Abstract
It is well established that cytochrome c is released from mitochondria when the permeability transition (PT) of this organelle is induced by Ca2+. Our previous study showed that valinomycin also caused the release of cytochrome c from mitochondria but without inducing this PT (Shinohara, Y., Almofti, M. R., Yamamoto, T., Ishida, T., Kita, F., Kanzaki, H., Ohnishi, M., Yamashita, K., Shimizu, S., and Terada, H. (2002) Permeability transition-independent release of mitochondrial cytochrome c induced by valinomycin. Eur. J. Biochem. 269, 5224–5230). These results indicate that cytochrome c may be released from mitochondria with or without the induction of PT. In the present study, we examined the protein species released from valinomycin- and Ca2+-treated mitochondria by LC-MS/MS analysis. As a result, the proteins located in the intermembrane space were found to be specifically released from valinomycin-treated mitochondria, whereas those in the intermembrane space and in the matrix were released from Ca2+-treated mitochondria. These results were confirmed by Western analysis. Furthermore to examine how the protein release occurred, we examined the correlation between the species of released proteins and those of the abundant proteins in mitochondria. Consequently most of the proteins released from mitochondria treated with either agent were highly expressed proteins in mitochondria, indicating that the release occurred not selectively but in a manner dependent on the concentration of the proteins. Based on these results, the permeabilization effects of Ca2+ and valinomycin on the inner and outer mitochondrial membranes are discussed.
Mitochondria are well known as the organelle for energy conversion in all eukaryotes. This energy conversion, i.e. ATP synthesis, is performed by using the electrochemical gradient of H+ across the inner mitochondrial membrane. To enable effective energy conversion, the mitochondrial inner membrane is highly resistant to the permeation of solutes and ions. However, under certain conditions, such as in the presence of Ca2+ and inorganic phosphate, the permeability of this inner membrane is known to be markedly increased. This phenomenon is referred to as the permeability transition (PT)1 and is believed to result from the formation of a proteinaceous pore, referred to as the PT pore, which makes the inner membrane permeable to various solutes and ions smaller than 1.5 kDa (1–3). The physiological importance of the PT has long been uncertain; however, recent studies have revealed that the changes in the permeability of the inner mitochondrial membrane due to the induction of PT cause the release of cytochrome c into the cytosol and that the released cytochrome c then triggers subsequent steps of programmed cell death, which is known as apoptosis (4–6). Thus, the PT is considered to be one of the major regulatory steps of apoptosis. However, the questions as to how the PT is induced and how cytochrome c is released accompanied by the induction of PT have remained unanswered.
To characterize the features of the mitochondrial PT and to understand the mechanism underlying the release of cytochrome c from mitochondria, investigators have studied the effects of various agents on this organelle. As a result, the PT and the release of cytochrome c were found to be induced not only by Ca2+ but also by other agents (7–9). We also found that copper-o-phenanthroline (10), metal ions (11), and cyanine dyes (12, 13) induced this PT and the release of cytochrome c from mitochondria. Furthermore we reported that valinomycin, known as a potassium-selective ionophore, also induces the release of cytochrome c from mitochondria but without the induction of PT (14). This finding indicated that cytochrome c could be released from mitochondria in two different manners: one with the induction of PT and the other without it. To understand how cytochrome c is released from mitochondria, it is very important to know what protein species are released from mitochondria concomitant with the release of cytochrome c. To address these questions, in the present study we used a mass spectrometry (LC-MS/MS system)-based proteome analysis approach, which allowed us to identify the protein species present in a limited amount of protein samples. Using proteomics techniques, we examined the protein species released from mitochondria treated with valinomycin or with Ca2+, and we discuss our findings on the status of inner and outer mitochondrial membranes treated with these agents.
EXPERIMENTAL PROCEDURES
Materials—
Valinomycin (code V-0627) was purchased from Sigma. Maleimide-activated keyhole limpet hemocyanin (code 77606) was obtained from Pierce. The ECL kit (code RPN2106), Pharmalyte (code 17-0456-01), and anti-rabbit IgG conjugated with peroxidase (code NA934) were obtained from GE Healthcare. Polyclonal antibody against Smac/DIABLO (code SA-219) came from BIOMOL Research Laboratories, Inc.
Preparation of Mitochondria from Rat Liver—
Liver mitochondria were isolated as described previously (15) with the following modification to purify them. Briefly the liver obtained from normal male Wistar rat was minced in +EDTA medium (250 mm sucrose, 2 mm Tris-Cl, 1 mm EDTA, pH 7.4) and then homogenized at low speed in a chilled Potter-Elvehjem homogenizer. This homogenate was subsequently centrifuged for 5 min at 800 × g at 4 °C to remove nuclei, erythrocytes, unbroken liver cells, and debris after which approximately three-quarters of the supernatant was transferred to new tubes and centrifuged under the same conditions. The resulting supernatant was centrifuged for 10 min at about 6800 × g. The obtained crude mitochondrial pellet thus obtained was resuspended in +EDTA medium and centrifuged under the same conditions. The resulting pellet was resuspended and centrifuged at 17,400 × g for 10 min, and the subsequent pellet was resuspended and centrifuged under the same conditions except that −EDTA medium (250 mm sucrose, 2 mm Tris-Cl, pH 7.4) was used. The protein concentration of the final mitochondrial suspension was determined by the Biuret method with bovine serum albumin used as the standard.
Measurements of Turbidity of Mitochondrial Suspension—
The turbidity of mitochondrial suspensions was measured by monitoring the absorbance at 540 nm with a Shimadzu spectrophotometer, model UV-3000.
Transmission Electron Microscopic Analysis of Mitochondrial Configuration—
Mitochondrial configurations were analyzed by transmission electron microscopy according to the method reported previously (14).
Preparation of the Protein Samples Released from Mitochondria—
Mitochondria were suspended in 22 ml of +Pi medium (200 mm sucrose, 10 mm potassium phosphate buffer, pH 7.4) supplemented with 10 mm succinate (plus 0.5 μg of rotenone/mg of protein) as a respiratory substrate to make their final protein concentration 0.7 mg of protein/ml. After the addition of 2.5 μm valinomycin or 100 μm Ca2+, they were incubated at 25 °C for 4 min and then centrifuged at 17,400 × g for 10 min. The obtained supernatants were centrifuged once more. The resulting supernatants were further centrifuged at 230,000 × g for 60 min at 4 °C. The proteins present in the supernatants were precipitated by the addition of trichloroacetic acid or acetone. The obtained precipitates were solubilized with 160 μl of 1% SDS.
Preparation of Antibodies—
Polyclonal antibodies were raised by injection of synthetic peptides corresponding to parts of various mitochondrial proteins into adult New Zealand White rabbits as described in our previous study (14). The amino acid sequences of the peptides used as immunogens are summarized in Table I. These peptides were conjugated with keyhole limpet hemocyanin, then emulsified with Freund's adjuvant, and injected into the rabbits. Total blood was obtained 7 days after the final booster shot and was kept at room temperature for 1 h and then overnight at 4 °C. The blood clot was removed by centrifugation at 3,000 × g for 10 min at 4 °C, and the resulting supernatant was used as antiserum without further purification.
Table I.
Amino acid sequences of the peptides used for the preparation of antibodies used in this study
| Antibody against | Accessiona | Locb | Amino acid sequencec | Regiond |
|---|---|---|---|---|
| Adenylate kinase 2 | P29410 | IMS | TVKQAEMLDDLMDKRKEKLDC | 104–123 |
| Adenine nucleotide translocase | Q09073 | IM | VQHASKQISAEKQYKGIIDC | 38–57 |
| Apoptosis-inducing factor | Q9JM53 | IMS | NRMPIARKIIKDGEQHEDLC | 582–600 |
| Cytochrome c | P62898 | IMS | HTVEKGGKHKTGPNLHGLFC | 19–37 |
| 3-Hydroxymethylglutaryl-CoA synthase 2 | P22791 | M | NQREQFYHKVNFSPPGDTSNC | 465–484 |
| Ornithine carbamoyltransferase | P00481 | M | KGYEPDPNIVKLAEQYAKENC | 221–240 |
| Sulfite oxidase | Q07116 | IMS | SEESYSHWQRRDYKGFSPSVC | 389–408 |
| Voltage-dependent anion channel | Q9Z2L0 | OM | FQLHTNVNDGTEFGGSIYQKVC | 178–198 |
Accession number used in UniProt.
Location of the protein in mitochondria is shown by the following abbreviations: IM, inner membrane; IMS, intermembrane space; M, matrix; and OM, outer membrane.
Except for adenine nucleotide translocase, C-terminal cysteine residues were artificially introduced to enable conjugation with maleimide-activated hemocyanin.
Regions of individual peptides in the entire proteins are shown.
Protein Detection by Western Blotting—
Proteins obtained as described above were solubilized in extraction buffer (12.5 mm Tris, pH 6.8, containing 1% (w/v) SDS, 10% (w/v) glycerol, 1% (w/v) dithiothreitol, and 0.05% (w/v) bromphenol blue). SDS-PAGE was performed in 10% (w/v) and 20% (w/v) acrylamide gels essentially as described previously (14). After transfer of proteins in the gel to nitrocellulose membranes, the membranes were soaked for 1 h in TS buffer (20 mm sodium phosphate buffer, pH 7.4, containing 0.05% (w/v) Tween 20, 150 mm NaCl, and 5% (w/v) skim milk). The blocked membranes were then incubated with antibodies against individual mitochondrial proteins for 1 h. Antibodies against adenylate kinase 2, adenine nucleotide translocase, cytochrome c, 3-hydroxymethylglutaryl-CoA synthase 2, ornithine carbamoyltransferase, and sulfite oxidase were used at a 1000-fold dilution, and those against Smac/DIABLO and voltage-dependent anion channel were used at an 800- and 5000-fold dilution, respectively, with TS buffer containing 5% (w/v) skim milk as the diluent. After washing with TS buffer, the membranes were incubated with secondary antibody (anti-rabbit IgG conjugated with peroxidase; 2000-fold diluted with TS buffer) for 1 h. Specific binding of antibodies was visualized by the use of ECL reagents and subsequent exposure to x-ray film (Fuji Photo Film).
Mass Spectrometry and Protein Identification—
After SDS-PAGE, discrete protein bands were excised from the CBB-stained gel. In-gel digestion with trypsin was carried out as described previously (16). Then to facilitate the extraction of digested peptides from the gel pieces, the gel pieces were first swelled by the addition of 70 μl of 25 mm NH4HCO3 and then shrunk by addition of 210 μl of acetonitrile. Subsequently a similar course of the extraction of digested peptides by swelling/shrinkage of gel pieces was performed in 5% (v/v) formic acid and acetonitrile, and the obtained supernatants were collected. After the tryptic peptides had been air-dried, they were resuspended in 5% acetonitrile, 0.1% formic acid and injected into a CapLC system (Waters). They were first preconcentrated on a 300-μm × 5-mm C18 PepMap100 precolumn (LC Packings) and then separated for 40 min at a flow rate of 200 nl/min on a 75-μm × 15-cm C18 PepMap100 column (LC Packings) with a linear gradient of 5–60% acetonitrile containing 0.1% formic acid. The eluted peptides were subjected to mass spectrum analysis using a Q-TOF Ultima instrument (MicroMass) directly coupled to a nano-LC system. Spectrum analysis was performed by using the survey scanning mode of MassLynx 4.0 (Waters). In this mode, each spectrum was scanned for less than 5 s. For the selection condition of the precursors in the MS/MS analysis, a mode enabling repeated selection of certain precursor ions was used. Other parameters were set at default values. The obtained peak lists were subjected to a database search using the MASCOT server 1.905 (Matrix Science, London, UK) and a UniProt database (release date, January 9, 2007; 252,616 sequences and 92,372,123 residues). Search parameters were as follow: Rattus species, trypsin specificity, one allowed missed cleavage, carbamidomethylation fixed modification, methionine oxidation variable modification, precursor ion mass tolerance of 1.0 Da, and fragment ion mass tolerance of ±0.67 Da.
In these processes, if a certain protein was identified by more than two peptides showing ion scores higher than 23, which is a statistically significant ion score (expectation value, p < 0.05), this result was cited without further manual validation. However, if a protein was identified by only one peptide showing a score higher than 23, the MASCOT suggestion was manually inspected, and at least three consecutive y- or b-ions with a significant signal-to-background ratio were required for the identification. The intracellular locations of individual proteins were determined from their annotations in UniProt or based on the published literature.
RESULTS
Preparation of Protein Samples Released from Mitochondria—
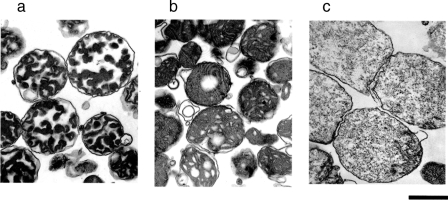
For preparation of samples of proteins released from mitochondria, the use of highly purified and functionally intact mitochondria is essential. Thus, we adopted the more stringent conditions of differential centrifugation for isolation of mitochondria from rat liver as described under “Experimental Procedures.” Our previous study using mitochondria prepared by the general protocol showed that valinomycin and Ca2+ caused a decrease in the turbidity of mitochondrial suspensions, but the membrane structures of the treated mitochondria were markedly different between the two treatments (14). Thus, we first examined whether the mitochondria prepared under the above higher stringency condition of centrifugation showed the reported response to valinomycin and Ca2+. As shown in Fig. 1, when 2.5 μm valinomycin was added to mitochondria, the turbidity of the mitochondrial suspension decreased. The addition of 100 μm Ca2+ also caused a decrease in the turbidity of mitochondrial suspensions that was more substantial than that caused by 2.5 μm valinomycin. The decrease in turbidity induced by Ca2+ was suppressed by 1 μm cyclosporin A, which is known to be a specific inhibitor of the PT (data not shown). When the configurations of mitochondria treated with valinomycin or with Ca2+ were examined by transmission electron microscopy (Fig. 2), no massive swelling or complete disappearance of the inner membrane structure was observed with valinomycin-treated mitochondria, whereas Ca2+ caused massive swelling and disappearance of the inner membrane structure, which are generally observed in PT-induced mitochondria (7, 10–14). Thus, we concluded that the mitochondria prepared in the present study showed essentially the same properties as those used in the previous study (14).
Fig. 1.
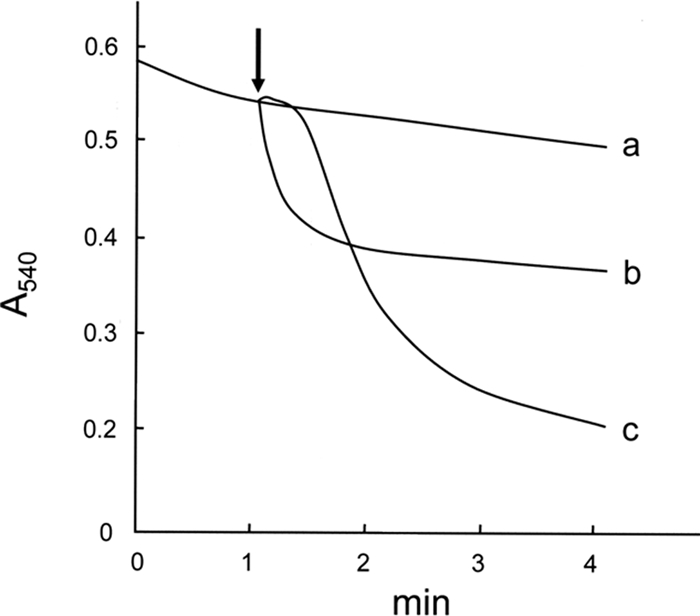
Effects of valinomycin and Ca2+ on the turbidity of mitochondrial suspensions. Time courses of the turbidity change in non-treated mitochondrial suspension (trace a) and suspensions of mitochondria treated with 2.5 μm valinomycin (trace b) or with 100 μm Ca2+ (trace c) are shown. Mitochondria were suspended to make a final protein concentration of 0.7 mg/ml in +Pi medium supplemented with 10 mm succinate (plus 0.5 μg of rotenone/mg of protein) as a respiratory substrate, and absorbance of this suspension at 540 nm was monitored. After incubation at 25 °C for 1 min, each agent was then added to the mitochondrial suspension at the time indicated by the arrow. A typical result of three independent runs is shown.
Fig. 2.
Effects of valinomycin and Ca2+ on mitochondrial configuration. The transmission electron microscopic appearance of non-treated mitochondria (a), valinomycin-treated mitochondria (b), and Ca2+-treated mitochondria (c) is shown. After treatment of mitochondria as described in the legend of Fig. 1, the mitochondria were promptly harvested and subjected to the fixation processes. The bar under photograph “c” indicates 1 μm. Of ∼30 randomly selected sections observed for each condition, typical images are shown.
To prepare samples of the proteins released from mitochondria, we first centrifuged mitochondria treated with 100 μm Ca2+ or with 2.5 μm valinomycin under the standard conditions (15,000 × g for 10 min at 4 °C). However, the supernatant thus obtained was not applicable for subsequent analysis due to remarkable contamination by non-pelleted mitochondria (data not shown). To settle this issue of contamination, we applied high speed centrifugation (230,000 × g for 1 h at 4 °C) to the reaction mixtures. With this application, contamination of the supernatant with whole mitochondria was significantly reduced (data not shown). To examine whether the supernatant samples prepared by this procedure were suitable for subsequent analyses, we subjected these samples to Western blotting (Figs. 2 and 3) using antibodies against cytochrome c, adenine nucleotide translocase, and voltage-dependent anion channel (VDAC). None of these proteins were detected in the supernatant of non-treated mitochondria (lane A). In the supernatant of valinomycin-treated and Ca2+-treated mitochondria (lanes B and C, respectively), cytochrome c, which is known to be released from mitochondria treated with these agents, was detected, whereas neither adenine nucleotide translocase nor VDAC, which are both located in the inner and outer membranes, was detected. The doublet bands of VDAC observed with whole mitochondrial protein reflect its isoforms (17). The results of this immunoassay showed that these supernatant samples were suitable as samples containing proteins released from mitochondria.
Fig. 3.
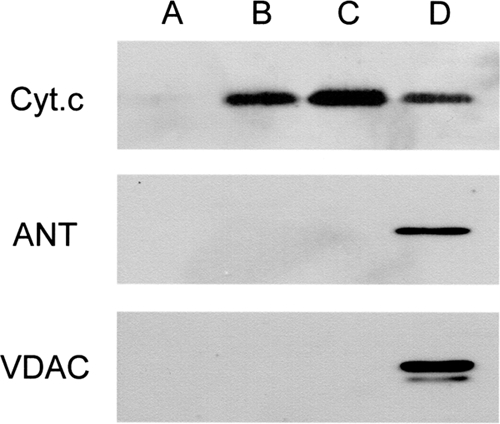
Quality check of the prepared protein samples by immunoblotting against various proteins. For lanes A–D, supernatants (15 μl/lane) of non-treated, valinomycin-treated, and Ca2+-treated mitochondria or whole proteins of rat liver mitochondria (5 μg/lane), respectively, were subjected to SDS-PAGE. After separation, the proteins were transferred to nitrocellulose membranes. Membranes thus prepared were first blocked with medium containing 5% skim milk and then incubated with specific antibodies against cytochrome c (Cyt.c), adenine nucleotide translocase (ANT), or VDAC. The immunoreactive proteins bands were visualized by using the ECL kit. A typical result of three independent runs is shown.
Identification by LC-MS/MS Analysis of the Proteins Released from Valinomycin- and Ca2+-treated Mitochondria—
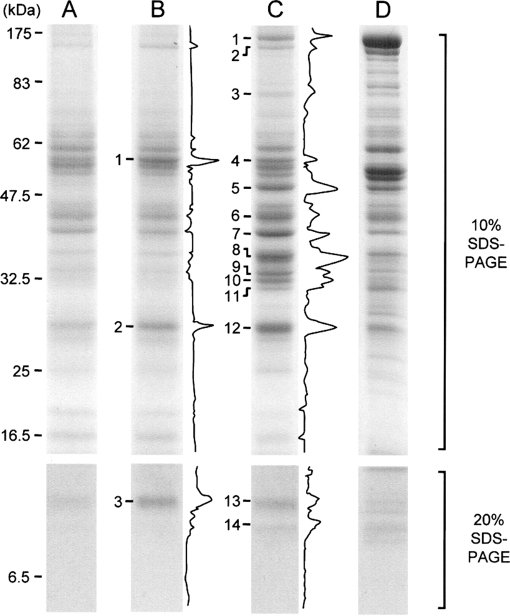
Next to elucidate the protein species present in the supernatant samples, we subjected these samples to SDS-PAGE followed by CBB staining (Fig. 4). SDS-PAGE was performed by using 10 and 20% polyacrylamide gels containing SDS to enable high resolution analysis of proteins covering a wide range of molecular weights. Although their staining intensities with CBB were faint, several protein bands were still detected in the samples of the supernatant of non-treated mitochondria; these bands were considered to be proteins from mitochondria broken during the preparation and from non-mitochondrial compartments (lane A). The band pattern of the supernatant prepared from valinomycin-treated mitochondria was similar to that from the non-treated mitochondria, whereas that from the Ca2+-treated mitochondria was apparently different (lanes B and C, respectively). For identification of the bands of proteins specifically released from mitochondria by valinomycin or Ca2+, the band densities in individual lanes were measured by using an image analyzer, and differential analysis against the densitogram for the non-treated mitochondria was performed. The differential densitograms, obtained by the subtraction of the densitogram of lane A from that of lane B or C, are shown at the right margins of lanes B and C in Fig. 4. From these densitograms, protein bands 1–3 in lane B (B1–3) and 1–14 in lane C (C1–14) were identified as those showing higher signal intensity in valinomycin- and Ca2+-treated mitochondria, respectively, than in non-treated mitochondria. These bands identified here were considered to contain the proteins released from mitochondria treated with each agent. The bands were cut out, the proteins present in each band were subjected to in-gel digestion with trypsin, and then the peptide mixtures obtained were analyzed by LC-MS/MS followed by a database search. In addition, the gel pieces in lane A corresponding to bands B1–3 and C1–14 were also cut out and analyzed as the background. Besides the above described bands, all of the visible bands were also analyzed as shown in supplemental Fig. S1 and Tables SI and SII.
Fig. 4.
SDS-PAGE analysis of proteins present in the supernatants of treated and non-treated mitochondria. For lanes A–D, supernatants (15 μl/lane) of non-treated, valinomycin-treated, and Ca2+-treated mitochondria or whole proteins of rat liver mitochondria (5 μg/lane), respectively, were subjected to SDS-PAGE. Upper and lower photographs represent the results of SDS-PAGE using 10 or 20% acrylamide gel, respectively. Numbers shown at the left margin of lane A represent the size of proteins used as molecular mass markers and their migration position. Densitograms shown at the right margins of lanes B and C represent the differential densitograms obtained by the subtraction of the densitogram of lane A from that of lane B or lane C. The protein bands in lanes B and C numbered at their left margins were analyzed by LC-MS/MS. A typical result of three independent runs is shown.
As a result of LC-MS/MS analysis of protein bands B1–3, multiple proteins were identified for individual protein bands (Table II). As features of the detected protein species, the following three points are noteworthy. First, all of the identified proteins were water-soluble proteins, and no membrane proteins were included. Second, the molecular masses of the identified proteins were almost consistent with those expected from mobilities of the analyzed protein bands with the following few exceptions. For example, carbamoyl-phosphate synthase (166 kDa) was identified in band B1, showing the mobility of ∼60 kDa; however, this band was assumed to be that of the degraded protein because this protein has been reported to be detectable as multiple fragments in immunoblotting analysis of rat liver mitochondria (18). Hydroxymethylglutaryl-CoA synthase (57 kDa) identified in band B2, showing the mobility of ∼30 kDa, was also considered to have been detected at that position for the same reason. Third, non-mitochondrial proteins such as hemoglobin or fatty acid-binding protein were observed in band B3. The reason for these ectopic proteins in this fraction is uncertain.
Table II.
LC-MS/MS analysis of proteins released from valinomycin-treated mitochondria
Protein species present in bands B1–3 (supernatant of valinomycin-treated mitochondria) were determined by LC-MS/MS analysis. Those present in the gel pieces in lane A (supernatant of non-treated mitochondria) corresponding to the positions of bands B1–3 were also determined as backgrounds and are shown in the column labeled “Non-treated.” AC, UniProt accession number.
| Band no. in lane B | Identified protein
|
Number of detected peptidesc (%)d
|
||||
|---|---|---|---|---|---|---|
| Name | AC | Molecular massa | Locb | Non-treated | Valinomycin-treated | |
| 1 | Sulfite oxidase | Q07116 | 55 | IMS | 18 (38) | 14 (70)e |
| Dihydrolipoyl dehydrogenase | Q6P6R2 | 55 | M | 12 (25) | 0 (0) | |
| Glutamate dehydrogenase 1 | P10860 | 62 | M | 10 (21) | 3 (15) | |
| Carbamoyl-phosphate synthase | P07756 | 166 | M | 8 (17) | 3 (15) | |
| 2 | Enoyl-CoA hydratase | P14604 | 32 | IMS | 15 (39) | 7 (26) |
| Adenylate kinase isoenzyme 2 | P29410 | 27 | M | 11 (29) | 15 (56)e | |
| Electron transfer flavoprotein subunit β | Q68FU3 | 28 | M | 4 (11) | 4 (15) | |
| 3,2-trans-Enoyl-CoA isomerase | P23965 | 32 | M | 3 (6) | 1 (3) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 2 (5) | 0 (0) | |
| 3 | Ribonuclease UK114 | P52759 | 14 | Mt, Cy | 4 (24) | 4 (15) |
| Cytochrome c | P62898 | 12 | IMS | 4 (24) | 12 (46)e | |
| Hemoglobin subunit β -1 | P02091 | 12 | Bl | 3 (18) | 3 (12) | |
| Fatty acid-binding protein, liver | P02692 | 12 | Cy | 3 (18) | 7 (27) | |
| Hemoglobin subunit α | P01946 | 12 | Bl | 3 (18) | 0 (0) | |
In kDa.
Location of the individual proteins is shown by following abbreviations: Bl, red blood cell; Cy, cytosol; IMS, intermembrane space of mitochondria; M, mitochondrial matrix; Mt, whole mitochondria.
Number of peptides derived from each protein identified by LC-MS/MS analysis.
Relative proportions of the peptide number derived from the protein to the total number of the peptides detected in each protein band are shown.
Peptides (their number and proportion to the total number of peptides) showing predominantly higher proportions in valinomycin-treated mitochondria than in the non-treated mitochondria are underlined.
We next determined the protein species specifically released from mitochondria by valinomycin treatment by comparing protein species identified from each protein band detected in the supernatant of valinomycin-treated mitochondria with those from the corresponding band in non-treated mitochondria. The rationale used for this determination is as follows. If the content of the peptides derived from the specific proteins in a certain gel piece of valinomycin-treated mitochondria was much higher than that in the non-treated mitochondria, those proteins could be considered as proteins selectively released from mitochondria. On the contrary, if that in valinomycin-treated mitochondria was similar to that in non-treated mitochondria, all proteins identified in the band could be considered to be unselectively released from mitochondria. As shown in the results of the LC-MS/MS analysis of each band (Table II), the proportions of the number of the peptides derived from sulfite oxidase, adenylate kinase 2, and cytochrome c to the total number of the peptides detected in bands B1–3 of valinomycin-treated mitochondria were 70, 56, and 46%, respectively (shown with underlines in Table II), which are about 2 times higher than the corresponding data for non-treated mitochondria (38, 29, and 24%, respectively). Therefore, we concluded that the protein species of sulfite oxidase, adenylate kinase 2, and cytochrome c were specifically released from mitochondria by valinomycin. This conclusion on the specific release of cytochrome c by valinomycin accords well with the result of the immunological analysis shown in Fig. 3. Sulfite oxidase, adenylate kinase 2, and cytochrome c are all known to be located in the mitochondrial intermembrane space, suggesting that valinomycin caused the release of proteins residing there.
Multiple protein species were also identified in the protein bands of Ca2+-treated mitochondria (Table III). Similar to the protein samples of valinomycin-treated mitochondria, the molecular weights of most of the identified proteins of Ca2+-treated mitochondria were consistent with the mobility of the analyzed bands. Non-mitochondrial hemoglobin and fatty acid-binding protein were also detected in band C13. To clarify the proteins specifically released from mitochondria by Ca2+, the proportions of peptides derived from each protein to all peptides detected in each band were compared with those in the corresponding gel pieces for the non-treated mitochondria. As a result, unlike the bands for the valinomycin-treated mitochondria, in most of the protein bands except for bands C4, C6, and C13, the proportions of peptides derived from each protein were not markedly different from those in the corresponding gel pieces for the non-treated mitochondria. Therefore, Ca2+ was concluded to cause the nonspecific release of all identified mitochondrial proteins in these protein bands. These protein species included not only proteins located in the intermembrane space such as adenylate kinase 2 (band C12) but also those situated in the matrix such as malate dehydrogenase (band C9), electron transfer flavoprotein subunit α (band C11), and enoyl-CoA hydratase (band C12).
Table III.
LC-MS/MS analysis of the proteins released from Ca2+-treated mitochondria
Protein species present in bands C1–14 (supernatant of Ca2+-treated mitochondria) were determined by LC-MS/MS analysis. Those present in the gel pieces in lane A (supernatant of non-treated mitochondria) corresponding to the positions of bands C1–14 were also determined as backgrounds and are shown in the column designated as “Non-treated.” AC, UniProt accession number; PH, pleckstrin homology.
| Band no. in lane C | Identified protein
|
Number of detected peptidesc (%)d
|
||||
|---|---|---|---|---|---|---|
| Name | AC | Molecular massa | Locb | Non-treated | Ca2+-treated | |
| 1 | Carbamoyl-phosphate synthase | P07756 | 166 | M | 31 (65) | 61 (94) |
| Aspartate aminotransferase | P00507 | 48 | M | 7 (15) | 0 (0) | |
| Catalase | P04762 | 60 | Pr | 4 (8) | 1 (2) | |
| Aldehyde dehydrogenase | P11884 | 57 | M | 2 (4) | 0 (0) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | Mt | 2 (4) | 1 (2) | |
| Malate dehydrogenase | P04636 | 36 | M | 0 (0) | 1 (2) | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 2 (4) | 0 (0) | |
| 2 | Carbamoyl-phosphate synthase | P07756 | 166 | M | 50 (84) | 58 (93) |
| Pyruvate carboxylase | P52873 | 130 | M | 5 (8) | 0 (0) | |
| Bile acid CoA:amino acid N-acyltransferase | Q63276 | 47 | Pr | 3 (5) | 1 (2) | |
| Catalase | P04762 | 60 | Pr | 2 (3) | 3 (5) | |
| 3 | Carbamoyl-phosphate synthase | P07756 | 166 | M | 18 (60) | 22 (42) |
| Dimethylglycine dehydrogenase | Q63342 | 96 | Mt | 6 (20) | 14 (26) | |
| Aspartate aminotransferase | P00507 | 48 | M | 4 (13) | 1 (2) | |
| Aconitate hydratase | Q9ER34 | 86 | Mt | 2 (7) | 16 (30) | |
| 4 | Sulfite oxidase | Q07116 | 55 | IMS | 18 (38) | 11 (69) |
| Dihydrolipoyl dehydrogenase | Q63342 | 55 | M | 12 (25) | 2 (13) | |
| Glutamate dehydrogenase 1 | P10860 | 62 | M | 10 (21) | 0 (0) | |
| Carbamoyl-phosphate synthase | P07756 | 166 | M | 8 (17) | 1 (6) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 0 (0) | 1 (6) | |
| Methylmalonate-semialdehyde dehydrogenase | Q02253 | 58 | M | 0 (0) | 1 (6) | |
| 5 | Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 24 (58) | 18 (100) |
| Carbamoyl-phosphate synthase | P07756 | 166 | M | 6 (15) | 0 (0) | |
| Fumarate hydratase | P14408 | 55 | Mt | 5 (12) | 0 (0) | |
| Acyl-coenzyme A oxidase 1 | P07872 | 75 | Pr | 4 (10) | 0 (0) | |
| Retinoid-inducible serine carboxypeptidase | Q920A6 | 51 | S | 2 (5) | 0 (0) | |
| 6 | Long chain-specific acyl-CoA dehydrogenase | P15650 | 48 | M | 23 (37) | 11 (35) |
| 3-Ketoacyl-CoA thiolase | P13437 | 42 | Mt | 16 (25) | 12 (39) | |
| Acetyl-CoA acetyltransferase | P17764 | 45 | Mt | 7 (11) | 1 (3) | |
| Aspartate aminotransferase | P00507 | 48 | M | 5 (8) | 5 (17) | |
| Medium chain-specific acyl-CoA dehydrogenase | P08503 | 47 | M | 5 (8) | 1 (3) | |
| Cathepsin D | P24268 | 45 | Ly | 4 (6) | 1 (3) | |
| Fumarylacetoacetase | P25093 | 46 | ? | 3 (5) | 0 (0) | |
| 7 | Aspartate aminotransferase | P00507 | 48 | M | 32 (51) | 37 (86) |
| 3-Ketoacyl-CoA thiolase A | P13437 | 44 | Pr | 11 (17) | 0 (0) | |
| Short chain-specific acyl-CoA dehydrogenase | P15651 | 45 | M | 10 (16) | 3 (7) | |
| α-Methylacyl-CoA racemase | P70473 | 42 | Pr, Mt | 6 (10) | 3 (7) | |
| Cathepsin D | P24268 | 45 | Ly | 2 (3) | 0 (0) | |
| PH domain leucine-rich repeat protein phosphatase | Q9WTR8 | 186 | Cy | 1 (2) | 0 (0) | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 1 (2) | 0 (0) | |
| 8 | Ornithine carbamoyltransferase | P00481 | 40 | M | 21 (50) | 23 (64) |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 13 (31) | 13 (36) | |
| Arginase-1 | P07824 | 35 | Cy | 4 (10) | 0 (0) | |
| Carbamoyl-phosphate synthase | P07756 | 166 | M | 3 (7) | 0 (0) | |
| Malate dehydrogenase | P04636 | 36 | M | 3 (7) | 0 (0) | |
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 | 36 | Cy | 1 (2) | 0 (0) | |
| 9 | Malate dehydrogenase | P04636 | 36 | M | 22 (58) | 34 (76) |
| Hydroxyacyl-CoA dehydrogenase | P22791 | 35 | M | 8 (21) | 5 (11) | |
| l-Lactate dehydrogenase A chain | P04642 | 37 | Cy | 4 (11) | 0 (0) | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 3 (8) | 0 (0) | |
| Cathepsin Z | NP_899159e | 35 | Ly | 1 (3) | 1 (2) | |
| Peroxisomal trans-2-enoyl-CoA reductase | Q9WVK3 | 33 | Pr | 0 (0) | 3 (7) | |
| 10 | Hydroxyacyl-Co A dehydrogenase | P22791 | 35 | M | 16 (52) | 9 (40) |
| Electron transfer flavoprotein subunit α | P13803 | 35 | M | 9 (29) | 4 (17) | |
| Malate dehydrogenase | P04636 | 36 | M | 2 (6) | 0 (0) | |
| 3-Mercaptopyruvate sulfurtransferase | P97532 | 33 | Cy, Mt | 2 (6) | 1 (4) | |
| Cathepsin Z | NP_899159e | 35 | Ly | 1 (3) | 1 (4) | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 1 (3) | 0 (0) | |
| Peroxisomal trans-2-enoyl-CoA reductase | Q9WVK3 | 33 | Pr | 0 (0) | 4 (17) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 0 (0) | 2 (9) | |
| Hydroxymethylglutaryl-CoA lyase | P97519 | 35 | M | 0 (0) | 1 (4) | |
| 11 | Electron transfer flavoprotein subunit aα | P13803 | 35 | M | 15 (26) | 20 (40) |
| 3-Mercaptopyruvate sulfurtransferase | P97532 | 33 | Cy, Mt | 11 (19) | 10 (20) | |
| Hydroxyacyl-Co A dehydrogenase | P22791 | 35 | M | 9 (16) | 3 (6) | |
| 2,4-Dienoyl-CoA reductase | Q64591 | 36 | Mt | 7 (12) | 7 (14) | |
| Δ3,5-Δ2,4-Dienoyl-CoA isomerase | Q62651 | 36 | Mt, Cy | 5 (9) | 1 (2) | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | 5 (9) | 2 (4) | |
| Betaine-homocysteine S-methyltransferase | O09171 | 45 | Cy | 3 (5) | 0 (0) | |
| Retinoid-inducible serine carboxypeptidase | Q920A6 | 51 | S | 2 (3) | 0 (0) | |
| Cathepsin Z | NP_899159e | 35 | Ly | 1 (2) | 0 (0) | |
| 3-Hydroxyisobutyrate dehydrogenase | P29266 | 36 | Mt | 0 (0) | 5 (10) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 0 (0) | 2 (4) | |
| 12 | Enoyl-CoA hydratase | P14604 | 32 | M | 15 (43) | 14 (26) |
| Adenylate kinase isoenzyme 2 | P29410 | 27 | IMS | 11 (31) | 17 (32) | |
| Electron transfer flavoprotein subunit β | Q68FU3 | 28 | M | 4 (11) | 10 (19) | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | 2 (6) | 0 (0) | |
| 3,2-trans-Enoyl-CoA isomerase | P23965 | 32 | M | 3 (9) | 11 (21) | |
| Adenylate kinase isoenzyme 4 | Q9WUS0 | 25 | M | 0 (0) | 1 (2) | |
| 13 | Ribonuclease UK114 | P52759 | 14 | Mt, Cy | 4 (24) | 6 (20) |
| Cytochrome c | P62898 | 12 | IMS | 4 (24) | 13 (45) | |
| Hemoglobin subunit β -1 | P02091 | 12 | Bl | 3 (18) | 2 (7) | |
| Fatty acid-binding protein, liver | P02692 | 12 | Cy | 3 (18) | 8 (28) | |
| Hemoglobin subunit α | P01946 | 12 | Bl | 3 (18) | 0 (0) | |
| 14 | 10-kDa heat shock protein | P26772 | 11 | M | 6 (75) | 14 (83) |
| Betaine-homocysteine S-methyltransferase | O09171 | 45 | Cy | 2 (25) | 2 (11) | |
In kDa.
Location of the individual proteins is shown by the following abbreviations: Bl, Red blood cell; Cy, cytosol; IMS, intermembrane space; Ly, lysosome; M, matrix; Mt, mitochondria; Pr, peroxisome; ?, unknown.
Number of peptides derived from each protein identified by LC-MS/MS analysis.
Relative proportions of the peptide number derived from the protein to the total number of the peptides detected in each protein band are shown.
NCBI accession number.
In protein bands C4 and C13 of Ca2+-treated mitochondria, the proportions of peptides derived from sulfite oxidase and cytochrome c were dominantly higher than those in non-treated mitochondria, respectively; and hence these two protein species were considered to be released from the Ca2+-treated mitochondria. As for the reason why the content of sulfite oxidase in band C4 was much higher for Ca2+-treated mitochondria than for non-treated mitochondria, we offer the following interpretation: Ca2+ treatment increases the permeability of the outer mitochondrial membrane even for proteins of the size of sulfite oxidase (50 kDa) but increases that of the inner mitochondrial membrane only for proteins smaller than sulfite oxidase. These differential effects of Ca2+ on the permeability of outer and inner mitochondrial membrane may result in the specific release of sulfite oxidase in band C4. We consider this interpretation to be reasonable because the staining intensities of protein bands smaller than this size in lane C in Fig. 4 were much stronger than those in lane D, but those of protein bands larger than this size in lane C were much weaker than those in lane D. As for the specific increase in cytochrome c in band C13 in Ca2+-treated mitochondria, high contents of non-mitochondrial proteins such as hemoglobin or fatty acid-binding protein in this fraction may be one of the major reasons causing the observed results. Other visible protein bands in the case of Ca2+-treated mitochondria showing signal intensities similar to those of the corresponding gel pieces in non-treated mitochondria mainly contained non-mitochondrial proteins such as catalase and Cu,Zn-superoxide dismutase, which were suspected to be contaminants arising during the preparation of the mitochondria (supplemental Tables SI and SII). Consequently our data suggest that, unlike valinomycin, Ca2+ causes the release of not only proteins located in the intermembrane space (e.g. adenylate kinase 2) but also those residing in the matrix (e.g. malate dehydrogenase).
Immunological Confirmation of the Protein Release from Valinomycin- and Ca2+-treated Mitochondria—
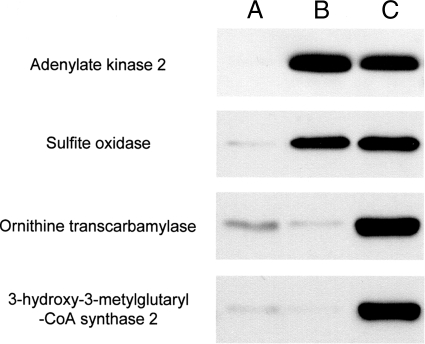
To reconfirm the results obtained from the above proteomics analysis, we also carried out immunological analysis. For this, we first raised antibodies against sulfite oxidase, adenylate kinase 2, hydroxymethylglutaryl-CoA synthase, and ornithine carbamoyltransferase: the former two are located in the intermembrane space, and the latter two are located in the matrix. These four proteins were the main proteins identified by LC-MS/MS analysis of bands B1, B2, C5, and C8, respectively. We used these antibodies for Western blot analysis of the supernatant samples. As shown in Fig. 5, adenylate kinase 2 and sulfite oxidase were detected in the supernatant prepared from mitochondria treated with valinomycin or with Ca2+, whereas ornithine carbamoyltransferase and hydroxymethylglutaryl-CoA synthase were detected in that from the organelles treated with Ca2+ but not in that from those treated with valinomycin. These results obtained by immunological analysis accord well with those found by our proteomics analysis.
Fig. 5.
Immunoblot confirmation of release of proteins from mitochondria. Lanes A–C represent the supernatants of mitochondria used in the experiments whose results are given in Fig. 3 or 4 (15 μl/lane). Proteins in these samples were subjected to immunoblotting under the same experimental conditions as indicated in the legend of Fig. 3. Adenylate kinase 2, sulfite oxidase, ornithine carbamoyltransferase, and 3-hydroxymethylglutaryl-CoA synthase 2 proteins present in individual samples were detected with their specific antibodies. A typical result of three independent runs is shown.
Analysis of the Selectivity of the Protein Release from Mitochondria—
To understand how proteins are released from mitochondria by valinomycin or Ca2+ treatment, it is important to reveal whether proteins are released from mitochondria in a selective manner or in a non-selective manner. We next tried to elucidate this problem by using proteome analysis. If the protein release occurs in an unselective manner, the released proteins would be those abundantly present in mitochondria because they would be expected to leak from mitochondria according to their expression or concentration levels. Conversely if the protein release occurs in a selective manner, the released proteins would not always necessarily be highly expressed proteins. To examine these possibilities, we first examined the abundantly expressed proteins in mitochondria. For this, all proteins of rat liver mitochondria were subjected to SDS-PAGE (Fig. 4, lane D), and then the gel was cut into pieces of ∼5-mm vertical thickness. The proteins included in these gel pieces were analyzed by using LC-MS/MS. The proteins identified here by LC-MS/MS can be roughly considered to be those abundantly expressed in mitochondria because the efficiency of identification of a protein by LC-MS/MS is generally considered to correlate coarsely with the amount of the protein. Table IV shows the 111 species of proteins identified by LC-MS/MS analysis of whole mitochondrial proteins. Mann and co-workers (19) identified ∼700 species of proteins in the proteome analysis of mitochondria isolated from rat various tissues. Thus, the proteins described in Table IV can be regarded as a part of all of the mitochondrial proteins and may be considered to be abundantly expressed in rat liver mitochondria. Actually the proteins known to be abundantly expressed in the liver mitochondria, such as carbamoyl-phosphate synthase 1 (18), ATP synthase subunit (20), phosphate carrier (21), and voltage-dependent anion channel 1 (17), appear in Table IV. We next examined whether the proteins revealed to be released from valinomycin- and Ca2+-treated mitochondria were also present in Table IV. As a result, 92% of the proteins released from mitochondria by valinomycin or by Ca2+ were present in Table IV. Therefore, only the protein species abundantly present in mitochondria were found to be released by valinomycin or Ca2+ treatment, and so we may conclude that the release of these proteins from valinomycin- and Ca2+-treated mitochondria occurred in an unselective manner.
Table IV.
LC-MS/MS profile of proteins expressed in mitochondria
The whole mitochondrial proteins obtained from gel pieces cut out from Fig. 4, lane D were subjected to LC-MS/MS analysis.
| Protein name | Aca | Molecular massb | Locc | Released |
|---|---|---|---|---|
| Carbamoyl-phosphate synthetase 1 | P07756 | 166 | M | Ca2+ |
| Pyruvate carboxylase | P52873 | 130 | M | |
| 2-Oxoglutarate dehydrogenase E1 component | Q5XI78 | 117 | M | |
| Ladybird homeobox corepressor 1 | P84551 | 101 | N | |
| Dimethylglycine dehydrogenase | Q63342 | 96 | Mt | |
| Glycerol-3-phosphate acyltransferase | P97564 | 95 | OM | |
| Aconitate hydratase | Q9ER34 | 86 | Mt | |
| Trifunctional enzyme subunit α | Q64428 | 83 | M | |
| NADH-ubiquinone oxidoreductase 75-kDa subunit | Q66HF1 | 80 | IM | |
| Peroxisomal multifunctional enzyme type 2 | P97852 | 80 | Pr | |
| Long-chain fatty acid CoA ligase 1 | P18163 | 79 | OM | |
| Peroxisomal bifunctional | P07896 | 79 | Pr | |
| Carnitine O-palmitoyltransferase 2 | P18886 | 75 | IM | |
| Stress-70 protein | P48721 | 74 | Mt | |
| Chaperone-activity of bc1 complex-like | Q5BJQ0 | 73 | Mt | |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit | Q920L2 | 73 | IM | |
| Very long chain-specific acyl-CoA dehydrogenase | P45953 | 71 | IM | |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | Q6UPE1 | 69 | IM | |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | P08461 | 68 | M | |
| Apoptosis-inducing factor 1 | Q9JM53 | 67 | IMS | |
| Carboxylesterase 3 | NP_579829e | 62 | ER | |
| Glutamate dehydrogenase 1 | P10860 | 62 | M | Ca2+ |
| 60-kDa heat shock protein | P63039 | 61 | M | |
| ATP synthase subunit α | P15999 | 60 | IM | |
| Catalase | P04762 | 60 | Pr | |
| UDP-glucuronosyltransferase 1-1 | Q64550 | 60 | Mic | |
| Amine oxidase (flavin-containing) B | P19643 | 59 | OM | |
| Propionyl-CoA carboxylase β chain | P07633 | 59 | M | |
| Seryl-tRNA synthetase, cytoplasmic | Q6P799 | 59 | Cy | |
| Alanine-glyoxylate aminotransferase 2 | Q64565 | 58 | Mt | |
| Methylmalonate-semialdehyde dehydrogenase | Q02253 | 58 | M | Ca2+ |
| Aldehyde dehydrogenase | P11884 | 57 | M | |
| Hydroxymethylglutaryl-CoA synthase | P22791 | 57 | M | Ca2+ |
| Protein-disulfide isomerase | P04785 | 57 | ER | |
| 4-Aminobutyrate aminotransferase | P50554 | 57 | M | |
| Amyloid β A4 precursor protein binding family B member 3 | O35827 | 56 | ? | |
| ATP synthase subunit β | P10719 | 56 | IM | |
| Dihydrolipoyl dehydrogenase | Q6P6R2 | 55 | M | Ca2+ |
| Fumarate hydratase | P14408 | 55 | Mt, Cy | |
| Neural Wiskott-Aldrich syndrome protein | O08816 | 55 | Cy, N | |
| Sulfite oxidase | Q07116 | 55 | IMS | Ca2+, val |
| Ubiquinol-cytochrome-c reductase complex core protein 1 | Q68FY0 | 54 | IM | |
| Trifunctional enzyme subunit β | Q60587 | 52 | M | |
| Acyl-CoA thioesterase 2 | O55171 | 50 | M | |
| 2-Oxoisovalerate dehydrogenase subunit α | P11960 | 50 | M | |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | Q01205 | 49 | Mt | |
| Ornithine aminotransferase | P04182 | 49 | M | |
| Aspartate aminotransferase | P00507 | 48 | M | Ca2+ |
| Kynurenine/α-aminoadipate aminotransferase | Q64602 | 48 | Mt | |
| Long chain-specific acyl-CoA dehydrogenase | P15650 | 48 | M | |
| Short/branched chain-specific acyl-CoA dehydrogenase | P70584 | 48 | M | |
| Ubiquinol-cytochrome-c reductase complex core protein 2 | P32551 | 48 | IM | |
| Bile acid CoA:amino acid N-acyltransferase | Q63276 | 47 | Pr | |
| Isovaleryl-CoA dehydrogenase | P12007 | 47 | M | |
| Medium chain-specific acyl-CoA dehydrogenase | P08503 | 47 | M | Ca2+ |
| Acetyl-CoA acetyltransferase | P17764 | 45 | Mt | Ca2+ |
| Betaine-homocysteine S-methyltransferase | O09171 | 45 | Cy | |
| Short-chain specific acyl-CoA dehydrogenase | P15651 | 45 | M | Ca2+ |
| α-Methylacyl-CoA racemase | P70473 | 42 | Mt, Pr | Ca2+ |
| 3β-Hydroxysteroid dehydrogenase type 5 | P27364 | 42 | ER | |
| 3-Ketoacyl-CoA thiolase | P13437 | 42 | Mt | Ca2+ |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex subunit 10 | Q561S0 | 41 | M | |
| Ornithine carbamoyltransferase | P00481 | 40 | M | Ca2+ |
| Phosphate carrier | P16036 | 40 | IM | |
| d-β-Hydroxybutyrate dehydrogenase | P29147 | 39 | M | |
| Proto-oncogene tyrosine-protein kinase FER | P09760 | 37 | ? | |
| Δ3,5-Δ2,4-Dienoyl-CoA isomerase | Q62651 | 36 | Mt, Pr | Ca2+ |
| Malate dehydrogenase | P04636 | 36 | M | Ca2+ |
| Sideroflexin-1 | Q63965 | 36 | Mt membr | |
| 2,4-Dienoyl-CoA reductase | Q64591 | 36 | Mt | Ca2+ |
| 3-Hydroxyisobutyrate dehydrogenase | P29266 | 36 | Mt | Ca2+ |
| Electron transfer flavoprotein subunit α | P13803 | 35 | M | Ca2+ |
| Hydroxyacyl-CoA dehydrogenase | Q9WVK7 | 35 | M | Ca2+ |
| Hydroxymethylglutaryl-CoA lyase | P97519 | 35 | M | Ca2+ |
| Uricase | P09118 | 35 | Pr | |
| Tricarboxylate transport protein | P32089 | 34 | IM | |
| Carnitine/acylcarnitine carrier | P97521 | 33 | IM | |
| Peroxisomal trans-2-enoyl-CoA reductase | Q9WVK3 | 33 | Pr | Ca2+ |
| Prohibitin-2 | Q5XIH7 | 33 | IM, Cy, N | |
| Thiosulfate sulfurtransferase | P24329 | 33 | M | Ca2+ |
| 3-Mercaptopyruvate sulfurtransferase | P97532 | 33 | Mt, Cy | Ca2+ |
| Enoyl-CoA hydratase | P14604 | 32 | M | Ca2+ |
| 3,2-trans-Enoyl-CoA isomerase | P23965 | 32 | M | Ca2+ |
| Voltage-dependent anion channel 1 | Q9Z2L0 | 31 | OM | |
| Voltage-dependent anion channel 3 | Q9R1Z0 | 31 | OM | |
| ATP synthase γ chain | P35435 | 30 | IM | |
| Prohibitin | P67779 | 30 | IM | |
| Ubiquinol-cytochrome-c reductase iron-sulfur subunit | NP_001008888e | 30 | IM | |
| ATP synthase B chain | P19511 | 29 | IM | |
| Electron transfer flavoprotein subunit β | Q68FU3 | 28 | M | Ca2+ |
| ES1 protein homolog | P56571 | 28 | Mt | |
| NADH dehydrogenase (ubiquinone) flavoprotein 2 | P19234 | 28 | IM | |
| Adenylate kinase isoenzyme 2 | P29410 | 27 | IMS | Ca2+, val |
| 3-Hydroxyacyl-CoA dehydrogenase type-2 | O70351 | 27 | Mt | |
| Cytochrome c oxidase subunit 2 | P00406 | 26 | IM | |
| Serum amyloid P component | P23680 | 26 | S | Ca2+ |
| Aldehyde dehydrogenase family 7 member A1 | Q64057 | 25 | ? | |
| Glutathione S-transferase Kappa 1 | P24473 | 25 | M | |
| Superoxide dismutase (manganese) | P07895 | 25 | M | |
| ATP synthase O subunit | Q06647 | 23 | IM | |
| Peptidyl-prolyl cis-trans isomerase | P29117 | 22 | M | Ca2+ |
| Cytochrome c oxidase subunit 4 isoform 1 | P10888 | 20 | IM | |
| ATP synthase D chain | P31399 | 19 | IM | |
| ATP synthase δ chain | P35434 | 18 | IM | |
| Succinate dehydrogenase (ubiquinone) iron-sulfur protein | P21913 | 17 | IM | |
| Cytochrome c oxidase subunit 5A | P11240 | 16 | IM | |
| Cytochrome c oxidase subunit 5B | P12075 | 14 | IM | |
| Ribonuclease UK114 | P52759 | 14 | Mt, Cy, N | |
| Cytochrome c | P62898 | 12 | IMS | Ca2+, val |
| ATP synthase e chain | P29419 | 8 | IM | |
| ATP synthase protein 8 | P11608 | 8 | IM |
Accession number from UniProt.
In kDa.
Location of the individual proteins is shown by the following abbreviations: Cy, cytosol; ER, endoplasmic reticulum; IM, inner membrane; IMS, intermembrane space; M, matrix; Mic, microsome; Mt, mitochondria; Mt membr, mitochondrial membrane; N, nuclei; OM, outer membrane; Pr, peroxisome; S, secretase; ?, unknown.
The proteins revealed to be released from valinomycin-treated or Ca2+-treated mitochondria are shown as val and Ca2+, respectively.
NCBI accession number.
DISCUSSION
In this study, we performed proteome analysis of the proteins released from valinomycin- and Ca2+-treated mitochondria. Until now, there was no report on proteomics studies on the proteins released from valinomycin-treated mitochondria. On the contrary, several studies reported the results of proteome analysis of the proteins released from PT-induced mitochondria. Patterson et al. (22) performed such an analysis of the proteins released from mitochondria with PT induced by atractyloside. Different from the results of our present study on the proteins released from mitochondria with PT induced by Ca2+ in which no membrane protein was detected as stated above, they detected three inner membrane proteins such as electron transfer flavoprotein α-subunit precursor in the released proteins. As was shown in our Fig. 4, the supernatant of non-treated mitochondria also showed numerous proteins, and complete elimination of proteins reflecting this background was difficult. Nevertheless Patterson et al. (22) simply applied the supernatant of mitochondria treated with atractyloside to the LC-MS/MS analysis without taking the background into account. For this reason, the samples used as the proteins released from mitochondria in their study seemed to be contaminated by a number of non-released proteins. To identify the proteins specifically released from PT-induced mitochondria, differential analysis between samples prepared from PT-induced mitochondria and from non-treated mitochondria, as performed in this study, is essential. Recently to eliminate the background proteins, Teilum et al. (23) prepared the released protein samples by using mitochondria immobilized on a sponge-like material, cryogel monoliths. As a result of the proteome analysis of thus prepared samples, they showed 68 proteins released from Ca2+-treated mitochondria. Of these, 30 proteins were also identified in the present study; however, two important proteins, i.e. adenylate kinase 2 and sulfite oxidase, which were released from either valinomycin-treated or Ca2+-treated mitochondria, were not identified in their study. The exact reason why these two proteins were not identified in their study is uncertain. One possible explanation for this discrepancy is that immobilization of mitochondria on the cryogel monoliths affects the property of the samples. Further careful examinations are necessary to understand the reason causing such differences. The two above cited studies were performed to enable effective screening of novel apoptosis-inducible proteins and to develop a novel method to prepare released protein samples, respectively, and selectivity of proteins released from mitochondria was not discussed.
In this study, we demonstrated that valinomycin caused the specific release of proteins located in the intermembrane space in an unselective manner. This result agrees with that of a previous study showing immunologically that not only cytochrome c but also adenylate kinase 2 is released from valinomycin-treated mitochondria (24). The exact mechanisms governing how these proteins located in the intermembrane space were released are unclear. However, valinomycin is thought to cause the specific rupture of the outer membrane at least under the experimental conditions used because the intermembrane space proteins having high molecular mass, such as sulfite oxidase (55 kDa), were also released.
Moreover we also demonstrated that treatment of mitochondria with Ca2+ caused the release of proteins located not only in the intermembrane space but also in the matrix. This finding indicates that both outer membrane and inner membranes are ruptured in Ca2+-treated mitochondria. Possible rupture of the outer mitochondrial membrane accompanied by induction of PT has been proposed by multiple researchers (8, 25, 26). These conclusions were mainly made based on morphological analysis of PT-induced mitochondria by electron microscopy or tomography (27–29). In the present study, during the configuration analysis of Ca2+-treated mitochondria by electron microscopy (Fig. 2), we also observed mitochondria with a ruptured outer membrane (data not shown). Thus, rupture of the outer mitochondrial membrane in PT-induced mitochondria was confirmed by both proteomics and structural analyses.
On the contrary, rupture of the inner mitochondrial membrane in PT-induced mitochondria has hardly ever been reported. Pfeiffer and co-workers (30) reported that the electrophoresis pattern of the proteins released from Ca2+-treated mitochondria was similar to that of the soluble protein fraction of mitochondria. This result may indicate the release of matrix proteins from PT-induced mitochondria, but analysis of these protein bands at the molecular level was not performed. In another report, possible rupture of the inner membrane of PT-induced mitochondria was proposed (31), but no experimental data supporting this statement were presented. In the present study, by using proteomics and immunological techniques, we showed that the proteins located in the matrix space could also be released by induction of PT. By the increased permeability of the mitochondrial inner membrane caused by Ca2+, proteins smaller than 50 kDa seemed to be easily released as evident from the staining intensities of the released proteins (Fig. 4C) and from the results showing the specific release of sulfite oxidase (band C4 in Table III).
It has been well established that the PT pore allows the mitochondrial inner membrane to be permeable to solutes or ions smaller than 1.5 kDa. This contradicts with our finding that matrix proteins bigger than 1.5 kDa such as ornithine carbamoyltransferase (40 kDa) were released from Ca2+-treated mitochondria. This contradiction may be interpreted as follows. The inner membrane of the Ca2+-treated mitochondria becomes permeable to solutes or ions smaller than 1.5 kDa by opening of the PT pores, and the inner membrane of some of these mitochondria becomes permeable to macromolecules probably by rupture following PT. Up to now, the mitochondrial apoptosis inducers have been considered to be present in the intermembrane space. However, the finding that matrix proteins are also released from PT-induced mitochondria suggests that the novel apoptosis inducers may be present not only in the intermembrane space but also in the mitochondrial matrix.
In this study, apoptosis-inducible proteins besides cytochrome c, such as caspase 9, AIF, Smac/DIABLO, endonuclease G, and Omi/HtrA2 (32–37), were not identified in the supernatants by proteomics analysis, but Smac/DIABLO was detected in the supernatant of valinomycin- or Ca2+-treated mitochondria by immunological analysis (supplemental Fig. S2). The main reason for the difficulty in observation of these proteins is possibly their low abundance. Actually proteins other than AIF were not observed by the proteome analysis of the whole mitochondrial proteins (Table IV). AIF is considered to be expressed more abundantly than the other apoptotic proteins (Table IV); however, it was not observed in the supernatants of Ca2+- or valinomycin-treated mitochondria (Tables II and III), indicating that AIF is not released from valinomycin- or Ca2+-treated mitochondria. Results on the localization of AIF in valinomycin-treated mitochondria are in accord with the results reported by Gogvadze et al. (24). On the contrary, two studies reported the release of AIF from PT-induced mitochondria by Ca2+ (38, 39). The exact reason for the discrepant behaviors of AIF in PT-induced mitochondria is uncertain. Possibly its distribution in PT-induced mitochondria may significantly dependent upon the experimental conditions used because AIF is also reported to be anchored to the inner mitochondrial membrane (40, 41).
In summary, we performed proteome analysis of the proteins released from valinomycin- and Ca2+-treated mitochondria. As a result, we succeeded in demonstrating differences in protein release from mitochondria treated with valinomycin or with Ca2+, suggesting differential permeabilization effects of Ca2+ and valinomycin on the inner and outer mitochondrial membranes. These findings are important to understand how the release of mitochondrial proteins is achieved.
Footnotes
Published, MCP Papers in Press, February 14, 2009, DOI 10.1074/mcp.M800377-MCP200
The abbreviations used are: PT, permeability transition; AIF, apoptosis-inducing factor; VDAC, voltage-dependent anion channel; CBB, Coomassie Brilliant Blue; DIABLO, direct IAP-binding protein with low pI.
This work was supported by research funding from the “knowledge cluster initiative” of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and by Grant-in-aid for Scientific Research 17076011 from the MEXT of Japan.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Gunter, T. E., and Pfeiffer, D. R. ( 1990) Mechanisms by which mitochondria transport calcium. Am. J. Physiol. Cell Physiol. 258, C755–786 [DOI] [PubMed] [Google Scholar]
- 2.Zoratti, M., and Szabò, I. ( 1995) The mitochondrial permeability transition. Biochim. Biophys. Acta 1241, 139–176 [DOI] [PubMed] [Google Scholar]
- 3.Bernardi, P., Colonna, R., Costantini, P., Eriksson, O., Fontaine, E., Ichas, F., Massari, S., Nicolli, A., Petronilli, V., and Scorrano, L. ( 1998) The mitochondrial permeability transition. Biofactors 8, 273–281 [DOI] [PubMed] [Google Scholar]
- 4.Crompton, M. ( 1999) The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 341, 233–249 [PMC free article] [PubMed] [Google Scholar]
- 5.Kroemer, G., and Reed, J. C. ( 2000) Mitochondrial control of cell death. Nat. Med. 6, 513–519 [DOI] [PubMed] [Google Scholar]
- 6.Bernardi, P., Petronilli, V., Di Lisa, F., and Forte, M. ( 2001) A mitochondrial perspective on cell death. Trends Biochem. Sci. 26, 112–117 [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer, D. R., Gudz, T. I., Novgorodov, S. A., and Erdahl, W. L. ( 1995) The peptide mastoparan is a potent facilitator of the mitochondrial permeability transition. J. Biol. Chem. 270, 4923–4932 [DOI] [PubMed] [Google Scholar]
- 8.Morin, D., Barthélémy, S., Zini, R., Labidalle, S., and Tillement, J. P. ( 2001) Curcumin induces the mitochondrial permeability transition pore mediated by membrane protein thiol oxidation. FEBS Lett. 495, 131–136 [DOI] [PubMed] [Google Scholar]
- 9.Zhu, Y., Xu, H., and Huang, K. ( 2002) Mitochondrial permeability transition and cytochrome c release induced by selenite. J. Inorg. Biochem. 90, 43–50 [DOI] [PubMed] [Google Scholar]
- 10.Shinohara, Y., Bandou, S., Kora, S., Kitamura, S., Inazumi, S., and Terada, H. ( 1998) Cationic uncouplers of oxidative phosphorylation are inducers of mitochondrial permeability transition. FEBS Lett. 428, 89–92 [DOI] [PubMed] [Google Scholar]
- 11.Almofti, M. R., Ichikawa, T., Yamashita, K., Terada, H., and Shinohara, Y. ( 2003) Silver ion induces a cyclosporine a-insensitive permeability transition in rat liver mitochondria and release of apoptogenic cytochrome C. J. Biochem. 134, 43–49 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto, T., Tachikawa, A., Terauchi, S., Yamashita, K., Kataoka, M., Terada, H., and Shinohara, Y. ( 2004) Multiple effects of DiS-C3(5) on mitochondrial structure and function. Eur. J. Biochem. 271, 3573–3579 [DOI] [PubMed] [Google Scholar]
- 13.Deleted in proof
- 14.Shinohara, Y., Almofti, M. R., Yamamoto, T., Ishida, T., Kita, F., Kanzaki, H., Ohnishi, M., Yamashita, K., Shimizu, S., and Terada, H. ( 2002) Permeability transition-independent release of mitochondrial cytochrome c induced by valinomycin. Eur. J. Biochem. 269, 5224–5230 [DOI] [PubMed] [Google Scholar]
- 15.Myers, D. K., and Slater, E. C. ( 1957) The enzymic hydrolysis of adenosine triphosphate by liver mitochondria. Biochem. J. 67, 558–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto, T., Yamada, A., Watanabe, M., Yoshimura, Y., Yamazaki, N., Yoshimura, Y., Yamauchi, T., Kataoka, M., Nagata, T., Terada, H., and Shinohara, Y. ( 2006) VDAC1, having a shorter N-terminus than VDAC2 but showing the same migration in an SDS-polyacrylamide gel, is the predominant form expressed in mitochondria of various tissues. J. Proteome Res. 5, 3336–3344 [DOI] [PubMed] [Google Scholar]
- 18.Ozaki, M., Terada, K., Kanazawa, M., Fujiyama, S., Tomita, K., and Mori, M. ( 1994. –1995) Enzyme-linked immunosorbent assay of carbamoylphosphate synthetase I: plasma enzyme in rat experimental hepatitis and its clearance. Enzyme Protein 48, 213–221 [DOI] [PubMed] [Google Scholar]
- 19.Forner, F., Foster, L. J., Campanaro, S., Valle, G., and Mann, M. ( 2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 20.Izquierdo, J. M., Luis, A. M., and Cuezva, J. M. ( 1990) Postnatal mitochondrial differentiation in rat liver. Regulation by thyroid hormones of the β-subunit of the mitochondrial F1-ATPase complex. J. Biol. Chem. 265, 9090–9097 [PubMed] [Google Scholar]
- 21.Fiermonte, G., Dolce, V., and Palmieri, F. ( 1998) Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem. 273, 22782–22787 [DOI] [PubMed] [Google Scholar]
- 22.Patterson, S. D., Spahr, C. S., Daugas, E., Susin, S. A., Irinopoulou, T., Koehler, C., and Kroemer, G. ( 2000) Mass spectrometric identification of proteins released from mitochondria undergoing permeability transition. Cell Death Differ. 7, 137–144 [DOI] [PubMed] [Google Scholar]
- 23.Teilum, M., Hansson, M. J., Dainiak, M. B., Månsson, R., Surve, S., Elmér, E., Onnerfjord, P., and Mattiasson, G. ( 2006) Binding mitochondria to cryogel monoliths allows detection of proteins specifically released following permeability transition. Anal. Biochem. 348, 209–221 [DOI] [PubMed] [Google Scholar]
- 24.Gogvadze, V., Robertson, J. D., Enoksson, M., Zhivotovsky, B., and Orrenius, S. ( 2004) Mitochondrial cytochrome c release may occur by volume-dependent mechanisms not involving permeability transition. Biochem. J. 378, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Heiden, M. G., and Thompson, C. B. ( 1999) Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis? Nat. Cell Biol. 8, E209–E216 [DOI] [PubMed] [Google Scholar]
- 26.Green, D. R., and Kroemer, G. ( 2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 27.Vander Heiden, M. G., Chandel, N. S., Williamson, E. K., Schumacker, P. T., and Thompson, C. B. ( 1997) Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91, 627–637 [DOI] [PubMed] [Google Scholar]
- 28.Kwong, J., Choi, H. L., Huang, Y., and Chan, F. L. ( 1999) Ultrastructural and biochemical observations on the early changes in apoptotic epithelial cells of the rat prostate induced by castration. Cell Tissue Res. 298, 123–136 [DOI] [PubMed] [Google Scholar]
- 29.Frey, T. G., Renken, C. W., and Perkins, G. A. ( 2002) Insight into mitochondrial structure and function from electron tomography. Biochim. Biophys. Acta 1555, 196–203 [DOI] [PubMed] [Google Scholar]
- 30.Igbavboa, U., Zwizinski, C. W., and Pfeiffer, D. R. ( 1989) Release of mitochondrial matrix proteins through a Ca2+-requiring, cyclosporin-sensitive pathway. Biochem. Biophys. Res. Commun. 161, 619–625 [DOI] [PubMed] [Google Scholar]
- 31.Garrido, C., Galluzzi, L., Brunet, M., Puig, P. E., Didelot, C., and Kroemer, G. ( 2006) Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 13, 1423–1433 [DOI] [PubMed] [Google Scholar]
- 32.Susin, S. A., Lorenzo, H. K., Zamzami, N., Marzo, I., Snow, B. E., Brothers, G. M., Mangion, J., Jacotot, E., Costantini, P., Loeffler, M., Larochette, N., Goodlett, D. R., Aebersold, R., Siderovski, D. P., Penninger, J. M., and Kroemer, G. ( 1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446 [DOI] [PubMed] [Google Scholar]
- 33.Verhagen, A. M., Ekert, P. G., Pakusch, M., Silke, J., Connolly, L. M., Reid, G. E., Moritz, R. L., Simpson, R. J., and Vaux, D. L. ( 2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102, 43–53 [DOI] [PubMed] [Google Scholar]
- 34.Du, C., Fang, M., Li, Y., Li, L., and Wang, X. ( 2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42 [DOI] [PubMed] [Google Scholar]
- 35.Davies, A. M., Hershman, S., Stabley, G. J., Hoek, J. B., Peterson, J., and Cahill, A. ( 2003) A Ca2+-induced mitochondrial permeability transition causes complete release of rat liver endonuclease G activity from its exclusive location within the mitochondrial intermembrane space. Identification of a novel endo-exonuclease activity residing within the mitochondrial matrix. Nucleic Acids Res. 31, 1364–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martins, L. M., Iaccarino, I., Tenev, T., Gschmeissner, S., Totty, N. F., Lemoine, N. R., Savopoulos, J., Gray, C. W., Creasy, C. L., Dingwall, C., and Downward, J. ( 2002) The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277, 439–444 [DOI] [PubMed] [Google Scholar]
- 37.Tang, X., Gao, J., Chen, J., Fang, F., Wang, Y., Dou, H., Xu, Q., and Qian, Z. ( 2005) Inhibition by ursolic acid on calcium-induced mitochondrial permeability transition and release of two proapoptotic proteins. Biochem. Biophys. Res. Commun. 337, 320–324 [DOI] [PubMed] [Google Scholar]
- 38.Yuste, V. J., Moubarak, R. S., Delettre, C., Bras, M., Sancho, P., Robert, N., d’Alayer, J., and Susin, S. A. ( 2005) Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 12, 1445–1448 [DOI] [PubMed] [Google Scholar]
- 39.Arnoult, D., Parone, P., Martinou, J. C., Antonsson, B., Estaquier, J., and Ameisen, J. C. ( 2002) Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J. Cell Biol. 159, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uren, R. T., Dewson, G., Bonzon, C., Lithgow, T., Newmeyer, D. D., and Kluck, R. M. ( 2005) Mitochondrial release of pro-apoptotic proteins. Electrostatic interactions can hold cytochrome c but not Smac/DIABLO to mitochondrial membranes. J. Biol. Chem. 280, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 41.Muñoz-Pinedo, C., Guío-Carrión, A., Goldstein, J. C., Fitzgerald, P., Newmeyer, D. D., and Green, D. R. ( 2006) Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc. Natl. Acad. Sci. U. S. A. 103, 11573–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]