Abstract
The processes that drive the evolution of snake venom variability, particularly the role of diet, have been a topic of intense recent research interest. Here, we test whether extensive variation in venom composition in the medically important viper genus Echis is associated with shifts in diet. Examination of stomach and hindgut contents revealed extreme variation between the major clades of Echis in the proportion of arthropod prey consumed. The toxicity (median lethal dose, LD50) of representative Echis venoms to a natural scorpion prey species was found to be strongly associated with the degree of arthropod feeding. Mapping the results onto a novel Echis phylogeny generated from nuclear and mitochondrial sequence data revealed two independent instances of coevolution of venom toxicity and diet. Unlike venom LD50, the speed with which venoms incapacitated and killed scorpions was not associated with the degree of arthropod feeding. The prey-specific venom toxicity of arthropod-feeding Echis may thus be adaptive primarily by reducing venom expenditure. Overall, our results provide strong evidence that variation in snake venom composition results from adaptive evolution driven by natural selection for different diets, and underscores the need for a multi-faceted, integrative approach to the study of the causes of venom evolution.
Keywords: snake venom, adaptation, diet, coevolution, Echis
1. Introduction
Variation in venom composition is ubiquitous among venomous snakes, occurring at all taxonomic and biological levels (Chippaux et al. 1991). Although it is generally accepted that the primary function of snake venom is to facilitate immobilization and/or digestion of prey, the extent to which adaptive processes drive the evolution of snake venom diversity has been widely debated. Several authors have supported an ‘overkill’ hypothesis of venom evolution, which postulates that, due to the apparent high toxicity of many snake venoms and the large doses injected, variation in venom composition is unlikely to be subject to natural selection for lethality to prey, and that venom diversity largely results from neutral evolutionary processes (Sasa 1999a,b; Mebs 2001). By contrast, other authors argue that snake venom composition is subject to strong natural selection, and that venom diversity results from adaptation to specific diets (e.g. Daltry et al. 1996; Wüster et al. 1999; Kordiš & Gubenšek 2000).
Well-documented instances of resistance to envenoming in prey species (Poran et al. 1987; Heatwole & Poran 1995; Biardi et al. 2006) demonstrate the potential of selection for increased venom toxicity in snakes, and the possibility that reciprocal coevolutionary ‘arm's races’ may occur between snakes and their prey. Further indication that venom composition is subject to natural selection comes from evidence that venom production is metabolically costly, representing a trade-off between the metabolic costs of venom synthesis and increasing foraging efficiency (McCue 2006). This contradicts the assumption that snakes inject many orders of magnitude more venom into prey than the lethal dose required, which is further challenged by evidence that at least some snakes ‘meter’ the amount of venom injected depending upon prey size (Hayes et al. 1995). The selective consequences of the metabolic cost of venom production are also demonstrated by an example of adaptive venom loss following an evolutionary shift to a diet of fish eggs in the sea snake Aipysurus eydouxii (Li et al. 2005).
Other studies have highlighted apparent coincident variation in venom composition and diet as evidence of adaptive venom evolution. Geographical variation in venom composition, as revealed by electrophoretic profiles or mass spectrometry, has been shown to be correlated with geographical variation in diet in some pitvipers (Daltry et al. 1996; Creer et al. 2003). At the molecular level, venom-coding genes show extremely high rates of sequence evolution and an excess of non-synonymous over synonymous substitutions, suggesting that selection drives rapid toxin diversification by favouring mutations that alter protein structure (Kordiš & Gubenšek 2000; Lynch 2007).
While compositional and molecular analyses of snake venoms provide evidence of adaptation, the functional significance of these adaptations remains unknown. Ultimately, this can only be tested by measuring the effects of venom on natural prey (Sasa 1999b; Wüster et al. 1999). Naturalistic prey models have been used to investigate the adaptive significance of ontogenetic venom variation, revealing instances of ontogenetic shifts in prey preference accompanied by corresponding shifts in prey-specific venom toxicity (Mackessy 1988; Andrade & Abe 1999; Mackessy et al. 2006). Previous studies have also compared venom toxicities to natural prey between related snake species with variable diets, and have typically found the venom of a given species to be more toxic to its preferred prey than that of congeners with different diets (Jorge da Silva & Aird 2001; Starkov et al. 2007). These results are certainly consistent with the hypothesis that natural selection for different diets has driven the evolution of interspecific venom variation. However, without knowledge of where evolutionary shifts in diet and venom composition have occurred during the evolutionary history of these species, the alternative hypothesis of phylogenetic constraint cannot be rejected: similarity in venom characteristics and diet may simply be the result of common ancestry rather than selection.
To provide a more robust test of hypotheses on the causes of variation in venom composition, approaches are required which examine venom characteristics and diet within a phylogenetic framework. This requires a group of closely related venomous snake species, some of which feed on radically different prey types with different physiology. The saw-scaled vipers (Viperidae: Echis) fulfil these criteria. This genus of small vipers is distributed throughout much of Africa north of the Equator, the Arabian Peninsula, India and Sri Lanka (Wüster et al. 1997), and represents the most medically significant group of snakes in several parts of this distribution (Warrell et al. 1977). Previous studies document considerable interspecific variation in venom composition (Schaeffer 1987), which is further supported by a lack of antivenom cross-reactivity against envenoming by different Echis species (Gillissen et al. 1994). Great variation in diet is also reported within the genus. Some species consume predominantly vertebrate prey (Tsairi & Bouskila 2004), while others prefer arthropods such as scorpions and centipedes (Gasperetti 1988; Revault 1996). Arthropods can be considered atypical viper prey (Shine et al. 1998), thus arthropod feeding most probably represents a derived condition arising at some point during the diversification of Echis. We hypothesized that this shift to physiologically distinct, well-defended and potentially dangerous arthropod prey would impose considerable selective pressure, resulting in adaptive evolution of venom composition for increased toxicity to arthropods.
We tested our hypothesis of adaptive venom evolution by integrating evidence of phylogeny, venom composition, diet and venom activity. This involved reconstructing phylogenetic relationships between the major clades of Echis and verifying that variation in venom composition occurs among them. Determining the extent of arthropod versus vertebrate feeding for each clade then allowed the inference of evolutionary shifts in diet occurring during the diversification of Echis. Finally, to investigate the functional significance of variation in Echis venom composition, the toxicity of representative venoms from each Echis clade was measured against a naturalistic arthropod prey model, the scorpion Scorpio maurus. Use of a phylogenetic framework allows an assessment of the extent to which dietary shifts are associated with adaptive venom evolution for increased toxicity to novel prey types, and a more rigorous test of the factors driving snake venom evolution than previous studies.
2. Material and methods
(a) Phylogenetic analysis
Although the intrageneric taxonomy of Echis is currently in a state of flux (Wüster et al. 1997), ongoing molecular phylogenetic work (W. Wüster & C. E. Pook 2008, unpublished data) has shown that previously described species (Cherlin 1990) form four monophyletic species complexes: (i) Echis carinatus group, including E. carinatus and Echis multisquamatus; (ii) Echis pyramidum group, including E. pyramidum, Echis leucogaster and Echis khosatzkii; (iii) Echis ocellatus group, including E. ocellatus and Echis jogeri; and (iv) Echis coloratus group, including E. coloratus and Echis omanensis. The phylogenetic relationships among the four major species groups were reconstructed using a single representative of each: E. carinatus sochureki (Sharjah, United Arab Emirates); E. pyramidum leakeyi (Baringo, Kenya); E. ocellatus (northern Cameroon); and E. coloratus (Israel). Corresponding sequences for the vipers Cerastes cerastes (Egypt) and Bitis arietans (Agadir, Morocco) were included as outgroup taxa (Wüster et al. 2008).
Total DNA was extracted from tissue (ventral scale clipping) or blood samples using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich). Parts of four mitochondrial genes and one nuclear gene were amplified using the polymerase chain reaction (PCR) and sequenced. These were cytochrome b (cytb), NADH dehydrogenase subunit 4 (ND4), 12s rRNA, 16s rRNA and the nuclear recombination activating gene 1 (RAG1). Details of PCR protocols, primers and reaction conditions are provided in the electronic supplementary material.
All protein-coding gene sequences were aligned manually. 12s and 16s rRNA sequences were aligned using ClustalW (Thompson et al. 1994), with later corrections by eye. Protein-coding genes were translated into amino acid sequences and checked for unexpected indels or stop codons to detect the possible presence of non-coding copies or pseudogenes (Zhang & Hewitt 1996).
Phylogenetic analysis was carried out using Bayesian inference (BI) methods. We first partitioned the data into eleven biologically relevant partitions: all protein-coding genes were partitioned separately into first, second and third codon positions, and 12s and 16s were also treated as separate partitions. The most appropriate model of sequence evolution for each partition under the Akaike information criterion was estimated using MrModeltest v. 2.2 (Nylander 2004). The final BI analysis, using MrBayes v. 3.1 (Ronquist & Huelsenbeck 2003), involved four parallel runs of five million generations, each with one cold and three heated chains. The number of generations prior to burn-in was determined through visual inspection of a plot of likelihood scores against generation number.
Previous analyses have suggested that E. coloratus may form the sister taxon to the remaining Echis (Lenk et al. 2001). To determine whether our data significantly rejected this phylogenetic hypothesis, we calculated the proportion of post-burn-in Bayesian trees consistent with it by filtering the trees in PAUP* v. 4b10 (Swofford 2002). The hypothesis was rejected if fewer than 5 per cent of post-burn-in trees were consistent with it.
(b) Electrophoresis of venom
To examine whether variation in venom composition exists among the major Echis species groups, we produced electrophoretic venom profiles for the same representative Echis taxa used for phylogenetic analysis, in addition to the outgroup taxa B. arietans and C. cerastes. The snakes (E. c. sochureki, United Arab Emirates; E. p. leakeyi, Kenya; E. ocellatus, Nigeria; E. coloratus, Egypt; B. arietans, Ghana; and C. cerastes, Egypt) were maintained in the herpetarium at the Liverpool School of Tropical Medicine. Venoms were extracted from at least 10 specimens of each species, pooled, lyophilized and stored at 4°C. The phosphate buffered saline-reconstituted venom samples were mixed with an equal volume of reduced protein-loading buffer (1 mg ml−1), boiled for 10 min and loaded onto a 15 per cent reduced SDS–PAGE gel and electrophoresis performed at 200 constant voltage for 57 min. The fractionated venom proteins were visualized by staining with Coomassie blue.
(c) Dietary analysis
To determine the extent and variation of arthropod feeding among the major Echis species groups, preserved museum specimens (E. carinatus group n=22; E. pyramidum group n=65; E. ocellatus group n=46; and E. coloratus group n=99) were dissected and the stomach and hindgut contents examined. Intact prey items were identified visually. Partially digested prey remnants were examined using a dissecting microscope (×10–40 magnification), and identified by the criteria shown in the electronic supplementary material. Very small arthropods or exoskeleton fragments were not recorded owing to the possibility of secondary ingestion via the gut contents of the actual prey (Creer et al. 2002). The snout–vent length (SVL) of each specimen was also measured.
Chi-squared tests were used to test for significant differences in the proportion of arthropods and vertebrates consumed between the four species groups. ANOVA was used to test for significant differences in SVL between snakes that had consumed arthropods and those that had consumed vertebrates in each species group. Analyses were performed using SPSS v. 12.
(d) Venom toxicity to scorpions
The scorpion S. maurus (Egypt) was selected as a suitable naturalistic arthropod prey species with which to compare Echis venom toxicities, as this species inhabits semi-arid and arid environments (Prendini et al. 2003) that are also typical for Echis, and has a very large distribution sympatric with several Echis species. The body mass of scorpions ranged from 0.6 to 2.7 g.
Venom toxicities were measured for each of the representative Echis taxa used in phylogenetic analysis and venom electrophoresis: E. p. leakeyi (Kenya); E. c. sochureki (Pakistan); E. ocellatus (Nigeria); and E. coloratus (Saudi Arabia). In order to infer the ancestral level of arthropod-specific venom toxicity that existed before the evolutionary shift to arthropod feeding, the venom toxicity of the vertebrate-feeding outgroup taxon B. arietans (Ghana) was also measured. Lyophilized venoms were obtained as described for venom electrophoresis, and dissolved in insect Ringer's solution to achieve specific concentrations used in toxicity tests. Venoms were administered to scorpions by injection into the lateral posterior pre-abdomen, through the pleural membrane.
Initially, median lethal doses (LD50) were determined by injecting groups of scorpions with varying venom doses and recording mortality after 24 hours. The range of doses tested and the number of scorpions in each group (n) are shown in table 1. All doses were mass (μg g−1 body weight) and volume (0.05 ml g−1 body weight) adjusted. Additionally, to test the possibility of mortality due to the injection procedure, control groups were injected with 0.05 ml g−1 (n=3) and 0.1 ml g−1 (n=3) of insect Ringer's solution only, representing, respectively, the same volume and double the volume used in venom injections. LD50 was calculated by probit analysis using Minitab v. 15. Dose was the stimulus and venom the factor. Response was in event (mortality)/trial (n) format. Weibull distribution was assumed, as this provided the best fit to the data (Pearson Χ232=19.15, p=0.692; deviance Χ232=9.41, p=0.994). The test for equal slopes indicated that the assumption of equal regression slopes among venoms was not violated (Χ42=8.09, p=0.088).
Table 1.
Scorpion mortality 24 h after venom injection. (Total group size (n) is shown in parentheses.)
| venom | dose (μg g−1) | |||||||
|---|---|---|---|---|---|---|---|---|
| 2.5 | 10 | 13 | 25 | 50 | 125 | 250 | 1000 | |
| E. p. leakeyi | 0 (5) | 2 (5) | 5 (5) | 5 (5) | 5 (5) | — | 2 (2) | 2 (2) |
| E. c. sochureki | 0 (5) | 2 (5) | 5 (5) | 5 (5) | 5 (5) | — | 2 (2) | 2 (2) |
| E. ocellatus | — | 0 (5) | — | 1 (5) | 4 (5) | — | 5 (5) | 2 (2) |
| E. coloratus | — | 0 (5) | — | — | 1 (5) | 1 (5) | 5 (5) | 2 (2) |
| B. arietans | — | 0 (5) | — | — | 0 (5) | 5 (5) | 5 (5) | 2 (2) |
As the standard LD50 is of questionable biological relevance (a snake requires a much greater probability of prey death in a much shorter duration), we also investigated the time taken for scorpions to become incapacitated (defined as inability to self-right when turned on to its back) and die (defined as complete lack of movement when touched) following a biologically realistic venom dose. We selected 3 mg g−1 as an appropriate dose, as this represents two-thirds of the average total venom yield (9 mg dry weight, R. A. Harrison 2007, personal observation) from a medium-sized E. ocellatus for the envenomation of a scorpion with a mass of 2 g. Times to incapacitation and death were recorded to the nearest minute. Five scorpions received each Echis venom (n=5 per venom) and four received B. arietans venom (n=4). ANOVA with post hoc tests (Tukey) was used to test for significant differences between venoms in the time to scorpion incapacitation and death. Analyses were performed using SPSS v. 12.
To test whether these doses of venom constitute a biologically realistic representation of natural predator–prey interactions between Echis and scorpions, we conducted feeding trials, in which captive E. c. sochureki (Sharjah, United Arab Emirates; n=2) were offered live S. maurus. Snakes were placed in a transparent enclosure and left undisturbed for a minimum of 24 hours before a scorpion was introduced. Behaviour was recorded using a video camera located outside the enclosure, and the time between scorpion envenomation and onset of feeding measured using the video camera time counter.
3. Results
(a) Phylogenetic analysis
A total of 3145 bp of sequence (cytb: 789 bp; ND4: 644 bp; 12s rRNA: 404 bp; 16s rRNA: 472 bp; and RAG1: 836 bp) was aligned. A 24 bp section of 16s and a 48 bp section of 12s were excluded due to alignment difficulties. No unexpected indels, frameshifts or non-sense codons were detected in protein-coding gene sequences. Nucleotide sequences were deposited in GenBank (accession numbers: EU852294–852305 and EU852312–852329). The final dataset contained 738 variable sites, of which 339 were parsimony informative.
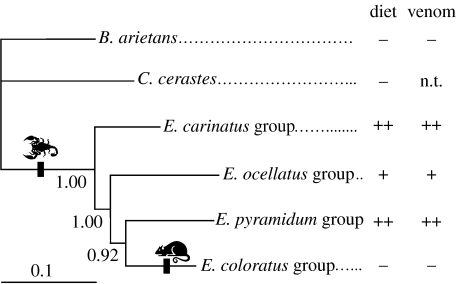
Models of sequence evolution for data partitions used in the BI analysis are shown in the electronic supplementary material. Examination of likelihood plots revealed that stationarity was reached after approximately 50 000 generations, but as a ‘safety margin’ we discarded the first million generations. The BI tree with Bayesian posterior probabilities is shown in figure 1, and yielded the topology (carinatus (ocellatus (coloratus, pyramidum))). The monophyly of the clade consisting of E. coloratus, E. pyramidum and E. ocellatus was supported by high Bayesian posterior probability values, but support for the monophyly of E. pyramidum and E. coloratus fell just short of statistical significance. Out of 32 004 post-burn-in BI trees, only 4 (0.012%) supported the position of E. coloratus as the sister taxon of the remaining Echis. We therefore consider this hypothesis rejected.
Figure 1.
Bayesian phylogeny of the major Echis species groups, inferred using a single representative taxon from each. Bayesian posterior probabilities are shown by relevant nodes. Columns to the right indicate the proportion of arthropod prey consumed (diet) and venom toxicity to scorpions (venom):++, high; +, moderate; −, low; nt, not tested. Instances of dietary shifts in prey type accompanied by coevolution of venom activity are indicated by vertical bars along branches.
(b) Electrophoresis of venom
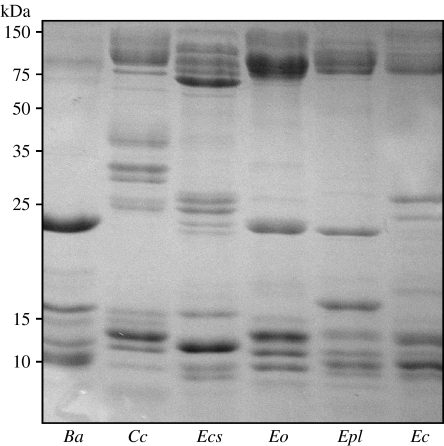
Subjecting venom samples to reduced SDS–PAGE (figure 2) revealed considerable variation in venom protein composition between representatives of each major Echis species group, which was comparable with that found between Echis and the more distantly related vipers B. arietans and C. cerastes. From this preliminary analysis, there was no obvious association between venom protein profile and diet.
Figure 2.
Venom protein profiles; venom from B. arietans (Ba), C. cerastes (Cc), E. c. sochureki (Ecs), E. ocellatus (Eo), E. p. leakeyi (Epl) and E. coloratus (Ec) were fractionated using 15% reduced SDS–PAGE.
(c) Dietary analysis
In total, 152 identifiable prey items were recovered. Chi-squared tests showed significant differences between Echis species groups in the proportion of arthropods consumed (Χ32=28.54, p<0.001). Arthropods constitute more than half the diets of the E. carinatus and E. pyramidum groups. The E. ocellatus group is less reliant on arthropod prey, and the E. coloratus group feeds almost exclusively on vertebrates (figure 3; more detailed dietary data are provided in the electronic supplementary material). In all species groups, the mean SVL of snakes that consumed arthropods was less than those that had consumed vertebrates, the difference being significant in the E. coloratus group (F=8.25, d.f.=1, p=0.007), the E. ocellatus group (F=7.60, d.f.=1, p=0.009) and the E. pyramidum group (F=9.93, d.f.=1, p=0.003), but not in the E. carinatus group (F=0.49, d.f.=1, p=0.49). However, there was extensive overlap in SVL between snakes that had consumed arthropods and those that had consumed vertebrates in all Echis species groups, indicating that no clear ontogenetic shift in diet occurs in Echis, and both vertebrates and arthropods are consumed at all life stages (see the electronic supplementary material).
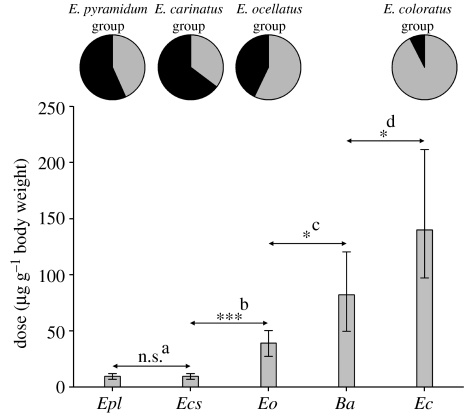
Figure 3.
Scorpion LD50 for Echis and B. arietans venoms. Species abbreviations are consistent with figure 2. Error bars show 95% confidence intervals. Pairwise statistical comparisons are shown by asterisks (*=p<0.05; ***=p<0.001; n.s., not significant) and superscripts (a: Z=0.0, p=1.0; b: Z=3.48, p<0.001; c: Z=2.47, p=0.014; d: Z=2.02, p=0.043). The pie charts above show the proportion of arthropods (black portions) and vertebrates (grey portions) consumed by each Echis species group.
(d) Venom toxicity to scorpions
Scorpion mortality for the doses tested is shown in table 1. Control experiments with insect Ringer solution only resulted in zero mortality. Probit analysis revealed that E. p. leakeyi and E. c. sochureki had the most scorpion-lethal venoms (lowest LD50), followed by, in order of diminishing toxicity; E. ocellatus, B. arietans and E. coloratus. Significant differences in LD50 were evident in all pairwise venom comparisons, except between E. p. leakeyi and E. c. sochureki (figure 3).
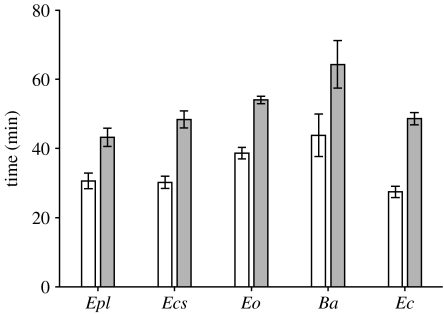
Following administration with a biologically realistic venom dose, mean times to scorpion incapacitation (27–43 min) and death (43–64 min) were longer than expected (figure 4). ANOVA revealed significant differences in both the time to incapacitation (F=5.49, d.f.=4, p=0.004) and death (F=5.80, d.f.=4, p=0.003) between the five venoms. Post hoc tests revealed significant differences between B. arietans venom and that of E. c. sochureki (incapacitation p=0.028 and death p=0.022), E. coloratus (incapacitation p=0.007 and death p=0.024) and E. p. leakeyi (incapacitation p=0.035 and death p=0.002). There were, however, no significant differences in either the mean time to scorpion incapacitation or death between any of the four Echis venoms, or between E. ocellatus venom and B. arietans venom.
Figure 4.
Mean time to scorpion incapacitation (white bars) and death (grey bars) following a biologically realistic venom dose. Species abbreviations are consistent with figure 2. Error bars show standard error.
During feeding trials, successful feeding was observed on three occasions. Snakes delivered a rapid strike to the scorpion and then quickly released it. One or two subsequent bites were delivered to the scorpion in all trials. These were spaced at variable time intervals (3–40 min between bites), but never delivered in rapid succession. Feeding did not commence until shortly after the scorpion stopped moving completely. The mean time from the initial bite to commencement of feeding was 43.1 min (s.e.=2.4 min; 3 trials; n=2 snakes), which is comparable with the time to scorpion death following a biologically realistic E. c. sochureki venom dose (figure 4), supporting our assumption that the dose selected was representative of natural predatory behaviour.
4. Discussion
The results provide strong evidence for the role of natural selection in shaping venom composition in the genus Echis. Electrophoresis of representative venoms verified the existence of considerable variation in venom protein composition between the four major Echis species groups (figure 2). Examination of stomach and hindgut contents also confirmed that significant variation occurs between species groups in the proportion of arthropod prey consumed. Measuring the effects of representative venoms to a naturalistic arthropod prey model reveals a strong association between venom toxicity (in terms of LD50) and diet: venoms from the frequent arthropod-feeding E. carinatus group and E. pyramidum group are significantly more toxic to scorpions than venoms from the occasional arthropod-feeding E. ocellatus group, which in turn is significantly more toxic than the venoms from the vertebrate-feeding E. coloratus group and the more distantly related B. arietans (figure 3). Comparing scorpion LD50 values with those reported for mice (E. p. leakeyi 0.7 μg g−1, E. c. sochureki 1.9 μg g−1, E. ocellatus 0.41 μg g−1 and E. coloratus 1.25 μg g−1—Theakston & Reid 1983) indicates that the relative lethality ranking of the Echis venoms is different for scorpions than for mice. This demonstrates that the increased scorpion toxicity associated with the venoms of arthropod-feeding Echis is prey specific, rather than simply the result of a venom that is generally more lethal. A similar association between diet and arthropod-specific venom toxicity is reported among the Eurasian vipers, Vipera (Pelias group), which also show marked interspecific variation in the amount of arthropod prey consumed (Starkov et al. 2007). Increased venom toxicity to natural prey species is also well known in vertebrate-feeding snakes (Jorge da Silva & Aird 2001; Pawlak et al. 2006; Mackessy et al. 2006).
Mapping the degree of arthropod feeding and venom lethality to scorpions onto the phylogeny of the major Echis species groups further reinforces the hypothesis of a coevolutionary relationship between venom and diet (figure 1). Both outgroup taxa are, like the majority of vipers (Mallow et al. 2003), vertebrate predators, suggesting that vertebrate feeding is the ancestral viper condition. The low scorpion toxicity of B. arietans venom (figure 3) similarly supports an ancestral condition of low arthropod venom toxicity. Within Echis, arthropod-feeding species groups are paraphyletic with respect to the vertebrate-feeding E. coloratus group, a relationship strongly supported by high Bayesian posterior probability values. The most parsimonious scenario for the evolution of arthropod feeding and venom specificity (figure 1) is thus that arthropod feeding arose at the base of the Echis radiation, and was accompanied by adaptive venom evolution for increased arthropod toxicity. Subsequently, a reversal to vertebrate feeding occurred in the E. coloratus group, whose venom characteristics reverted to the ancestral condition of low arthropod toxicity. Application of a phylogenetic approach therefore reveals not one but two independent instances of dietary shifts in prey type accompanied by coevolution of venom activity, a conclusion that could not be inferred from venom toxicity data alone. The fact that E. ocellatus is intermediate in both diet composition and scorpion lethality of its venom (figure 3) further supports the hypothesis of a coevolutionary relationship.
In contrast to venom LD50, the speed of scorpion incapacitation and death following a biologically realistic venom dose is not associated with the degree of arthropod feeding (figure 4). Indeed, the venom of vertebrate-feeding E. coloratus caused the most rapid incapacitation. This suggests that increased scorpion toxicity of the venoms of frequent arthropod-feeding Echis is unlikely to result from selection for greater speed of prey immobilization or death. Instead, as venom is metabolically expensive to produce (McCue 2006), increased venom toxicity of arthropod-feeding Echis could be adaptive in terms of metabolic energy savings, as less venom is required to achieve the same effect. Furthermore, as venom yield in snakes decreases exponentially with SVL (Mackessy 1988), increased venom toxicity may also allow juvenile Echis to produce and store sufficient venom reserves to adequately subdue their arthropod prey, which, based on murine LD50 values, requires a larger venom dose than for vertebrate prey of an equivalent size.
Another notable outcome of our investigations was the long-time periods taken for venoms to incapacitate and kill scorpions following a biologically realistic venom dose (figure 4) as well as during feeding trials. This was unexpected, particularly considering that the dose selected represents 574, 85, 34 and 21 times the scorpion LD50 for E. c. sochureki and E. p. leakeyi, E. ocellatus, B. arietans and E. coloratus venom, respectively, and a large proportion of the total venom stores of an averaged sized E. ocellatus. This provides new information on the feeding ecology of arthropod-feeding Echis. Scorpions and other arthropod prey may have to be tracked for extended periods following envenomation, increasing exposure to snake predators and the probability of prey loss; a scenario further supported by our observations that E. c. sochureki releases prey following envenomation, rather than holding it in the mouth as documented in some viper species (Wüster et al. 2005; Starkov et al. 2007).
In conclusion, this study provides strong evidence of adaptive evolution of venom composition in the genus Echis. The observation of increased lethality of venom for arthropod prey is strengthened by the phylogenetic approach, which documents two independent instances of coevolution of diet and venom. Our results also suggest that the principal driver for increased arthropod-specific lethality may not be the need for rapid killing times, but selection for venom economy. These conclusions underscore the need for a broad, integrative approach to the study of the causes and correlates of the evolution of venom composition. In particular, the use of natural prey items is essential to illuminate the interaction between venom and diet, and this interaction may be considerably more complex than suggested by the reliance on standard LD50 tests in much of the literature. The genus Echis, with its extreme variation in diet composition, is shown to be a promising model for further studies on the basis and mechanisms for adaptive evolution of venom composition in snakes.
Acknowledgements
Procedures involving animals were approved by the Bangor University Animal Ethics Committee.
We thank Damien Egan, Boaz Shacham and Tomáš Mazuch, as well as Franck Principaud and Yvon Doljanski at Latoxan, for providing tissue samples. For access to specimens for dietary analysis, we thank Colin J. McCarthy (Natural History Museum, London), Andreas Schmitz (Muséum d'Histoire Naturelle de Genève), Harold Voris (Field Museum of Natural History, Chicago), Jens Vindum (California Academy of Science, San Francisco), Wolfgang Böhme (Zoologisches Museum Alexander Koenig, Bonn), Robert Wilson (National Museum of Natural History, Washington DC), Ivan Ineich (Muséum National d'Histoire Naturelle, Paris), Natalia Ananjeva (Zoological Museum, St Petersburg) and Mark-Oliver Rödel (Würzburg University/Humboldt Museum, Berlin). For laboratory assistance and logistical support, we thank Wendy Grail, Ann Pennell, Rachel Inman and Dylan Davenport. For his expert herpetological assistance, we thank Paul Rowley. For reading of the final proof, we thank Faye Barlow. This research was funded by the Leverhulme Trust (grant number F/00 174/I, to W.W.).
Supplementary Material
PCR conditions and primer information; criteria for prey identification
Models of sequence evolution; detailed dietary data; SVL comparisons
References
- Andrade D.V., Abe A.S. Relationship of venom ontogeny and diet in Bothrops. Herpetologica. 1999;55:200–204. [Google Scholar]
- Biardi J.E., Chien D.C., Coss R.G. California ground squirrel (Spermophilus beecheyi) defences against rattlesnake venom digestive and hemostatic toxins. J. Chem. Ecol. 2006;31:2501–2518. doi: 10.1007/s10886-005-7610-1. doi:10.1007/s10886-005-7610-1 [DOI] [PubMed] [Google Scholar]
- Cherlin V.A. Taxonomic review of the snake genus Echis (Viperidae). II. An analysis of taxonomy and descriptions of new forms. Proc. Zool. Inst. Leningrad. 1990;207:193–223. [Google Scholar]
- Chippaux J.P., Williams V., White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. doi:10.1016/0041-0101(91)90116-9 [DOI] [PubMed] [Google Scholar]
- Creer S., Chou W.-H., Malhotra A., Thorpe R.S. Offshore insular variation in the diet of the Taiwanese bamboo viper Trimeresurus stejnegeri (Schmidt) Zool. Sci. 2002;19:907–931. doi: 10.2108/zsj.19.907. doi:10.2108/zsj.19.907 [DOI] [PubMed] [Google Scholar]
- Creer S., Malhotra A., Thorpe R.S., Stocklin R., Favreau P., Chou W.-H. Genetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing. J. Mol. Evol. 2003;56:317–329. doi: 10.1007/s00239-002-2403-4. doi:10.1007/s00239-002-2403-4 [DOI] [PubMed] [Google Scholar]
- Daltry J.C., Wüster W., Thorpe R.S. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. doi:10.1038/379537a0 [DOI] [PubMed] [Google Scholar]
- Gasperetti J. The snakes of Arabia. Fauna Saudi Arabia. 1988;9:169–450. [Google Scholar]
- Gillissen A., Theakston D.J., Barth J., May B., Krieg M., Warrell D.A. Neurotoxicity, haemostatic disturbances and haemolytic anaemia after a bite by a Tunisian saw-scaled or carpet viper (Echis ‘pyramidum’ complex): failure of antivenom treatment. Toxicon. 1994;32:937–944. doi: 10.1016/0041-0101(94)90372-7. doi:10.1016/0041-0101(94)90372-7 [DOI] [PubMed] [Google Scholar]
- Hayes W.K., Lavin-Murcio P., Kardong K.V. Northern Pacific rattlesnakes (Crotalus viridis oreganus) meter venom when feeding on prey of different sizes. Copeia. 1995;1995:337–343. doi:10.2307/1446896 [Google Scholar]
- Heatwole H., Poran N.S. Resistances of sympatric and allopatric eels to sea snake venoms. Copeia. 1995;1995:136–147. doi:10.2307/1446808 [Google Scholar]
- Jorge da Silva N., Jr, Aird S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. 2001;128:425–456. doi: 10.1016/s1532-0456(00)00215-5. doi:10.1016/S1096-4959(00)00347-X [DOI] [PubMed] [Google Scholar]
- Kordiš D., Gubenšek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261:43–52. doi: 10.1016/s0378-1119(00)00490-x. doi:10.1016/S0378-1119(00)00490-X [DOI] [PubMed] [Google Scholar]
- Lenk P., Kalyabina S., Wink M., Joger U. Evolutionary relationships among the true vipers (Reptilia: Viperidae) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2001;19:94–104. doi: 10.1006/mpev.2001.0912. doi:10.1006/mpev.2001.0912 [DOI] [PubMed] [Google Scholar]
- Li M., Fry B.G., Manjunatha Kini R. Eggs-only diet: its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii) J. Mol. Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. doi:10.1007/s00239-004-0138-0 [DOI] [PubMed] [Google Scholar]
- Lynch V.J. Inventing an arsenal: adaptive evolution and neofunctionalisation of venom phospholipase A2 genes. BMC Evol. Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. doi:10.1186/1471-2148-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy S.P. Venom ontogeny in the Pacific rattlesnakes Crotalus viridis helleri and Crotalus v. oreganus. Copeia. 1988;1988:92–101. doi:10.2307/1445927 [Google Scholar]
- Mackessy S.P., Sixberry N.M., Heyborne W.H., Fritts T. Venom of the brown tree snake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon. 2006;47:537–548. doi: 10.1016/j.toxicon.2006.01.007. doi:10.1016/j.toxicon.2006.01.007 [DOI] [PubMed] [Google Scholar]
- Mallow D., Ludwig D., Nilson G. Krieger; Malabar, FL: 2003. True vipers: natural history and toxinology of old world vipers. [Google Scholar]
- Mebs D. Toxicity in animals. Trends in evolution? Toxicon. 2001;39:87–96. doi: 10.1016/s0041-0101(00)00155-0. doi:10.1016/S0041-0101(00)00155-0 [DOI] [PubMed] [Google Scholar]
- McCue M.D. Cost of producing venom in three North American pitviper species. Copeia. 2006;2006:818–825. doi:10.1643/0045-8511(2006)6[818:COPVIT]2.0.CO;2 [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University. Program distributed by the author; Uppsala, Sweden: 2004. MrModeltest v. 2. [Google Scholar]
- Pawlak J., et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J. Biol. Chem. 2006;281:29 030–29 041. doi: 10.1074/jbc.M605850200. doi:10.1074/jbc.M605850200 [DOI] [PubMed] [Google Scholar]
- Poran N.S., Coss R.G., Benjamini E. Resistance of California ground squirrels (Spermophilus beecheyi) to the venom of the northern Pacific rattlesnake (Crotalus viridis oreganus): a study of adaptive variation. Toxicon. 1987;25:767–777. doi: 10.1016/0041-0101(87)90127-9. doi:10.1016/0041-0101(87)90127-9 [DOI] [PubMed] [Google Scholar]
- Prendini L., Crowe T.M., Wheeler W.C. Systematics and biogeography of the family Scorpionidae (Chelicerata: Scorpiones) with a discussion on phylogenetic methods. Invert. Systemat. 2003;17:185–259. doi:10.1071/IS02016 [Google Scholar]
- Revault P. Ecology of Echis ocellatus and peri-urban bites in Ouagadougou. Toxicon. 1996;34:144. doi:10.1016/0041-0101(96)83655-5 [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sasa M. Diet and snake venom evolution: can local selection alone explain intraspecifc venom variation? Toxicon. 1999a;37:249–252. doi: 10.1016/s0041-0101(98)00121-4. doi:10.1016/S0041-0101(98)00121-4 [DOI] [PubMed] [Google Scholar]
- Sasa M. Reply. Toxicon. 1999b;37:259–260. doi: 10.1016/s0041-0101(98)00121-4. doi:10.1016/S0041-0101(98)00123-8 [DOI] [PubMed] [Google Scholar]
- Schaeffer R.C., Jr Heterogeneity of Echis venoms from different sources. Toxicon. 1987;25:1343–1346. doi: 10.1016/0041-0101(87)90012-2. doi:10.1016/0041-0101(87)90012-2 [DOI] [PubMed] [Google Scholar]
- Shine R., Branch W.R., Harlow P.S., Webb J.K. Reproductive biology and food habits of horned adders, Bitis caudalis (Viperidae), from southern Africa. Copeia. 1998;1998:391–401. doi:10.2307/1447433 [Google Scholar]
- Starkov V.G., Osipov A.V., Utkin Y.N. Toxicity of venoms from vipers of Pelias group to crickets Gryllus assimilis and its relation to snake entomophagy. Toxicon. 2007;49:995–1001. doi: 10.1016/j.toxicon.2007.01.010. doi:10.1016/j.toxicon.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 2002. PAUP*—phylogenetic analysis using parsimony (*and other methods) Beta version 4.0b10. [Google Scholar]
- Theakston R.D.G., Reid H.A. Development of simple standard assay procedures for the characterization of snake venoms. Bull. World Health Organ. 1983;61:949–956. [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsairi H., Bouskila A. Ambush site selection of a desert snake (Echis coloratus) at an oasis. Herpetologica. 2004;60:13–23. doi:10.1655/20-47 [Google Scholar]
- Warrell D.A., Davidson N.McD, Greenwood B.M., Ormerod L.D., Pope H.M., Watkins B.J., Prentice C.R.M. Poisoning by bites of the saw-scaled or carpet viper (Echis carinatus) in Nigeria. Q. J. Med. 1977;46:33–62. [PubMed] [Google Scholar]
- Wüster W., Golay P., Warrell D.A. Synopsis of recent developments in venomous snake systematics. Toxicon. 1997;35:319–340. doi: 10.1016/s0041-0101(96)00152-3. doi:10.1016/S0041-0101(96)00152-3 [DOI] [PubMed] [Google Scholar]
- Wüster W., Daltry J.C., Thorpe R.S. Can diet explain intraspecific venom variation? Reply to Sasa. Toxicon. 1999;37:253–258. doi: 10.1016/s0041-0101(98)00121-4. doi:10.1016/S0041-0101(98)00248-7 [DOI] [PubMed] [Google Scholar]
- Wüster W., Duarte M.R., Salomão M.D.G. Morphological correlates of incipient arboreality and ornithophagy in island pitvipers, and the phylogenetic position of Bothrops insularis. J. Zool. Lond. 2005;266:1–10. doi:10.1017/S0952836904006247 [Google Scholar]
- Wüster W., Peppin L., Pook C.E., Walker D.E. A nesting of vipers: phylogeny, historical biogeography and patterns of diversification of the Viperidae (Squamata: Serpentes) Mol. Phylogenet. Evol. 2008;49:445–459. doi: 10.1016/j.ympev.2008.08.019. doi:10.1016/j.ympev.2008.08.019 [DOI] [PubMed] [Google Scholar]
- Zhang D.-X., Hewit G.M. Nuclear integrations: challenges for mitochondrial DNA markers. Trends Ecol. Evol. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. doi:10.1016/0169-5347(96)10031-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR conditions and primer information; criteria for prey identification
Models of sequence evolution; detailed dietary data; SVL comparisons