated the production of several secreted proteins. Some of the proteins secreted by the flagellar export apparatus could be subunits of the flagellum shed by the bacterium during growth under laboratory conditions, as is the case for flagellin (4, 5). Another flagellar protein that is known to be secreted is the negative regulator FlgM (22). Other flagellum proteins exported by this system that could be shed into the extracellular environment include those that form the hook and the filament components. However, in the absence of any proteolytic degradation, these proteins would account for no more than 9 of the more than 14 extracellular proteins detected, indicating that some have other functions.
One protein that is not involved in motility is the virulence-associated phospholipase YplA, which was shown in this study to be secreted by the flagellar export apparatus. YplA is related to another extracellular phospholipase, PhlA, produced by S. liquefaciens, which may be transported by a similar mechanism (23). This suggests this type of protein transport is common among many bacteria. The function of the other nonflagellum-associated proteins identified in this study remains to be elucidated. Certainly, these other proteins also may affect bacterial–host interactions. Secretion of YplA by Y. enterocolitica required genes that are known to compose the flagellum basal body and hook. Secretion did not require additional flagellum components such as the filament because YplA was secreted by an fliA mutant. For activity, YplA must be exported because phospholipase activity could not be detected for strains that were demonstrated to produce YplA but were defective for its transport out of the cell. The absence of YplA activity in secretion-defective strains was probably a result of proteolysis, which may occur in any situation in which the protein does not associate efficiently with components of the flagellar secretion apparatus. Rapid degradation, in this case, is probably an essential feature for bacterial viability because YplA accumulation in the cytoplasmic compartment could be deleterious since it may expose intracellular phospholipids to uncontrolled degradation.
The homology between flagellar basal body subunits and other type III secretion apparatus subunits indicates YplA secretion occurs by a type III mechanism. Based on the DNA sequence of its structural gene, secreted YplA should be transported without being modified and have a molecular mass of 35.5 kDa. The apparent molecular mass for secreted YplA was consistent with this prediction and the same as the apparent molecular mass of the full-length species of YplA detected in whole cells.
Demonstration of dual functions for the flagellar export machinery in flagellum biogenesis and protein secretion reinforces the hypothesis that this system has a common evolutionary origin with the contact-dependent type III secretion systems. The overlapping functions between these systems for protein secretion will allow future studies of type III secretion to take advantage of the wealth of biochemical and genetic tools already developed for the study of flagellum biosynthesis. In Y. enterocolitica and Salmonella typhimurium there are several different type III secretion systems (including the flagellar export apparatus) that appear to be dedicated to the transport of a specific set of proteins (3, 24). It is probable that other bacteria similarly will produce many type III secretion systems. Bacteria therefore must coordinate the expression and specificity of these different secretion systems to maintain appropriate control over physiological responses to environmental cues.
A role for motility in bacterial–host interactions has been suggested in many instances but has not been clarified (25). Previous studies have focused specifically on the role of flagella in these interactions. However, the results presented here indicate components of the bacterial flagellum may have a role in the secretion of proteins not specifically related to motility but that influence bacterial–host interactions. Indeed, for Y. enterocolitica, mutations in fliA that eliminate production of a mature flagellum had no obvious effect on host colonization (26). However, a fliA mutant still produces a flagellum basal body that is competent for secretion, suggesting that these strains still may be able to express essential virulence factors in vivo.
Acknowledgments
We thank Andrew Darwin and Briana Young for critical review of this manuscript, Scott Minnich for valuable discussions, and Michael Givskov for providing strains and plasmids. This work was supported by National Institutes of Health Grants AI27342 and AI01230 to V.L.M. and 5 T AI07172-17 to G.M.Y.
ABBREVIATIONS
- T medium
1% (wt/vol) tryptone
- IPTG
isopropyl β-d-thiogalactopyranoside
References
- 1.Wandersman C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 955–966. [Google Scholar]
- 2.Galan J E, Sansonetti P J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2757–2773. [Google Scholar]
- 3.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacNab R M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 123–145. [Google Scholar]
- 5.Aizawa S. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 6.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 7.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Infect Immunol. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 9.Young, G. M., Smith, M., Minnich, S. A. & Miller, V. L. (1999) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 10.Kapatral V, Olson J W, Pepe J C, Miller V L, Minnich S A. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 11.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 12.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 13.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Blum H, Beier H, Gross H J. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 16.Givskov M, Olsen L, Molin S. J Bacteriol. 1988;170:5855–5862. doi: 10.1128/jb.170.12.5855-5862.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 18.Young G M, Miller V L. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 19.Pepe J C, Badger J L, Miller V L. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 20.Givskov M, Eberl L, Christiansen G, Benedik M J, Molin S. Mol Microbiol. 1995;16:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 21.Fauconnier A, Allaoui A, Campos A, Van Elsen A, Cornelis G, Bollen A. Microbiology. 1997;143:3461–3471. doi: 10.1099/00221287-143-11-3461. [DOI] [PubMed] [Google Scholar]
- 22.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 23.Givskov M, Molin M. Mol Microbiol. 1993;8:229–242. doi: 10.1111/j.1365-2958.1993.tb01567.x. [DOI] [PubMed] [Google Scholar]
- 24.Cornelis G R, Wolf-Watz H. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 25.Ottemann K M, Miller J F. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 26.Iriarte M, Stainier I, Mikulskis A V, Cornelis G R. J Bacteriol. 1995;177:2299–2304. doi: 10.1128/jb.177.9.2299-2304.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]