Abstract
Fetal growth restriction (FGR) is a common disorder in which a fetus is unable to achieve its genetically determined potential size. High concentrations of insulin-like growth factor-binding protein-1 (IGFBP-1) have been associated with FGR. Phosphorylation of IGFBP-1 is a mechanism by which insulin-like growth factor-I (IGF-I) bioavailability can be modulated in FGR. In this study a novel strategy was designed to determine a link between IGF-I affinity and the concomitant phosphorylation state characteristics of IGFBP-1 phosphoisoforms. Using free flow electrophoresis (FFE), multiple IGFBP-1 phosphoisoforms in amniotic fluid were resolved within pH 4.43–5.09. The binding of IGFBP-1 for IGF-I in each FFE fraction was determined with BIAcore biosensor analysis. The IGF-I affinity (K) for different IGFBP-1 isoforms ranged between 1.12e−08 and 4.59e−07. LC-MS/MS characterization revealed four phosphorylation sites, Ser(P)98, Ser(P)101, Ser(P)119, and Ser(P)169, of which Ser(P)98 was new. Although the IGF-I binding affinity for IGFBP-1 phosphoisoforms across the FFE fractions did not correlate with phosphopeptide intensities for Ser(P)101, Ser(P)98, and Ser(P)169 sites, a clear association was recorded with Ser(P)119. Our data demonstrate that phosphorylation at Ser119 plays a significant role in modulating affinity of IGFBP-1 for IGF-I. In addition, an altered profile of IGFBP-1 phosphoisoforms was revealed between FGR and healthy pregnancies that may result from potential site-specific phosphorylation. This study provides a strong basis for use of this novel approach in establishing the linkage between phosphorylation of IGFBP-1 and FGR. This overall strategy will also be broadly applicable to other phosphoproteins with clinical and functional significance.
The insulin-like growth factor (IGF)1 axis plays an important role in human fetal growth and development. Insulin-like growth factor-binding protein-1 (IGFBP-1) is a major IGF-binding protein in amniotic fluid (AF) (1, 2). The physiological role of IGFBP-1 is considered to be highly dependent on its differential phosphorylation (3–5). Phosphorylation of IGFBP-1 increases its affinity for IGF-I (6), suggesting that IGFBP-1 may modulate the action of IGF-I specifically with respect to fetal and placental growth (4, 7).
AF is a dynamic and complex biofluid and reflects the physiological status of the developing fetus (8). Fetal growth restriction (FGR) is a condition in which a fetus is unable to achieve its genetically determined potential size. The concentration of total IGFBP-1 is increased in FGR (9–12). Multiple phosphorylated species of IGFBP-1 have been detected during healthy pregnancy in both maternal circulation and in AF throughout gestation (1, 13, 14). Several studies have considered the clinical implications of IGFBP-1 phosphorylation, focusing on correlating variable ratios of high to low concentrations of IGFBP-1 phosphoisoforms with fetal outcome in FGR pregnancies (15–19). Although phosphorylation of IGFBP-1 has since been suggested to be critical, the predictive or functional value of IGFBP-1 phosphorylation in FGR is still not clear. The inconsistency in measurements of variable degrees of IGFBP-1 phosphorylation by ELISAs has resulted in inconclusive findings (20).
IGFBP-1 phosphoisoforms have been characterized previously using conventional methods (1, 13, 14). IGFBP-1, although relatively abundant in AF, still represents less than 0.01% of the total protein content (21). With restricted volumes available from clinical samples, isolation of functional IGFBP-1 phosphoisoforms using traditional approaches (13, 22, 23) is challenging. Our goal is to obtain a comprehensive understanding of the clinical and functional implications of IGFBP-1 phosphorylation in human FGR pregnancies. We developed an efficient, reproducible, and entirely liquid-based native IEF separation technology based on free flow electrophoresis (FFE) (24) to facilitate characterization of both the state of phosphorylation and IGF-I binding kinetics of variably phosphorylated IGFBP-1 isoforms in a clinical sample. As a prerequisite to application of this approach clinically, we also evaluated representative AF samples to determine whether or not differential patterns of IGFBP-1 phosphorylation exist in FGR and whether these changes could be attributed to augmentation of IGF-I affinity in the disease.
MATERIALS AND METHODS
FFE reagents were obtained from BD Diagnostics as BD™ FFE kits, IEF Native DSE (depletion-separation-enrichment) (separation range, pH 3–10) and IEF Native pH 4–6. H2SO4 and NaOH were purchased from Riedl-de-Haen (St. Louis, MO). All chemicals for gel electrophoresis and MS analysis were of electrophoresis or analytical grade. Anti-human anti-IGFBP-1 polyclonal antibody was a kind gift from Dr. R. Baxter from the Kolling Institute of Medical Research, Sydney, Australia, and recombinant human IGF-I (rIGF-I) was from Dr. George Bright from Tercica Inc., San Francisco, CA. Anti-human anti-IGFBP-1 monoclonal antibody (mAb 6303) was obtained from Medix Biochemica (Kauniainen, Finland). Protease inhibitor mixture (Complete Mini) was purchased from Roche Diagnostics. Alkaline phosphatase (calf intestinal) was obtained from Sigma-Aldrich. The ELISA kit for IGFBP-1 was from Diagnostic Systems Laboratories (Webster, TX) and Titansphere TiO (titanium dioxide (TiO2) was from GL Sciences Inc. (Tokyo, Japan). Biotin labeling for rIGF-I was performed using biotin EZ-Link NHS-LC from Pierce. BIAcore analysis was performed using the BIAcore X instrument (BIAcore, Inc., Piscataway, NJ).
Sample Collection
The AF sample used in FFE separation was obtained at delivery from a healthy pregnant woman for which no consent was necessary. Other samples used in this study involved AF from FGR pregnancies and gestational age-matched controls (n = 6 each) for which consent was obtained from the donors at the time of delivery as described previously (25). All AF samples were collected applying a protease inhibitor mixture. An aliquot of sample used in FFE was also collected without protease inhibitors to test the stability of IGFBP-1. All samples were filtered, centrifuged, and flash frozen at −80 °C until used.
Sample Preparation for FFE Separation
The AF sample was thawed and centrifuged (Beckman centrifuge JA 10 rotor) at 8000 rpm (both at 4 °C) to remove any debris. Prior to FFE separation, IGFBP-1 in AF was isolated using a salting out procedure (26) by adding solid ammonium sulfate (0–75% saturation) to the clarified AF sample. The sample was then centrifuged at 13,000 rpm for 1 h (Beckman model J2-21 centrifuge, JA 20 rotor). The pellet was collected and dissolved in 20 mm NaPO4 buffer, pH 7.0. Desalting of the samples was performed by dialysis (12,000–14,000 molecular weight cutoff, Spectrum Laboratories Inc.) using the same buffer (20 mm NaPO4 buffer, pH 7.0) at 4 °C followed by distilled water for additional 48 h. The resulting sample was spun at 13,000 rpm, and the clear supernatant consisting of IGFBP-1 (referred to as the enriched IGFBP-1) was stored at −80 °C until use. Total protein concentration was determined using the BCA protein assay kit (Pierce).
Free Flow Electrophoresis
FFE separations were conducted in the IEF mode (IEF-FFE) using a BD FFE System (BD Diagnostics) as described previously (27). Separation of proteins in the enriched AF sample was performed under two different conditions using either the FFE kit IEF Native DSE for lower resolution separation across the pH range of 3–10 or the FFE kit IEF Native pH 4–6 for a narrow range pH gradient providing a higher resolution separation (see Fig. 2, i and ii). In the case of the DSE protocol, the medium contained 10% glycerol in aqueous solution in addition to the ampholytes used to create the pH gradient. The FFE separation was performed according to the manufacturer's protocols. In brief, the electrolyte anode circuit was 100 mm H2SO4, and the cathode circuit was 100 mm NaOH. The flow rates for the separation medium were set at 120 ml/h and for delivering the sample the rate was 3 ml/h. A Tecan (Maennedorf, Switzerland) liquid handling system equipped with a pH electrode was used to measure the pH value of each fraction. After FFE equilibration and pI marker test, the enriched AF sample (6 mg of total protein) was diluted 10× in the separation medium, and the sample was infused into the chamber for IEF-FFE. Electrophoresis was performed at 10 °C with a constant voltage (400 V). The separated sample was collected (3.5 ml/fraction) to a 96-well plate with a separation time of 1.5 h.
Fig. 2.
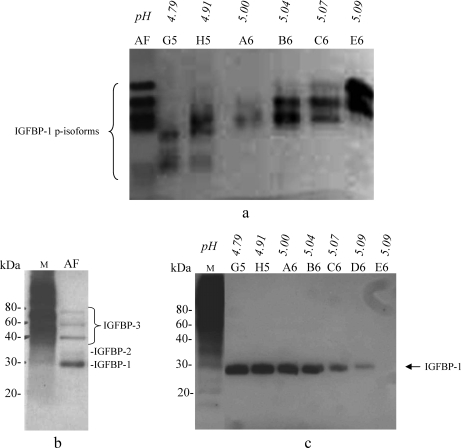
a, FFE pH gradients used for the separation of AF proteins. The flat regions below pH 2 and above pH 12 in the broad range pH gradient (DSE) (i) and below pH 3 and above pH 7 in the narrow range pH gradient (ii) represent the pH values of the anodic and cathodic stabilization media, respectively. Fractions (1–96) and their positions in the sample collection plates (A1–H12) are indicated. Sample preparation and FFE fractionation were performed as described in the text. The range of FFE fractions (C5–B7) used for the analysis of IGFBP-1 from the DSE separation is also indicated. b, SDS-PAGE with SilverQuest staining of FFE-fractionated AF. The distribution of total proteins is shown in fractions A4 to C10 (pH 1.6–12.4). FFE samples (10 μl/lane) were loaded. Lane M is the molecular mass marker, and Lane S is an aliquot of the enriched AF used in FFE separation. The perspective range of fractions for analysis of IGFBP-1 isoforms is indicated. c, immunoblot analysis of FFE fractions using mAb 6303. Lane M is the molecular mass marker, and E5 to D6 FFE fractions (20 μl/lane) (DSE separation) are indicated with respective pH values.
Alternatively FFE separation was performed for a narrow range separation of proteins using an IEF gradient from pH 4 to 6. The separation medium, in addition to other constituents as described for the DSE protocol, contained 0.2% hydroxypropyl methyl cellulose. Flow rates used were 60 and 1 ml/h, for medium and sample, respectively, to provide a separation of 4 mg of protein/h. In the narrow range FFE separation, AF (20 mg of protein) was diluted 5× prior to separation to avoid precipitation in the separation chamber. Electrophoresis was performed at 10 °C using 1350 V. Furthermore to avoid degradation of the separated samples, the FFE fractions were collected into plates prefilled with 500 μl of HBS EP buffer (10 mm HEPES, 150 mm NaCl, 3.4 mm EDTA, 0.005% surfactant P20, pH 7.4)/ml of FFE fraction.
Of the total fractions (n = 96) collected from FFE separations, selected fractions between A5 and C10 (from the 96-well plate) were analyzed for total protein separation by SDS-PAGE. The total IGFBP-1 concentrations in FFE fractions E5 to B7 were analyzed by ELISA (28). FFE fractions corresponding to pH 4.2–5.1 were selected for detection of IGFBP-1 by Western immunoblotting.
SDS-PAGE and Western Immunoblot Analysis
Total protein in samples was separated by 12% SDS-PAGE and stained either by Coomassie Blue or silver staining. For detection of IGFBP-1 by Western immunoblot analysis, proteins on gels were transferred onto PVDF membranes (0.45 μm) (Roche Diagnostics). In brief, for 1-D immunoblotting, 5% BSA was used as a blocking agent, and mAb 6303 (1:10,000 dilution) was used as the primary antibody. For 2-D immunoblot analyses, protein were separated on 2-D gels (25) using 7-cm IPG strips (pH 4–7), and IGFBP-1 isoforms were detected using IGFBP-1 polyclonal antibody (1:15,000 dilution). Mouse or rabbit HRP-conjugated antibodies (1:10,000 dilution) were used as secondary antibodies. Western markers, either low range protein standard (Invitrogen) or Western Magic HRP-conjugated protein marker, were used for size estimations. The ECL Plus reagent (GE Life Sciences) system and Eastman Kodak Co. X-Omat LS films were used for detection of proteins, and Alpha Innotech software 5.10 (Nonlinear Dynamics, Newcastle, UK) was used for imaging purposes. The phosphorylation state of IGFBP-1 phosphoisoforms was confirmed by alkaline phosphatase treatment prior to 2-D immunoblot analysis. The reactions were stopped by adding SDS-PAGE sample buffer. Dephosphorylation of IGFBP-1 for IGF-I binding kinetics was performed similarly except the reactions were terminated using EDTA (50 mm final concentration). The samples were immediately desalted and concentrated 10× using 10,000 molecular weight cutoff Centricon tubes (PALL Life Sciences, Ann Arbor, MI) and HBS EP buffer.
Native Western Immunoblot and Ligand Blot Analysis
For separation of IGFBP-1 in the native state, all samples were prepared for 1-D gels using the running buffer (50 mm Tris-HCl, pH 6.8, 25% glycerol, 1% Nonidet P-40, 0.005% bromphenol blue). Proteins transferred onto PVDF membrane (Roche Applied Science) were used for IGFBP-1 immunoblot analysis using mAb 6303. Additionally ligand blot analysis was performed using biotin-labeled rIGF-I (10 ng/ml). IGF-binding proteins were detected by using HRP-conjugated streptavidin (1:1000 dilution) and the ECL Plus reagent kit.
BIAcore Biosensor Interaction Using Surface Plasmon Resonance (SPR) Analysis
The interaction of IGFBP-1 with IGF-I was determined using the BIAcore X instrument using CM5 Sensor Chip™ overlaid with two flow cells. For immobilization, rIGF-I (10 mg/ml) was applied to the sample cell according to the manufacturer's protocol following amine coupling; the second flow cell was used as reference. Regeneration of the chip was performed using 30 μl of 10 mm glycine-HCl buffer, pH 2.0. Samples were obtained from two sets of FFE fractions. The unfractionated enriched AF was used for comparison. The kinetics measurements were performed in triplicate using a set of three dilutions. All FFE fractions used were desalted, concentrated 10×, and frozen at −20 °C until further use. Various concentrations of analyte (IGFBP-1) were injected for a 60-s association phase in both reference and sample cells at 50 μl/min.
Data Analysis
All binding curves were corrected for background by the subtraction of the signal obtained from the reference cell. Representative curves were generated for the association and the dissociation phases using the BIAevaluation software version 3.0 according to the 1:1 Langmuir binding model to fit kinetics (BIAcore). The data are represented as mean of three independent runs with S.D.
Phosphorylation of IGFBP-1 Using MS
In-solution Digestion—
The FFE fractions were selected, and aliquots with equal amount of IGFBP-1 were desalted and buffer exchanged using 10,000 molecular weight cutoff Centricon tubes at 4 °C with ammonium bicarbonate buffer, pH 8.0. The reduction and alkylation were carried out by treating samples (200 ng of IGFBP-1) with 10 mm dithiothreitol followed by 100 mm iodoacetamide. Subsequently proteins were digested first with Asp-N endoproteinase (Sigma) (25 ng/μl) at 37 °C overnight and then with sequencing grade trypsin (12.5 ng/μl) (Promega, Madison, WI) at 37 °C overnight. Digested samples were dried and stored at −80 °C.
Enrichment of IGFBP-1 Phosphopeptides Using TiO2—
IGFBP-1 phosphopeptides in the digested samples were enriched using TiO2 particles. In brief, the pellet with phosphopeptides was dissolved in 20 μl of loading buffer (80% ACN, 1% TFA) and incubated with 1 μl of TiO2 slurry (5 μm; 1 mg; 50% ACN) for 20 min at room temperature on a shaker. The solution and TiO2 particles were transferred to a pipette tip to which a piece of filter paper was inserted at its end to serve as a frit. The tip was placed in a microcentrifuge tube and centrifuged for 5 min. The particles were washed using 20 μl of loading buffer (50 mg/ml dihydroxybenzoic acid and 0.2% TFA in 40% ACN). The phosphopeptides were eluted using 20 μl of buffer (5% ammonium hydroxide, pH 11.0). To the receiving tube, 5 μl of 5% TFA was added prior to eluting the bound phosphopeptides. The samples were dried and reconstituted in 0.1% formic acid in water or in 50 mm EDTA in water prior to LC-MS or LC-MS/MS analysis.
IGFBP-1 Phosphoresidue Identification by MS—
The TiO2 enriched IGFBP-1 phosphopeptides prepared from each fraction (F5 to D6) were analyzed on a CapLC instrument (Waters, Milford, MA) coupled with a Q-TOF mass spectrometer (Global Ultima, Micromass, Manchester, UK) using a C18 precolumn and an analytical column (75 μm × 150 mm) (LC Packings, Amsterdam, Netherlands) with a flow rate of 300 nl/min. LC-MS/MS analysis was performed using a gradient elution and the data-dependent acquisition function (29). For determining the phosphopeptide concentration from each IGFBP-1 phosphoisoform, LC-MS analyses were carried out on individual FFE fractions (F6 to D6) using identical instrument settings. The selected ion chromatograms for different phosphopeptide peaks were plotted, and the spectra were summed. The intensities of the phosphopeptide peaks in the summed spectra were used for the semiquantitative determination of the relative amounts of the individual phosphopeptides in the samples.
LC-MS/MS spectra were processed to generate peak list files using the Maxnt 3 function in MassLynx software (Version 4.0) with default sets. The obtained peak list files were submitted for database search using PEAKS software (Bioinformatics Solution Inc., Waterloo, Ontario, Canada (version 4.2)) or Mascot (Matrix Science, Boston, MA) against Swiss-Prot database (version 51.3) for protein identification. Asp-N and/or trypsin were designated as proteases, and up to one missed cleavage was allowed. Taxonomy was set to Homo sapiens only. Peptide m/z tolerance was set to 0.15, and the peptide fragment ion tolerance was set to 0.1 Da. Carbamidomethylation on cysteine residues was included as a fixed modification, and oxidation of methionine and phosphorylation of serine/threonine/tyrosine residues were selected as a variable modifications. In total 15,347 sequences of human were searched from 250,296 sequences in the database. Score/expectation value of 20/1 was set for accepting a sequence identified for an MS/MS spectrum. However, all phosphopeptides identified were manually inspected to verify that the majority of high abundance peaks were y or b sequence ions or y − H2O/H3PO4 or b − H2O/H3PO4 ions when appropriate. For all phosphopeptides, their phosphorylation sites were verified manually.
RESULTS
Assessment of IGFBP-1 in Ammonium Sulfate-fractionated AF—
AF shares the distinctive characteristics of human biofluids such as a high dynamic range in protein abundance and high salt concentration. Salting out using ammonium sulfate fractionation (0–75%) was essential for removal of some interfering proteins and the high salt concentration in AF prior to FFE separation. Rather than conventional albumin depletion (30, 31), the salting out procedure proved to be efficient in elimination of albumin to minimize masking effects during FFE separation. Based on the molecular mass marker, although albumin (55–60-kDa band) was still detectable in the concentrated sample (Fig. 1a, Lane 2), the relative intensity of the band (30 kDa) corresponding to IGFBP-1 was nonetheless increased. Immunoblot analysis using the crude and the enriched AF samples (Fig. 1b, Lanes 1 and 2) confirmed the 30-kDa band as IGFBP-1. The sample also showed an enhanced band (20 kDa) corresponding to the proteolyzed IGFBP-1 fragment. To monitor the stability of IGFBP-1, AF sample (100 ng of IGFBP-1) collected without protease inhibitors as described above was incubated at 4 °C for up to 30 h. The samples with and without protease inhibitors were analyzed by immunoblot analysis using mAb 6303. The AF sample without protease inhibitors showed degradation (data not shown), whereas the sample with inhibitors resulted in only one additional band (20 kDa) (Fig. 1b, shown by an arrow), suggesting that the proteolysis may be endogenous.
Fig. 1.
Enrichment and detection of IGFBP-1 in AF. a, SDS gel (12% PAGE) with silver staining. Lane 1, crude AF; Lane 2, enriched AF (total protein, 28 μg/lane). An enrichment of IGFBP-1 (∼30 kDa) was achieved with a reduction in band (∼60 kDa) intensity for albumin (Lane 2). b, immunoblot analysis using anti-IGFBP-1 mAb 6303. Lane 1, crude AF; Lane 2, enriched AF (total protein, 8 μg) showing intact IGFBP-1 and its proteolytic fragment. c–e, 2-D immunoblot analysis using anti IGFBP-1 polyclonal antibody. c, crude AF; d, enriched AF (∼25 μg of total protein) with multiple IGFBP-1 phosphoisoforms represented by the numbered spots in each panel. e, AF sample following alkaline phosphatase treatment with dephosphorylated IGFBP-1. Lane M, molecular mass markers.
Multiple IGFBP-1 phosphoisoforms in AF samples are represented by several spots on 2-D immunoblotting (Fig. 1, c and d). The crude AF showed one spot (spot 1) toward pH 4.0 (20-kDa range) corresponding to the proteolyzed IGFBP-1 fragment. An additional six spots (spots 2–7) detected in the 30–35-kDa region corresponded to the intact IGFBP-1 phosphoisoforms. Similarly six spots (spots 4–9) were observed for the intact IGFBP-1 with the enriched AF sample (Fig. 1d). In addition, three distinct spots (spots 1–3) in this sample were detected for the 20-kDa fragment (Fig. 1d) possibly because of enrichment of the fragmented peptide. Alkaline phosphatase treatment (Fig. 1e) confirmed multiple spots as a result of different pI values of the phosphoisoforms. It should be noted that detection of IGFBP-1 on 2-D immunoblots required the use of IGFBP-1 polyclonal antibody because mAb 6303 (used in 1-D immunoblotting) was not efficient in detection of IGFBP-1 in the sample following 2-D gel analysis.
Total Protein Distribution in FFE Fractions—
The FFE procedure using a broad pH gradient provided resolution of proteins in the pH range ∼3–10 (Fig. 2a, i). The DSE pH gradient includes a plateau of 10 fractions where the pH gradient is extremely shallow covering the pH range 5.0–5.1. This plateau enabled an efficient focusing of the abundant albumin at its pI value during separation. Total protein distribution in selected fractions from a total of 96 fractions collected following FFE separation is shown Fig. 2b. As compared with the enriched AF (Lane S), the FFE-fractionated samples show a high resolution separation of several individual proteins. FFE fractions marked within the pH 4.2–5.1 range (C5 to B7) were selected for the investigation of IGFBP-1.
In addition, a higher resolution separation of the proteins and of IGFBP-1 was achieved by using the narrow range IEF gradient (pH 4–6; Fig. 2a, ii) of the same enriched AF (data not shown). FFE fractions were similarly selected for the analysis of IGFBP-1.
Total IGFBP-1 Distribution in FFE Fractions—
Total IGFBP-1 in individual FFE fractions E5 to F6 (3.5 ml/each) estimated by ELISA showed an overall 79% recovery as shown in Table I. The separation of total IGFBP-1 in FFE fractions was also confirmed qualitatively using immunoblot analysis. Because distribution of IGFBP-1 in FFE fractions was based on differences in pI values (pH 4.43–5.09), individual FFE fractions were considered representative of specific IGFBP-1 phosphoisoforms. IGFBP-1 phosphoisoforms in FFE fractions (Fig. 2c) from acidic to basic pH (E5 to F6) represent potentially highly phosphorylated to the less phosphorylated isoforms (Lanes E6 and F6 are not shown). At the acidic end, fraction E5 was co-fractionated with the proteolytic 20-kDa fragment (Fig. 2c). No attempts were made to evaluate the number of IGFBP-1 phosphoisoforms separated by FFE. However, an overall similarity could be discerned with multiple IGFBP-1 phosphoisoforms separated using both gel-free FFE fractionation (Fig. 2c) and 2-D gel-based immunoblotting of the unfractionated AF samples (Fig. 1d).
Table I.
Fractionation of human amniotic fluid for separation of IGFBP-1 phosphoisoforms using FFE
Ammonium sulfate-fractionated amniotic fluid sample (enriched AF; 6 mg of total protein) was diluted (10×) in the FFE separation medium and fractionated by FFE in IEF mode (DSE, pH 3–10). Fractions (3.5 ml each) were collected in a 96-well plate. The yield is the sum of total IGFBP-1 concentration measured by ELISA in individual FFE fractions (E5 to F6). FFE fractionation resulted in approximately 79% recovery of IGFBP-1 from the enriched AF sample.
| Samples | Sample volume | Total protein | Total protein | IGFBP-1 | Total IGFBP-1 | -Fold enrichment (IGFBP-1) |
|---|---|---|---|---|---|---|
| ml | mg/ml | mg | μg/ml | μg | ||
| Crude AF | 5.1 | 1.8 | 9.0 (100%) | 4.1 | 20.9 (100%) | 1.0 |
| Ammonium sulfate-fractionated AF (enriched AF) | 0.3 | 19.0 | 5.7 (64.5%) | 47.5 | 14.3 (58.4%) | 1.6 |
| FFE fractions (E5 to F6) | 49.0 | 4.7 (42.4%) | 11.3 (54.5%) | 1.9 |
Native immunoblot analysis of the crude AF sample (Fig. 3a, Lane AF) showed five discernible variants of IGFBP-1 denoted by four closely associated intense bands and a fainter lower band. Shown on the blot (Fig. 3a) are also the FFE fractions G5 to E6. Native immunoblot analysis confirmed that the distribution of IGFBP-1 in FFE fractions (G5 to E6 (F6, not shown)) was due to the relative state of IGFBP-1 phosphorylation. IGFBP-1 phosphoisoforms were detectable as two to three bands in each lane. The ascending shifts in bands from lower to higher pH with small differences in pH between the fractions were likely due to minute variability in the IGFBP-1 phosphorylation. Multiple bands also represent the partial separation of or overlapping IGFBP-1 phosphoisoforms. Qualitatively differences in the intensities of the bands in FFE fractions B6, C6, and E6 (Fig. 3a) suggest predominance of some phosphoisoforms over the others.
Fig. 3.
IGFBP-1 phosphoisoform separation using native gel electrophoresis. a, immunoblot analysis using mAb 6303 combined with native gel analysis. Lane AF, crude AF (∼50 ng of total IGFBP-1); Lanes G5–E6, equal volumes (30 μl/lane) of the FFE fractions loaded showing multiple bands. In fractions G5 to E6 variable intensities of the double bands indicate low to high abundance of IGFBP-1 phosphoisoforms (p-isoforms) that are partially overlapping in the individual fractions. b and c, ligand blot analysis using biotinylated rIGF-I showing crude AF (b, Lane AF; 5 μl) and individual FFE fractions (c, Lanes G5–E6; 5 μl). Several IGF-I-binding proteins were identified in AF by their molecular mass, whereas only IGFBP-1 was detected in the FFE fractions. Lane M, molecular mass markers.
IGF-I Ligand Blot Analysis—
Ligand blot analysis performed using crude AF in Fig. 3b demonstrated that IGFBP-1 was the major IGF-binding protein. Additional faint bands (Lane AF) in the sample represent low levels of IGFBP-3 and some other IGFBPs in the crude AF. In FFE fractions G5 to E6, only IGFBP-1 was detected (Fig. 3c). The qualitative data in this study confirm that FFE separation resulted in highly selective enrichment of specifically IGFBP-1 and removal of other IGFBPs due to differences in pI values (32). Because no other IGF-I-binding proteins were detectable under our experimental conditions, further purification was considered not to be necessary in the kinetic measurements of IGFBP-1.
IGF-I Binding Kinetics Using SPR Analysis—
BIAcore data (Table II) depict the kinetics of IGFBP-1/IGF-I binding in the isoforms separated into individual FFE fractions with high reproducibility. Superimposition of IGF-I binding kinetics data (Table II) from FFE fractions of the broad range separation DSE protocol with the narrow range FFE separation (pH 4–6) (data not shown) depicted matching patterns and reproducibility of the procedure.
Table II.
BIAcore biosensor-derived kinetics and equilibrium constants for interaction of IGFBP-1 phosphoisoform binding to immobilized rIGF-I
BIAcore analysis of individual samples was performed in triplicate. Selected FFE fractions shown with their corresponding pH represent the pI values of the IGFBP-1 phosphoisoforms in each fraction.
| FFE fraction containing IGFBP-1 phosphoisoforms | pH | ka | kd | KA (mean) | KD (mean) | S.D. |
|---|---|---|---|---|---|---|
| 1/ms | 1/s | 1/m | m | |||
| E5 | 4.43 | 1.33e+04 | 5.67e−04 | 2.35e+07 | 4.26e−08 | 3.98e−09 |
| F5 | 4.65 | 1.34e+04 | 5.40e−04 | 2.48e+07 | 4.03e−08 | 1.15e−10 |
| G5 | 4.79 | 3.23e+04 | 7.50e−04 | 4.28e+07 | 2.34e−08 | 6.08e−10 |
| H5 | 4.91 | 3.88e+05 | 5.20e−03 | 7.46e+07 | 1.34e−08 | 1.53e−09 |
| H5 AKPa | 1.11e+03 | 6.17e−03 | 1.79e+05 | 5.58e−06 | ||
| A6 | 5.00 | 1.34e+04 | 5.00e−04 | 2.69e+07 | 3.72e−08 | 1.00e−09 |
| A6 AKPa | 7.04e+03 | 4.79e−03 | 1.47e+06 | 6.80e−07 | ||
| B6 | 5.04 | 8.26e+02 | 4.20e−05 | 2.00e+07 | 5.01e−08 | 1.53e−09 |
| C6 | 5.07 | 3.88e+05 | 4.36e−03 | 8.93e+07 | 1.12e−08 | 5.77e−11 |
| C6 AKPa | 3.07e+03 | 2.36e−03 | 1.30e+06 | 7.70e−07 | ||
| D6 | 5.09 | 2.40e+03 | 1.08e−03 | 2.21e+06 | 4.52e−07 | 5.29e−09 |
| E6 | 5.09 | 2.40e+03 | 1.09e−03 | 2.20e+06 | 4.54e−07 | 5.86e−09 |
| F6 | 5.09 | 2.40e+03 | 1.10e−03 | 2.18e+06 | 4.59e−07 | 1.00e−09 |
| Unfractionated AF | 2.48e+03 | 1.21e−03 | 2.05e+06 | 4.88e−07 | 1.37e−11 |
AKP represents alkaline phosphatase-treated samples.
In comparison with enriched AF (KD 4.88e−07), the affinity of IGFBP-1 isoforms for IGF-I separated in the acidic fractions E5 toward A6 (pH 4.43–5.00) was higher than those toward the basic fractions (A6 to F6). Whereas a decrease in IGF-I binding affinity in fractions A6 and B6 (pH 5.00 and 5.04) followed the expected pattern, in C6 (pH 5.07) an intermittently higher affinity suggests a more complex interplay between phosphorylation and IGF-I binding. A relative -fold change in IGF-I binding affinity (KD) in FFE fractions E5 to D6 as compared with the most basic fraction E6 (listed in Table II) is shown in Table III. The highest affinities for IGF-I, 1.34e−08 and 1.12e−08 (Table II), were detected in fractions H5 (pH 4.91) and C6 (pH 5.07), respectively. Dephosphorylation following alkaline phosphatase treatment of IGFBP-1 phosphorylation (Table II) reverted the affinity for IGF-I and confirmed that the greater IGF-I binding affinities were due to the state of phosphorylation.
Table III.
Relative changes in IGF-I binding affinity (KD) (BIAcore data, Table II) and corresponding phosphopeptide peak intensities (LC-MS) detected from individual FFE fractions
| FFE fraction containing IGFBP-1 phosphoisoforms | -Fold change in binding affinity of IGFBP-1 phosphoisoforms for IGF-I (KD)a | Ratio of the peak intensity for each phosphopeptide to the intensity of Ser(P)101 (LC-MS)b
|
|||
|---|---|---|---|---|---|
| Ser(P)101 | Ser(P)98 and Ser(P)101 | Ser(P)119 | Ser(P)169 | ||
| F5 (pH 4.65) | 11.3 | 1,090 (1.00) | Not detected | 2,290 (2.10) | Not detected |
| G5 (pH 4.79) | 19.4 | 2,500 (1.00) | 476 (0.19) | 13,100 (5.24) | 941 (0.38) |
| H5 (pH 4.91) | 33.9 | 5,370 (1.00) | 1,880 (0.35) | 30,000 (5.59) | Not detected |
| A6 (pH 5.00) | 12.2 | 8,250 (1.00) | 1,830 (0.22) | 26,600 (3.22) | Not detected |
| B6 (pH 5.04) | 9.1 | 5,430 (1.00) | 989 (0.18) | 25,600 (4.71) | Not detected |
| C6 (pH 5.07) | 40.5 | 9,980 (1.00) | 1,190 (0.12) | 47,900 (4.80) | 475 (0.05) |
| D6 (pH 5.09) | 1.0 | 9,520 (1.00) | 976 (0.10) | 14,900 (1.57) | Not detected |
-Fold change in IGFBP-1 affinity (KD values for rIGF-I are illustrated in Table II) for rIGF-1 in FFE fractions E5 to C6 relative to fraction D6.
Corresponding phosphopeptide intensities (LC-MS) for Ser98, Ser101, Ser119, and Ser169 for IGFBP-1 phosphoisoforms separated in fractions E5 to D6. The relative -fold change in phosphopeptide peak intensity for each site compared with Ser(P)101 intensity (Ser(P)101 has been shown to alter the affinity for IGF-I (37)) is represented in parentheses.
Representative biosensorgrams for kinetics of IGFBP-1 binding to immobilized rIGF-I are shown in Fig. 4: a represents the phosphoisoforms separated in the most acidic fraction, b represents the most alkaline fraction, and c represents the unfractionated enriched AF. The kinetic pattern in fraction E5 (pH 4.43) indicates a slight bulk shift (Fig. 4a) due to the presence of the fragmented peptide with the intact IGFBP-1.
Fig. 4.
BIAcore biosensor interaction of immobilized rIGF-I with IGFBP-1 displaying the association and dissociation phases of concentration-dependent IGFBP-1 binding with rIGF-I. a–c, IGFBP-1 phosphoisoform(s) present in FFE fractions E5 (pH 4.43) and F6 (pH 5.09) and in the enriched AF, respectively. The kinetics analysis shows differences between dissociation phases for the phosphoisoforms separated at the lower pH (4.91) compared with the higher pH (5.09) or the unfractionated AF.
Major Sites of IGFBP-1 Phosphorylation—
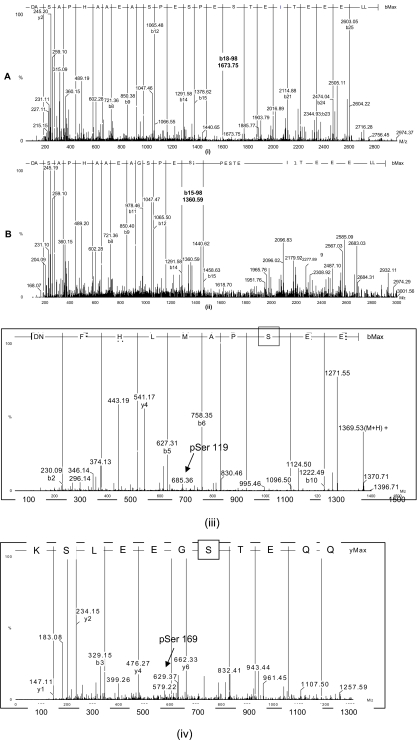
Phosphorylation of IGFBP-1 at Ser101, Ser119, and Ser169 sites has been reported earlier (33). Following Asp-N and trypsin digestion of individual FFE fractions containing different IGFBP-1 phosphoisoforms together with TiO2 enrichment of phosphopeptides, the amino acid sequences for the above three phosphopeptides were confirmed by LC-MS/MS: Ser(P)101: DASAPHAAEAGSPESPEpS101TEITEEELL (where pS is phosphoserine); m/z 949.73; charge state, +3; Ser(P)119: DNFHLMAPpS119EE; 685.30 m/z; charge state, +2; and Ser(P)169: AQETpS169GEEISK; m/z 629.78; charge state, +2. In addition, a doubly phosphorylated peptide with Ser(P)98 together with Ser(P)101 (DASAPHAAEAGSPEpS98PEpS101TEITEEELL; m/z 976.42; charge state, +3) was also detected. This doubly phosphorylated peptide was identified using a modified protocol (29) where addition of EDTA to the sample increases the detection sensitivity of multiply phosphorylated peptides. The LC-MS/MS spectra using FFE fraction C6 with a peak at m/z 949.73 for the singly phosphorylated peptide at Ser101 and at m/z 976.42 for Ser98 and Ser101 are shown in Fig. 5a (i and ii). The MS spectra with the observed b and y ion series allowed assignment of both the phosphopeptide sequence and the phosphorylation site. We did not obtain a spectrum that corresponds to a peptide with a singly phosphorylated site at Ser98. The spectra for phosphorylated peptide with Ser(P)119 (685.30 m/z; charge state, +2) and Ser(P)169 (m/z 629.78; charge state, +2) identified in the same fraction (C6) are shown in Fig. 5b (iii and iv).
Fig. 5.
Representative LC-MS/MS ion spectra of IGFBP-1 phosphopeptides identified in FFE fraction C6. The peptide sequences with highlighted IGFBP-1 phosphorylation sites are also indicated for each spectrum. Shown is the deconvoluted spectra of the parent ion at m/z 949.73, representing the singly phosphorylated Ser101 (Ser(P)101 with the peptide sequence in (i) and m/z 976.42 representing the doubly phosphorylated peptide (Ser(P)101 and Ser(P)98) in (ii). Both ions were observed as triply charged ions. In spectrum (i), intense b ions confirm the amino acid sequence of the peptide. The observed b18 ion at m/z 1771.68 and the b18 − 98 ion at m/z 1673.75 that is derived from b18 ion with a loss of H3PO4 confirmed phosphorylation on the Ser101 residue. In spectrum (ii), the precursor ion is 80 Da heavier than the ion in spectrum (i), indicating additional phosphorylation. The observed b15 ion at m/z 1458.63 and the b15 − 98 ion at m/z 1360.59 confirmed phosphorylation on Ser98 in addition to Ser101. Also are shown representative LC-MS/MS ion spectra of IGFBP-1 phosphopeptides identified with Ser(P)119, 685.30 m/z, charge state +2 (iii) and Ser(P)169, m/z 629.78, charge state +2 in (iv) using an aliquot of FFE sample C6.
LC-MS analyses were performed for selected fractions (F5 to D6) with an equal amount of total IGFBP-1 to estimate the peak intensity for each phosphorylated peptide. The differences in the recoveries of phosphopeptide upon TiO2 enrichment and the LC-MS are summarized in Table III. Because the intensities of each phosphopeptide are largely dependent on their sequence, the concentration of different phosphopeptides could not be directly compared with their intensities. However, the intensities of the same phosphopeptide from different fractions were comparable, and these intensities reflected the relative amount of the phosphopeptide in the specific fraction.
To determine the phosphorylation status of IGFBP-1 phosphoisoforms in individual FFE fractions, the phosphopeptide intensities in the corresponding fractions were matched with the affinity of IGFBP-1 for IGF-I (KD values; Table II). Although the IGF-I binding affinity across fractions F5 to D6 did not correlate with the intensities for Ser(P)101, Ser(P)98, and Ser(P)169 peptides, it linked well with the Ser(P)119 phosphopeptide as shown in Table III. These data suggest that Ser119 phosphorylation plays a significant role in modulating affinity of IGFBP-1 for IGF-I. A more precise quantitative analysis of these four phosphopeptides requires isotope labeling, which is the goal of a subsequent study.
IGFBP-1 Phosphoisoforms in AF from FGR and Control Pregnancies—
We applied 2-D immunoblotting to distinguish IGFBP-1 phosphoisoforms in representative control and gestational age-matched FGR samples (Fig. 6). Although as expected biological variability in IGFBP-1 isoforms was evident within each group, a lower number of spots corresponding to IGFBP-1 phosphoisoforms were obvious in the controls. In contrast, not only relatively greater numbers of phosphoisoforms were detected in the FGR samples but also higher intensities of specific spots. Additionally the acidic phosphoisoforms (toward pH 4) in FGR samples migrated further toward the acidic end with an upward shift due to a potentially greater phosphorylation (Fig. 6). Qualitatively the differences observed between the control and FGR groups (Fig. 6, a and b) suggest further evaluation to determine whether the variations are site-specific and affect IGF-I affinity. In addition, this study provides a strong basis for use of FFE in studying the clinical and functional role of IGFBP-1 phosphorylation in FGR.
Fig. 6.
2-D immunoblot analysis of IGFBP-1 phosphoisoforms in crude AF. Samples (total protein, 25 μg/AF sample) were analyzed using anti-IGFBP-1 polyclonal antibody. a, normal (healthy) (n = 6); b, FGR pregnancies (n = 6). The patterns of spots representing multiple IGFBP-1 phosphoisoforms were compared visually.
DISCUSSION
Phosphorylation of IGFBP-1 mediates acute metabolic regulation of IGF-I bioavailability; hence it is significant in FGR. The current assessments for molecular characterization of IGFBP-1 phosphorylation are inadequate in establishing its association with FGR. We report a novel approach that enables acquisition of IGF-I binding kinetics and complementary phosphorylation state characteristics of IGFBP-1 phosphoisoforms. Vital to the strategy was the precise separation of IGFBP-1 phosphoisoforms based on their pI values. Greater IGF-I affinity of IGFBP-1 in FFE fractions correlated with higher intensities for Ser(P)119 but not for Ser(P)101, Ser(P)169, or Ser(P)98 phosphopeptides. Our data demonstrate that Ser(P)119 plays an important role in altering the affinity of IGFBP-1 for IGF-I. Furthermore we also demonstrated for the first time differences in profiles of IGFBP-1 phosphoisoforms between FGR and healthy pregnancies. Our novel strategy may successfully identify IGFBP-1 phosphoisoforms characteristic of the disease.
Non- and phosphorylated isoforms of IGFBP-1 have been detected in AF using conventional approaches (13, 14). In this study, we introduced salting out (26) as a novel procedure for efficiently desalting and concentrating IGFBP-1 in a biofluid with abundant albumin. Rather than albumin depletion (31) or using ultrafiltration devices (34, 35), ammonium sulfate fractionation of AF (enriched AF) resulted in comparable yield. Using ammonium sulfate-fractionated enriched AF, FFE fractionation effectively separated several functional IGFBP-1 phosphoisoforms with minute differences in pI values (36). Subsequently overlaps in IGFBP-1 phosphoisoforms were expected, but overall differentially phosphorylated IGFBP-1 isoforms devoid of other IGF-I-binding proteins were separated. Direct assessment of IGF-I binding kinetics for IGFBP-1 phosphoisoforms was therefore feasible without further purification.
Multisite modifications on a protein constitute a complex regulatory mechanism in modulating protein functions in vivo (37). In our study, we detected phosphorylation of IGFBP-1 at Ser101, Ser119, and Ser169 residues as reported previously (33). With incorporation of phosphopeptide enrichment using LC-MS/MS technology, we also identified phosphorylation at Ser98 in the non-conserved central domain of IGFBP-1. In understanding the pathophysiological basis of a disease, explicit functional correlations of specific post-translational modifications are vital in evaluating the precise role of isoforms (38, 39). It is recognized that Ser(P)101 is a unique site on IGFBP-1 that is functionally significant in IGF-I binding (33). Using IGFBP-1 secreted by HepG2 cells, however, we recently confirmed higher phosphorylation at multiple sites to be linked with increased IGF-I affinity; additionally a newly identified Ser(P)98 site was hyperphosphorylated uniquely in hypoxic stress (40). Although phosphorylation of Ser101 may be critical, the increased Ser(P)119 phosphorylation associated with higher affinity for IGF-I currently detected in amniotic fluid is highly relevant.
IGFBP-1, -3, and -5 are all phosphorylated; however, phosphorylation of only IGFBP-1 elicits distinct changes on IGF-I binding affinity (3). The crystal structure of the C-terminal domain of IGFBP-1 has been elucidated previously (41, 42). This study proposes that a change in the conformation of the C terminus may be induced by hyperphosphorylation of IGFBP-1 at Ser(P)119 or other additional residues identified that could alter IGF-I affinity. Further structural-functional studies for evaluations of the multiple phosphorylation events and use of synthetic phosphopeptides should verify such a phenomenon more explicitly.
Previous clinical studies have shown the levels of total IGFBP-1 to increase in FGR (9–12). The physiological significance of alteration in the phosphorylation state of IGFBP-1 specifically during healthy pregnancy is also well recognized (4, 13). We report for the first time greater numbers and altered profiles of IGFBP-1 phosphoisoforms in pathological (FGR) pregnancies compared with the healthy. The data from this study provide a rationale for mapping the phosphorylation sites and the degree of phosphorylation to determine the linkage between IGFBP-1 phosphorylation and FGR.
In conclusion, we have characterized IGFBP-1 phosphoisoforms with respect to IGF-I binding and correlated binding affinity with individual phosphorylation states of IGFBP-1 phosphoisoforms. Such an approach to study the significance of IGFBP-1 phosphorylation clinically has not been achievable so far. This study establishes a framework on which biological fluids/clinical samples could be used in ascertaining correlations of IGFBP-1 phosphorylation with fetal outcomes as yet ambiguous in FGR (16, 17, 19, 43). The current strategy is also broadly pertinent for other clinically and functionally relevant proteins regulated by phosphorylation.
Acknowledgments
We gratefully acknowledge Dr. Gillis Lajoie, Director, Biological Mass Spectrometry Laboratory (BMSL), University of Western Ontario, London, Ontario, Canada for interest, insight, and invaluable discussions throughout the project. We thank Dr. Suya Liu (BMSL) for intellectual and technical contributions in conducting mass spectral analysis. We also express our thanks to Sylvia Katzer for interest and time in diligently proofreading the manuscript. Samad Shah, recipient of a Natural Sciences and Engineering Research Council of Canada undergraduate student research award, contributed toward obtaining preliminary data as a trainee in M. B. Gupta's laboratory.
Footnotes
Published, MCP Papers in Press, February 3, 2009, DOI 10.1074/mcp.M800571-MCP200
The abbreviations used are: IGF, insulin-like growth factor; AF, amniotic fluid; DSE, depletion-separation-enrichment; FGR, fetal growth restriction; FFE, free flow electrophoresis; IGFBP, insulin-like growth factor-binding protein; rIGF-I, recombinant human IGF-I; TiO2, titanium dioxide; mAb, monoclonal antibody; 1-D, one-dimensional; 2-D, two-dimensional; HRP, horseradish peroxidase.
REFERENCES
- 1.Martina, N. A., Kim, E., Chitkara, U., Wathen, N. C., Chard, T., and Giudice, L. C. ( 1997) Gestational age-dependent expression of insulin-like growth factor-binding protein-1 (IGFBP-1) phosphoisoforms in human extraembryonic cavities, maternal serum, and decidua suggests decidua as the primary source of IGFBP-1 in these fluids during early pregnancy. J. Clin. Endocrinol. Metab. 82, 1894–1898 [DOI] [PubMed] [Google Scholar]
- 2.Rajaram, S., Baylink, D. J., and Mohan, S. ( 1997) Insulin-like growth factor-binding proteins in serum and other biological fluids: Regulation and functions. Endocr. Rev. 18, 801–831 [DOI] [PubMed] [Google Scholar]
- 3.Coverley, J. A., and Baxter, R. C. ( 1997) Phosphorylation of insulin-like growth factor binding proteins. Mol. Cell. Endocrinol. 128, 1–5 [DOI] [PubMed] [Google Scholar]
- 4.Gibson, J. M., Aplin, J. D., White, A., and Westwood, M. ( 2001) Regulation of IGF bioavailability in pregnancy. Mol. Hum. Reprod. 7, 79–87 [DOI] [PubMed] [Google Scholar]
- 5.Yu, J., Iwashita, M., Kudo, Y., and Takeda, Y. ( 1998) Phosphorylated insulin-like growth factor (IGF)-binding protein-1 (IGFBP-1) inhibits while non-phosphorylated IGFBP-1 stimulates IGF-I-induced amino acid uptake by cultured trophoblast cells. Growth Horm. IGF Res. 8, 65–70 [DOI] [PubMed] [Google Scholar]
- 6.Jones, J. I., D'Ercole, A. J., Camacho-Hubner, C., and Clemmons, D. R. ( 1991) Phosphorylation of insulin-like growth factor (IGF)-binding protein 1 in cell culture and in vivo: Effects on affinity for IGF-I. Proc. Natl. Acad. Sci. U. S. A. 88, 7481–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, J. M., Westwood, M., Lauszus, F. F., Klebe, J. G., Flyvbjerg, A., and White, A. ( 1999) Phosphorylated insulin-like growth factor binding protein 1 is increased in pregnant diabetic subjects. Diabetes 48, 321–326 [DOI] [PubMed] [Google Scholar]
- 8.Tisi, D. K., Liu, X. J., Wykes, L. J., Skinner, C. D., and Koski, K. G. ( 2005) Insulin-like growth factor II and binding proteins 1 and 3 from second trimester human amniotic fluid are associated with infant birth weight. J. Nutr. 135, 1667–1672 [DOI] [PubMed] [Google Scholar]
- 9.Giudice, L. C., de Zegher, F., Gargosky, S. E., Dsupin, B. A., de las Fuentes, L., Crystal, R. A., Hintz, R. L., and Rosenfeld, R. G. ( 1995) Insulin-like growth factors and their binding proteins in the term and preterm human fetus and neonate with normal and extremes of intrauterine growth. J. Clin. Endocrinol. Metab. 80, 1548–1555 [DOI] [PubMed] [Google Scholar]
- 10.Maures, T. J., and Duan, C. ( 2002) Structure, developmental expression, and physiological regulation of zebrafish IGF binding protein-1. Endocrinology 143, 2722–2731 [DOI] [PubMed] [Google Scholar]
- 11.Huang, S. T., Vo, K. C., Lyell, D. J., Faessen, G. H., Tulac, S., Tibshirani, R., Giaccia, A. J., and Giudice, L. C. ( 2004) Developmental response to hypoxia. FASEB J. 18, 1348–1365 [DOI] [PubMed] [Google Scholar]
- 12.Kajimura, S., Aida, K., and Duan, C. ( 2005) Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc. Natl. Acad. Sci. U. S. A. 102, 1240–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koistinen, R., Angervo, M., Leinonen, P., Hakala, T., and Seppala, M. ( 1993) Phosphorylation of insulin-like growth factor-binding protein-1 increases in human amniotic fluid and decidua from early to late pregnancy. Clin. Chim. Acta 215, 189–199 [DOI] [PubMed] [Google Scholar]
- 14.Westwood, M., Gibson, J. M., Davies, A. J., Young, R. J., and White, A. ( 1994) The phosphorylation pattern of insulin-like growth factor-binding protein-1 in normal plasma is different from that in amniotic fluid and changes during pregnancy. J. Clin. Endocrinol. Metab. 79, 1735–1741 [DOI] [PubMed] [Google Scholar]
- 15.Bankowski, E., Sobolewski, K., Palka, J., and Jaworski, S. ( 2004) Decreased expression of the insulin-like growth factor-I-binding protein-1 (IGFBP-1) phosphoisoform in pre-eclamptic Wharton's jelly and its role in the regulation of collagen biosynthesis. Clin. Chem. Lab. Med. 42, 175–181 [DOI] [PubMed] [Google Scholar]
- 16.Loukovaara, M., Leinonen, P., Teramo, K., Nurminen, E., Andersson, S., and Rutanen, E. M. ( 2005) Effect of maternal diabetes on phosphorylation of insulin-like growth factor binding protein-1 in cord serum. Diabet. Med. 22, 434–439 [DOI] [PubMed] [Google Scholar]
- 17.Bhatia, S., Faessen, G. H., Carland, G., Balise, R. L., Gargosky, S. E., Druzin, M., El-Sayed, Y., Wilson, D. M., and Giudice, L. C. ( 2002) A longitudinal analysis of maternal serum insulin-like growth factor I (IGF-I) and total and nonphosphorylated IGF-binding protein-1 in human pregnancies complicated by intrauterine growth restriction. J. Clin. Endocrinol. Metab. 87, 1864–1870 [DOI] [PubMed] [Google Scholar]
- 18.Kajantie, E., Dunkel, L., Rutanen, E. M., Seppala, M., Koistinen, R., Sarnesto, A., and Andersson, S. ( 2002) IGF-I, IGF binding protein (IGFBP)-3, phosphoisoforms of IGFBP-1, and postnatal growth in very low birth weight infants. J. Clin. Endocrinol. Metab. 87, 2171–2179 [DOI] [PubMed] [Google Scholar]
- 19.Fowler, D., Albaiges, G., Lees, C., Jones, J., Nicolaides, K., and Miell, J. ( 1999) The role of insulin-like growth factor binding protein-1 phosphoisoforms in pregnancies with impaired placental function identified by Doppler ultrasound. Hum. Reprod. 14, 2881–2885 [DOI] [PubMed] [Google Scholar]
- 20.Khosravi, J., Krishna, R. G., Bodani, U., Diamandi, A., Khaja, N., Kalra, B., and Kumar, A. ( 2007) Immunoassay of serine-phosphorylated isoform of insulin-like growth factor (IGF) binding protein (IGFBP)-1. Clin. Biochem. 40, 86–93 [DOI] [PubMed] [Google Scholar]
- 21.Rutanen, E. M., and Pekonen, F. ( 1991) Assays for IGF binding proteins. Acta Endocrinol. ( Copenh.) 124, Suppl. 2, 70–73 [PubMed] [Google Scholar]
- 22.Busby, W. H., Hossenlopp, P., Binoux, M., and Clemmons, D. R. ( 1989) Purified preparations of the amniotic fluid-derived insulin-like growth factor-binding protein contain multimeric forms that are biologically active. Endocrinology 125, 773–777 [DOI] [PubMed] [Google Scholar]
- 23.Westwood, M., Gibson, J. M., and White, A. ( 1997) Purification and characterization of the insulin-like growth factor-binding protein-1 phosphoform found in normal plasma. Endocrinology 138, 1130–1136 [DOI] [PubMed] [Google Scholar]
- 24.Nissum, M., and Foucher, A. L. ( 2008) Analysis of human plasma proteins: a focus on sample collection and separation using free-flow electrophoresis. Expert Rev. Proteomics 5, 571–587 [DOI] [PubMed] [Google Scholar]
- 25.Gupta, M. B., Seferovic, M. D., Liu, S., Gratton, R. J., Doherty-Kirby, A., Lajoie, G. A., and Han, V. K. M. ( 2006) Altered proteome profiles in maternal plasma in pregnancies with fetal growth restrictions. Clin. Proteomics 2, 169–184 [Google Scholar]
- 26.Baijal-Gupta, M., Fraser, J. E., Clarke, M. W., Xuan, J. W., and Finkelman, M. A. ( 1996) A new scalable purification procedure for prostatic secretory protein (PSP94) from human seminal plasma. Protein Expr. Purif. 8, 483–488 [DOI] [PubMed] [Google Scholar]
- 27.Moritz, R. L., and Simpson, R. J. ( 2005) Liquid-based free-flow electrophoresis-reversed-phase HPLC: a proteomic tool. Nat. Methods 2, 863–873 [DOI] [PubMed] [Google Scholar]
- 28.Khosravi, M. J., Diamandi, A., and Mistry, J. ( 1997) Immunoassay of insulin-like growth factor binding protein-1. Clin. Chem. 43, 523–532 [PubMed] [Google Scholar]
- 29.Liu, S., Zhang, C., Campbell, J. L., Zhang, H., Yeung, K. K., Han, V. K., and Lajoie, G. A. ( 2005) Formation of phosphopeptide-metal ion complexes in liquid chromatography/electrospray mass spectrometry and their influence on phosphopeptide detection. Rapid Commun. Mass Spectrom. 19, 2747–2756 [DOI] [PubMed] [Google Scholar]
- 30.Moritz, R. L., Clippingdale, A. B., Kapp, E. A., Eddes, J. S., Ji, H., Gilbert, S., Connolly, L. M., and Simpson, R. J. ( 2005) Application of 2-D free-flow electrophoresis/RP-HPLC for proteomic analysis of human plasma depleted of multi high-abundance proteins. Proteomics 5, 3402–3413 [DOI] [PubMed] [Google Scholar]
- 31.Seferovic, M. D., Krughkov, V., Pinto, D., Han, V. K., and Gupta, M. B. ( 2008) Quantitative 2-D gel electrophoresis-based expression proteomics of albumin and IgG immunodepleted plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 865, 147–152 [DOI] [PubMed] [Google Scholar]
- 32.Weber, M. M., Spottl, G., Gossl, C., and Engelhardt, D. ( 1999) Characterization of human insulin-like growth factor-binding proteins by two-dimensional polyacrylamide gel electrophoresis and western ligand blot analysis. J. Clin. Endocrinol. Metab. 84, 1679–1684 [DOI] [PubMed] [Google Scholar]
- 33.Jones, J. I., Busby, W. H., Jr., Wright, G., and Clemmons, D. R. ( 1993) Human IGFBP-1 is phosphorylated on 3 serine residues: effects of site-directed mutagenesis of the major phosphoserine. Growth Regul. 3, 37–40 [PubMed] [Google Scholar]
- 34.Harper, R. G., Workman, S. R., Schuetzner, S., Timperman, A. T., and Sutton, J. N. ( 2004) Low-molecular-weight human serum proteome using ultrafiltration, isoelectric focusing, and mass spectrometry. Electrophoresis 25, 1299–1306 [DOI] [PubMed] [Google Scholar]
- 35.Hornig, M., Goodman, D. B., Kamoun, M., and Amsterdam, J. D. ( 1998) Positive and negative acute phase proteins in affective subtypes. J. Affect. Disord. 49, 9–18 [DOI] [PubMed] [Google Scholar]
- 36.Halligan, B. D., Ruotti, V., Jin, W., Laffoon, S., Twigger, S. N., and Dratz, E. A. ( 2004) ProMoST (protein modification screening tool): a web-based tool for mapping protein modifications on two-dimensional gels. Nucleic Acids Res. 32, W638–W644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johannessen, M., and Moens, U. ( 2007) Multisite phosphorylation of the cAMP response element-binding protein (CREB) by a diversity of protein kinases. Front. Biosci. 12, 1814–1832 [DOI] [PubMed] [Google Scholar]
- 38.Yang, X. J. ( 2005) Multisite protein modification and intramolecular signaling. Oncogene 24, 1653–1662 [DOI] [PubMed] [Google Scholar]
- 39.Ubersax, J. A., and Ferrell, J. E., Jr. ( 2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 40.Seferovic, M. D., Ali, R., Kamei, H., Liu, S., Khosravi, J. M., Nazarian, S., Han, V. K., Duan, C., and Gupta, M. B. ( 2009) Hypoxia and leucine deprivation induce human insulin-like growth factor binding protein-1 hyperphosphorylation and increase its biological activity. Endocrinology 150, 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sitar, T., Popowicz, G. M., Siwanowicz, I., Huber, R., and Holak, T. A. ( 2006) Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 103, 13028–13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sala, A., Capaldi, S., Campagnoli, M., Faggion, B., Labo, S., Perduca, M., Romano, A., Carrizo, M. E., Valli, M., Visai, L., Minchiotti, L., Galliano, M., and Monaco, H. L. ( 2005) Structure and properties of the C-terminal domain of insulin-like growth factor-binding protein-1 isolated from human amniotic fluid. J. Biol. Chem. 280, 29812–29819 [DOI] [PubMed] [Google Scholar]
- 43.Kajantie, E. ( 2003) Insulin-like growth factor (IGF)-I, IGF binding protein (IGFBP)-3, phosphoisoforms of IGFBP-1 and postnatal growth in very-low-birth-weight infants. Horm. Res. 60, Suppl. 3, 124–130 [DOI] [PubMed] [Google Scholar]