Abstract
Mitochondria are a vital component of eukaryotic cells with functions that extend beyond energy production to include metabolism, signaling, cell growth, and apoptosis. Their dysfunction is implicated in a large number of metabolic, degenerative, and age-related human diseases. Therefore, it is important to characterize and understand the mitochondrion. Many experiments have attempted to define the mitochondrial proteome, resulting in large and complex data sets that are difficult to analyze. To address this, we developed a new public resource for the storage and investigation of this mitochondrial proteomics data, called MitoMiner, that uses a model to describe the proteomics data and associated biological information. The proteomics data of 33 publications from both mass spectrometry and green fluorescent protein tagging experiments were imported and integrated with protein annotation from UniProt and genome projects, metabolic pathway data from Kyoto Encyclopedia of Genes and Genomes, homology relationships from HomoloGene, and disease information from Online Mendelian Inheritance in Man. We demonstrate the strengths of MitoMiner by investigating these data sets and show that the number of different mitochondrial proteins that have been reported is about 3700, although the number of proteins common to both animals and yeast is about 1400, and membrane proteins appear to be underrepresented. Furthermore analysis indicated that enzymes of some cytosolic metabolic pathways are regularly detected in mitochondrial proteomics experiments, suggesting that they are associated with the outside of the outer mitochondrial membrane. The data and advanced capabilities of MitoMiner provide a framework for further mitochondrial analysis and future systems level modeling of mitochondrial physiology.
Mitochondria have a varied and critical role in many aspects of eukaryotic metabolism and are implicated in a large number of metabolic, degenerative, and age-related human diseases, including cancer and aging itself (1–4). About 1500 different proteins are estimated to be present in the mammalian mitochondrion (5), and many of these proteins are tissue and development state-specific (6), but despite intense interest in this organelle, the mitochondrial proteome has yet to be fully defined and characterized. Efforts to identify mitochondrial proteins and their post-translational modifications (7, 8) from proteomics studies of purified mitochondrial organelles to in-depth analyses of protein complexes have resulted in the publication of various data sets. The number, size, and complexity of these data sets coupled with a lack of common standards for proteomics data are a major challenge to their use and integration with resources such as the public protein databases. However, understanding the mitochondrial proteome and modeling mitochondrial physiology and molecular pathology at a systems level needs a fully defined and searchable catalog of mitochondrial proteins that is cross-referenced with relevant data.
Ten Web-accessible resources are available currently that store data on the mitochondrial proteome (Table I). Among these, there is a large variation in the number of data sets included, the way the data are stored, and the sophistication of the query interface. Each resource has its own strengths and weaknesses, but some limitations are common. First, many do not appear to be actively maintained. Although their experimental data remains valid, it has been integrated with information from public databases that is subject to revision, which undermines confidence in the resource. This emphasizes that even small resources can become difficult to maintain without careful design. Second, many resources are limited to a single species or have no protein homology data, which hinders cross-species comparisons and using orthology to annotate related proteins. Third, many resources do not cite experimental references for individual proteins. Yet provenance is needed to assess whether a protein has been identified correctly as mitochondrial. Fourth, the sophistication of the query interfaces varies considerably. For some, the data are presented as a text file with queries limited to a single identifier, whereas others use relational databases, which allow greater flexibility in the number of searchable fields as well as to constrain attributes. A few resources have query interfaces with multiple options and constraints that are combined to build complex queries. However, their flexibility and ease of use could be improved.
Table I.
Species and evidence types cataloged in public databases reporting the mitochondrial localization of proteins
HMPDb, human mitochondrial protein database; AMPDb, Arabidopsis mitochondrial protein database; AMPP, Arabidopsis mitochondrial proteome project; ORMD, organelle map database; YMP, yeast mitochondrial proteome database; YDPM, yeast deletion project and proteomics of mitochondria database.
| Database | Speciesa
|
Evidenceb | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hs | Mm | Rn | Dm | Ce | Nc | Sc | At | |||
| MitoP2 | + | + | − | − | − | + | + | − | M, G, A | 61 |
| MiGenes | + | + | + | + | + | − | + | − | A | 62 |
| MitoResc | + | + | + | + | + | − | − | − | A | 63 |
| MitoProteome | + | − | − | − | − | − | − | − | M, A | 64 |
| HMPDb | + | − | − | − | − | − | − | − | A | |
| AMPDb | − | − | − | − | − | − | − | + | M, A | 65 |
| AMPP | − | − | − | − | − | − | − | + | M | 66 |
| ORMD | − | + | − | − | − | − | − | − | M, G | 29 |
| YMP | − | − | − | − | − | − | + | − | M | 38 |
| YDPM | − | − | − | − | − | − | + | − | M | 40 |
| MitoMiner | + | + | + | + | + | − | + | − | M, G, A | |
Species: Hs, H. sapiens; Mm, M. musculus; Rn, R. norvegicus; Dm, D. melanogaster; Ce, Caenorhabditis elegans; Nc, Neurospora crassa; Sc, S. cerevisiae; and At, A. thaliana.
Evidence type reported for mitochondrial protein localization: identification from mass spectrometry of purified mitochondria (M), localization from GFP tagging (G), or curated annotation from public databases and literature (A).
MitoRes includes metazoan species from UniProt.
Given the limitations of the other resources, we developed a new public resource for the storage and analysis of data about the mitochondrial proteome, called MitoMiner. The foundation of this resource is a model that describes cellular localization by GFP1 tagging and mass spectrometry of purified organelles as well as associated biological information and that formalizes the relationship among these different data. In developing MitoMiner, we addressed the four major limitations common to other resources. First, to ease the long term maintenance and continuity of the MitoMiner infrastructure beyond the original developers, we built it using the InterMine data warehouse (9) rather than develop a bespoke system. InterMine is easier to maintain by being an open source system with documentation, tutorials, and an active user and development community. To ease maintenance of the data, the underlying data sources in MitoMiner can be updated with minimal manual intervention by using automated Perl scripts, and we aim for the resource to be updated every 4–6 months. Furthermore new types of data sources can be added by extending the data model and then using InterMine to generate a new relational database schema. Data files in an XML format that is compatible with the new schema can then be easily loaded. Second, the model is not species-specific, and MitoMiner currently includes data sets from six species. Furthermore by incorporating protein orthology in the model it is possible to compare data among these species. Third, with regard to data provenance, MitoMiner records all the evidence for the classification of each individual protein as mitochondrial. This creates a comprehensive provenance for each protein, and a user can evaluate the evidence for the cellular localization of a protein and use this as a constraint in queries. Fourth, InterMine provides a user-friendly query interface for simple data browsing and querying as well as powerful and flexible methods to facilitate complex analyses incorporating multiple resources and search constraints.
We demonstrate the advantages of defining a data model and the variety of data imported in MitoMiner by using the flexible query interface of the InterMine system to report (i) the annual growth in the number of studies and the mitochondrial proteins they identify, (ii) the number of proteins (of a particular species) that are annotated as mitochondrial or have experimental evidence of mitochondrial localization, (iii) the evidence for the mitochondrial localization of proteins in metabolic pathways, and (iv) the union, intersection, and subtraction of mitochondrial proteins among data sets from different studies or organisms. When all the mitochondrial data currently loaded were considered, about 3700 different proteins have been reported as mitochondrial, and about 1400 proteins are common to yeast and animals. Combining the data from multiple studies showed that the identification of transmembrane proteins remains difficult and that these proteins are likely to be underrepresented in the data. Furthermore some cytosolic proteins, such as those of glycolysis, may be co-localized with the mitochondrion through interactions with the outer mitochondrial membrane. The analyses also highlighted known differences in the mitochondrial physiology of organisms, such as fermentation in yeast and apoptosis in animals.
EXPERIMENTAL PROCEDURES
Database Architecture of MitoMiner—
MitoMiner was built using the InterMine open source data warehouse system (9), and version 11.0 was installed and configured. The functionality of InterMine relies upon an object model that describes each data type, its attributes, and the relationships among these different data types, which are defined by the use of shared identifiers. The core object model of InterMine includes definitions of genes, proteins, publications, and the Gene Ontology (10). This object model was extended to incorporate data types and attributes for describing cellular localization, protein homology, metabolic pathways, genetic phenotypes, and post-translational modifications as well as GFP targeting and mass spectrometry data. The MitoMiner object model was not normalized as it was designed for optimal query performance and ease of navigation in the InterMine Query Builder. The relational database schema of MitoMiner was generated automatically from the object model by the InterMine system.
Public Sources of Data Used in MitoMiner—
MitoMiner was populated with data downloaded from the Web sites of several public resources. To allow the cross-referencing and integration of data, protein identifiers in all data sets were unified to UniProt (11) accession numbers by using the on-line conversion tools of the Mouse Genome Informatics (MGI) (12) for proteins from Mus musculus and the Protein Information Resource (PIR) ID program (13) for other species. In many cases a protein was mapped to more than one UniProt identifier because when using these programs separate entries for fragments, isoforms, and duplicates can be associated with the original identifier.
The literature was searched with PubMed for publications that reported large scale data sets on the mitochondrial localization of proteins. Each data set of these publications was downloaded and imported into Microsoft Excel. Recorded from each publication were the type of experiment, tissues or cell lines from which proteins had been isolated, and the PubMed identifier. Recorded for each protein of the mass spectrometry data sets were, where available, the original protein identifier, subcellular location, sequence of identified peptides, sequence coverage, and the experimental techniques that had been used for the purification, separation, and identification of the protein. If the original protein identifier could not be mapped to a UniProt primary accession number by PIR ID or MGI, then the protein was compared with proteins in UniProt by using BLASTP (14). If there was a significant match, then the UniProt primary accession number was assigned to the protein. Those proteins without a significant match were discarded. By using the PIR ID and the MGI identifier conversion tools, the evidence of mitochondrial localization for a protein was linked to many of the UniProt entries representing it. Identifiers of proteins encoded in the mitochondrial genome of organisms were taken from the Organelle database of the European Molecular Biology Laboratory-European Bioinformatics Institute and used to annotate the appropriate proteins in MitoMiner.
The source of protein sequences, related features, and annotation was UniProt (11). All UniProt entries were downloaded for the six species with mitochondrial localization data sets. The literature citations in each UniProt entry were retrieved from PubMed by using an InterMine parser. Additional Gene Ontology annotation on the biological process, metabolic function, and cellular component of each protein was taken from UniProt (15) and individual genome projects of M. musculus (12), Rattus norvegicus (16), Drosophila melanogaster (17), and Saccharomyces cerevisiae (18).
Finally lists of human genes and the descriptions of their associated disease phenotypes were taken from OMIM (19), the definitions of groups of homologous proteins were taken from HomoloGene (20), and data on the reactions, enzymes, and compounds of metabolic pathways were taken from KEGG (21). The EC numbers of proteins in UniProt were used to define the cross-reference between proteins and metabolic pathways.
Import of Data into MitoMiner—
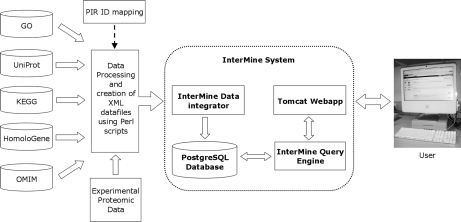
The data files for UniProt and Gene Ontology were loaded into MitoMiner by using InterMine parsers. The other data sources were converted into XML data files compatible with the MitoMiner object model by using Perl scripts that use BioPerl (22) modules, and then these were loaded into MitoMiner. These scripts were designed to allow the sources to be updated quickly and with minimal manual intervention. A simplified data flow for MitoMiner is shown in Fig. 1.
Fig. 1.
The process of extracting data from external public resources for loading and integration into MitoMiner. Data were downloaded from public sources, and the identifiers among the different sources were mapped by using either the MGI or PIR identifier mapping tools. XML-formatted data and configuration files were created from the data sources by using Perl scripts. The XML files were loaded into an underlying PostgreSQL database by the InterMine system. InterMine gives access to the data through a configurable Web application (Webapp) by using Apache Tomcat. GO, Gene Ontology.
Data Queries and Analysis in MitoMiner—
InterMine provides access to the data by using Apache Tomcat to create a configurable Web interface. This interface allows sophisticated cross-resource queries to be created using the integral Query Builder that are executed using the InterMine query engine. With the exception of analyses involving BLAST searches and DAVID functional classifications (23), the analyses reported in the results were done using queries written with the Query Builder.
The UniProt database contains redundancy as the same protein can be represented by multiple entries. Therefore the number of UniProt entries reported as mitochondrial in MitoMiner is not the same as the number of mitochondrial proteins. This redundancy was reduced by incorporating HomoloGene into MitoMiner and using it to cluster duplicate entries. However, it should be noted that HomoloGene does cluster some highly similar paralogs. The number of HomoloGene clusters was given for analyses that reported the number of proteins, unless stated otherwise, for example, when the number of proteins with evidence for mitochondrial localization was evaluated. To prevent double counting, proteins were excluded from analyses if they were not members of a HomoloGene cluster as many of these were fragments that were of insufficient size to have been clustered with their corresponding full-length counterparts. HomoloGene was also used in MitoMiner to identify orthologous proteins among different species. As HomoloGene appeared to be too stringent in its criteria for homology for some analyses, more distant orthologs were identified by using BLASTP with an expect value cutoff of 10−35. The BLAST searches were done on lists of proteins exported in FASTA format from MitoMiner.
Customization and Deployment of MitoMiner—
Queries considered to meet the most common requirements of users were written by using the integral Query Builder tool of InterMine. These template queries were made available on the relevant data category Web pages, as well as together on a single searchable Web page. The user interface of the InterMine Web application was customized, and the service was deployed.
Functional Analysis and Classification of Proteins—
To determine which Gene Ontology annotation terms were significantly overrepresented (p < 0.001) in lists of proteins compared with a background population, the DAVID Functional Annotation Clustering tool (23) was used; it uses a modified version of Fisher exact p value. The DAVID analyses were done using lists of UniProt identifiers exported from MitoMiner. If the list contained identifiers from more than one species, then the identifiers from each species were analyzed separately.
RESULTS
The User Interface of MitoMiner—
MitoMiner is publicly accessible. For ease of navigation in the Web interface, the data in MitoMiner were divided into separate data categories, and these are available from the MitoMiner home page. The data categories are mass spectrometry data, GFP tagging data, homology information (from HomoloGene), protein annotation (from UniProt and others), metabolic pathways (from KEGG), proteomics publications (from PubMed), and genetic phenotypes and disease (from OMIM). The data categories organize and provide background information on their source, access to bulk data sets, relevant template queries, and pertinent starting points for the Query Builder. For example, the protein data category page (Fig. 2) provides (i) what protein annotation is available and from where it was taken, (ii) the option to download all proteins that have experimental evidence of mitochondrial localization, and (iii) template queries for the most common searches with regard to protein annotation, such as show all proteins of a particular species that have experimental evidence of mitochondrial localization. Report pages specify the information in the database related to entries of a data category and provide cross-references to the relevant entries in external public resources. For example, the report page of a protein (Fig. 3) lists attributes of the protein and a link to the entry at the UniProt Web site as well as tabulating and cross-referencing the available data on GFP tagging, mass spectrometry, publications, tissue distribution, orthology in HomoloGene, functional annotation in the Gene Ontology, and phenotypic information in OMIM.
Fig. 2.
The Web page of the data category for proteins. This page describes the data (top left) and provides bulk download options (top right) and template queries that perform common searches (bottom).
Fig. 3.
The protein report page for the oxysterol-binding protein homolog 7 (Osh7p) of S. cerevisiae (UniProt accession number P38755). This page shows properties of the protein, cross-references to other data including the UniProt entry, and the evidence for its mitochondrial localization from GFP tagging and mass spectrometry. Osh7p is not annotated as mitochondrial in UniProt, the Saccharomyces Genome Database, or the Gene Ontology.
InterMine provides a Quick Search option, and this is the simplest way to query MitoMiner on an identifier or description in UniProt, KEGG, or OMIM. A search returns a list of entries that match the search term from which individual report pages can be selected. The Quick Search text box is available from the main menu bar, which is present at the top of all pages of MitoMiner, and wild cards can be used to broaden the search.
Advanced querying of data in MitoMiner is provided by the InterMine Query Builder. Exceptionally flexible queries can be created by using Boolean logic to combine constraints on the attributes of any data type defined in the model. In addition, the Query Builder specifies the data fields that are shown in the results table and in what order. Any identifiers in the results table, such as a UniProt accession number, are linked to the report page for that entry, and the results can be exported in a variety of formats, including Microsoft Excel. For example, the query “show all proteins that are present in fatty acid metabolism (KEGG pathway 00071) in human and that have either mass spectrometry or GFP experimental evidence for mitochondrial localization” was built in the Query Builder starting from the data category of KEGG metabolic pathways and incorporating the constraint for mitochondrial localization of proteins via the EC cross-reference (Fig. 4). To execute the query, the InterMine system coordinated the required integration of data from UniProt, experimental proteomics, and metabolic pathways. The results confirmed that many human proteins of fatty acid metabolism, as defined in KEGG, have experimental evidence of mitochondrial localization (Fig. 5).
Fig. 4.
The Web page of the Query Builder for building bespoke queries. The Web page has three components: the model browser (top left) from which data classes and attributes of the object model are selected for inclusion in the query; the data classes included in the query, the constraints on their attributes, and the Boolean logic used to combine them (top right); and the data columns to display and sort the output of the results (bottom). The query displayed is to “show all proteins that are present in KEGG pathway 00071 (fatty acid metabolism) in H. sapiens and have evidence of mitochondrial localization by either mass spectrometry or GFP experiments.”
Fig. 5.
An example of a Web page for the results for a query. The query was “show all proteins that are present in KEGG pathway 00071 (fatty acid metabolism) in H. sapiens and have evidence of mitochondrial localization by either mass spectrometry or GFP tagging experiments.” Data in columns can be sorted or summarized, the columns can be moved or hidden, identifiers can be saved to a list, and the results can be exported in different formats.
To fulfill the most common user queries of data in MitoMiner, the Query Builder was used to create template queries. These are accessed from either the MitoMiner home page or data category pages (Fig. 2). A template query includes constraints on one or more attributes of the data model, such as specifying a particular organism or metabolic pathway (Fig. 6). To make this straightforward for a user, example attributes together with descriptions are provided. The collection of template queries can be searched using the Quick Search by changing the selection menu from identifiers to templates. Users can further modify template queries by using the Query Builder, and this provides a good introduction to using the Query Builder facility.
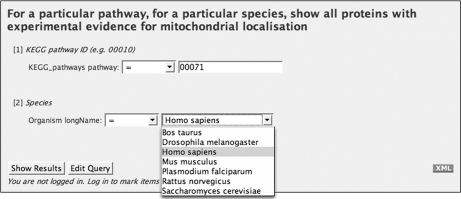
Fig. 6.
A Web page for a predefined template query. The query is show all proteins that are present in KEGG pathway 00071 (fatty acid metabolism) in H. sapiens and have evidence of mitochondrial localization by either mass spectrometry or GFP experiments. Before the query is run, the user specifies the KEGG pathway by its identifier and picks the desired organism from a drop-down box.
Two additional features of InterMine are accessed from the menu bar: Lists and MyMine. The List feature allows a list of objects, such as UniProt identifiers, to be uploaded and then used as a constraint in a compatible template query or in the Query Builder. Several public lists were created: the proteins in the major respiratory complexes of oxidative phosphorylation and the MitoCarta lists of mitochondrial proteins for Homo sapiens and M. musculus (6). The MyMine feature provides a personal account, with a username and password, that stores queries and results so that they can be accessed, exported, or modified later. A set of results saved to the InterMine system can be combined, intersected, or subtracted from any other saved result set of the same type to generate a list that is the product of several different queries.
Study of the Mitochondrial Proteome by Using MitoMiner—
We identified 33 publications reporting large scale data sets on the mitochondrial localization of proteins. Thirty publications described proteins determined from mass spectrometry of purified mitochondria cell fractions (8, 24–49), three publications had used GFP tagging (50–52), and three had used both techniques (6, 53, 54). These publications included 13 data sets from H. sapiens, eight from M. musculus, eight from S. cerevisiae, four from R. norvegicus, one from Bos taurus, and one from D. melanogaster. The data from all these publications were imported into MitoMiner.
To assess progress in determining the mitochondrial proteome, we used MitoMiner to find the annual growth in data and publications on the mitochondrial localization of proteins from the 33 publications. The increase in the number of proteins (evaluated as HomoloGene clusters) reported in the publications closely matched the growth in the number of publications (Fig. 7). By 2008, the total number of proteins reported across all species was 3672. In 2008, the increase in the number of mitochondrial proteins was 220, although Pagliarini et al. (6) had published a study that identified about 1100 mitochondrial proteins. The proportion of proteins with experimental evidence of mitochondrial localization that had the “transmembrane” keyword in their UniProt entry increased from 18% in 2002 to 21% in 2008. Of the 3672 proteins, 1506 (41%) had been annotated as mitochondrial by either the Gene Ontology or UniProt, 1326 (36%) had not been annotated as mitochondrial but had been annotated in another subcellular location, and 840 (23%) had no annotation for subcellular localization.
Fig. 7.
The cumulative increase in the number of (i) proteins reported as mitochondrial (gray bars) and (ii) publications (black line) over the last 10 years using the 33 publications on mitochondrial localization included in MitoMiner. Protein redundancy was removed by using HomoloGene to merge orthologs and duplicate proteins. The number of transmembrane proteins (dark gray) as annotated in UniProt that have been found is about 20% of the total (light gray).
We used the capabilities of MitoMiner to investigate the similarities and differences of two of the largest and most recent mass spectrometry data sets: those of Pagliarini et al. (6) and Kislinger et al. (35). Applying the HomoloGene clustering, 1746 different proteins were identified in the Kislinger et al. (35) publication, and 905 were identified in the Pagliarini et al. (6) study. The number of transmembrane proteins was 323 (18.5%) in the Kislinger et al. (35) data set, and the number of transmembrane proteins was 204 (22.5%) in the Pagliarini et al. (6) data set. The overlap between these two data sets was 392 proteins.
To determine whether some proteins were identified more often than others by mass spectrometry, we queried MitoMiner to determine the number of publications in which proteins had been reported. The distribution showed scale-free characteristics with approximately half of the proteins identified in one publication and about 30% reported in three or more publications (Fig. 8). Ninety-five proteins had been reported as mitochondrial in four or more mass spectrometry publications but had not been reported as mitochondrial in either UniProt or their respective genome database. For example, the oxysterol-binding protein homolog 7 (Osh7p) from S. cerevisiae (UniProt accession number P38755) had been identified as localizing to the mitochondrion in four mass spectrometry publications (39, 40, 42, 46) as well as by GFP tagging (50) (Fig. 3).
Fig. 8.
The frequency distribution (gray bars) and cumulative frequency distribution (black line) for the number of publications reporting a protein as mitochondrial by mass spectrometry. Protein redundancy was removed by using HomoloGene to combine orthologs and duplicate proteins. Proteins that have been identified in 10 or more studies are grouped as “10+.”
As the techniques used in identifying mitochondrial proteins can be sensitive to the properties of the proteins such as hydrophobicity, we used MitoMiner to investigate in how many publications the subunits of well known mitochondrial proteins had been identified by GFP tagging or mass spectrometry. For the subunits of the mitochondrial F-type ATP synthase in M. musculus and their orthologs, the subunits of the F1 subcomplex generally had been identified in fewer studies than those of the Fo subcomplex (Table II). In particular the transmembrane subunits were represented poorly. For example in M. musculus, the hydrophobic a and c subunits had been identified in a single mass spectrometry study, whereas the hydrophilic catalytic α and β subunits had been identified by GFP tagging and in many mass spectrometry studies. Among the 46 mammalian members of the mitochondrial transporter family (55), all except eight had been reported by mass spectrometry in at least one publication, although the majority had been reported only by Pagliarini et al. (6). The proteins without evidence from a mass spectrometry study were uncoupling protein 2, mitoferrin 1, Graves disease carrier protein, glutamate carrier 2, SLC25A38, SLC25A39, SLC25A41, and SLC25A43.
Table II.
Experimental evidence for the mitochondrial localization of the subunits of the FoF1 ATP synthase in M. musculus and in other species
| Subunit | UniProt ID | GFP tagging study
|
No. of mass spectrometry publications
|
No. of TM helicesb | ||
|---|---|---|---|---|---|---|
| M. musculus | Other speciesa | M. musculus | Other speciesa | |||
| a | P00848 | − | − | 1 | 8 | 6 |
| b | Q9CQQ7 | − | + | 5 | 13 | 0 |
| cc | P48202 | − | − | 1 | 2 | 2 |
| cd | P56383 | − | − | 0 | 0 | 2 |
| ce | P56384 | − | − | 0 | 1 | 2 |
| d | Q9DCX2 | − | + | 5 | 18 | 0 |
| e | Q06185 | − | + | 3 | 7 | 0 |
| f | P56135 | − | + | 5 | 9 | 0 |
| g | Q9CPQ8 | − | + | 4 | 14 | 0 |
| F6 | P97450 | − | − | 5 | 5 | 0 |
| 8 (A6L) | P03930 | − | − | 1 | 4 | 1 |
| 6.8 kDa | P56379 | − | − | 2 | 3 | 0 |
| DAPIT | Q78IK2 | − | + | 2 | 7 | 1 |
| α | Q03265 | + | + | 7 | 18 | 0 |
| β | P56480 | + | + | 7 | 19 | 0 |
| γ | Q91VR2 | + | + | 6 | 14 | 0 |
| δ | Q9D3D9 | − | − | 6 | 11 | 0 |
| ε | P56382 | − | + | 4 | 9 | 0 |
| OSCP | Q9DB20 | − | + | 5 | 15 | 0 |
Species were H. sapiens, R. norvegicus, B. taurus, D. melanogaster, and S. cerevisiae.
Number of transmembrane helices (TM) in the subunit as reported in UniProt.
Isoform 1 of subunit c.
Isoform 2 of subunit c.
Isoform 3 of subunit c.
We estimated the size of mitochondrial proteomes in each of the six species by using MitoMiner to combine the data from mass spectrometry, GFP tagging, and annotation. M. musculus had the largest number of proteins reported as mitochondrial when the results of its mass spectrometry studies were combined followed by H. sapiens and then S. cerevisiae (Table III). However, the number of proteins annotated as mitochondrial in UniProt or genome databases was about the same for these three species. Only S. cerevisiae had a large number of mitochondrial proteins identified from GFP tagging studies. Next we inferred the mitochondrial localization of proteins by considering the evidence from their orthologs by using HomoloGene. This identified many more proteins in each species as mitochondrial (Table III). Finally to estimate the size of the mitochondrial proteome in each species, we combined the proteins with direct evidence and annotation with those inferred from orthologs (Table III). H. sapiens and M. musculus each had about 3000 proteins that had been reported as mitochondrial, whereas S. cerevisiae had about 1500. Included in this list were proteins that are not usually considered mitochondrial but had experimental evidence for mitochondrial localization, such as the proteins from the core pathway of glycolysis.
Table III.
Numbers of non-redundant proteins (by using HomoloGene) described as mitochondrial in six species
| Species | No. of proteins in species
|
No. with orthologous proteins in other speciese
|
Totalf | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GFPa | Mass specb | Annotatedc | Combinedd | GFP | Mass spec | Annotated | Combined | ||
| H. sapiens | 142 | 1037 | 853 | 1551 | 243 | 2465 | 1109 | 2701 | 3025 |
| M. musculus | 52 | 2411 | 942 | 2626 | 327 | 1436 | 1063 | 1896 | 3124 |
| R. norvegicus | 0 | 533 | 413 | 754 | 207 | 1700 | 665 | 1835 | 1907 |
| B. taurus | 0 | 18 | 415 | 416 | 207 | 1483 | 649 | 1595 | 1602 |
| D. melanogaster | 0 | 37 | 236 | 255 | 271 | 1407 | 713 | 1520 | 1576 |
| S. cerevisiae | 574 | 1004 | 922 | 1196 | 46 | 584 | 251 | 620 | 1461 |
GFP, number of proteins determined by GFP tagging and microscopy as having mitochondrial localization.
Mass spec, number of proteins determined by using mass spectrometry of purified mitochondrial fractions.
Annotated, number of proteins annotated by UniProt, genome databases, or the Gene Ontology as mitochondrial.
Combined, number of proteins described as mitochondrial by combining evidence from GFP, mass spectrometry, and annotation.
Number of proteins that have an ortholog (as defined by HomoloGene) described as mitochondrial in the other five species.
Number of proteins described as mitochondrial by combining direct evidence and that inferred from orthologs.
To determine the mitochondrial proteins that were common among H. sapiens, M. musculus, and S. cerevisiae, we assessed the overlap in the mitochondrial proteomes we had calculated. Many of the proposed mitochondrial proteins of H. sapiens, M. musculus, and S. cerevisiae were orthologous using the relationships defined in HomoloGene (Fig. 9). Over 90% of the mitochondrial proteins from H. sapiens and M. musculus were orthologous, whereas about 50% of the proteins of S. cerevisiae were orthologous to proteins of H. sapiens and M. musculus. However, among the proteins that were considered to be non-orthologous between H. sapiens and M. musculus according to HomoloGene, there were examples of known orthologs. Therefore, more distant orthologies than defined by HomoloGene were identified by comparing all the proteins using BLASTP with an expect value cutoff of 10−35. Subsequently the number of proteins unique to S. cerevisiae fell to 562, whereas the number of orthologs present in all three species rose to 1393, and those in both H. sapiens and M. musculus rose to 3330 (close to 99% of the reported proteins). Of the remaining species-specific proteins in H. sapiens and M. musculus, several had an expect value slightly greater than that used for the BLASTP cutoff, and a couple had orthologs in RefSeq, but these sequences were missing from UniProt. The remaining singletons were all small proteins under 100 amino acids in length, and some were annotated as hypothetical.
Fig. 9.
The numbers of orthologous proteins among three mitochondrial proteomes. A protein was assigned as mitochondrial by either experimental evidence (mass spectrometry or GFP tagging) or annotation or by the mitochondrial localization of an ortholog. Main numbers were calculated by using HomoloGene to determine redundancy and orthology among proteins. Numbers in parentheses were calculated by using BLAST (with a threshold of 10−35 for the expect score) to define orthologs.
The DAVID Functional Annotation Clustering tool Web service was used to determine statistically significant (p < 0.001) biological process terms from the Gene Ontology among the set of proteins that were (i) common to all three species (using the proteins of S. cerevisiae to calculate the p values), (ii) specific to S. cerevisiae, and (iii) shared by H. sapiens and M. musculus (using the proteins of H. sapiens to calculate the p values) but not S. cerevisiae. There were many statistically significant but uninformative annotation terms among each set of proteins such as “biosynthetic process” (312 proteins; p < 10−26). However, among the set of proteins common to all three species, there were more specific Gene Ontology annotations reported by DAVID, including carboxylic acid metabolic process (136 proteins; p < 10−18), cofactor metabolic process (87 proteins; p < 10−15), tRNA aminoacylation (29 proteins; p < 10−12), mitochondrial transport (36 proteins; p < 10−10), amino acid metabolic process (80 proteins; p < 10−9), tricarboxylic acid cycle (18 proteins; p < 10−9), mitochondrial organization and biogenesis (60 proteins; p < 10−8), glycolysis (16 proteins; p < 10−4), lipid metabolic process (71 proteins; p < 10−3), and ubiquinone metabolic process (eight proteins; p < 10−3). Although the proteins specific to S. cerevisiae had many of the terms reported for the common group, there were also statistically significant Gene Ontology biological process terms that were absent from H. sapiens and M. musculus, including branched chain family amino acid biosynthetic process (nine proteins; p < 10−4) and fermentation (nine proteins; p < 10−3). Likewise for the proteins specific to H. sapiens and M. musculus, the term “apoptotic program” (30 proteins; p < 10−12) as well as the cellular component term “respiratory chain complex I” (38 proteins; p < 10−33) were significantly overrepresented and absent from S. cerevisiae.
DISCUSSION
We developed MitoMiner as a public resource to define and characterize the mitochondrial proteome by integrating the results of published experiments determining mitochondrial proteins with associated biological information. This experimental evidence is generally not included in the public sequence databases because of its size, complexity, and high false positive rate (6). MitoMiner differs from other mitochondrial proteome resources (Table I) by using a model to describe these data that is implemented in the open source InterMine data warehouse and allowed the integration of data from multiple sources as well as cross-resource queries. We incorporated data sets from 33 publications on the mitochondrial localization of proteins by either GFP tagging or mass spectrometry of purified mitochondria together with data and annotation on protein function, protein homology, metabolic pathways, genetic phenotypes, and post-translational modifications. Additional information was captured from these experimental studies such as the techniques used for purification, separation, and identification, providing a level of detail that is not present in any other mitochondrial resource. This extra information is accessible on a per entry basis and creates a comprehensive provenance that allows a user to directly evaluate the experimental evidence for the localization of a protein. The unique inclusion of metabolic pathway data gives a physiological context for a protein as well as being essential for supporting systems biology and modeling approaches. The mitochondrion is the site of many metabolic and bioenergetic pathways and is a prime candidate for the construction of metabolic models (60). These reconstructions are far from complete, and the integration of genomics and proteomics data with metabolic pathway data will allow for further refinement.
To demonstrate the unique features of MitoMiner, we analyzed the data of the 33 studies integrated in MitoMiner to address some general questions about the size and overlap of mitochondrial proteomes among species as well as progress in its definition by different technologies. The number of different mitochondrial proteins identified in these studies was calculated as about 3700 and appears to be reaching a plateau (Fig. 7), although only two studies were published after 2006. However, the study by Pagliarini et al. (6) in 2008 identified 1000 proteins that contributed 219 new proteins to the total number, suggesting that further studies are likely to identify new mitochondrial proteins. In particular, it has been estimated that a third of proteins encoded in genomes are membrane-bound (8). Yet until 2003, the identification of transmembrane proteins was relatively rare (Fig. 7); this may be due to the limitations of the two-dimensional electrophoresis method (8) that was commonly used to separate the protein fractions at that time. The proportion of transmembrane proteins identified increased steadily to 21% by 2008 as more GFP tagging studies were carried out and more sophisticated mass spectrometry techniques such as reverse phase chromatography were used. Note that the quoted proportions of transmembrane proteins may be inaccurate because of missing or incorrect annotation of transmembrane helices in UniProt. Mass spectrometry has been used in most mitochondrial proteomics studies because of its advantages for high throughput identification, but the isolation and purification of membrane proteins that are compatible with this method is difficult (8). Thus transmembrane proteins are likely to be underrepresented in these studies. Further sampling of the mitochondrial proteome, particularly using methods that can capture hydrophobic membrane proteins, is therefore likely to identify new mitochondrial proteins.
Only 392 proteins were shared between the two largest mass spectrometry data sets from Kislinger et al. (35) and Pagliarini et al. (6) on mouse mitochondria and represented about 22% of the Kislinger et al. (35) set and 43% of the Pagliarini et al. (6) set. This indicated that about 2200 different proteins are identified as mitochondrial by combining just these two studies. If the false discovery rates reported by Pagliarini et al. (6) are applied, then the total number falls to about 1770. This is higher than the 1500 different proteins estimated to be present in the mammalian mitochondrion (5). It is unlikely that these two studies have identified all the mitochondrial proteins particularly with regard to membrane proteins, and it suggests that the mitochondrial proteome could be much larger than current estimates.
We speculated that the greater the number of mass spectrometry studies reporting the mitochondrial localization of a protein the more likely it is that the protein is mitochondrial. We found that the majority of proteins reported in four or more publications are annotated in UniProt as mitochondrial. These may represent abundant soluble proteins that are components of ubiquitous pathways and processes and thus are more amenable to purification and identification by mass spectrometry. However, there were nearly 100 proteins identified in MitoMiner that are reported in four or more data sets and that are not annotated as mitochondrial in UniProt or their genome database. An example is the oxysterol-binding protein homolog 7 (Osh7p) from S. cerevisiae (Fig. 3) that has evidence from both GFP and mass spectrometry. As oxysterols generated in the mitochondria may play an important role in the maintenance of intracellular cholesterol homeostasis (56), the novel presence of an oxysterol-binding protein in the mitochondrion is plausible.
Infrequently observed proteins could be considered as contaminants from the cytosol and other organelles and thus could be discarded. However, many mitochondrial proteins are likely to be hydrophobic membrane proteins (8), to be of low abundance, or to be specific to particular development stages or tissues. These are features that make them difficult to isolate and detect using mass spectrometry. Thus, ignoring the nearly 1700 proteins that have been reported in only one mass spectrometry publication (Fig. 8) could result in a large number of genuine mitochondrial proteins being discounted. For example, the hydrophobic subunits of ATP synthase have poor experimental evidence for mitochondrial localization (Table II), in particular the hydrophobic c subunit, despite a very high abundance in the inner mitochondrial membrane. The same is true for members of the mitochondrial transporter family. Although all the proteins in these examples are annotated as mitochondrial in UniProt and the Gene Ontology and so would not be missing from a search in MitoMiner, this does not apply to novel proteins. MitoMiner could also be used to exclude proteins annotated as localizing to a subcellular location other than the mitochondrion in an attempt to reduce false positives. However, this would affect 36% of the total, many of which may be instances of dual localization (57) or proteins that function in the cytosol but are co-localized with the mitochondrion presumably by an interaction with the outside of the outer mitochondrial membrane. For example, searches using MitoMiner showed that all the core enzymes of the glycolytic pathway are regularly detected in mitochondrial proteomics determinations from animals and yeast, suggesting that they are structurally associated with the mitochondrion, as is reported for Arabidopsis thaliana (58), rather than a contaminant, although such interactions are disputed (29).
We estimated the size of the mitochondrial proteome in different species by combining the direct evidence and that inferred from orthologs (Table III). The total number of proteins reported for M. musculus and H. sapiens was about 3300, which was much higher than previous estimates (5). Prior estimates have been calculated using a limited number of data sets, so it was perhaps unsurprising that the combination of results from 33 publications in MitoMiner, which use a range of experimental techniques, was more likely to identify a wider range of the mitochondrial proteome than a single technique used in isolation. However, this combination also increases the likelihood of false positives, which for mass spectrometry studies of purified organelles have been estimated to be as high as 40% (6). We did not eliminate any proteins that had been identified from only a few peptide sequences or had inadequate sequence coverage, although the Query Builder can be used to apply filters for these. For example, it would be possible to reject any proteins reported with sequence coverage of less than 25% in a single mass spectrometry study, although few studies record this level of detail currently.
We investigated the overlap between the mitochondrial proteomes of three species (Fig. 9) and found that a substantial proportion of the mitochondrial proteome is common among them. This is expected given the universal role of the mitochondrion in energy production, metabolism, its own biogenesis and maintenance, and its shared evolutionary origin. This was confirmed by the Gene Ontology terms, found by using DAVID, that describe the functions of these common proteins. However, S. cerevisiae had nearly 600 proteins that were not present in H. sapiens and M. musculus, whereas H. sapiens and M. musculus shared nearly 2000 mitochondrial proteins that were not present in S. cerevisiae. Some of these differences probably arise from an expansion or reduction in the number of genes participating in common metabolic pathways. But analysis confirmed the presence in S. cerevisiae of proteins for processes that are lacking in H. sapiens and M. musculus, such as fermentation and branched chain amino acid biosynthesis. The functional annotation of the proteins specific to H. sapiens and M. musculus, compared with S. cerevisiae, identified proteins that were involved in NADH:ubiquinone oxidoreductase (complex I), which is lacking from S. cerevisiae, and apoptosis. The role of apoptosis in yeast remains controversial (59); however, in multicellular organisms it is an extremely important and well regulated process for programmed cell death that is mediated by the mitochondrion. Thus, functional analysis of the mitochondrial proteomes confirmed the different processes occurring in the mitochondria of species from different kingdoms because of either newly evolved functions or the loss of function in one kingdom compared with another.
MitoMiner is a new mitochondrial proteome resource that combines experimental data from 33 published studies with protein annotation from UniProt and genome projects, metabolic pathway data from KEGG, homology relationships from HomoloGene, and disease phenotypes from OMIM. This unique combination allows for further analysis of the mitochondrial proteome and provides the foundation of systems level evaluation of mitochondrial physiology and metabolism. Our preliminary analysis suggests that the mitochondrial proteome is much larger than previous estimates, although distinguishing between contaminants and hydrophobic, low abundance, or dual localized proteins remains challenging.
A public version of the MitoMiner resource is accessible from the website of the MRC Mitochondrial Biology Unit.
Acknowledgments
We thank Richard Smith, Kim Rutherford, and Gos Micklem of the FlyMine project for assistance with the InterMine system and John Walker and Ian Fearnley for valuable suggestions on proteomics.
Footnotes
Published, MCP Papers in Press, February 9, 2009, DOI 10.1074/mcp.M800373-MCP200
The abbreviations used are: GFP, green fluorescent protein; KEGG, Kyoto Encyclopedia of Genes and Genomes; OMIM, Online Mendelian Inheritance in Man; XML, Extensible Markup Language; MGI, Mouse Genome Informatics; PIR, Protein Information Resource; ID, identifier; BLAST, Basic Local Alignment Search Tool; DAVID, Database for Annotation, Visualization, and Integrated Discovery.
This work was supported by the Medical Research Council, UK.
REFERENCES
- 1.DiMauro, S., and Schon, E. A. ( 2003) Mitochondrial respiratory-chain diseases. N. Eng. J. Med. 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- 2.Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A. T., Bruder, C. E., Bohlooly, Y. M., Gidlof, S., Oldfors, A., Wibom, R., Tornell, J., Jacobs, H. T., and Larsson, N. G. ( 2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 [DOI] [PubMed] [Google Scholar]
- 3.Fliss, M. S., Usadel, H., Caballero, O. L., Wu, L., Buta, M. R., Eleff, S. M., Jen, J., and Sidransky, D. ( 2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287, 2017–2019 [DOI] [PubMed] [Google Scholar]
- 4.Wallace, D. C. ( 2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor, S. W., Fahy, E., and Ghosh, S. S. ( 2003) Global organellar proteomics. Trends Biotechnol. 21, 82–88 [DOI] [PubMed] [Google Scholar]
- 6.Pagliarini, D. J., Calvo, S. E., Chang, B., Sheth, S. A., Vafai, S. B., Ong, S. E., Walford, G. A., Sugiana, C., Boneh, A., Chen, W. K., Hill, D. E., Vidal, M., Evans, J. G., Thorburn, D. R., Carr, S. A., and Mootha, V. K. ( 2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, J., Fearnley, I. M., Skehel, J. M., Runswick, M. J., Shannon, R. J., Hirst, J., and Walker, J. E. ( 2005) The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 4, 693–699 [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J., Fearnley, I. M., and Walker, J. E. ( 2006) Definition of the mitochondrial proteome by measurement of molecular masses of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 103, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyne, R., Smith, R., Rutherford, K., Wakeling, M., Varley, A., Guillier, F., Janssens, H., Ji, W., McLaren, P., North, P., Rana, D., Riley, T., Sullivan, J., Watkins, X., Woodbridge, M., Lilley, K., Russell, S., Ashburner, M., Mizuguchi, K., and Micklem, G. ( 2007) FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8, R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. ( 2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, C. H., Apweiler, R., Bairoch, A., Natale, D. A., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Mazumder, R., O'Donovan, C., Redaschi, N., and Suzek, B. ( 2006) The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bult, C. J., Eppig, J. T., Kadin, J. A., Richardson, J. E., and Blake, J. A. ( 2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36, D724–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu, C. H., Yeh, L. S., Huang, H., Arminski, L., Castro-Alvear, J., Chen, Y., Hu, Z., Kourtesis, P., Ledley, R. S., Suzek, B. E., Vinayaka, C. R., Zhang, J., and Barker, W. C. ( 2003) The Protein Information Resource. Nucleic Acids Res. 31, 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. ( 1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camon, E., Magrane, M., Barrell, D., Lee, V., Dimmer, E., Maslen, J., Binns, D., Harte, N., Lopez, R., and Apweiler, R. ( 2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32, D262–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twigger, S. N., Shimoyama, M., Bromberg, S., Kwitek, A. E., and Jacob, H. J. ( 2007) The Rat Genome Database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res. 35, D658–D662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson, R. J., Goodman, J. L., and Strelets, V. B. ( 2008) FlyBase: integration and improvements to query tools. Nucleic Acids Res. 36, D588–D593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong, E. L., Balakrishnan, R., Dong, Q., Christie, K. R., Park, J., Binkley, G., Costanzo, M. C., Dwight, S. S., Engel, S. R., Fisk, D. G., Hirschman, J. E., Hitz, B. C., Krieger, C. J., Livstone, M. S., Miyasato, S. R., Nash, R. S., Oughtred, R., Skrzypek, M. S., Weng, S., Wong, E. D., Zhu, K. K., Dolinski, K., Botstein, D., and Cherry, J. M. ( 2008) Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 36, D577–D581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A., and McKusick, V. A. ( 2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler, D. L., Barrett, T., Benson, D. A., Bryant, S. H., Canese, K., Chetvernin, V., Church, D. M., DiCuccio, M., Edgar, R., Federhen, S., Geer, L. Y., Kapustin, Y., Khovayko, O., Landsman, D., Lipman, D. J., Madden, T. L., Maglott, D. R., Ostell, J., Miller, V., Pruitt, K. D., Schuler, G. D., Sequeira, E., Sherry, S. T., Sirotkin, K., Souvorov, A., Starchenko, G., Tatusov, R. L., Tatusova, T. A., Wagner, L., and Yaschenko, E. ( 2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35, D5–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., Katayama, T., Araki, M., and Hirakawa, M. ( 2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stajich, J. E., Block, D., Boulez, K., Brenner, S. E., Chervitz, S. A., Dagdigian, C., Fuellen, G., Gilbert, J. G., Korf, I., Lapp, H., Lehvaslaiho, H., Matsalla, C., Mungall, C. J., Osborne, B. I., Pocock, M. R., Schattner, P., Senger, M., Stein, L. D., Stupka, E., Wilkinson, M. D., and Birney, E. ( 2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. ( 2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 24.Alonso, J., Rodriguez, J. M., Baena-Lopez, L. A., and Santaren, J. F. ( 2005) Characterization of the Drosophila melanogaster mitochondrial proteome. J. Proteome Res. 4, 1636–1645 [DOI] [PubMed] [Google Scholar]
- 25.Carroll, J., Altman, M. C., Fearnley, I. M., and Walker, J. E. ( 2007) Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc. Natl. Acad. Sci. U. S. A. 104, 14330–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Da Cruz, S., Xenarios, I., Langridge, J., Vilbois, F., Parone, P. A., and Martinou, J. C. ( 2003) Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 27.Devreese, B., Vanrobaeys, F., Smet, J., Van Beeumen, J., and Van Coster, R. ( 2002) Mass spectrometric identification of mitochondrial oxidative phosphorylation subunits separated by two-dimensional blue-native polyacrylamide gel electrophoresis. Electrophoresis 23, 2525–2533 [DOI] [PubMed] [Google Scholar]
- 28.Forner, F., Foster, L. J., Campanaro, S., Valle, G., and Mann, M. ( 2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 29.Foster, L. J., de Hoog, C. L., Zhang, Y., Zhang, Y., Xie, X., Mootha, V. K., and Mann, M. ( 2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 30.Fountoulakis, M., Berndt, P., Langen, H., and Suter, L. ( 2002) The rat liver mitochondrial proteins. Electrophoresis 23, 311–328 [DOI] [PubMed] [Google Scholar]
- 31.Fountoulakis, M., and Schlaeger, E. J. ( 2003) The mitochondrial proteins of the neuroblastoma cell line IMR-32. Electrophoresis 24, 260–275 [DOI] [PubMed] [Google Scholar]
- 32.Fukada, K., Zhang, F., Vien, A., Cashman, N. R., and Zhu, H. ( 2004) Mitochondrial proteomic analysis of a cell line model of familial amyotrophic lateral sclerosis. Mol. Cell. Proteomics 3, 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaucher, S. P., Taylor, S. W., Fahy, E., Zhang, B., Warnock, D. E., Ghosh, S. S., and Gibson, B. W. ( 2004) Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J. Proteome Res. 3, 495–505 [DOI] [PubMed] [Google Scholar]
- 34.Jiang, X. S., Dai, J., Sheng, Q. H., Zhang, L., Xia, Q. C., Wu, J. R., and Zeng, R. ( 2005) A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase. Mol. Cell. Proteomics 4, 12–34 [DOI] [PubMed] [Google Scholar]
- 35.Kislinger, T., Cox, B., Kannan, A., Chung, C., Hu, P., Ignatchenko, A., Scott, M. S., Gramolini, A. O., Morris, Q., Hallett, M. T., Rossant, J., Hughes, T. R., Frey, B., and Emili, A. ( 2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 36.Lescuyer, P., Strub, J. M., Luche, S., Diemer, H., Martinez, P., Van Dorsselaer, A., Lunardi, J., and Rabilloud, T. ( 2003) Progress in the definition of a reference human mitochondrial proteome. Proteomics 3, 157–167 [DOI] [PubMed] [Google Scholar]
- 37.McDonald, T., Sheng, S., Stanley, B., Chen, D., Ko, Y., Cole, R. N., Pedersen, P., and Van Eyk, J. E. ( 2006) Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell. Proteomics 5, 2392–2411 [DOI] [PubMed] [Google Scholar]
- 38.Ohlmeier, S., Kastaniotis, A. J., Hiltunen, J. K., and Bergmann, U. ( 2004) The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J. Biol. Chem. 279, 3956–3979 [DOI] [PubMed] [Google Scholar]
- 39.Pflieger, D., Le Caer, J. P., Lemaire, C., Bernard, B. A., Dujardin, G., and Rossier, J. ( 2002) Systematic identification of mitochondrial proteins by LC-MS/MS. Anal. Chem. 74, 2400–2406 [DOI] [PubMed] [Google Scholar]
- 40.Prokisch, H., Scharfe, C., Camp, D. G., II, Xiao, W., David, L., Andreoli, C., Monroe, M. E., Moore, R. J., Gritsenko, M. A., Kozany, C., Hixson, K. K., Mottaz, H. M., Zischka, H., Ueffing, M., Herman, Z. S., Davis, R. W., Meitinger, T., Oefner, P. J., Smith, R. D., and Steinmetz, L. M. ( 2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabilloud, T., Kieffer, S., Procaccio, V., Louwagie, M., Courchesne, P. L., Patterson, S. D., Martinez, P., Garin, J., and Lunardi, J. ( 1998) Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: toward a human mitochondrial proteome. Electrophoresis 19, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 42.Reinders, J., Zahedi, R. P., Pfanner, N., Meisinger, C., and Sickmann, A. ( 2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 43.Rezaul, K., Wu, L., Mayya, V., Hwang, S. I., and Han, D. ( 2005) A systematic characterization of mitochondrial proteome from human T leukemia cells. Mol. Cell. Proteomics 4, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Romero, C., Lopez-Armada, M. J., and Blanco, F. J. ( 2006) Mitochondrial proteomic characterization of human normal articular chondrocytes. Osteoarthritis Cartilage 14, 507–518 [DOI] [PubMed] [Google Scholar]
- 45.Scheffler, N. K., Miller, S. W., Carroll, A. K., Anderson, C., Davis, R. E., Ghosh, S. S., and Gibson, B. W. ( 2001) Two-dimensional electrophoresis and mass spectrometric identification of mitochondrial proteins from an SH-SY5Y neuroblastoma cell line. Mitochondrion 1, 161–179 [DOI] [PubMed] [Google Scholar]
- 46.Sickmann, A., Reinders, J., Wagner, Y., Joppich, C., Zahedi, R., Meyer, H. E., Schonfisch, B., Perschil, I., Chacinska, A., Guiard, B., Rehling, P., Pfanner, N., and Meisinger, C. ( 2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 100, 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, S. W., Fahy, E., Zhang, B., Glenn, G. M., Warnock, D. E., Wiley, S., Murphy, A. N., Gaucher, S. P., Capaldi, R. A., Gibson, B. W., and Ghosh, S. S. ( 2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 [DOI] [PubMed] [Google Scholar]
- 48.Xie, J., Techritz, S., Haebel, S., Horn, A., Neitzel, H., Klose, J., and Schuelke, M. ( 2005) A two-dimensional electrophoretic map of human mitochondrial proteins from immortalized lymphoblastoid cell lines: a prerequisite to study mitochondrial disorders in patients. Proteomics 5, 2981–2999 [DOI] [PubMed] [Google Scholar]
- 49.Zahedi, R. P., Sickmann, A., Boehm, A. M., Winkler, C., Zufall, N., Schonfisch, B., Guiard, B., Pfanner, N., and Meisinger, C. ( 2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. ( 2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 51.Kumar, A., Agarwal, S., Heyman, J. A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., Cheung, K. H., Miller, P., Gerstein, M., Roeder, G. S., and Snyder, M. ( 2002) Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozawa, T., Sako, Y., Sato, M., Kitamura, T., and Umezawa, Y. ( 2003) A genetic approach to identifying mitochondrial proteins. Nat. Biotechnol. 21, 287–293 [DOI] [PubMed] [Google Scholar]
- 53.Calvo, S., Jain, M., Xie, X., Sheth, S. A., Chang, B., Goldberger, O. A., Spinazzola, A., Zeviani, M., Carr, S. A., and Mootha, V. K. ( 2006) Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 38, 576–582 [DOI] [PubMed] [Google Scholar]
- 54.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., Patterson, N., Lander, E. S., and Mann, M. ( 2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 55.Palmieri, F. ( 2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pfluegers Arch. Eur. J. Physiol. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 56.Ren, S., Hylemon, P., Zhang, Z. P., Rodriguez-Agudo, D., Marques, D., Li, X., Zhou, H., Gil, G., and Pandak, W. M. ( 2006) Identification of a novel sulfonated oxysterol, 5-cholesten-3β,25-diol 3-sulfonate, in hepatocyte nuclei and mitochondria. J. Lipid Res. 47, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 57.Regev-Rudzki, N., and Pines, O. ( 2007) Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. BioEssays 29, 772–782 [DOI] [PubMed] [Google Scholar]
- 58.Giege, P., Heazlewood, J. L., Roessner-Tunali, U., Millar, A. H., Fernie, A. R., Leaver, C. J., and Sweetlove, L. J. ( 2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng, W. C., Leach, K. M., and Hardwick, J. M. ( 2008) Mitochondrial death pathways in yeast and mammalian cells. Biochim. Biophys. Acta 1783, 1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vo, T. D., and Palsson, B. O. ( 2007) Building the power house: recent advances in mitochondrial studies through proteomics and systems biology. Am. J. Physiol. 292, C164–C177 [DOI] [PubMed] [Google Scholar]
- 61.Andreoli, C., Prokisch, H., Hortnagel, K., Mueller, J. C., Munsterkotter, M., Scharfe, C., and Meitinger, T. ( 2004) MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res. 32, D459–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basu, S., Bremer, E., Zhou, C., and Bogenhagen, D. F. ( 2006) MiGenes: a searchable interspecies database of mitochondrial proteins curated using gene ontology annotation. Bioinformatics 22, 485–492 [DOI] [PubMed] [Google Scholar]
- 63.Catalano, D., Licciulli, F., Turi, A., Grillo, G., Saccone, C., and D'Elia, D. ( 2006) MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa. BMC Bioinformatics 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cotter, D., Guda, P., Fahy, E., and Subramaniam, S. ( 2004) MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 32, D463–D467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heazlewood, J. L., and Millar, A. H. ( 2005) AMPDB: the Arabidopsis Mitochondrial Protein Database. Nucleic Acids Res. 33, D605–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H. P. ( 2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710 [PMC free article] [PubMed] [Google Scholar]