Abstract
DivIVA is a conserved protein in Gram-positive bacteria and involved in various processes related to cell growth, cell division and spore formation. DivIVA is specifically targeted to cell division sites and cell poles. In Bacillus subtilis, DivIVA helps to localise other proteins, such as the conserved cell division inhibitor proteins, MinC/MinD, and the chromosome segregation protein, RacA. Little is known about the mechanism that localises DivIVA. Here we show that DivIVA binds to liposomes, and that the N terminus harbours the membrane targeting sequence. The purified protein can stimulate binding of RacA to membranes. In mutants with aberrant cell shapes, DivIVA accumulates where the cell membrane is most strongly curved. On the basis of electron microscopic studies and other data, we propose that this is due to molecular bridging of the curvature by DivIVA multimers. This model may explain why DivIVA localises at cell division sites. A Monte-Carlo simulation study showed that molecular bridging can be a general mechanism for binding of proteins to negatively curved membranes.
Keywords: Bacillus subtilis , cell division, DivIVA, membrane curvature, phospholipids
Introduction
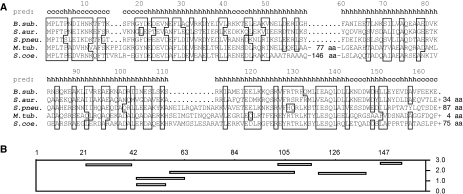
DivIVA is conserved in Gram-positive bacteria. Secondary structure predictions show that the protein mainly forms α-helices (Figure 1A), and as DivIVA has some sequence similarity with tropomyosin, it is assumed that it forms coiled coils (Edwards et al, 2000). Biochemical and electron microscopic studies have shown that the purified protein forms multimers and assembles into large ordered lattices (Stahlberg et al, 2004). Despite the sequence conservation, the functional role of this protein varies between different bacterial species. In Bacillus subtilis, mutations in divIVA result in elongated cells that occasionally divide aberrantly near existing cell poles to produce minicells (Cha and Stewart, 1997). Fluorescence microscopy studies have shown that DivIVA is located at mid-cell during cell division and at matured cell poles, and that it is responsible for the polar localisation of the division inhibitor, MinC/MinD (Edwards and Errington, 1997; Marston et al, 1998). The first step in cell division in most bacteria is the polymerisation of the tubulin-like protein, FtsZ, into a ring-like structure (the Z-ring) onto which the cytokinesis apparatus assembles. In rod-shaped bacteria, MinC/MinD prevents polymerisation of FtsZ close to cell poles (Margolin, 2001; Hale and de Boer, 2002). When MinC/MinD is delocalised, as a consequence of inactive DivIVA, cell division is largely inhibited and, in addition, cells can divide aberrantly close to cell poles, producing small anucleate minicells. DivIVA also plays an important role during sporulation. Incorporation of DNA into the polar prespore compartment is achieved by anchoring one chromosome copy to the distal pole of the prespore compartment. One of the proteins involved in this process is RacA, and DivIVA is responsible for the polar localisation of RacA (Ben-Yehuda et al, 2003; Wu and Errington, 2003).
Figure 1.
Amino-acid alignment of DivIVA homologues (A) from the following: B. subtilis (B. sub.), S. aureus (S. aur.), S. pneumoniae (S. pneu.), M. tuberculosis (M. tub.) and S. coelicolor (S. coe.). Homologues and similar amino acids are boxed. The length of the C termini that extend beyond the B. subtilis DivIVA sequence is indicated. Presented above the sequence is the secondary structure prediction for B. subtilis DivIVA: c, coiled; h, helix. (B) Amphipathic helix prediction from the LOCATE program. The amino-acid positions are indicated and the Y axis shows the hydrophobic moments of the putative amphipathic helices.
DivIVA homologues are also present in Gram-positive cocci such as Streptococcus pneumoniae and Staphylococcus aureus (Figure 1A), although these bacteria do not contain a Min system and do not sporulate. In S. pneumoniae, deletion of divIVA results in a severe cell division defect, and two-hybrid experiments suggested that DivIVA interacts with a number of cell division proteins (Fadda et al, 2003). It is surprising that in S. aureus, a divIVA deletion shows no apparent phenotype (Pinho and Errington, 2004). In the filamentous branching actinomycete, Streptomyces coelicolor DivIVA has an important function in tip growth and branching (Flardh, 2003; Hempel et al, 2008). Recently, it was suggested that a cellulose-synthase-like protein, involved in tip growth, may interact with DivIVA (Xu et al, 2008). In the non-sporulating, non-branching actinomycetes, such as Corynebacterium glutamicum and Mycobacterium tuberculosis, DivIVA is also required for polar growth (Kang et al, 2008; Letek et al, 2008). In all bacteria investigated so far, DivIVA localises to division sites and/or polar regions. It is therefore assumed that DivIVA functions as a scaffold that helps to localise other proteins to specific regions within the cell.
Despite the importance of DivIVA for cell morphology in such a diversity of Gram-positive bacteria, little is known about the mechanism responsible for the localisation of DivIVA. An earlier mutagenesis study showed that mutations in the conserved N terminus of B. subtilis DivIVA destroy its polar localisation (Perry and Edwards, 2004). These mutations likely block interactions of the protein with a specific polar target. As the target might be a membrane-localised protein, we examined the affinity of DivIVA for isolated cell membranes. It is surprising that it appeared that DivIVA has a general affinity for the phospholipid fraction of the cell membrane. Using cell shape mutants, we show that DivIVA preferably binds to strongly curved cell membranes. On the basis of electron microscopy (EM) results and earlier published data, we propose a general model for binding of proteins to negatively curved membranes.
Results
DivIVA binds to membranes
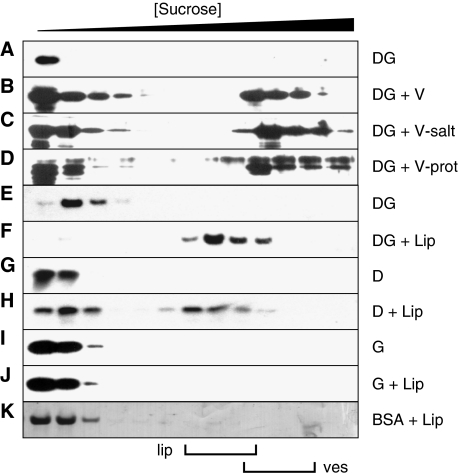
A simple explanation for the localisation of DivIVA would be that this protein is targeted by another protein; possibly a membrane protein. To examine this, we isolated cell membranes from a B. subtilis strain lacking DivIVA (B. subtilis 3310) and mixed these membranes with purified DivIVA-GFP. This mixture was then loaded onto a sucrose step gradient. After centrifugation, fractions from the gradient were analysed by western blotting using a GFP-specific antibody. As shown in Figure 2, the addition of cell membranes resulted in shifting a part of the DivIVA-GFP fraction towards the bottom of the gradient, where the membranes accumulated (Figure 2 B). When no membranes were present, DivIVA-GFP remained in the top fraction of the sucrose gradient (Figure 2A). The reaction mixture contained an excess of BSA (1 mg/ml), suggesting that the interaction of DivIVA-GFP with the membranes was specific. This was further supported by an experiment in which the binding buffer and gradient contained a high concentration of salt (0.5 M NaCl, Figure 2C).
Figure 2.
DivIVA-membrane interactions analysed with sucrose density gradient centrifugation. Gradient fractions were analysed by western blotting using GFP- or DivIVA-specific antibodies. The top fractions (low density) to the bottom fractions (high density) run from left to right. DivIVA-GFP (DG) was mixed with membrane vesicles (V; B) that were either incubated at high-salt concentrations (V-salt; C) or treated with Proteinase K (V-prot; D). Alternatively, purified DivIVA-GFP (DG), DivIVA (D) or GFP (G) was incubated with liposomes (Lip; F, H, and J). In all experiments, an excess of BSA (BSA) was present, which is indicated in the Coomassie staining of a blot (K). (A, E, G and I) The results of protein mixtures that contained no membrane vesicles or liposomes. Membranes vesicles (ves) and liposomes (lip) were clearly visible, and their position in the gradients is indicated below the figure.
To ascertain whether DivIVA-GFP interacted with membrane-bound proteins, we treated the membranes with Proteinase K. To our surprise, this treatment had no effect on the binding of DivIVA-GFP (Figure 2D). The amino-acid sequence of DivIVA does not show an apparent membrane-interacting domain. Nevertheless, the lack of any effect of Proteinase K suggested that DivIVA might interact with the lipid fraction of the cytoplasmic membrane. To test this, we examined the association of DivIVA with lipid vesicles constructed from pure phospholipids. As shown in Figure 2E and F, purified DivIVA-GFP bound efficiently to liposomes as well. It was possible that the GFP moiety somehow stimulated the interaction with phospholipids, therefore we purified wild-type DivIVA and repeated the experiments. Pure DivIVA (D) also showed a clear interaction with liposomes (Figure 2G and H). As a final control, we tested purified GFP. This protein (G) did not show any interaction with liposomes (Figure 2I and J). In the sucrose gradients that contained DivIVA or DivIVA-GFP, the liposomes appeared to aggregate. Clustering of liposomes by these proteins was in fact easily detectable by light microscopy (Supplementary data).
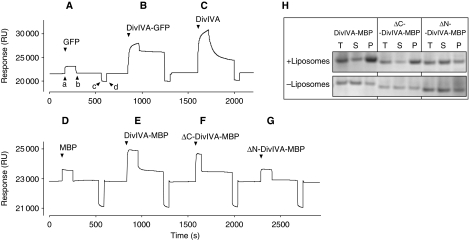
As an independent way to test the affinity of DivIVA for lipids, we turned to surface plasmon resonance (SPR). Liposomes were adsorbed onto an L1 sensor chip and purified GFP, DivIVA-GFP, or DivIVA was injected. As shown in the sensograms of Figure 3A, the addition of GFP yielded only a small response (a), likely due to a buffer effect given that the signal dropped back to the baseline when the GFP injection ended (b). In contrast, the magnitude of the response was strong when DivIVA-GFP was injected (Figure 3B), and the signal remained when the flow of DivIVA-GFP ceased, indicating that the fusion protein strongly interacted with the phospholipid membranes (the initial sharp decrease was due to buffer effects). Induction of DivIVA (Figure 3C) also resulted in a strong response and, together with the low off-rate, confirmed that this protein binds specifically to liposomes. Owing to the complex oligomerisation characteristics of DivIVA, it is difficult to deduce kinetic parameters from these sensograms. However, the difference in off-rates between DivIVA and DivIVA-GFP suggests that the GFP-tag influences membrane binding.
Figure 3.
Binding of purified GFP (A), DivIVA-GFP (B) and DivIVA (C) to liposomes adhered to a Biacore L1 sensor chip. Protein samples were injected (a), and after ∼2 min, followed by an injection of buffer alone (b). The chip was regenerated by a short injection (c and d) with 0.1 M NaOH solution. The flow rate was 30 μl/min, and protein concentrations were 3.1, 1.1 and 1.5 mg/ml for GFP, DivIVA-GFP and DivIVA, respectively. The response is given in artificial resonance units (RU). (D–G) SPR analysis of DivIVA deletions that were purified as MBP fusions. The C-terminal deletion (ΔC-DivIVA-MBP) lacks the last 20 amino acids of DivIVA and the N-terminal deletion (ΔN-DivIVA-MBP) lacks the first 40 amino acids of DivIVA. Protein concentrations were 0.4 mg/ml. (H) Sedimentation analyses of the N- and C-terminal DivIVA deletions in the presence and absence of liposomes. The total fraction, before centrifugation (T), and the supernatant (S) and pellet (P) fractions after centrifugation, was analysed by SDS–PAGE.
The N terminus of DivIVA contains the membrane-targeting domain
DivIVA contains several amphipathic helices that stimulate multimerisation by means of coiled-coil interactions (Muchova et al, 2002; Rigden et al, 2008). However, amphipathic helices can also interact with lipid bilayers, and, for example, the cell division proteins FtsA and MinD bind to membranes by means of amphipathic helices at their C termini (Szeto et al, 2002; Hu and Lutkenhaus, 2003; Pichoff and Lutkenhaus, 2005). Possibly, the association of DivIVA with membranes is mediated by an amphipathic helix as well. To rate the potential amphipathic helices in DivIVA, we used the program LOCATE (Jones et al, 1992). Figure 1B shows the outcome of this algorithm. The putative amphipathic helices with the highest scores span amino acids 22–41, 102–118, and 145–154 (Figure 1B). Of these three sequences, the helix at amino acids 102–118 seemed least likely, as this region showed less sequence conservation than the others (Figure 1A). To test whether the N- or C-terminally located amphipathic helices were required for membrane binding, we made several deletion constructs. Initial attempts using a DivIVA-GFP fusion resulted in inclusion bodies or very low protein yields. Therefore, we used maltose-binding protein (MBP) as a tag, as this protein is very soluble and easy to purify. As shown in the sensograms of Figure 3D, MBP does not bind to liposomes. However, when MBP was fused to DivIVA, the purified fusion protein did bind to the sensor chip (Figure 3E). Two deletion mutants were constructed, namely ΔN-DivIVA-MBP and ΔC-DivIVA-MBP, in which the N-terminal 40 amino acids or the last 20 amino acids of DivIVA are lacking. Gel filtration experiments showed that neither deletion interfered with the oligomeric state of the protein (Supplementary data). As shown in the sensograms of Figure 3F and G, the absence of the predicted C-terminal amphipathic helix did not affect binding to liposomes, whereas the absence of the predicted N-terminal amphipathic helix abolished binding. The fusion to MBP reduced aggregation of DivIVA and this made it possible to test membrane binding using a simple sedimentation protocol (Figure 3H). Furthermore, membrane binding was abolished by the N-terminal deletion but not by the C-terminal deletion. When we looked at the effect on liposomes with light microscopy, it was clear that the N-terminal deletion abolished clustering of liposomes, whereas purified ΔC-DivIVA-MBP was still able to aggregate liposomes (data not shown). These results suggest that the N-terminal amphipathic helix is the membrane-targeting domain.
The first 60 amino acids of DivIVA are sufficient for membrane binding
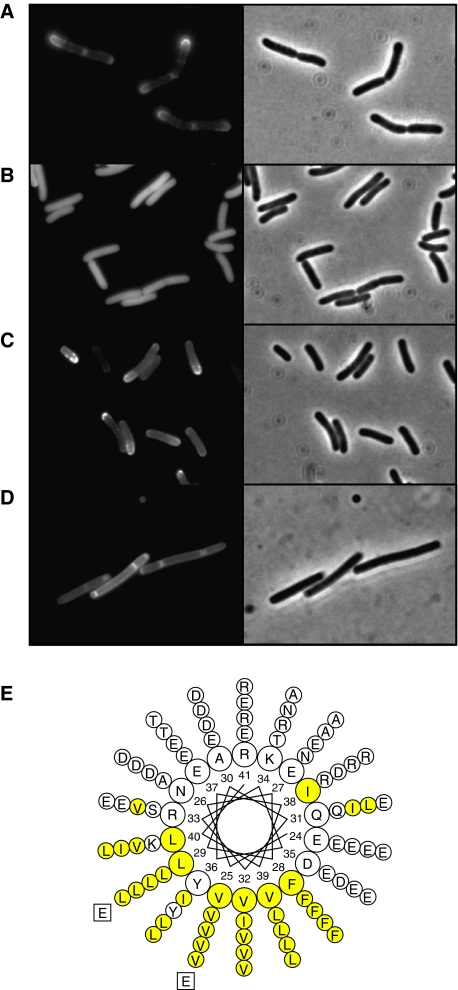
As described earlier (Edwards et al, 2000), DivIVA fused to GFP accumulates at the polar periphery of Escherichia coli cells (Figure 4A). We used this as an assay to further analyse the membrane-targeting domain. Short amino-acid stretches of the N terminus of DivIVA were fused to GFP, and the fusion proteins were expressed in E. coli. As the N-terminal amphipathic helix spans amino acids 22–41 (Figure 4E), we fused the first 40 or 50 amino acids of DivIVA to GFP. Both fusions resulted in a diffuse fluorescence signal in E. coli (Figure 4B, and data not shown). Western blot analysis showed that the fusions were stable; hence, the diffuse GFP signal was not a consequence of proteolytic cleavage (data not shown). However, when the first 60 amino acids of the N terminus were fused to GFP, a peripheral fluorescence signal appeared (Figure 4C). Expression of this fusion protein in a B. subtilis strain lacking divIVA resulted in a peripheral fluorescence signal as well (Figure 4D), suggesting that the N terminus contains the membrane-binding domain. However, when this truncation was purified and mixed with liposomes, no clustering was observed (data not shown). To test whether the large amphipathic helix within the N-terminal 60 amino-acid domain is involved in membrane binding, we changed one of the hydrophobic amino acids into a hydrophilic glutamate residue (Figure 4E). When valine 25 was mutated into a glutamate, a completely diffuse fluorescence signal was observed in E. coli. The same occurred when leucine 29 was mutated into a glutamate (data not shown, results comparable to Figure 4B). Western blot analysis indicated that these point mutations did not affect the stability of the fusion protein in E. coli (data not shown).
Figure 4.
Fluorescence microscopy of cells expressing different DivIVA-GFP deletion constructs: (A) E. coli cells expressing full-length DivIVA fused to GFP, (B) E. coli cells expressing amino acids 1–40 of DivIVA fused to GFP, (C) E. coli cells expressing amino acids 1–60 of DivIVA fused to GFP and (D) B. subtilis cells expressing amino acids 1–60 of DivIVA fused to GFP (expression was induced by xylose). The latter strain (B. subtilis LH60) contains a deletion of the wild-type divIVA and minCD genes, which results in polar division and minicells. (E) Helical wheel projection of the amphipathic helix located at amino acids 22–41 of DivIVA. Hydrophobic residues are marked in yellow. The inner circle represents the amino-acid residues of DivIVA from B. subtilis. The amino-acid residues of S. aureus, S. pneumoniae, M. turberculosis and S. coelicolor DivIVA are depicted from the second (inner) to the fifth (outer) circle, respectively. Mutations in hydrophobic residues (V25E or L29E) are indicated.
In E. coli, the MinCD proteins oscillate between the cell poles in less than a minute (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999). The fluorescent signals in Figure 4A and C are unequally distributed between the cell poles, which could indicate oscillation of the GFP-fusion proteins, as well. However, time-lapse experiments of DivIVA-GFP-expressing E. coli cells show no oscillation of the fluorescence signal, but indicate that DivIVA-GFP continues to accumulate as the cell poles age (Supplementary data). This is possibly due to the inert nature of cell poles compared with the continuously growing lateral wall (Lindner et al, 2008).
EM analyses of DivIVA-membrane interactions
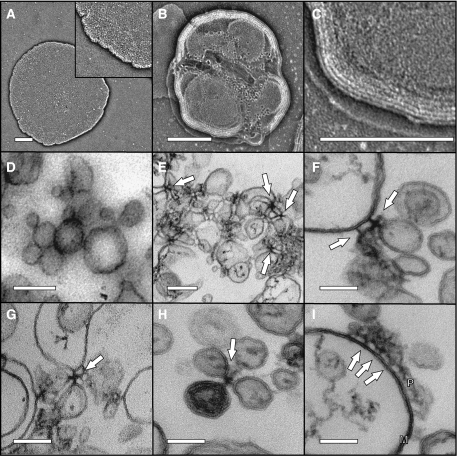
There are several examples of peripheral membrane proteins that cause deformation of phospholipid bilayers, MinD is one such protein (Hu et al, 2002). Whether the binding of DivIVA influences the shape of lipid membranes was tested using EM. We first examined liposome–DivIVA mixtures by negative staining EM. As shown in Figure 5A–C, the presence of DivIVA seemed to result in liposomes stacked on top of each other, which gives a multilayered appearance (Figure 5C). Some ruffled edges can be observed on the top layers (Figure 5B), but this could be an artifact due to drying and was not observed in isolated liposomes. As the stacking of liposomes in negative staining EM obscured the observation of a possible deformation of liposomes, we fixed the samples and prepared them for thin section transmission electron microscope (TEM). Furthermore, the clustering of liposomes by DivIVA was apparent (Figure 5E). Often the DivIVA complexes that held liposomes together appeared as cruciform-like structures (Figure 5E–H). Figure 5I shows part of a large liposome onto which a cluster of DivIVA molecules is attached via short stalks. Despite the clear association between DivIVA and liposomes, the TEM micrographs showed no indication that DivIVA deforms lipid membranes.
Figure 5.
Electron micrographs of negatively stained liposomes (A), and liposomes mixed with DivIVA (B, C). Liposomes were extruded using a 400-nm pore filter. Inset in panel A is a 2 × enlargement. Panel C is a 3 × enlargement of panel B, to show the layered structure due to stacking of liposomes. Scale bars=200 nm. Electron micrographs of thin sections of liposomes (D), and liposomes mixed with DivIVA (E–I). Liposomes were extruded using a 100-nm pore filter. Arrows indicate cruciform-like structures. (I) Part of a large liposome to which a cluster of DivIVA proteins (P) is bound; arrows point to DivIVA stalks attached to the membrane (M). Scale bars=200 nm in panels D and E, and 100 nm in panels F–I.
DivIVA accumulation at negatively curved membranes
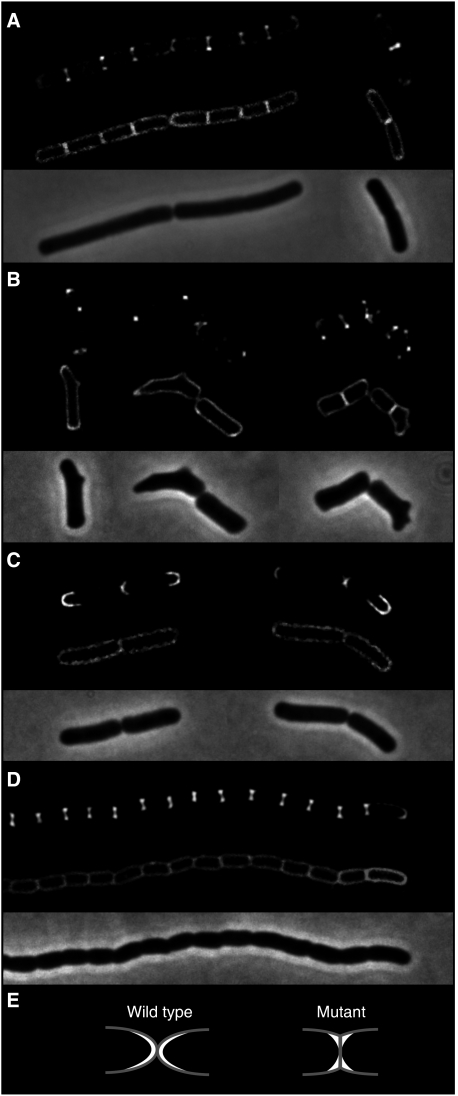
In B. subtilis, the brightest DivIVA-GFP signal appears at cell division sites in a ring-like pattern that does not follow the constriction of the Z-ring (Harry and Lewis, 2003). This is also illustrated in Figure 6A. In this case, we used deconvolution of a Z-stack of images to increase the resolution. At the site of cell division (bands across cells in the membrane image), DivIVA-GFP gives a dumbbell-shaped fluorescence signal that is indicative of a ring-like accumulation of the fusion protein. Interestingly, this is also the region where the membrane is most strongly curved. So, although DivIVA itself does not induce curves in lipid membranes, it might be that the protein has a preference for strongly curved membrane regions. There is currently no established method to test for binding to negatively curved membranes. As an alternative, we looked at the localisation of DivIVA-GFP in aberrantly shaped cells. First, we re-examined an old observation that suggested that DivIVA-GFP tends to accumulate at protrusions formed in cells that contain an mreB mutation (Hamoen and Errington, 2003). These protrusions form strongly negative curved membranes, and indeed, as shown in Figure 6B, the strongest GFP signals colocalised with the bulges in the deformed mreB mutants. Thus, the preference for negatively curved membranes does not seem to be restricted to cell division sites. To test this further, we analysed the situation in E. coli. In wild-type E. coli cells, DivIVA-GFP accumulates primarily at the poles and forms clear fluorescent arcs that follow the curvature of the poles (Figure 6C). There are E. coli mutants that are devoid of murein hydrolases necessary to hydrolyse the peptidoglycan of the division septum. These mutants cannot separate after division and form long chains of cells (Heidrich et al, 2001). A consequence of this is that the poles of the cells are not round but flat, and that a relative sharply curved membrane is generated at the transition from a pole to a lateral wall (see model in Figure 6E). Expressing DivIVA-GFP in such an E. coli mutant resulted in strong dumbbell-shaped fluorescence signals at the fused cell poles, as shown in Figure 6D; thus, an indication for a ring-like accumulation pattern. Hence, also in E. coli, DivIVA seems to accumulate at sites where the cytoplasmic membrane is most strongly curved. When we expressed the 60 amino-acid N-terminal domain of DivIVA (fused to GFP) in this E. coli mutant, the fluorescent signal marked the cell membrane, and no dumbbell-shaped accumulation was observed (data not shown).
Figure 6.
Localisation of DivIVA-GFP in cells of different shape: (A) wild-type B. subtilis cells, (B) compilation of several deformed B. subtilis mreB mutant cells (B. subtilis 3292), (C) wild-type E. coli cells and (D) E. coli murein hydrolase mutant cells (E. coli MHD63). To increase the resolution, deconvoluted images of Z-stacks are shown. The upper panels show GFP fluorescence, the middle panels show fluorescent membrane stain and the lower panels show phase-contrast images. (E) A schematic representation of the poles of wild-type E. coli cells and the murein hydrolase mutant (grey). DivIVA-GFP is indicated in white.
DivIVA recruits RacA to the membrane
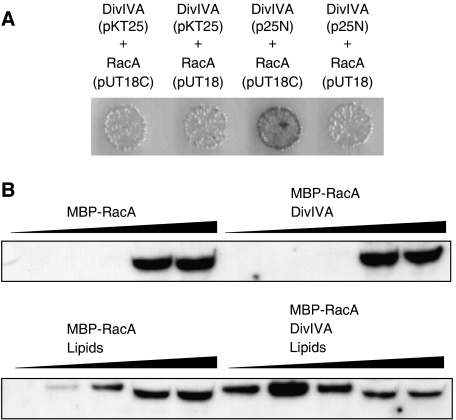
DivIVA is required for the localisation of proteins, but so far there is no biochemical data showing a direct interaction between DivIVA and other proteins. We were unable to detect any effect on the lipid-binding affinity of purified MinD by DivIVA (data not shown). Very recently, it was reported that MinJ (YvjD), a previously unknown transmembrane protein, is required for MinD localisation (Bramkamp et al, 2008; Patrick and Kearns, 2008), which explains our failure to show a direct interaction between DivIVA and MinD in vitro. Another potential target for DivIVA interaction is the chromosome segregation protein, RacA (Ben-Yehuda et al, 2003). We first used a bacterial two-hybrid assay to obtain more evidence for a direct interaction between DivIVA and RacA. Such a test gave a negative outcome in case of MinD and DivIVA (data not shown). We tested different combinations of both C- and N-terminal fusions, and low- and high-copy vectors (Supplementary data), and found a positive interaction between a DivIVA-adenylate cyclase T25 fragment fusion on a low-copy plasmid and an adenylate cyclase T18 fragment-RacA fusion on a high-copy plasmid (Figure 7A). Encouraged by this result, we purified RacA as a fusion with MBP. We used density gradient flotation experiments to test whether DivIVA would stimulate binding of RacA to lipid membranes. MBP-RacA was mixed with DivIVA and liposomes, and loaded at the bottom of a sucrose gradient. A high concentration of BSA (0.5 mg/ml) was present to ensure specificity. After centrifugation, fractions were loaded onto a protein gel and analysed by western blotting using RacA-specific antibodies (Figure 7B). In the absence of DivIVA, a small amount of RacA could be detected in the phospholipid fractions. However, the presence of DivIVA led indeed to a substantial increase in the amount of RacA in the lipid fraction.
Figure 7.
Bacterial two-hybrid interaction assay (A). divIVA and racA were cloned in different expression vectors, and the combinations screened for adenylate cyclase activity (blue colonies). A positive interaction was observed with a DivIVA-adenylate cyclase T25 fragment on the low-copy plasmid p25-N and an adenylate cyclase T18 fragment-RacA fusion on the high-copy plasmid pUT18C. pKT25: low-copy plasmid for N-terminal adenylate cyclase T25 fragment fusion, p25-N: low-copy plasmid for C-terminal adenylate cyclase fusion, pUT18C: high-copy plasmid for N-terminal adenylate cyclase T18 fragment fusion, pUT18: high-copy plasmid for C-terminal adenylate cyclase T18 fragment fusion. The effect of DivIVA on the binding of MBP-RacA to liposomes (B). MBP-RacA (6.4 ng) and DivIVA (3.6 μg) were mixed with liposomes (90 μg) in a sucrose-containing buffer, and loaded at the bottom of a sucrose gradient. After centrifugation, gradients where sampled in five fractions (top fractions (low density) to bottom fractions (high density) run from left to right). Liposomes floated to the two top fractions and where clearly visible. Gradient fractions were analysed by western blotting using RacA-specific antibodies.
Discussion
DivIVA plays a structural role in the development of Gram-positive bacteria by recruiting proteins to cell division sites and cell poles. How this process operates is unknown. Here, we show that the localisation of DivIVA to the cell periphery involves a direct interaction with the lipid bilayer, and that the protein can stimulate the binding of other proteins, such as RacA, to the lipid membrane. In addition, we show that DivIVA accumulates where the cytoplasmic membrane is most strongly curved. Very recently, it was shown that the B. subtilis protein, SpoVM, uses the curvature of membranes as a cue for its localisation (Ramamurthi et al, 2009). SpoVM is a small peripheral membrane protein that is involved in the spore coat assembly. The protein accumulates only at the spore membrane and not at the cytoplasmic membrane of the mother cell. SpoVM makes this distinction by having a stronger affinity for the positive curvature of the spore membrane compared with the negative curvature of the mother cell membrane. Hence, SpoVM is an example of a protein that prefers the convex side of curved membranes, whereas DivIVA is an example of a protein that prefers the concave side.
A deletion analysis of DivIVA showed that the first 60 amino acids are sufficient for membrane binding. The amino-acid alignment of different DivIVA molecules (Figure 1A) suggests that these 60 amino acids form a distinct domain, as DivIVA sequences from both M. tuberculosis and S. coelicolor contain large non-conserved inserts immediately after this region. Amino acids 22–41 are predicted to form a long amphipathic helix (Figure 4E). Such amphipathic helices form the basis for coiled-coil interactions; however, an extensive mutagenesis study of DivIVA from Enterococcus faecalis showed that the N-terminal domain is not required for oligomerisation (Rigden et al, 2008). There are also several examples of amphipathic helices that bind specifically to lipid bilayers (Szeto et al, 2003; Pichoff and Lutkenhaus, 2005). Therefore, the 20 amino-acid long amphipathic helix at the N terminus of DivIVA would be a good candidate for the membrane-targeting sequence. In fact, the hydrophobic and hydrophilic phases of this amphipathic helix are highly conserved in different DivIVA species (Figure 4E). Replacing some of the hydrophobic residues in this helix abolished the interaction with the cell membrane.
Despite the fluidity of the lipid bilayer, phospholipids are not homogenously distributed in the cytoplasmic membrane (Fishov and Woldringh, 1999; Mileykovskaya and Dowhan, 2000; Kawai et al, 2004). Using fluorescent dyes with preference for certain lipids, it was shown that the concentration of cardiolipin (CP) and phosphatidylethanolamine (PE) is increased at the septal and polar regions of B. subtilis (Kawai et al, 2004; Nishibori et al, 2005). Given that DivIVA binds to lipids, a striking possibility would be that the localisation of this protein depends on the localisation of certain lipid molecules. The synthesis of CP or PE can be blocked in B. subtilis without clear adverse effects on growth (Kawai et al, 2004; Nishibori et al, 2005). We have deleted genes involved in the biogenesis of these lipids but found no effect on the localisation of DivIVA (Supplementary data). We have also introduced a mutation that blocks the synthesis of another major lipid component, phosphatidylglycerol (Kobayashi et al, 2003), but again the septal and polar localisation of DivIVA remained undisturbed (Supplementary data). It seems therefore less likely that DivIVA binds a specific lipid species that would accumulate at cell division sites and cell poles.
Both in B. subtilis and in E. coli, DivIVA tends to accumulate at sites where the membrane is most strongly curved. As the two bacteria are evolutionary rather distinct, it seems that this tendency could be an intrinsic property of DivIVA. The Bar domain is a typical example of a protein structure that binds specifically to curved membranes. This conserved domain is found in proteins that are involved in vesicle formation and membrane remodelling (Peter et al, 2004). Bar domains bind to curved membranes by forming crescent-shaped dimers that fit the curvature. In fact, when Bar domains are added to liposomes, they deform the membrane, which results in long tubular-shaped liposomes (Peter et al, 2004). It should be mentioned that Bar domains bind to the outside of vesicles, thus to positively curved membranes, whereas DivIVA is enriched at negatively curved membranes. Nevertheless, if DivIVA binds to curved membranes in a similar manner, it seems likely that DivIVA deforms spherical liposomes, as the lipid bilayer will try to wrap around curved DivIVA complexes. However, our EM data showed no indication for membrane deformation. We have also tested whether the affinity of DivIVA depends on the diameter, thus curvature, of liposomes, but we found no differences (Supplementary data). It seems that the accumulation of DivIVA is not a consequence of a possible curved shaped of DivIVA multimers.
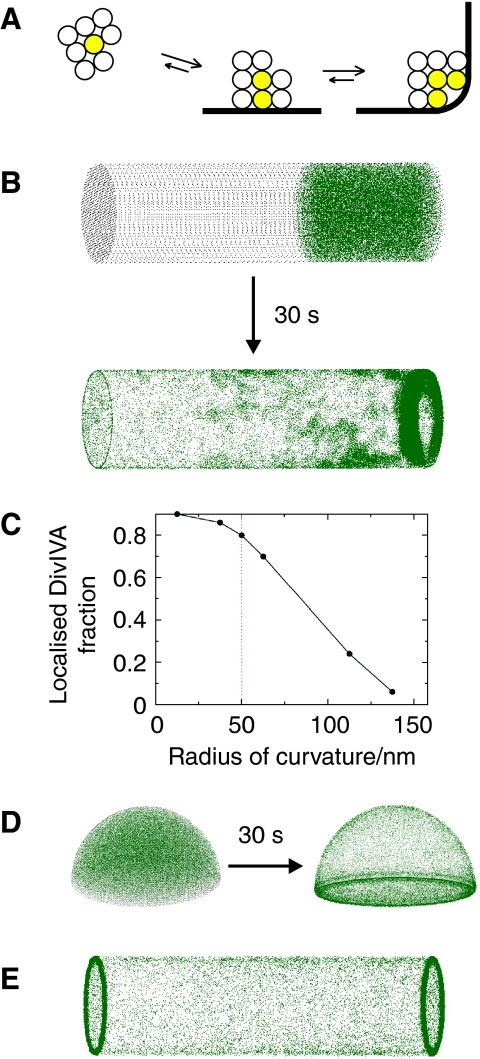
In one of the most detailed studies on DivIVA oligomerisation, it was shown that DivIVA forms doggy bone-like structures of ∼22.4 nm in length, which assemble into large lattices (Stahlberg et al, 2004). These doggy bones appear to be composed of 6–8 DivIVA subunits. In the TEM pictures of Figure 5, the crucifix- and stalk-like protein structures span on average 25 nm, which is almost the same size as doggy bones. These are remarkably large structures that in length surpass the diameter of ribosomes (∼20 nm). Possibly, this is one of the key factors that contribute to binding of DivIVA to negatively curved membranes. We propose that (i) the mutual interaction between DivIVA oligomers (doggy bones), (ii) their affinity for membranes and (iii) their large size stabilises DivIVA clusters by ‘bridging' opposing membranes. This ‘molecular bridging' is schematically depicted in Figure 8A. For simplicity, the DivIVA oligomers are depicted as spheres. The spheres can interact with each other and with the cell membrane. Three different situations are depicted: a cluster of eight spheres that freely floats in the cytoplasm, the same cluster but bound to the lateral (flat) cell membrane and a cluster that is localised in the corner formed by a curved membrane. The spheres at the periphery of these clusters (transparent spheres) have fewer interactions and are more prone to detach and diffuse away. Binding of the cluster to the membrane decreases the exposed surface and stabilises the cluster, and this is most noticeable when the membrane is strongly curved. In fact, the transition from the lateral to the perpendicular membrane region can be rather gradual, as the spheres will bridge the groove by forming a cluster. This model could explain the accumulation of DivIVA in a ring-like formation at the site of cell division, without the need for either intrinsic curvature of the protein, binding to specific lipid molecules or the presence of other proteins.
Figure 8.
Schematic presentation of the molecular bridging model for the formation of stable DivIVA clusters at negatively curved cell membranes (A). DivIVA oligomers are indicated as spheres that form a free-floating cluster (left), a cluster that attaches to the cell membrane (middle) and a cluster that fills the corner of a curved membrane (right). Oligomers that can detach and diffuse away are transparent, and oligomers that are enclosed are indicated in yellow (see main text for more details). (B) Monte-Carlo simulation of 200 spheres that diffuse freely in a cylindrical volume, representing a rod-like bacterium. The pictures show the distribution over 25k iterations. The spheres depicting DivIVA oligomers (green) have a radius of 12.5 nm, and the dimensions of the cylinder are 4 × 1 μm (length × diameter). The curvature of the membranes at the transition from lateral wall of the cylinder to the sides has a radius of 50 nm. In this simulation, the spheres can make eight contacts, with two membrane contacts maximal (by weighing a membrane interaction as four contacts). Epp and Epm were 3.5 kBT and 5.5 kBT. To reduce cpu time, we started with an asymmetric distribution (the simulation took 5 days on a dual-core Intel processor). (C) Localisation of 12.5 nm spheres in relation to the curvature of the membrane. The simulation conditions were the same as in panel B. To quantify the localisation, we define a cutoff distance, typically 150 nm, and assume that a sphere is localised at the high curvature region when it is within this cutoff distance. (D) Monte-Carlo simulation of 200 spheres diffusing in a hemispheric volume with a diameter of 1 μm, representing a dividing coccoid. The same constants were used as in panel B except that in this case the spheres can make only four contacts, and there are no restrains on the number of membrane contacts. (E) Monte-Carlo simulation of rod-shaped structures. The simulation conditions were the same as in panel B except that only 100 rods were used to limit cpu time.
The relative simplicity of the molecular bridging model meant that we could apply a whole-cell Monte-Carlo simulation to test it. The Monte-Carlo algorithm employed attempts to move one molecule at a time, by a small amount in a three-dimensional space. The new interaction energy experienced by the particle is then computed and compared with its old energy, and the move is subsequently accepted or rejected according to the standard Metropolis test (Allen and Tildesley, 1989; Binder and Heermann, 2002). This procedure is used to guide the system to the correct Boltzmann distribution in equilibrium. Provided that the moves are small enough and that the acceptance probability remains high, it has been shown that this Monte-Carlo dynamics corresponds well with Brownian or molecular dynamics (Whitelam and Geissler, 2007). In our simulation, DivIVA oligomers (doggy bones) are represented as diffusing spheres with a 12.5-nm radius. The bacterial cell is represented as a cylinder with a length of 4 μm and a diameter of 1 μm. The lateral membrane and the membranes at the sides (ends) of the cylinder are joined by a smooth region with variable radius of curvature Rc. The spheres experience a mutual interaction energy of Vpp, and are also attracted to the membrane via an interaction potential Vpm (see Supplementary data for details). For simplicity, the two potentials have been chosen to have constant magnitude (Epp and Epm, respectively) over their interaction ranges, which we take to be equal to rint for both interactions. The magnitude of Epp and Epm was chosen in the range 1.5–6 kBT (equivalent to 1–4 kcal/mol), which is in the range of typical weak protein–protein attractions. Figure 8B shows an example of a simulation. The simulation lasted 30 million iterations, which roughly corresponds to a 30-s time interval (see Supplementary data on how to map simulation into real time). In this specific case, the spheres were allowed to make no more than eight interactions, which is a reasonable assumption considering the large lattices that are formed by doggy bones (Stahlberg et al, 2004). According to our TEM data, the DivIVA structures seem to bridge 2–4 membranes. Therefore, we also limited the possible interactions of the spheres to two membrane contacts in this simulation. The membrane curvature at the cylinder ends has a radius of 50 nm, which is roughly the measure of curvature found at cell division sites in B. subtilis (Supplementary data). Clearly, under these simulation conditions, the spheres accumulate at the ends of the cylinder, where the membrane is most strongly curved. Thus, the cooperative binding enables the spheres to accumulate at curved membranes that have a considerably larger radius than the radius of the spheres themselves. Figure 8C shows that this accumulation depends on the measure of curvature in a highly nonlinear way. This might explain why the localisation of DivIVA-GFP at division sites of E. coli is not as distinct as that of B. subtilis. In contrast to B. subtilis, E. coli does not make a cross-wall when dividing, but division occurs by constriction of the whole lateral wall. As a consequence, the curvature of membranes at E. coli division sites is less pronounced. When DivIVA-GFP is expressed in the fission yeast, Schizosaccharomyces pombe, the protein accumulates at division sites as well (Edwards et al, 2000). Although S. pombe cells are much larger than B. subtilis cells, EM images of S. pombe show a sharp angle between the septum and the lateral wall (Sipiczki and Bozsik, 2000; Osumi et al, 2006), and we estimated a curvature with radius of about 60 nm (Supplementary data), which is close to the radius measured for B. subtilis division sites.
We have repeated the Monte-Carlo simulations with different parameters, binding conditions and number of spheres, and a more detailed description of these simulations can be found in the Supplementary data. In most cases, a comparable result was achieved indicating that the postulated molecular bridging is a relatively robust mechanism for the localisation of proteins to negatively curved membranes. As a proof of principle, we have also simulated molecular bridging in a hemispherical cell, in analogy with dividing cocci. To show the general principle of the mechanism, we allowed this time only four contact sites, and no constrains in protein–lipid interactions. As shown in Figure 8D, a clear localisation in the equatorial ring is observed that is reminiscent of the localisation of DivIVA in S. pneumoniae and S. aureus cells (Pinho and Errington, 2004; Fadda et al, 2007).
It should be emphasised that we have made several approximations in our modelling, and caution should be exercised in translating these simulation results into the cellular reality. One of the main shortcomings of simulating spherical particles is that the binding sites are not defined at fixed positions on the surface. In case of the DivIVA doggy bones, it seems that only the ends are forming the contacts. In an attempt to account for this, we modelled doggy bones as a rigid rod-shaped stack of four smaller spheres, whereby the top and bottom spheres make the interactions either with other doggy bones or with the membrane (see Supplementary data for details). Using these rod-shaped structures in the Monte-Carlo simulation resulted in a clear accumulation of protein at the edges of the cylinder (Figure 8E), and it appears that in this case, the localisation would persist to an even larger radius of curvature (Supplementary data). Although more research is required to confirm molecular bridging as a mechanism for DivIVA localisation, it will be interesting to see whether the principles can be applied to other proteins that accumulate at negatively curved membranes.
Materials and methods
General methods
A detailed description of strains, plasmid construction, protein purification and bacterial two-hybrid assay can be found in the Supplementary data.
Membrane and liposome preparations
B. subtilis cell membranes were isolated based on Henstra et al (1996). An overnight culture of B. subtilis 3310, lacking divIVA (divIVA∷Tet, minCD∷Km) (Edwards and Errington, 1997), was used to inoculate 500 ml LB medium. The culture was grown at 37°C to the end-log phase. Cells were harvested, washed in ice-cold Wash buffer (20 mM Tris–HCl pH 8, 200 mM NaCl) and stored at −80°C. Cells were resuspended in 10 ml ice-cold KPi buffer (50 mM KPi pH 7.5, 5 mM MgCl2, 0.5 mM DTT) containing PMSF, protease inhibitor (Complete Mini, Roche), Benzonase (Merck) and RNase. The cells were broken using French Press, which produces inside-out vesicles, and cell wall debris was removed with a 2 × low-spin centrifugation (SW51, 10k r.p.m., 10 min, 4°C). The supernatant was carefully collected with a pipette, and membranes were gathered following two subsequent high spins (SW51, 35k r.p.m., 1 h, 4°C). The membrane pellet was resuspended in 0.4 ml KPi buffer without DTT, aliquoted, frozen in liquid N2 and stored at −80°C.
Liposomes were prepared as described by Avanti Polar Lipids. E. coli polar lipid extract (Avanti Polar Lipids) dissolved in chloroform was desiccated in a rotary evaporator, followed by vacuum excitation (4 h). Lipids were resuspended in 50 mM Tris-acetate (pH 7.5) by vigorous vortexing (20 min), and sonication (2 × 5 min) in a bath sonicator. Aliquots were covered with Argon and stored at −80°C. For interaction studies, liposomes were mixed with the specified buffer and freeze-thawed several times followed by extrusion through a 0.1- or 0.4-μm filter (Avanti Polar Lipids).
Density gradient centrifugations and sedimentation experiments
For interaction studies with cell membrane preparations, a step gradient of 10, 20, 30, and 70% sucrose was used. After centrifugation, membranes were visible at the 30–70% sucrose interface. When liposomes were used, a step gradient of 5, 10 and 20% sucrose was used. After centrifugation, liposomes were visible at the 10–20% sucrose interface. Centrifugation was carried out for 2 h (25k r.p.m., 25°C) in a Beckman Rotor SW50.1/55 using 0.8 ml tubes (5 × 41 mm) and suitable adapters. Purified proteins (approximately 0.1–0.5 mg/ml) and membranes (1 mg/ml liposomes) were mixed in 50 μl binding buffer (20 mM Tris–HCl pH 8, 5 mM MgCl2, 200 mM KCl, 1 mM DTT, 1 mg/ml BSA) and incubated for 30 min at 30°C before loading onto sucrose gradients. The sucrose gradients were prepared using the binding buffer. After centrifugation, the gradients were fractionated in 50 μl samples that were analysed by western blotting using GFP- or DivIVA-specific antisera.
Sedimentation experiments were performed in the same binding buffer as used for density gradient experiments with the omission of BSA. Purified proteins (0.2 mg/ml) were mixed with liposomes (1 mg/ml) and after incubation for 30 min, the samples were centrifuged (Beckman TL-100 rotor, 80k r.p.m., 15 min, 30°C). Pellet and supernatant fractions were analysed by SDS–PAGE and Coomassie staining.
For the RacA interaction experiments, density gradient flotation was used. MBP-RacA was mixed with or without liposomes and DivIVA in the presence of 0.5 mg/ml BSA and 20% sucrose in buffer (20 mM Tris–HCl pH 8, 200 mM KCl, 2 mM MgCl2 and 0.2 mM DTT). Before adding MBP-RacA, the mixtures were preincubated for 15 min at room temperature. After 1 h incubation at room temperature, the solutions were loaded into 0.8 ml centrifuge tubes (5 × 41 mm). Further, 100 μl of 15 sucrose, 10 sucrose, 5 sucrose, and 0% sucrose in the buffer was loaded. Gradients were centrifuged at 25k r.p.m. at 30°C for 2 h in a Beckman Optima MAX Ultracentrifuge using an MLS 50 rotor. After centrifugation, the gradients were sampled in five fractions, which were analysed by western blotting using RacA-specific antibodies.
SPR
SPR measurements were carried out on a Biacore 2000 system (Biacore AB) using L1 chips at 25°C. Loading of liposomes on the L1 chip was carried out as described (Anderluh et al, 2005). An L1 chip was washed 3 × 1 min wash with isopropanol, 50 mM NaOH (3:2 (v/v)) at a flow rate of 30 μl/min. Liposomes (1 mg/ml prepared in the running buffer and extruded through a 0.1-μm pore filter) were loaded in 15 min at a flow rate of 2 μl/min, and excess lipid was washed 3 × 1 min with injections of 100 mM NaOH at 30 μl/min. The integrity of the lipid layer was checked using a 1-min injection of BSA (0.1 mg/ml). Protein samples were first dialysed for 3 h in the running buffer (20 mM Tris–HCl pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM EDTA) before injection. A 1-min injection of 100 mM NaOH at 30 μl/min was used to disrupt lipid–protein interactions.
Fluorescence microscopy
For fluorescence microscopy, the divIVA-gfp region of plasmid pSG1612 (Edwards et al, 2000) was isolated (XhoI × SpeI) and cloned into pSG1154 (Lewis and Marston, 1999), resulting in plasmid pDG7. This plasmid replicates in E. coli and integrates in B. subtilis. The divIVA-gfp fusion includes the divIVA promoter, which is also active in E. coli. The different deletions were made by PCR using pDG7 plasmid as a template (pDG13=DivIVa aa 1–40 -GFP, primers LH16 and GFP7, pDG15=DivIVA aa 1–60 -GFP, primers LH20 and GFP7). The point mutations were made by PCR using pDG15 a as template (pDG23=V25E, primers LH110 and LH111, pDG26=L29E, primers LH112 and LH113). Cultures were grown in PAB at 30°C. Samples were taken at exponential growth, and mounted onto microscope slides coated with a thin layer of 1.5% agarose. Membranes were stained with Nile Red or FM5-95. Images were taken with a Zeiss Axiovert 200M coupled to a CoolsnapHQ CCD camera, and using Metamorph imaging software (Universal Imaging). For deconvolution, about 15–20 Z-stacks were obtained with 0.1- to 0.2-μm intervals. Two-dimensional deconvolution (nearest neighbour) calculations were performed using Metamorph with the following settings: filter size 9, background subtraction 97% and result scale intensity of 2.
EM
Liposomes (0.1 mg/ml) with and without DivIVA (0.01 mg/ml) were incubated in the binding buffer (20 mM Tris–HCl pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM EDTA). Aliquots of liposomes with and without DivIVA were loaded onto glow-discharged formvar-coated 400-mesh copper grids and negatively stained with 1% (wt/v) aqueous uranyl acetate. Excess stain was removed by blotting with a filter paper, and the grids were air-dried. For thin section, TEM samples were fixed for 40 min on ice in a freshly prepared mixture of 1% glutaraldehyde, 1% osmium tetraoxide in 100 mM phosphate buffer (pH 7.0). The pelleted samples were washed in distilled water and sequentially treated with 1% aqueous tannic acid (1 h at 23°C) followed by 1% aqueous uranyl acetate (1 h at 4°C), dehydrated and embedded in Agar 100 resin. The thin sections were viewed in a Zeiss Omega 912 electron microscope equipped with an in column 2K Proscan CCD camera.
Supplementary Material
Supplementary Information
Acknowledgments
We thank the members of the Centre for Bacterial Cell Biology for helpful advice and discussions, and in particular Robyn Emmins for help with the bacterial two-hybrid assays. Richard Losick kindly provided RacA antibodies, and Waldemar Vollmer E. coli strains. Simon Cockell is acknowledged for his support with initial modelling attempts. This research was supported by an EMBO Long-Term Fellowship, a Wellcome Trust Research Career Development Fellowship (LWH), a grant from the UK Biotechnology and Biological Science Research Council (JE and LWH) and a grant from the Engineering and Physical Sciences Research Council (DM).
References
- Allen MP, Tildesley DJ (1989) Computer Simulation of Liquids. Oxford: Oxford University Press [Google Scholar]
- Anderluh G, Besenicar M, Kladnik A, Lakey JH, Macek P (2005) Properties of nonfused liposomes immobilized on an L1 Biacore chip and their permeabilization by a eukaryotic pore-forming toxin. Anal Biochem 344: 43–52 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 [DOI] [PubMed] [Google Scholar]
- Binder K, Heermann D (2002) Monte Carlo Simulation in Statistical Physics: An Introduction, 4th edn. Berlin: Springer-Verlag. [Google Scholar]
- Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J (2008) A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol 70: 1556–1569 [DOI] [PubMed] [Google Scholar]
- Cha JH, Stewart GC (1997) The divIVA minicell locus of Bacillus subtilis. J Bacteriol 179: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24: 905–915 [DOI] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J (2000) Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J 19: 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O (2003) Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol 185: 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda D, Santona A, D'Ulisse V, Ghelardini P, Ennas MG, Whalen MB, Massidda O (2007) Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J Bacteriol 189: 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishov I, Woldringh CL (1999) Visualization of membrane domains in Escherichia coli. Mol Microbiol 32: 1166–1172 [DOI] [PubMed] [Google Scholar]
- Flardh K (2003) Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49: 1523–1536 [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PA (2002) ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol 184: 2552–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Errington J (2003) Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J Bacteriol 185: 693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ, Lewis PJ (2003) Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol Microbiol 47: 37–48 [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Holtje JV (2001) Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41: 167–178 [DOI] [PubMed] [Google Scholar]
- Hempel AM, Wang SB, Letek M, Gil JA, Flardh K (2008) Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol 190: 7579–7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstra SA, Tolner B, ten Hoeve Duurkens RH, Konings WN, Robillard GT (1996) Cloning, expression, and isolation of the mannitol transport protein from the thermophilic bacterium Bacillus stearothermophilus. J Bacteriol 178: 5586–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J (2002) Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA 99: 6761–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34: 82–90 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2003) A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47: 345–355 [DOI] [PubMed] [Google Scholar]
- Jones MK, Anantharamaiah GM, Segrest JP (1992) Computer programs to identify and classify amphipathic alpha helical domains. J Lipid Res 33: 287–296 [PubMed] [Google Scholar]
- Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN (2008) Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154: 725–735 [DOI] [PubMed] [Google Scholar]
- Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K (2004) Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol 186: 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Débarbouille M, Dervyn E, Deuerling E et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letek M, Ordonez E, Vaquera J, Margolin W, Flardh K, Mateos LM, Gil JA (2008) DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol 190: 3283–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227: 101–110 [DOI] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F (2008) Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA 105: 3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W (2001) Spatial regulation of cytokinesis in bacteria. Curr Opin Microbiol 4: 647–652 [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J (1998) Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12: 3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W (2000) Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol 182: 1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchova K, Kutejova E, Scott DJ, Brannigan JA, Lewis RJ, Wilkinson AJ, Barak I (2002) Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology 148: 807–813 [DOI] [PubMed] [Google Scholar]
- Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K (2005) Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol 187: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi M, Konomi M, Sugawara T, Takagi T, Baba M (2006) High-pressure freezing is a powerful tool for visualization of Schizosaccharomyces pombe cells: ultra-low temperature and low-voltage scanning electron microscopy and immunoelectron microscopy. J Electron Microsc (Tokyo) 55: 75–88 [DOI] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB (2008) MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol 70: 1166–1179 [DOI] [PubMed] [Google Scholar]
- Perry SE, Edwards DH (2004) Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol Microbiol 54: 1237–1249 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55: 1722–1734 [DOI] [PubMed] [Google Scholar]
- Pinho MG, Errington J (2004) A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol Lett 240: 145–149 [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R (2009) Geometric cue for protein localization in a bacterium. Science 323: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden MD, Baier C, Ramirez-Arcos S, Liao M, Wang M, Dillon JA (2008) Identification of the coiled-coil domains of Enterococcus faecalis DivIVA that mediate oligomerization and their importance for biological function. J Biochem 144: 63–76 [DOI] [PubMed] [Google Scholar]
- Sipiczki M, Bozsik A (2000) The use of morphomutants to investigate septum formation and cell separation in Schizosaccharomyces pombe. Arch Microbiol 174: 386–392 [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Kutejova E, Muchova K, Gregorini M, Lustig A, Muller SA, Olivieri V, Engel A, Wilkinson AJ, Barak I (2004) Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol 52: 1281–1290 [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Habrukowich CL, King GF (2003) The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem 278: 40050–40056 [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF (2002) Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA 99: 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam S, Geissler PL (2007) Avoiding unphysical kinetic traps in Monte Carlo simulations of strongly attractive particles. J Chem Phys 127: 154101. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2003) RacA and the Soj–Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol 49: 1463–1475 [DOI] [PubMed] [Google Scholar]
- Xu H, Chater KF, Deng Z, Tao M (2008) A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in streptomyces. J Bacteriol 190: 4971–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information