Abstract
Tamoxifen resistance is a major cause of death in patients with recurrent breast cancer. Current clinical factors can correctly predict therapy response in only half of the treated patients. Identification of proteins that are associated with tamoxifen resistance is a first step toward better response prediction and tailored treatment of patients. In the present study we intended to identify putative protein biomarkers indicative of tamoxifen therapy resistance in breast cancer using nano-LC coupled with FTICR MS. Comparative proteome analysis was performed on ∼5,500 pooled tumor cells (corresponding to ∼550 ng of protein lysate/analysis) obtained through laser capture microdissection (LCM) from two independently processed data sets (n = 24 and n = 27) containing both tamoxifen therapy-sensitive and therapy-resistant tumors. Peptides and proteins were identified by matching mass and elution time of newly acquired LC-MS features to information in previously generated accurate mass and time tag reference databases. A total of 17,263 unique peptides were identified that corresponded to 2,556 non-redundant proteins identified with ≥2 peptides. 1,713 overlapping proteins between the two data sets were used for further analysis. Comparative proteome analysis revealed 100 putatively differentially abundant proteins between tamoxifen-sensitive and tamoxifen-resistant tumors. The presence and relative abundance for 47 differentially abundant proteins were verified by targeted nano-LC-MS/MS in a selection of unpooled, non-microdissected discovery set tumor tissue extracts. ENPP1, EIF3E, and GNB4 were significantly associated with progression-free survival upon tamoxifen treatment for recurrent disease. Differential abundance of our top discriminating protein, extracellular matrix metalloproteinase inducer, was validated by tissue microarray in an independent patient cohort (n = 156). Extracellular matrix metalloproteinase inducer levels were higher in therapy-resistant tumors and significantly associated with an earlier tumor progression following first line tamoxifen treatment (hazard ratio, 1.87; 95% confidence interval, 1.25–2.80; p = 0.002). In summary, comparative proteomics performed on laser capture microdissection-derived breast tumor cells using nano-LC-FTICR MS technology revealed a set of putative biomarkers associated with tamoxifen therapy resistance in recurrent breast cancer.
Tamoxifen is an antiestrogenic agent that has been widely and successfully used in the treatment of breast cancer over the past decades (1). Tamoxifen targets and inhibits the estrogen receptor-α, which is expressed in ∼70% of all primary breast tumors and is known to be important in the development and course of the disease. When diagnosed at an early stage, adjuvant systemic tamoxifen therapy can cure ∼10% of the patients (1). In recurrent disease, ∼50% of patients have no benefit from tamoxifen (intrinsic resistance). From the other half of patients who initially respond to therapy with an objective response (OR)1 or no change (NC), a majority eventually develop progressive disease (PD) due to acquired tamoxifen resistance (2, 3). With the markers available to date we can insufficiently predict therapy response. Therefore, identification of new biomarkers that can more effectively predict response to treatment and that can potentially function as drug targets is a major focus of research.
The search for new biomarkers has been enhanced by the introduction of microarray technology. Gene expression studies have resulted in a whole spectrum of profiles for e.g. molecular subtypes, prognosis, and therapy prediction in breast cancer (4–10). Corresponding studies at the protein level are lagging behind because of immature technology. However, protein-level information is crucial for the functional understanding and the ultimate translation of molecular knowledge into clinical practice, and proteomics technologies continue to progress at a rapid pace.
Proteomics studies reported so far have mainly been performed with breast cancer cell lines using either two-dimensional gel electrophoresis (11–14) or LC-MS for protein separation (15–17). However, it is known that the proteomic makeup of a cultured cell is rather different from that of a tumor cell surrounded by its native microenvironment (18). Furthermore cell lines lack the required follow-up information for answering important clinical questions. In addition, tumor tissues in general and breast cancer tissues in particular are very heterogeneous in the sense that they harbor many different cell types, such as stroma, normal epithelium, and tumor cells. LCM technology has emerged as an ideal tool for selectively extracting cells of interest from their natural environment (19) and has therefore been an important step forward in the context of genomics and proteomics cancer biomarker discovery research. LCM-derived breast cancer tumor cells have been used for comparative proteomics analyses in the past using both two-dimensional gel electrophoresis (20, 21) and LC-MS (22). This has resulted in the identification of proteins involved in breast cancer prognosis (21) and metastasis (20, 22). Although these studies demonstrated that proteomics technology has advanced to the level where it can contribute to biomarker discovery, major drawbacks, such as large sample requirements (42–700 μg) and low proteome coverage (50–76 proteins), for small amounts of starting material (∼1 μg) persist. Because clinical samples are often available in limited quantities, in-depth analysis of minute amounts of material (<1 μg) necessitates advanced technologies with sufficient sensitivity and depth of coverage.
Recently we demonstrated the applicability of nano-LC-FTICR MS in combination with the accurate mass and time (AMT) tag approach for proteomics characterization of ∼3,000 LCM-derived breast cancer cells (23). This study showed that proteome coverage was improved compared with conventional techniques. The AMT tag approach initially utilizes conventional LC-MS/MS measurements to establish a reference database of AMT tags specific for a particular proteome sample (e.g. breast cancer tissue). Each tag consists of a theoretical mass calculated from the peptide sequence, an LC normalized elution time (NET) value, and an indicator of quality. The AMT tag database serves as a “lookup table” for identifying peptides in subsequent quantitative LC-MS analyses. Substituting routine LC-MS/MS analyses (shotgun approach) with LC-FTICR MS analyses (AMT tag approach) significantly increases overall throughput and sensitivity while reducing sample requirements. Additionally quantitative intensity information related to the abundance of the protein can be discerned from these MS analyses (24). In the present study, we used the same strategy to analyze eight pools of tumor cells in duplicate or triplicate (resulting in 19 samples) derived from 51 fresh frozen primary invasive breast carcinomas that appeared to be either sensitive or resistant to tamoxifen treatment after recurrence. This work resulted in the identification of a putative protein profile associated with tamoxifen therapy resistance. In addition, the top discriminating protein of the putative profile, extracellular matrix metalloproteinase inducer (EMMPRIN), was validated in an independent patient cohort and was significantly associated with resistance to tamoxifen therapy and shorter time to progression upon tamoxifen treatment in recurrent breast cancer.
EXPERIMENTAL PROCEDURES
Patients and Tumor Tissues—
For the discovery phase of the study, 51 different fresh frozen primary breast cancer tissues from our liquid N2 tissue bank were used. Primary tumors were selected from patients that did not receive any systemic adjuvant hormonal therapy and were treated with the antiestrogen tamoxifen as first line therapy upon detection of recurrent breast cancer. Furthermore tumors were selected on the basis of positive estrogen receptor-α expression as assessed by ligand binding assay or enzyme-linked immunosorbent assay (≥10 fmol/mg of cytosolic protein). Tumor tissues were divided into two classes based on the type of response to tamoxifen therapy. 24 tumors were sensitive to tamoxifen therapy, showing either complete remission (CR) or partial remission (PR), and were assigned as OR. 27 tumors were resistant to therapy, showing an increase in tumor size, and were designated as PD. Clinical response was defined by standards of the International Union against Cancer criteria of tumor response (25). 20 of the above mentioned tumor tissues were selected for the verification study. Tissues were included based on their high tumor cell content of >70%. Tumor cell content was judged after hematoxylin/eosin stain of a separately cut 4-μm tissue section.
For immunohistochemical validation, a primary breast tissue microarray (TMA) containing 0.6-μm cores of formalin-fixed paraffin-embedded tumors was used. Within the TMA, there were 156 tumor tissues from patients that received tamoxifen as first line treatment upon recurrence. Median follow-up of patients alive after primary surgery was 103 months (range, 16–222 months) and 51 months after the onset of tamoxifen treatment (range, 9–136 months). Included patients showed CR, PR, PD, and NC of >6 and ≤6 months. Further patient and tumor characteristics are summarized in Table IV.
Table IV.
Patient characteristics
Patient and tumor characteristics for samples included in the validation set are shown.
ER, estrogen receptor α; PgR, progesterone receptor.
| Characteristics | Numbers | Median | Percent |
|---|---|---|---|
| Patients | 130 | 100 | |
| Age (years) | |||
| Primary surgery | 53.5 | ||
| Start first line | 56.5 | ||
| Menopausal status at start first line | |||
| Pre | 40 | 30.8 | |
| Post | 90 | 69.2 | |
| ER (fmol/mg protein) | 97 | ||
| PgR (fmol/mg protein) | 54.5 | ||
| Response | |||
| Clinical benefit: CR, PR, S.D. ≥ 6 months | 77 | 59.2 | |
| No clinical benefit: S.D. < 6 months, PD | 53 | 40.8 | |
| Dominant site of relapse | |||
| Local regional relapse | 15 | 11.5 | |
| Bone | 65 | 50.0 | |
| Other | 50 | 38.5 | |
| Disease-free interval (months) | |||
| ≤12 | 16 | 12.3 | |
| 12–36 | 59 | 45.4 | |
| ≥36 | 55 | 42.3 | |
| Nodal status | |||
| N0 | 62 | 47.7 | |
| N1–3 | 30 | 23.1 | |
| N > 3 | 34 | 26.2 | |
| Unknown | 4 | 3.1 | |
| Tumor size | |||
| ≤2 cm | 67 | 51.5 | |
| >2 cm | 63 | 48.5 | |
| Tumor grade | |||
| Poor | 46 | 35.4 | |
| Unknown | 52 | 40.0 | |
| Good/moderate | 32 | 24.6 |
This study was approved by the Medical Ethics Committee of the Erasmus Medical Center Rotterdam, The Netherlands (MEC 02.953) and was performed in accordance to the Code of Conduct of the Federation of Medical Scientific Societies in The Netherlands, and wherever possible we adhered to the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) (26).
Laser Capture Microdissection—
LCM was performed on 8-μm tissue cryosections that were fixed in ice-cold 70% ethanol and stained with hematoxylin as described previously (27). Briefly slides were washed in Milli-Q water, stained for 30 s in hematoxylin, washed again in Milli-Q water, subsequently dehydrated twice in 50, 70, 95, and 100% ethanol for 30 s each, and air-dried. Laser microdissection and pressure catapulting was performed directly after staining. Tumor epithelial cells were collected, using a P.A.L.M. LCM device, type P-MB (P.A.L.M. Microlaser Technologies AG, Bernried, Germany). From each cryosection an area of ∼500,000 μm2 that corresponds to ∼4,000 cells (area × slide thickness/1,000-μm3 cell volume) was collected in P.A.L.M. tube caps containing 10 μl of 0.1% RapiGest (Waters Corp., Milford, MA) and then spun down into 0.5-ml Eppendorf Protein LoBind tubes (Eppendorf, Hamburg, Germany). Collected cells were stored at −80 °C until further processing. Because we used small numbers of microdissected cells in this study, the protein concentration was typically below the detection limit of any protein assay. Hence the protein concentration for samples undergoing LC-MS analysis was estimated based on microdissected tissue area and extrapolations from protein assays performed on whole tissue lysates (i.e. ∼4,000 cells corresponds to ∼400 ng of total protein).
Sample Preparation—
Microdissected cell batches were pooled into OR and PD tumor groups (corresponding to ∼25,000 cells/pool) prior to sample preparation. Briefly cells were lysed by sonication directly in RapiGest solution using an Ultrasonic Disruptor Sonifier II (Model W-250/W-450, Branson Ultrasonics, Danbury, CT) for 1 min at 60% amplitude. Proteins were subsequently equilibrated for 2 min at 37 °C, denatured at 99 °C for 5 min, and processed for overnight trypsin digestion according to the instructions of the manufacturer using MS-grade porcine modified trypsin gold (Promega, Madison, WI) at a 1:20 (w/v) ratio as described previously (23). Digestion was stopped by incubation with 0.5% TFA at 37 °C for 30 min. Remaining cellular debris were spun down for 20 min at 10,600 × g, and supernatant was transferred to a new Eppendorf LoBind cup. Peptides were lyophilized and stored at −80 °C until further analysis. Prior to FTICR MS analysis, samples were reconstituted in 18 μl of NH4HCO3, vortexed briefly, and spun down again for 10 min at 10,600 × g to pellet any contaminating particulate material.
For the verification study, whole tissue lysates were prepared from 20 tumor tissues from which 6 × 4-μm cryosections per sample were cut. Tissue cryosections were placed in a Teflon container, frozen in liquid N2, and then pulverized in a frozen state in a microdismembrator (Braun Biotech International). The resulting powder was resuspended in 100 μl of 0.1% RapiGest. Cell lysis and trypsin digestion were performed as described above. Prior to trypsin digestion, a BCA protein assay (Pierce) was performed to determine protein concentration. From each total tissue sample, 50 μg of protein lysate was used for trypsin digestion at a trypsin:protein ratio of 1:50 (w/w) and further handled as described above.
Nano-LC-FTICR MS—
Nano-LC-FTICR MS was performed using a slightly modified procedure as described previously (23, 28). Each pooled sample was analyzed in triplicate by injecting 4 μl (equivalent to ∼5,500 cells or ∼550 ng) directly via a 3-μl sample loop onto a custom-built reversed-phase (RP) 80-cm × 50-μm-inner diameter fused silica capillary column (Polymicro Technologies, Phoenix, AZ) packed in house with 3-μm C18 particles (300-Å pore size; Jupiter, Phenomenex, Torrence, CA) and subjected to an applied pressure of 10,000 p.s.i. through a high pressure syringe pump (ISCO, Lincoln, NE). Flow rate over the column was ∼ 250 nl/min. After an injection period of 45 min, peptides were eluted from the column using a gradient from 100% mobile phase A (99.75% H2O, 0.2% acetic acid, 0.05% TFA) to ∼70% mobile phase B (90% acetonitrile, 9.9% H2O, 0.1% TFA) over a ∼200-min period. The nano-LC column outlet was coupled on line to a 7-tesla FTICR mass spectrometer through a nano-ESI emitter; 4,000 mass spectra were acquired in each LC-MS analysis using 0.3-s ion accumulation time and 50-μs gas pulse (29).
LC-MS/MS—
In the verification study, tryptic digests of 20 different whole tissue lysates (8 OR and 12 PD) were analyzed on a custom-built RPLC system via ESI utilizing an ion funnel (30) coupled to a ThermoFisher Scientific LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA). Separation was performed using a custom-made column (60 cm × 75-μm inner diameter) packed in house with Jupiter particles (C18 stationary phase, 5-μm particles, 300-Å pore size). The capillary RPLC system used for peptide separations has been described previously (23, 28). Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 100% acetonitrile. The column was equilibrated at 10,000 p.s.i. with 100% mobile phase A. A mobile phase selection valve was switched 50 min after injection to create a near exponential gradient as mobile phase B displaced mobile phase A in a 2.5-ml mixer. A split was used to provide an initial flow rate through the column of ∼400 nl/min. The column was coupled to the mass spectrometer using an in-house manufactured ESI interface with homemade 20-μm-inner diameter chemically etched emitters (31). The heated capillary temperature and spray voltage were 200 °C and 2.2 kV, respectively. Mass spectra were acquired for 80 min over the m/z range 400–2,000 at a resolving power of 100,000. An inclusion list with m/z values corresponding to peptide masses of 100 target proteins was used to select precursor ions. In cases when no targeted precursor ion was present, a maximum of six data-dependant LTQ tandem mass spectra were recorded for the most intense peaks in each survey mass spectrum.
Protein Identification and Quantitation—
FT mass spectra, acquired with the 7-tesla FTICR or LTQ-Orbitrap, were processed using ICR-2LS, Decon2LS (32), and VIPER v3.39 software developed in house (33). The output data files were visualized as two-dimensional displays of peptide monoisotopic mass versus LC elution time (i.e. spectrum number). Next MS peaks with similar measured neutral masses and LC elution times were clustered to form LC-MS features (or unique mass classes). LC elution times were converted into NET to make multiple LC-MS runs comparable (34). The assembled set of LC-MS features was then searched against the human mammary epithelial cell line AMT tag database (35), MCF-7 epithelial breast carcinoma cell line AMT tag database (36), and a composite database for a mixture of human mammary epithelial cells and MCF-7-c18, BT-474, MDA-231, and SKBR-3 breast cancer cell lines (37) using stringent filtering criteria: Xcorr ≥1.5, 2.7, and 3.3 for 1+, 2+, and 3+ fully tryptic peptides, respectively, and Xcorr ≥3.0, 3.7, and 4.5 for 1+, 2+, and 3+ partially tryptic peptides (with a minimum length of 6 amino acids), respectively, as reported previously (23). The LCMSWARP (liquid chromatography-based mass spectrometric warping and alignment of retention times of peptides) algorithm (38) was used to match LC-MS features to AMT tags. A tolerance window of mass measurement accuracy <6 ppm and NET error <0.025 was applied to ensure reliable peptide identification with false discovery rate of ≤10%. Identified peptides were coupled to their corresponding proteins using the human International Protein Index (IPI) databases, 2006 version 3.20 including 61,255 protein entries (discovery phase) and 2008 version 3.39 including 69,731 protein entries (verification phase), and in-house built Qrollup v2.2 software. Two or more constituent peptides were required to confidently identify a protein. In the case of proteins with multiple splice isoforms, these isoforms were only specifically listed if they were identified by at least one unique peptide (in addition to overlapping peptide sequences). For average abundance calculation, only highly abundant and, where possible, unique peptides were used. Protein names and descriptions were then converted to TrEMBL, NCBI (National Center for Biotechnology Information), and Swiss-Prot database formats. Protein information was retrieved from European Molecular Biology Laboratory-European Bioinformatics Institute databases. Proteins identified from all available AMT tag databases were assembled into a single list, giving rise to some redundancy. A final non-redundant protein list was generated using ProteinProphet software (SourceForge, Inc.). MS peak intensities were used as a measure of the relative peptide abundances. The mean abundance of the LC-MS features was used, and the relative abundances of constituent peptides were averaged to derive the relative abundance of the parent protein.
Tandem mass spectra acquired with the LTQ-Orbitrap were searched against the human IPI 2008 database using TurboSEQUEST v27. We used in-house developed DeconMSn software to correct the monoisotopic masses prior to generation of the dta files used for subsequent database search. Peptide sequences were considered confident with the following filtering criteria: Xcorr of 1.9, 2.2, and 3.75 for 1+, 2+, and ≥3+ peptides and ΔCn ≥ 0.1. We also applied the AMT tag strategy to identify peptides in survey mass spectra acquired with the LTQ-Orbitrap by matching the accurate masses and elution times against the composite breast cancer cell line AMT database. Peak intensities measured in high resolution survey spectra were used to retrieve relative abundance information as described above.
Immunohistochemistry—
Immunohistochemical validation was performed with an in-house prepared TMA. The TMA was established in close collaboration with a dedicated pathologist (M. A. d. B.) who evaluated all tissues for histology, grade, and Bloom and Richardson scoring (39). Tissue sections of 4 μm were stained overnight at 4 °C for EMMPRIN using a 1:100 diluted antibody directed against the C terminus of the protein (8D6, sc-21746, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Antigen retrieval was performed prior to antibody incubation for 40 min at 95 °C using DAKO retrieval solution, pH 6 (DakoCytomation, Carpinteria, CA) after which the slides were cooled down to room temperature. Staining was visualized using the anti-mouse EnVision+® System-HRP (DAB) (DakoCytomation) according to the instructions provided by the manufacturer. Scoring of immunostaining was performed by two independent observers who recorded both percentage of positive tumor cells and staining intensity.
Data Analysis and Statistics—
Relative abundance levels of all identified proteins in one sample were intra- and intersample normalized by log2 transformation using in-house developed MultiAlign software v1.1. Subsequently Z-score normalization was applied to each protein across the samples using the formula (value − mean)/standard deviation. Sample sets 1 and 2 were separately Z-score-normalized to correct for time and experimental variation. Normalized values were subjected to class comparison and prediction analysis using BRB-ArrayTools version 3.5.0 beta1 developed by Dr. Richard Simon and Amy Peng Lam. Class comparison involved finding differentially abundant proteins between therapy-sensitive (OR) and therapy-resistant (PD) tumors using a univariate two-sample t test with a significance threshold of 0.05. All data from sample sets 1 and 2 were combined to create a general list of differentially abundant proteins between OR and PD tumors and subjected to a Mann-Whitney Wilcoxon rank sum test performed with the STATA statistical package, release 10.0 (STATA, College Station, TX).
Hierarchical clustering of the data was performed using the OmniViz Desktop 3.8.0 package. For clustering, average linkage and the Euclidian similarity metric were used. Principal component analysis (PCA) was performed using Spotfire DecisionSite 8.1, version 14.3.
Kaplan-Meier survival analysis as a function of time to progression after the onset of first line tamoxifen treatment as well as correlation with response and other clinical parameters was performed using STATA. The primary end point for the Cox proportional hazard model was disease progression after the onset of tamoxifen treatment.
RESULTS
Protein Identification by Nano-LC-FTICR MS—
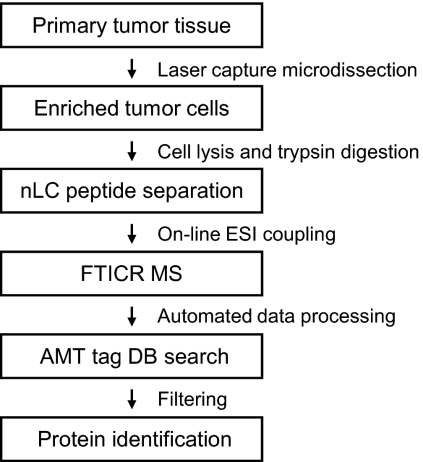
Large scale protein identification is a pivotal step in the discovery of a predictive protein profile. We have previously shown that nano-LC-FTICR MS coupled to AMT tag-based protein identification provides the sensitivity and proteome coverage required to achieve this goal (23). In the present study, we describe the clinical applicability of this approach by analyzing the proteome of eight pools of tumor cells procured by LCM from breast cancer tissues derived from 24 tamoxifen therapy-sensitive and 27 therapy-resistant patients. Fig. 1 summarizes our study design.
Fig. 1.
Experimental flow chart. All steps from sample preparation, MS analysis, and protein identification are described in the text. nLC, nano-LC; DB, database.
Tryptic peptides corresponding to ∼550 ng of protein lysate were analyzed using nano-LC-FTICR MS. Resulting data sets were visualized in a form of a two-dimensional plot, displaying monoisotopic mass versus spectrum number (NET) as shown in supplemental Fig. 1. On average ∼40,000 LC-MS features were detected in each analysis. These features were matched against previously established breast (cancer) cell line AMT tag databases. On average, ∼20% of LC-MS features matched with peptides in the database and were thus identified as illustrated in supplemental Fig. 1B.
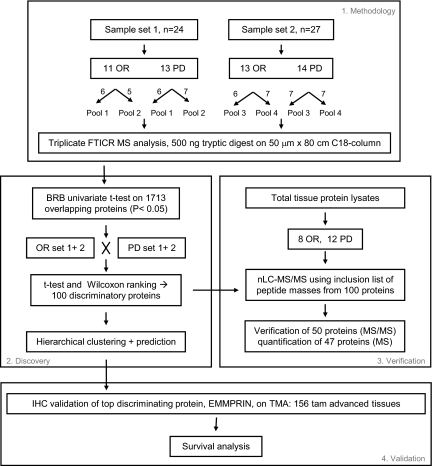
For this study, two sample sets were independently prepared and analyzed, using a different set of tumors, as shown in Fig. 2. Sample set 1 consisted of 24 tumors of which 11 were sensitive (OR) and 13 were resistant (PD) to tamoxifen treatment. Sample set 2 contained 27 tumors, 13 OR and 14 PD tissues. Microdissected cells were pooled to average sample heterogeneity and to enable triplicate analysis and were analyzed by nano-LC-FTICR MS. Replicate MS analyses, for which technical problems such as clogged tips were observed, were excluded from further data analysis, leaving 19 LC-MS data sets for further analysis (Table I). In total, 17,263 peptides corresponding to 2,556 proteins were identified through AMT tag database matching. Between the two sample sets 1,713 proteins, identified by 13,729 peptides, were identical, corresponding to an overlap of 67% (Table I). Protein abundance was computed by averaging intensities of the highly abundant peptides identified for the given protein and, where possible, using unique peptide sequences to account for multiple splice isoforms. It needs to be mentioned that it is difficult to correctly assess average protein abundance of highly homologous proteins that may have different abundance levels if these proteins are identified through identical peptides. In those cases, the additional use of unique peptide sequences may partly overcome this problem. Information on protein identification, such as filtering scores, assigned peptides and number of peptides used for abundance, mass and NET errors, and additional information is reported in supplemental Table S1. Normalized protein abundances for 1,713 proteins are displayed in supplemental Table S2.
Fig. 2.
Data analysis flow chart. Tryptic digests from two independently processed sample sets were analyzed in triplicate by nano-LC-FTICR. MS peak intensity-derived peptide/protein abundances were subjected to statistical analysis to determine differentially abundant proteins between OR and PD samples in both sample sets combined as well as in the two samples sets separately. Subsequently hierarchical clustering and class prediction was performed. nLC, nano-LC.
Table I.
FTICR MS summary
Peptide and protein information for LC-MS analyses that were used for further statistical analysis are summarized for sample set 1, set 2, the combined set, and the overlap.
| Data set | Tumor set 1 | Tumor set 2 | Total | Overlap sets 1 + 2 |
|---|---|---|---|---|
| Number of analyzed samples | 5 OR; 4 PD | 5 OR; 5 PD | 10 OR; 9 PD | |
| Total unique peptides | 14,933 | 16,059 | 17,263 | 13,729 |
| Total unique proteins | 1,998 | 2,271 | 2,556 | 1,713 (67%) |
Discovery of Tamoxifen Therapy Response-associated Proteins—
For the discovery of proteins that were associated with tamoxifen resistance, the 1,713 overlapping proteins were subjected to statistical analysis. The univariate two-sample t test from BRB-ArrayTools was used to search for differentially abundant proteins between OR and PD samples (Fig. 2). Protein abundances from all OR and PD samples from the two sample sets were analyzed together and compared with each other. The BRB analysis resulted in a list of 153 discriminating proteins using a significance threshold of p < 0.05 (the complete BRB analysis list is provided in supplemental Table S3). These 153 proteins were subsequently subjected to a Wilcoxon rank sum test, which narrowed the list down to 100 proteins with a p value <0.05. These 100 differentially abundant proteins were designated as a putative protein profile associated with the type of response to tamoxifen therapy. In this putative protein profile, 46 proteins had higher relative abundance in PD, and 54 had higher abundance in OR tissues. Protein information as well as OR:PD ratios and p values are listed in Table II in which the order and numbering of proteins corresponds to the order in Fig. 4. Our top discriminating protein in the putative protein profile was splice isoform 2 of basigin precursor (number 13 in Table II), also described in the literature as CD147 or EMMPRIN.
Table II.
Tamoxifen-response protein profile
Name, ratio, p value, IPI number, molecular mass, localization are given on the putative 100-protein profile. The order and numbering are identical to Fig. 4. EPH, ephrin; snRNA, small nucleolar RNA; GDNF, glial cell-derived neurotrophic factor.
| No.a | Protein description | Ratio of geometric means, OR:PD | Wilcoxon rank sum | IPI | Gene symbol | Molecular mass | Localizationb |
|---|---|---|---|---|---|---|---|
| kDa | |||||||
| 1 | EPH receptor B2 | 0.458 | 0.0057 | IPI00252979.7 | EPHB2 (ERK, EPTH3) | 110 | Membrane |
| 2 | Splice isoform 1 of protein kinase C and casein kinase substrate in neurons protein 2 | 0.538 | 0.0366 | IPI00027009.2 | PACSIN2 | 56 | Cytoplasm |
| 3 | 40 S ribosomal protein S4, X isoformc,d | 0.478 | 0.0412 | IPI00217030.5 | RPS4X (SCAR) | 29 | Ribosome |
| 4 | Calponin-2c | 0.478 | 0.0134 | IPI00015262.9 | CNN2 | 33.5 | Cytoskeleton |
| 5 | Calgranulin Bc,d | 0.483 | 0.0127 | IPI00027462.1 | S100A9 (CAGB, MRP14) | 13 | Cytoplasm |
| 6 | Anchor attachment protein 1 | 0.513 | 0.047 | IPI00021594.2 | GPAA1 | 68 | ER |
| 7 | Epididymal secretory protein E1 precursor | 0.54 | 0.0085 | IPI00301579.3 | NCP2 | 16.5 | Secreted protein |
| 8 | Pyrroline-5-carboxylate reductase 1c,d | 0.49 | 0.0274 | IPI00550882.2 | PYCR1 (P5CR1) | 33 | u |
| 9 | Nucleolar protein NOP5c | 0.453 | 0.0202 | IPI00006379.1 | NOP5 (HSPC120) | 60 | Nucleus |
| 10 | Annexin A8 | 0.394 | 0.0127 | IPI00218835.4 | ANXA8 (ANX8) | 37 | u |
| 11 | Lysyl-tRNA synthetasec,d | 0.539 | 0.0127 | IPI00014238.2 | KARS (KIAA0070) | 68 | Cytoplasm |
| 12 | Syntaxin 7 | 0.484 | 0.0338 | IPI00289876.2 | STX7 | 30 | Endosome |
| 13 | Splice isoform 2 of basigin precursore | 0.342 | 0.0004 | IPI00019906.1 | BSG (EMMPRIN, CD147) | 42 | Cell membrane |
| 14 | FLJ20625 protein | 0.457 | 0.0411 | IPI00016670.2 | FLJ20625 | 18 | u |
| 15 | Eukaryotic translation initiation factor 5 | 0.491 | 0.003 | IPI00022648.2 | EIF5 | 49 | Cytosol |
| 16 | Splice isoform 1 of Surfeit locus protein 4c,d | 0.449 | 0.0097 | IPI00005737.1 | SURF4 | 30 | ER |
| 17 | Splice isoform 1 of calumenin precursorc,d | 0.514 | 0.0222 | IPI00014537.1 | CALU | 38 | ER/Golgi |
| 18 | Coronin-1Ac,d | 0.507 | 0.0179 | IPI00010133.1 | CORO1A (CLIPINA) | 51 | Actin cytoskeleton |
| 19 | RAS-related protein RAB-10c | 0.516 | 0.0412 | IPI00016513.3 | RAB10 | 22.5 | Cell membrane |
| 20 | Splice isoform long of potential phospholipid-transporting ATPase IIA | 0.519 | 0.0221 | IPI00024368.1 | ATP9A (ATPIIA, KIAA0611) | 119 | Membrane |
| 21 | DNA replication licensing factor MCM2 | 0.528 | 0.0292 | IPI00184330.5 | MCM2 (BM28, CDCL1, KIAA0030) | 102 | Nucleus |
| 22 | Splice isoform long of splicing factor, proline- and glutamine-richc,d | 0.475 | 0.0179 | IPI00010740.1 | SFPQ (PSF) | 76 | Nucleus |
| 23 | Collagen-binding protein 2 precursorc,d | 0.534 | 0.0114 | IPI00032140.2 | SERPINH1 (SERPINH2, CBP2, HSP47, Colligin) | 46 | ER |
| 24 | Small nuclear ribonucleoprotein SM D2c,d | 0.495 | 0.0412 | IPI00017963.1 | SNRPD2 (Sm-D2) | 13.5 | Nucleus |
| 25 | 4F2 cell surface antigen heavy chainc,d | 0.546 | 0.05 | IPI00027493.1 | SLC3A2 (MDU1) | 58 | Membrane |
| 26 | Growth factor receptor-bound protein 7 | 0.483 | 0.038 | IPI00448767.3 | GRB7 | 60 | u |
| 27 | Copine I | 0.507 | 0.0221 | IPI00018452.1 | CPNE1 (CPN1) | 59 | u |
| 28 | Serum amyloid A protein precursor | 0.442 | 0.0055 | IPI00022368.1 | SAA1 (SAA2) | 13.5 | u |
| 29 | Ephrin type-A receptor 2 precursor | 0.503 | 0.0221 | IPI00021267.1 | EPHA1 (ECK) | 108 | Membrane |
| 30 | T-complex protein 1, η subunitc,d | 0.492 | 0.05 | IPI00018465.1 | CCT7 (TCP-1η, CCTH) | 59 | Cytoplasm |
| 31 | Guanine nucleotide-binding protein β subunit 4c,d | 0.53 | 0.0403 | IPI00012451.1 | GNB4 | 37 | u |
| 32 | Metalloprotease 1 | 0.494 | 0.022 | IPI00219613.3 | PITRM1 (hMP1) | 117 | Mitochondrion |
| 33 | C-1-Tetrahydrofolate synthase, cytoplasmicc | 0.485 | 0.0275 | IPI00218342.9 | MTHFD1 (MTHFC) | 101 | Cytoplasm |
| 34 | Predicted: septin 8 | 0.54 | 0.0179 | IPI00022082.4 | SEPT8 (KIAA0202) | 50 | u |
| 35 | Acetolactate synthase homolog | 0.465 | 0.0135 | IPI00549240.1 | OR10B1P (ILVBL) | 68 | u |
| 36 | Predicted: hypothetical protein XP_114317 | 0.511 | 0.0085 | IPI00145623.1 | RPL22L1 | 15–21 | u |
| 37 | Prefoldin subunit 6 | 0.512 | 0.0395 | IPI00005657.1 | PFDN6 (HKE2) | 14.5 | Cytosol |
| 38 | NADH-cytochrome b5 reductasec,d | 0.395 | 0.0071 | IPI00328415.8 | CYB5R3 (DIA1) | 34 | ER/mitochondrion/cytoplasm |
| 39 | Adenylate kinase isoenzyme 4, mitochondrial | 0.428 | 0.0036 | IPI00016568.1 | AK3L1(AK3, AK4) | 25 | Mitochondrion |
| 40 | Phosphoprotein enriched in astrocytes 15c | 0.481 | 0.0363 | IPI00014850.3 | PEA15 (PED) | 15 | Cytoplasm |
| 41 | Thioredoxin domain-containing protein 5 | 0.51 | 0.0496 | IPI00171438.2 | TXNDC5 (TLP46, ERp46) | 48 | ER |
| 42 | Coronin-1Bc,d | 0.425 | 0.0055 | IPI00007058.1 | CORO1B | 54 | Leading edge |
| 43 | Ephrin type-B receptor 3 precursor | 0.455 | 0.0077 | IPI00289329.1 | EPHB3 (ETK2, HEK2) | 110 | Membrane |
| 44 | RAB11 family-interacting protein 1Bc | 0.518 | 0.0191 | IPI00419433.1 | RAB11 FIP1(RCP) | 137 | Membrane |
| 45 | Splice isoform 1 of exocyst complex component SEC6 | 0.467 | 0.0231 | IPI00157734.2 | EXOC3 (SEC6, SEC6L1) | 87 | u |
| 46 | Splice isoform 1 of protein C20ORF116 precursor | 0.54 | 0.0266 | IPI00028387.3 | C20ORF116 | 36 | Secreted protein |
| 47 | Hypothetical protein DKFZP434E248 | 0.525 | 0.0221 | IPI00300094.5 | LSG1 | 75 | u |
| 48 | Adenylate kinase 2 isoform Ac,d | 2.036 | 0.0338 | IPI00215901.1 | AK2 (ADK2) | 26 | Mitochondrion |
| 49 | Trifunctional enzyme α subunit, mitochondrial precursorc,d | 2.105 | 0.0071 | IPI00031522.2 | HADHA (HADH) | 83 | Mitochondrion |
| 50 | Nucleosome assembly protein 1-like 1c,d | 1.847 | 0.0275 | IPI00023860.1 | NAP1L1 (NRP) | 45 | Nucleus |
| 51 | Secretory carrier-associated membrane protein 1 | 2.301 | 0.0055 | IPI00005129.6 | SCAMP1 | 40 | Membrane |
| 52 | Sphingosine-1-phosphate lyase 1d | 1.924 | 0.0135 | IPI00099463.2 | SGPL1 | 64 | ER membrane |
| 53 | Splice isoform 1 of glucosamine-fructose-6-phosphate aminotransferase (isomerizing) 1c,d | 2.377 | 0.0101 | IPI00217952.6 | GFPT1 (GFAT) | 79 | u |
| 54 | Ubiquinol-cytochrome c reductase iron-sulfur subunit, mitochondrial precursorc,d | 1.991 | 0.0236 | IPI00026964.1 | UQCRFS1 | 30 | Mitochondrion |
| 55 | U6 snRNA-associated SM-like protein LSM2 | 1.906 | 0.0394 | IPI00032460.3 | LSM2 (G7B) | 10 | Nucleus |
| 56 | Lisch protein, isoform 2 | 2.178 | 0.0084 | IPI00409640.1 | LSR (LISCH) | 71 | Membrane |
| 57 | Splice isoform 1 of epsin 4 | 2.385 | 0.0064 | IPI00291930.5 | CLINT1 (EPN4) | 68 | Cytoplasm |
| 58 | Endothelial protein C receptor precursor | 1.932 | 0.0178 | IPI00009276.1 | PROCR (EPCR) | 30 | Membrane |
| 59 | Annexin VI isoform 2c,d | 2.035 | 0.0114 | IPI00002459.3 | ANXA6 | 75 | u |
| 60 | Pyridoxine-5′-phosphate oxidase | 2.081 | 0.0238 | IPI00018272.3 | PNPO | 30 | u |
| 61 | Ectonucleotide pyrophosphatase/phosphodiesterase 1d | 2.087 | 0.0193 | IPI00184311.2 | ENPP1 (NPPS, PC1) | 105 | Membrane |
| 62 | Protein C20ORF178, charged multivesicular body protein 4bc,d | 1.997 | 0.0275 | IPI00025974.3 | CHMP4B (SHAX1) | 25 | Cytoplasm |
| 63 | Occludind | 1.848 | 0.0178 | IPI00003373.1 | OCLN | 59 | Membrane |
| 64 | Adipose most abundant gene transcript 2c | 2.099 | 0.0141 | IPI00020017.1 | APM2(C10ORF116) | 8 | u |
| 65 | Eukaryotic translation initiation factor 3 subunit 4 | 2.413 | 0.0062 | IPI00290460.3 | EIF3F | 36 | u |
| 66 | Hypothetical protein MGC5395c,d | 1.896 | 0.05 | IPI00031605.1 | AHNAK | 16 | u |
| 67 | Splice isoform 2 of methylcrotonoyl-CoA Carboxylase β chain, mitochondrial precursor | 1.902 | 0.0066 | IPI00294140.4 | MCCC2 (MCCB) | 58 | Mitochondrion |
| 68 | Tubulin β-3 chainc,d | 1.907 | 0.009 | IPI00013683.2 | TUBB3 (TUBB4) | 50 | u |
| 69 | KIAA2014 protein (formin-like protein 1) | 2.091 | 0.0236 | IPI00385874.4 | KIAA2014 | 117 | u |
| 70 | Hypothetical protein FLJ90697 | 2.348 | 0.0377 | IPI00329600.3 | u | ||
| 71 | Hypothetical protein, isoform 1 of protein CDV3 homolog | 1.986 | 0.0193 | IPI00014197.1 | CDV3 | 22–27 | u |
| 72 | ATP synthase oligomycin sensitivity conferral protein, mitochondrial precursorc,d | 2.153 | 0.0179 | IPI00007611.1 | ATP5O (ATPO) | 23 | Mitochondrion |
| 73 | Ubiquilin-2 | 1.843 | 0.0412 | IPI00409659.1 | UBQLN2 (PLIC2) | 66 | Cytoplasm/nucleus |
| 74 | Ubiquitin and ribosomal protein S27Ac,d | 2.273 | 0.0071 | IPI00179330.5 | RP27A | 18 | Ribosome |
| 75 | Tubulin α-1 chainc,d | 1.953 | 0.0222 | IPI00007750.1 | TUBA1 | 50 | u |
| 76 | ATP synthase α chain, mitochondrial precursorc,d | 1.947 | 0.0412 | IPI00440493.2 | ATP5O (ATPO) | 60 | Mitochondrion |
| 77 | Chaperonin containing TCP1, subunit 3c,d | 1.872 | 0.0412 | IPI00290770.2 | CCT3 | 60 | Cytoplasm |
| 78 | Nascent polypeptide-associated complex α subunitc,d | 2.159 | 0.0143 | IPI00023748.3 | NACA (HSD48) | 23 | Cytoplasm/nucleus |
| 79 | Emerin | 2.189 | 0.0178 | IPI00032003.1 | EMD | 29 | Nuclear inner membrane |
| 80 | Hypothetical protein KIAA0152c,d | 1.974 | 0.0412 | IPI00029046.1 | KIAA0152 | 32 | Membrane |
| 81 | Histone H1.5c,d | 2.533 | 0.05 | IPI00217468.2 | HIST1H1B (H1F5) | 23 | Nucleus |
| 82 | Cation channel TRPM4B | 2.03 | 0.0465 | IPI00294933.6 | TRPM4B (TRPM4) | 134 | Membrane |
| 83 | Calcyclinc,d | 2.122 | 0.0363 | IPI00027463.1 | S100A6 (CACY) | 10 | Cytoplasm/nucleus |
| 84 | Splice isoform 2 of GDNF family receptor α 1 precursor | 2.26 | 0.0184 | IPI00220291.1 | GFRA1 (GDNFRA, TRNR1) | 51 | Cell membrane |
| 85 | Complement component 1, Q subcomponent-binding protein, mitochondrial precursorc,d | 2.222 | 0.0274 | IPI00014230.1 | C1QBP (GC1QBP) | 31 | Mitochondrion |
| 86 | Chloride intracellular channel protein 4c,d | 2.023 | 0.0275 | IPI00001960.2 | CLIC4 | 29 | Cytoplasm/mitochondrion |
| 87 | Eukaryotic translation initiation factor 3 subunit 6c,d | 2.076 | 0.0178 | IPI00013068.1 | EIF3E (INT6) | 52 | Cytoplasm |
| 88 | Protein-disulfide isomerase A4 precursorc,d | 2.157 | 0.0275 | IPI00009904.1 | PDIA4 (ERP70) | 73 | ER |
| 89 | Hypothetical protein MGC5352c,d | 1.867 | 0.0394 | IPI00063242.3 | PGAM5 | 28 | u |
| 90 | Splice isoform 1 of polypeptide N-acetylgalactosaminyltransferase 3d | 2.229 | 0.0066 | IPI00004670.1 | GALNT3 | 73 | Golgi |
| 91 | OTTHUMP00000028732 (thioredoxin, mitochondrial precursor) | 1.823 | 0.0462 | IPI00017799.3 | TXN2 (TRX2) | 18–22 | Mitochondrion |
| 92 | Fatty acid-binding protein, epidermal | 2.203 | 0.0075 | IPI00007797.1 | FABP5 | 15 | Cytoplasm |
| 93 | Programmed cell death 6-interacting protein, PDCD6IP proteinc,d | 2.202 | 0.0199 | IPI00246058.3 | PDCD6IP (AIP1) | 97 | Cytoplasm |
| 94 | Ezrin-radixin-moesin-binding phosphoprotein 50c,d | 2.42 | 0.0025 | IPI00003527.3 | SLC9A3R1 (EBP50, NHERF1) | 39 | Intracytoplasmic membrane, actin cytoskeleton |
| 95 | Splice isoform 1 of ubiquitin thiolesterase protein | 1.974 | 0.05 | IPI00549574.2 | OTUB1 | u | |
| 96 | Endozepinec,d | 2.184 | 0.0211 | IPI00010182.3 | ACBP (DBI, EZ) | 10 | u |
| 97 | Phosphoribosylformylglycinamidine synthase | 2.221 | 0.0177 | IPI00004534.3 | PFAS (KIAA0361) | 145 | Cytoplasm |
| 98 | Histidine triad nucleotide-binding protein 1c | 1.853 | 0.0274 | IPI00239077.4 | HINT1 (PKCI1) | 14 | Cytoplasm/nucleus |
| 99 | BAG family molecular chaperone regulator-3 | 1.97 | 0.0175 | IPI00000644.3 | u | ||
| 100 | Exocyst complex component SEC8d | 1.856 | 0.0109 | IPI00059279.5 | EXOC4 (KIAA1699, SEC8) | 110 | u |
Numbering according to Fig. 4.
u, data unknown in database; ER, endoplasmic reticulum.
Presence verified in individual tumors by MS/MS.
Presence verified in individual tumor MS survey spectrum and quantified by AMT database match.
Validated by immunohistochemistry.
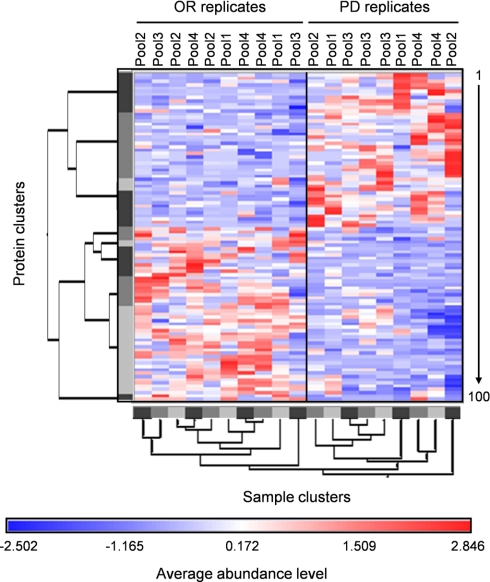
Fig. 4.
Hierarchical clustering of OR and PD samples. Red and blue colors indicate relative high and low protein abundance, respectively, and white equals median abundance. Gray bars represent sample and protein clusters. The length of the tree arms is inversely correlated with similarity. Proteins are listed vertically from top to bottom and numbered from 1 to 100 in the same order as in Table II.
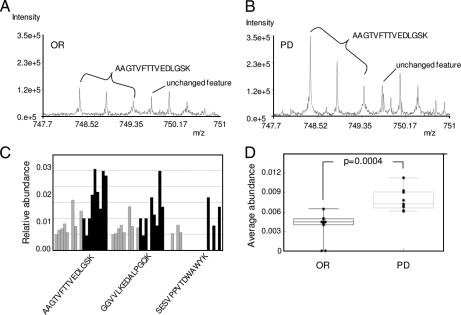
Multiple isoforms of EMMPRIN have been described that are identical in their C-terminal sequence but vary in length and sequence at the N-terminal part of the protein (EntrezGene 682). In our final, non-redundant protein list we report the identification of isoforms 1 and 2 by five and six peptides, respectively (supplemental Table S1). Only one of the six peptides (AAGTVFTTVEDLGSK) was unique for isoform 2. Isoform 1 is the longer variant of 385 amino acids, whereas isoform 2 lacks amino acids 24–139. Peptide AAGTVFTTVEDLGSK is uniquely positioned at the splice site in which the first two amino acids (AA) are positioned at residues 22 and 23 and the third amino acid (Gly) is positioned at residue 140 in the full-length sequence. Therefore, this peptide sequence is specific for isoform 2. The raw mass spectrum for EMMPRIN peptide AAGTVFTTVEDLGSK (Mr = 1,496.75 and m/z = 748.38) showed a 3-fold higher intensity for the PD sample (Fig. 3B) in comparison with the OR sample (Fig. 3A). The spectra also showed that there is no significant difference in peak intensity between OR and PD for the second feature appearing at m/z 749.76, suggesting that the observed difference in peak intensity for the AAGTVFTTVEDLGSK peptide is not an artifact introduced by e.g. loading differences. It needs to be mentioned, however, that we did not use single spectra to determine abundance ratios of peptides but LC-MS feature intensity, which is defined as a sum of intensities of all members of the unique mass class. Using LC-MS feature intensity, we investigated the relative abundance of three EMMPRIN peptides across all of the samples. The peptides AAGTVFTTVEDLGSK and GGVVLKEDALPGQK were present in virtually all samples and clearly showed a 2–3-fold increase in abundance in PD samples. SESVPPVTDWAWYK peptide was only present in a few samples but showed the same increase in PD (Fig. 3C). This increase in relative peptide abundance therefore correlated very well with the observed 2-fold increase of EMMPRIN at the protein level (Fig. 3D).
Fig. 3.
EMMPRIN differential peptide and protein abundance. Representative mass spectra of an LC-MS feature identified as EMMPRIN peptide AAGTVFTTVEDLGSK in OR (A) and PD (B) indicate a 3-fold increase in intensity for PD sample. C, relative abundance ratios of four EMMPRIN peptides in OR (gray) and PD (black) samples. D, average relative abundance of EMMPRIN protein in all OR and PD samples. p value was calculated using the Wilcoxon rank sum test. Box-Whisker plot in which each dot represents the value of a sample, and the error bars show the highest and lowest value. The line in the box represents the mean value.
To test the predictive power of the putative profile of 100 proteins within the two sample sets, supervised hierarchical clustering was performed, represented as a tree-shaped dendrogram (Fig. 4). Vertically the different proteins are listed numbered from 1 to 100 from top to bottom. Horizontally the different samples are listed. Based on their average relative abundances, OR and PD samples were effectively separated from each other as illustrated by the two main clusters in the dendrogram (Fig. 4). Separation of the samples was based on higher (red) and lower (blue) than median abundance of each protein within all samples. Furthermore the length of the dendrogram arms shows that some samples (replicates) show more similarity to each other than to the rest of the samples as expected. The order of the proteins numbered from 1 to 100 is identical to the order and numbering in Table II.
Similar results were obtained by PCA (supplemental Fig. 2). In the PCA complex information is reduced to three principal components, represented by the x, y, and z axes. Samples are visualized in a three-dimensional plot and cluster according to their relative protein abundance. From this PCA it is clear that, in this sample set, OR (green squares) and PD samples (red squares) were completely separated from each other based on their protein abundance profile.
To verify that individual peptides showed differential abundance similar to that of their corresponding proteins, we performed hierarchical clustering on all peptides corresponding to the putative 100-protein profile. As expected, clustering based on peptides resembled the results of protein clustering (data not shown).
Verification of Differential Protein Abundance—
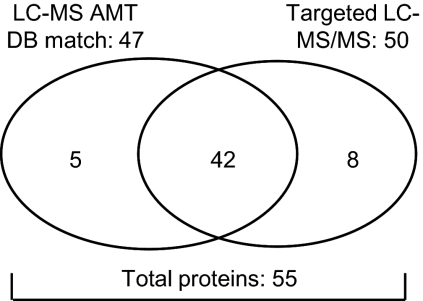
Our next goal was to verify the presence and abundance level of all profile proteins in separate tumor samples. Because we used pooled microdissected tumor cells for the discovery study, information on the single tumor level as well as the relation with clinical factors was lost. To verify our putative profile proteins, we performed targeted LC-MS/MS analyses using an inclusion list (supplemental Table S4) compiled from the m/z values of the peptides that corresponded to the 100 putative profile proteins. We prepared whole tissue protein lysates from tumors (eight OR and 12 PD) with a high tumor cell content (>70%) so that microdissection could be omitted. Using this approach, we identified and therefore verified the presence of 50 proteins from the inclusion list. In addition, peak intensities of survey mass spectra (on average ∼14,000 LC-MS features per sample) were used for quantitation. In this case, peptide identity was derived by matching LC-MS features from survey spectra to the composite breast cancer cell line AMT tag database. This resulted in the identification and quantitation of 47 target proteins of which 42 were also identified by MS/MS sequencing (Fig. 5). Overall a total of 55 proteins (50 by MS/MS sequencing and five additional by LC-MS feature (survey mass spectra) matching with the AMT database of the 100-putative protein list) were verified in an independent targeted LC-MS/MS experiment. The 47 proteins for which relative abundance was available were used in further analyses. Surprisingly the top discriminating protein in the original profile, EMMPRIN, was not identified through this targeted approach. Raw MS/MS data obtained for verified proteins and relative abundance ratios for verified proteins are listed in supplemental Tables S5 and S6, respectively.
Fig. 5.
Verification of putative profile proteins. Putative profile proteins were verified in non-microdissected tumor samples through targeted MS/MS. Peptide abundance information was retrieved from peak intensities of MS survey spectra. For protein identification MS survey spectra were matched with the AMT database (DB).
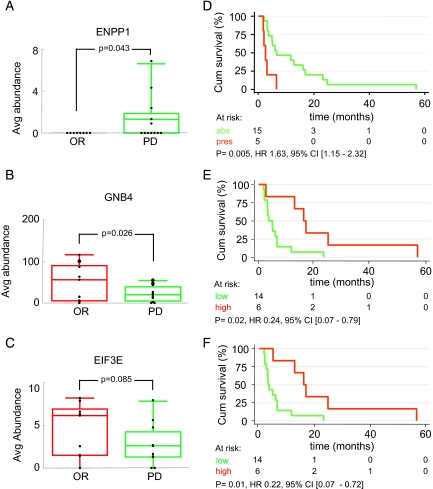
Relative abundances of the 47 verified proteins were statistically analyzed using either Wilcoxon rank sum or Student's t test depending on the outcome of a test for normality based on skewness and kurtosis. Three proteins, ectonucleotide phosphatase/phosphodiesterase 1 (ENPP1; number 61 in Table II), guanine nucleotide-binding protein β subunit 4 (GNB4; number 31 in Table II), and ubiquinol-cytochrome c reductase iron-sulfur subunit mitochondrial precursor (UQCRFS1; number 54 in Table II) were significantly differentially abundant between OR and PD with p values of 0.043 (Fig. 6A), 0.026 (Fig. 6B), and 0.036 (not shown), respectively (Table III). ENPP1 was not detected in any of the OR samples but in five of 12 PD samples (Fig. 6A), whereas GNB4 (Fig. 6B) and UQCRFS1 were higher in OR samples (Table III). In addition, eukaryotic translation initiation factor 3 subunit 6/E (EIF3E) (Fig. 6C), occludin (OCLN), splice isoform 1 of surfeit locus protein 4 (SURF4), thioredoxin domain-containing protein 5 precursor (TXNDC5), and ubiquitin and ribosomal protein S27A (RP27A) showed a trend toward differential abundance (0.05 < p < 0.1). Mean abundance and 95% confidence intervals (CIs) are listed in Table III. It needs to be mentioned that analysis groups for verification were rather small (eight OR versus 12 PD); thus the outcomes may change when more samples are analyzed in future studies. Subsequently relative abundance of all 47 verified proteins was coupled to clinical end points of patients. Of these 47 proteins, ENPP1, EIF3E, and GNB4 showed significant association with progression-free survival, whereas UQCRFS1 did not, although it did associate with response as described above. Kaplan-Meier analysis as a function of ENPP1 status showed that the presence of ENPP1 was significantly correlated with shorter progression-free survival after the start of tamoxifen treatment with a hazard ratio (HR) of 1.63 (95% CI, 1.15–2.32; p = 0.005) (Fig. 6D). Survival analyses as a function of EIF3E and GNB4 levels were performed after dividing the relative abundance levels into low + median versus high because low and median level survival curves were superimposable. High levels of EIF3E and GNB4 were significantly associated with prolonged progression-free survival with HRs of 0.22 (95% CI, 0.07–0.71; p = 0.01) (Fig. 6E) and 0.24 (95% CI, 0.07–0.79; p = 0.02) (Fig. 6F), respectively. In conclusion, we were able to associate high GNB4 and EIF3E levels with a favorable outcome and ENPP1 with an adverse outcome on tamoxifen therapy.
Fig. 6.
Clinical association of verified proteins. Differences in relative abundance ratios between OR (red) and PD (green) tumors for ENPP1 (A), GNB4 (B), and EIF3E (C) are shown. Shown is the Kaplan-Meier survival analysis of time to progression upon tamoxifen treatment for recurrent breast cancer patients according to LC-MS abundance levels. For ENPP1, absence (abs) (green line) and presence (pres) (red line) of abundance was compared (D). For GNB4 (E) and EIF3E (F) low abundance and medium abundance were grouped (green line) and compared with high abundance (red line). The number of patients at risk in each group is displayed together with the hazard ration, 95% confidence interval, and p value. Avg, average; Cum, cumulative; CI, confidence interval.
Table III.
Verified differentially abundant proteins
Shown are a subset of putative profile proteins verified in targeted MS/MS experiment with a p value <0.1.
| Protein description | Gene symbol | Higher in | Δ mean/median (95% CI) | p value |
|---|---|---|---|---|
| Guanine nucleotide-binding protein β subunit 4 | GNB4 | OR | −35.1 (−65.2 to −4.8) | 0.026 |
| Ubiquinol-cytochrome c reductase iron-sulfur subunit, mitochondrial precursor | UQCRFS1 | OR | −31.6 (−61.0 to −2.3) | 0.036 |
| Ectonucleotide pyrophosphatase/phosphodiesterase 1a | EPP1 | PD | 0 (0–1.3) | 0.043 |
| Thioredoxin domain-containing protein 5 precursora | TXNDC5 | PD | 2.8 (−0.02 to 20.3) | 0.081 |
| Eukaryotic translation initiation factor 3 subunit 6 | EIF3E | OR | −2.2 (−4.8 to 0.3) | 0.085 |
| Occludina | OCLN | OR | 0 (−1.6 to 0) | 0.087 |
| Splice isoform 1 of O15260 Surfeit locus protein 4 | SURF4 | PD | 3.7 (−0.8 to 8.3) | 0.098 |
| Ribosomal protein S27A | RP27A | OR | −168.1 (−376.3 to 40.2) | 0.100 |
Wilcoxon rank sum.
Validation of EMMPRIN and Association with Clinical End Points—
A pivotal step in the process of biomarker discovery is the validation of putative markers in independent patient cohorts and preferably by using a different methodology, such as using immunohistochemistry (IHC). In our case, validation was only performed for the top discriminating protein, EMMPRIN, because there are no appropriate antibodies available for ENPP1, EIF3E, and GNB4 or for any of the other differentially abundant proteins we discovered. The antibody we used in this study was directed against the C-terminal part of EMMPRIN and therefore recognizes all splice isoforms.
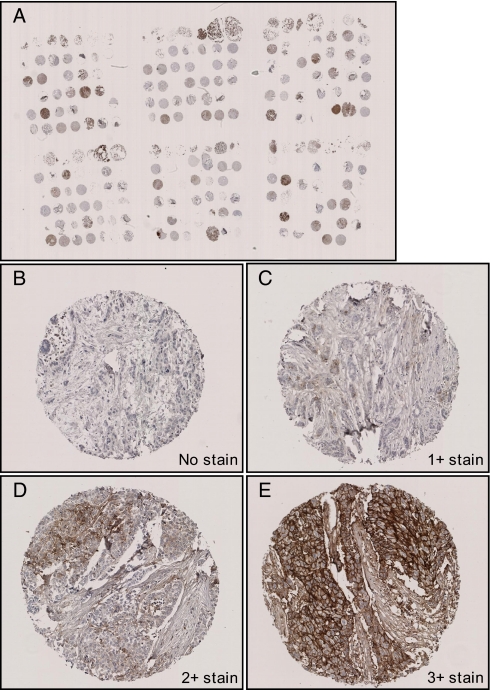
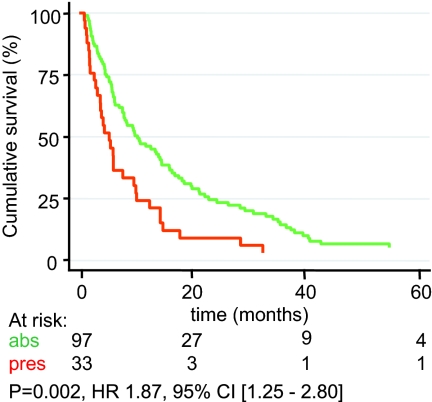
To independently validate differential EMMPRIN protein abundance between OR and PD patients, IHC was performed using our primary breast cancer TMA. Among the different tissues, there were 156 breast tumors of patients who received first line tamoxifen therapy after recurrence. This set of tumors had no overlap with the discovery set tumors. In total, 130 tumors showed reproducible IHC staining on the TMA when assays were performed in triplicate. Patient and tumor characteristics are described in Table IV. Different staining outcomes were categorized as undetectable, weak, medium, and strong membrane staining. Weak membrane staining, present in <10% of tumor cells, was scored as 1+. Medium membrane staining, present in 10–50% of tumor cells, was scored as 2+. Strong membrane staining, observed in >50% of tumor cells, was assigned score 3+ (Fig. 7). These scoring outcomes were subsequently related to clinical endpoints. We observed that none of the CR tumors displayed EMMPRIN staining, whereas highest EMMPRIN staining (3+) was observed in PD tumors (Table V). This finding, originally indicated using LC-MS-based technology, was thus confirmed by IHC. For comparison, we defined a “clinical benefit” group composed of tumors showing NC for >6 months, CR, and PR and a “no clinical benefit” group representing NC for ≤6 months and PD tumors. Absence of detectable EMMPRIN levels showed a significant clinical benefit with an odds ratio of 2.98 (95% CI, 1.32–6.73; p = 0.009). The presence of detectable EMMPRIN levels was more frequently observed in premenopausal women (X2 = 11.7; p < 0.001) and in patients with a shorter disease-free interval (X2 = 11.2; p = 0.004) defined as the time from primary diagnosis to recurrence (Table VI). In addition, Cox regression analysis showed that presence of EMMPRIN significantly correlated with shorter progression-free survival from the start of tamoxifen treatment (HR, 1.87; 95% CI, 1.25–2.80; p = 0.002) (Fig. 8). Thus, high EMMPRIN levels correlate with poor outcome on first line tamoxifen treatment.
Fig. 7.
Immunohistochemical staining of EMMPRIN. EMMPRIN immunohistochemical staining was performed on an independent sample set of 156 tissues using TMA. A, overview of TMA; B, negatively stained tissue; C, 1+ membrane stain; D, 2+ stain; E, 3+ stain. Overview picture was taken at 5× magnification; other pictures were taken at 100× magnification.
Table V.
IHC score of EMMPRIN
The average (Avg) EMMPRIN score in tumors grouped by therapy response is shown.
| Avgscore | CR | PR | NC > 6months | NC ≤ 6months | PD | Total |
|---|---|---|---|---|---|---|
| 0 | 4 | 20 | 40 | 8 | 25 | 97 |
| 1 | 0 | 4 | 6 | 3 | 12 | 25 |
| 2 | 0 | 0 | 2 | 0 | 4 | 6 |
| 3 | 0 | 0 | 1 | 0 | 1 | 2 |
| Total | 4 | 24 | 49 | 11 | 42 | 130 |
Table VI.
EMMPRIN correlation with clinical factors
EMMPRIN protein abundance correlated with menopausal status and disease-free interval is shown.
| EMMPRIN | n(%) | Menopausal status
|
Disease-free interval (months)
|
|||
|---|---|---|---|---|---|---|
| Pre (%) | Post (%) | ≤12 (%) | 12–36 (%) | ≥36 (%) | ||
| Absent | 97 (74.6) | 22 (55.0) | 75 (83.3) | 9 (56.3) | 39 (66.1) | 49 (89.1) |
| Present | 33 (25.4) | 18 (45.0) | 15 (16.7) | 7 (43.7) | 20 (34.9) | 6 (10.9) |
| Total | 130 (100) | 40 (30.7) | 90 (69.2) | 16 (12.3) | 59 (45.3) | 55 (42.3) |
| Pearson χ2 | 11.7 | 11.2 | ||||
| p value | <0.001 | 0.004 | ||||
Fig. 8.
Kaplan-Meier survival analysis. EMMPRIN abundance was measured by IHC using TMA and was correlated to time to progression after the onset of first line tamoxifen treatment. Absence (abs) of detectable EMMPRIN (green line) was compared with presence (pres) (1+, 2+, and 3+) of EMMPRIN staining (red line).
DISCUSSION
We performed a comparative proteomics study using nano-LC-FTICTR MS analyses of tamoxifen therapy-resistant and therapy-responsive tumor cells isolated from breast cancer tissue by LCM. This approach proved to be extremely powerful as exemplified by identification of several thousand unique proteins from sub-μg quantities of clinically relevant samples. These efforts resulted in the identification of a putative protein profile that is associated with the type of response to tamoxifen therapy. Furthermore we validated our top discriminating protein, EMMPRIN, in an independent patient cohort and confirmed its association with tamoxifen therapy resistance in recurrent breast cancer.
Protein Identification by Nano-LC-FTICR MS—
Many different proteomics technologies are available nowadays that all aid in the quest for cancer biomarkers. The method of choice will depend on the type of question asked, the type of material being investigated, and the availability of resources. Several studies have shown that the combination of dedicated nano-LC separation coupled to high end FT MS offers the best potential for in-depth analysis of limited sample quantity, which is usually the case with clinical material (23, 28, 36, 37, 40). In the present study, we used nano-LC-FTICR MS and a composite breast cancer cell line AMT tag database for the identification of peptides from as little as ∼550 ng of protein lysate. Overall we identified over 17,000 unique peptides corresponding to over 2,500 unique proteins, a significantly larger fraction of the proteome than attainable with more conventional proteomics techniques (20, 22). Furthermore we believe there is more to gain if a breast cancer tissue-specific AMT tag database becomes available. Although breast cancer cell lines represent aspects of normal and malignant breast tissue, it is well known that cultured cell lines have quite a distinct proteomic profile compared with primary cells or tissues. This was clearly demonstrated by Ornstein et al. (18) who compared proteomes of microdissected prostate tumor cells with proteomes of matching cell lines from the same patient. They showed that protein expression was strikingly altered in cultured cells, which had less than 20% proteins in common with uncultured cells (18). Therefore, it is very well possible that proteins involved in therapy resistance of breast tumors are not expressed in cell lines and thus are missing from the AMT tag database used in this study. To overcome this problem, we are currently constructing an AMT tag database from breast cancer tissues using a selection of tumors that have distinct phenotypic characteristics. A breast cancer tissue-specific AMT tag database will most likely increase the number of identified peptides (i.e. proteome coverage) in LC-MS analyses, thus increasing our chances of identifying relevant biomarkers. Proteome coverage could even be further improved using “smart MS/MS,” e.g. by fragmenting currently unidentified LC-MS features.
Discovery and Verification of Putative Tamoxifen Therapy Response-associated Proteins—
The putative protein profile described in this study consists of 100 proteins involved in a variety of biological processes. These proteins can be categorized into different functional classes, such as structural proteins, signaling proteins and kinases, metabolic enzymes, proteins involved in apoptosis, and others (see Table II). Several of the putative profile proteins (NAP1L1, pyridoxine-5′-phosphate oxidase, and UQCRFS1) have been previously associated with tamoxifen therapy resistance in breast cancer (41, 42) or chemotherapy resistance (SGPL1 and TUBB3) in vitro and in clinical specimens (43–45) and with aggressiveness of breast cancer (S100A6, S100A9, CLIC4, EBP50, and OCLN) (46–51).
Because the discovery of putative tamoxifen response-predictive proteins was performed in pooled samples, it was important to verify the presence and relative abundance of these proteins in each individual tumor tissue. Using a targeted MS/MS approach, we successfully identified 55 profile proteins in individual, non-microdissected tumor lysates and retrieved quantitative information for 47 of these proteins. Clearly 45 putative proteins were left unverified in individual tumor samples, including our top discriminating protein, EMMPRIN. The relatively low verification rate can be justified by the use of different samples and LC-MS platforms for the discovery and verification part of the study. Microdissected tumor cell lysates were analyzed by ultranarrow LC coupled to FTICR for discovery, whereas whole tissue lysates representing a mixture of cell types were analyzed by a standardized LC-MS/MS platform for verification. Nano-LC-FTICR analysis yielded an average of ∼40,000 LC-MS features, whereas LC-MS/MS Orbitrap analysis detected on average ∼14,000 LC-MS features. Therefore, the nano-LC-FTICR platform yielded ∼3× higher proteome coverage and, one can speculate, resulted in a similar improvement in sensitivity (i.e. limit of detection). Similarly we only used information on accurate mass in targeted MS/MS experiments because it was not possible to use NET information as an inclusion criterion with the software version available at the time. The addition of NET information as an inclusion criterion will most likely increase the success rate of target peptide identification through MS/MS in future studies using updated instrument control software. The compilation of these effects (i.e. LC-MS platform with lower overall sensitivity and inadequate targeted MS/MS strategy) resulted in a failure to confirm the identity of our top discriminating protein as EMMPRIN in the verification study.
Nevertheless the presence of 55 putative profile proteins was verified, and based on the abundance ratios, ENPP1, UQCRFS1, and GNB4 were confirmed to be significantly differentially abundant between OR and PD tumors. In addition ENPP1, EIF3E, and GNB4, were significantly associated with time to progression upon first line tamoxifen treatment of recurrent breast cancer. So far, no link between ENPP1 or GNB4 and breast cancer or response to tamoxifen has been described, although ENPP1 overexpression and polymorphisms have been repeatedly associated with insulin resistance and obesity (52, 53). Obesity is a risk factor for breast cancer (54), and insulin resistance may be linked to tamoxifen therapy resistance.
EIF3E protein expression has been shown to be significantly decreased in breast cancer, which was frequently associated with loss of heterozygosity at the Int-6/eIF3-p48 locus (55). EIF3E is ubiquitously expressed and highly conserved, and it encodes the p48 subunit of the translation initiation factor eIF3, also named INT6. In a multiplex tissue immunoblotting study by Traicoff et al. (56), EIF3E expression was determined in 124 breast cancer tissues. It was shown that breast tissues clustered according to high or low EIF3E expression, and this segregation was not dependent on tumor stage. Furthermore EIF3E expression positively correlated with tumor suppressors, such as p53, suggesting a function in the same signaling pathway (56). It was postulated that EIF3E has diverse functions in cell growth in addition to translation initiation, including tumor suppressive properties. This was particularly clearly shown in studies where truncation or knockdown of EIF3E induced angiogenesis and tumor formation (57, 58). This tumor-suppressive role correlates well with the elevated abundance of EIF3E in OR tumors and its contribution to prolonged progression-free survival upon tamoxifen treatment.
Validation of EMMPRIN—
The validation study was focused on our top discriminating protein, EMMPRIN, which is known to be involved in breast cancer and for which an appropriate antibody is conveniently available. EMMPRIN has been previously described to play a role in tumor cell invasion and metastasis (59). In particular, it acts through up-regulation of the urokinase-type plasminogen activator system, thereby promoting tumor cell invasion (60). In an immunohistochemical study using high density breast cancer tissue microarrays, it was shown that positive EMMPRIN staining correlated with various histopathological parameters, in particular with decreased tumor-specific survival in postmenopausal patients (61). EMMPRIN is up-regulated in many types of cancer (62), supporting the previous findings that the involvement of EMMPRIN in urokinase-type plasminogen activator deregulation may be a universal phenomenon in tumorigenesis and is not restricted to breast cancer. In addition, EMMPRIN has been recently shown to predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer (63). An IHC analysis in 101 advanced bladder cancer patients showed that high EMMPRIN expression strongly correlated with shorter survival time, in particular in patients with metastatic tumors, and that response to chemotherapy could also be predicted with an odds ratio of 4.41 (63). In our study, high expression of EMMRPIN was more frequently observed in PD than OR tumors, and it was significantly associated with an early tumor progression after the onset of first line tamoxifen treatment in recurrent breast cancer. Combining our results with previous findings, one can speculate that EMMPRIN-induced tumor aggressiveness may be the result of therapy resistance in general (i.e. tamoxifen and chemotherapy) and that this mechanism is not restricted to breast cancer.
Concluding Remarks—
In this study we demonstrated quantitative analysis of minute amounts of clinically relevant tumor tissues using ultrasensitive nano-LC-FTICR technology. These analyses have put forward a putative protein profile that may predict the outcome of response for tamoxifen therapy in breast cancer patients. Whether this profile as a whole is a good predictor for tamoxifen therapy response in a larger, independent group of patients and whether it is applicable to chemotherapy as well will be the subject of further investigations.
Acknowledgments
We thank Ron Moore for LC-MS/MS analyses and Angela Norbeck, Matthew Monroe, Nikola Tolić, and Samual Purvine for assistance in MS data management and analysis. Anita Trapman-Jansen and Renée Foekens are acknowledged for technical assistance with IHC and TMA. Portions of this research were performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, Richland, WA.
Footnotes
Published, MCP Papers in Press, March 27, 2009, DOI 10.1074/mcp.M800493-MCP200
The abbreviations used are: OR, objective response; AMT, accurate mass and time; BRB, biometric research branch; CI, confidence interval; CLIC4, chloride intracellular channel protein 4; CR, complete remission; EBP50, ezrin-radixin-moesin-binding phosphoprotein 50; EIF3E, eukaryotic translation initiation factor 3 subunit 6/E; EMMPRIN, extracellular matrix metalloproteinase inducer; GNB4, guanine nucleotide-binding protein β subunit 4; HR, hazard ratio; IHC, immunohistochemistry; IPI, International Protein Index; LCM, laser capture microdissection; NAP1L1, nucleosome assembly protein 1-like 1; NC, no change; NET, normalized elution time; OCLN, occluding; PCA, principal component analysis; PD, progressive disease; PR, partial remission; RP, reversed-phase; S100A6, calcyclin; S100A9, calgranulin B; SGPL1, sphingosine-1-phosphate lyase; TMA, tissue microarray; TUBB3, β-tubulin type 3; UQCRFS1, ubiquinol-cytochrome c reductase iron-sulfur subunit mitochondrial precursor; LTQ, linear trap quadrupole; ENPP1, ectonucleotide phosphatase/phosphodiesterase 1.
This work was supported, in whole or in part, by National Institutes of Health Grant RR18522 from the National Center for Research Resources. This work was also supported by the National Genomics Initiative/Netherlands Organization for Scientific Research (NWO).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) ( 2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–1717 [DOI] [PubMed] [Google Scholar]
- 2.Clarke, R., Liu, M. C., Bouker, K. B., Gu, Z., Lee, R. Y., Zhu, Y., Skaar, T. C., Gomez, B., O'Brien, K., Wang, Y., and Hilakivi-Clarke, L. A. ( 2003) Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene 22, 7316–7339 [DOI] [PubMed] [Google Scholar]
- 3.Massarweh, S., and Schiff, R. ( 2006) Resistance to endocrine therapy in breast cancer: exploiting estrogen receptor/growth factor signaling crosstalk. Endocr.-Relat. Cancer. 13, Suppl. 1, S15–S24 [DOI] [PubMed] [Google Scholar]
- 4.Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., Fluge, O., Pergamenschikov, A., Williams, C., Zhu, S. X., Lønning, P. E., Børresen-Dale, A. L., Brown, P. O., and Botstein, D. ( 2000) Molecular portraits of human breast tumours. Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 5.Sørlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Thorsen, T., Quist, H., Matese, J. C., Brown, P. O., Botstein, D., Eystein Lønning, P., and Børresen-Dale, A. L. ( 2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen, M. P., Foekens, J. A., van Staveren, I. L., Dirkzwager-Kiel, M. M., Ritstier, K., Look, M. P., Meijer-van Gelder, M. E., Sieuwerts, A. M., Portengen, H., Dorssers, L. C., Klijn, J. G., and Berns, E. M. ( 2005) Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J. Clin. Oncol. 23, 732–740 [DOI] [PubMed] [Google Scholar]
- 7.Wang, Y., Klijn, J. G., Zhang, Y., Sieuwerts, A. M., Look, M. P., Yang, F., Talantov, D., Timmermans, M., Meijer-van Gelder, M. E., Yu, J., Jatkoe, T., Berns, E. M., Atkins, D., and Foekens, J. A. ( 2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 8.Yang, F., Foekens, J. A., Yu, J., Sieuwerts, A. M., Timmermans, M., Klijn, J. G., Atkins, D., Wang, Y., and Jiang, Y. ( 2006) Laser microdissection and microarray analysis of breast tumors reveal ER-alpha related genes and pathways. Oncogene 25, 1413–1419 [DOI] [PubMed] [Google Scholar]
- 9.Smid, M., Wang, Y., Klijn, J. G., Sieuwerts, A. M., Zhang, Y., Atkins, D., Martens, J. W., and Foekens, J. A. ( 2006) Genes associated with breast cancer metastatic to bone. J. Clin. Oncol. 24, 2261–2267 [DOI] [PubMed] [Google Scholar]
- 10.Foekens, J. A., Atkins, D., Zhang, Y., Sweep, F. C., Harbeck, N., Paradiso, A., Cufer, T., Sieuwerts, A. M., Talantov, D., Span, P. N., Tjan-Heijnen, V. C., Zito, A. F., Specht, K., Hoefler, H., Golouh, R., Schittulli, F., Schmitt, M., Beex, L. V., Klijn, J. G., and Wang, Y. ( 2006) Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J. Clin. Oncol. 24, 1665–1671 [DOI] [PubMed] [Google Scholar]
- 11.Gehrmann, M. L., Hathout, Y., and Fenselau, C. ( 2004) Evaluation of metabolic labeling for comparative proteomics in breast cancer cells. J. Proteome Res. 3, 1063–1068 [DOI] [PubMed] [Google Scholar]
- 12.Malorni, L., Cacace, G., Cuccurullo, M., Pocsfalvi, G., Chambery, A., Farina, A., Di Maro, A., Parente, A., and Malorni, A. ( 2006) Proteomic analysis of MCF-7 breast cancer cell line exposed to mitogenic concentration of 17beta-estradiol. Proteomics 6, 5973–5982 [DOI] [PubMed] [Google Scholar]
- 13.Ou, K., Kesuma, D., Ganesan, K., Yu, K., Soon, S. Y., Lee, S. Y., Goh, X. P., Hooi, M., Chen, W., Jikuya, H., Ichikawa, T., Kuyama, H., Matsuo, E., Nishimura, O., and Tan, P. ( 2006) Quantitative profiling of drug-associated proteomic alterations by combined 2-nitrobenzenesulfenyl chloride (NBS) isotope labeling and 2DE/MS identification. J. Proteome Res. 5, 2194–2206 [DOI] [PubMed] [Google Scholar]
- 14.Chuthapisith, S., Layfield, R., Kerr, I. D., Hughes, C., and Eremin, O. ( 2007) Proteomic profiling of MCF-7 breast cancer cells with chemoresistance to different types of anti-cancer drugs. Int. J. Oncol. 30, 1545–1551 [PubMed] [Google Scholar]
- 15.Wu, S. L., Hancock, W. S., Goodrich, G. G., and Kunitake, S. T. ( 2003) An approach to the proteomic analysis of a breast cancer cell line (SKBR-3). Proteomics 3, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 16.Sandhu, C., Connor, M., Kislinger, T., Slingerland, J., and Emili, A. ( 2005) Global protein shotgun expression profiling of proliferating mcf-7 breast cancer cells. J. Proteome Res. 4, 674–689 [DOI] [PubMed] [Google Scholar]
- 17.Sarvaiya, H. A., Yoon, J. H., and Lazar, I. M. ( 2006) Proteome profile of the MCF7 cancer cell line: a mass spectrometric evaluation. Rapid Commun. Mass Spectrom. 20, 3039–3055 [DOI] [PubMed] [Google Scholar]
- 18.Ornstein, D. K., Gillespie, J. W., Paweletz, C. P., Duray, P. H., Herring, J., Vocke, C. D., Topalian, S. L., Bostwick, D. G., Linehan, W. M., Petricoin, E. F., 3rd, and Emmert-Buck, M. R. ( 2000) Proteomic analysis of laser capture microdissected human prostate cancer and in vitro prostate cell lines. Electrophoresis 21, 2235–2242 [DOI] [PubMed] [Google Scholar]
- 19.Emmert-Buck, M. R., Bonner, R. F., Smith, P. D., Chuaqui, R. F., Zhuang, Z., Goldstein, S. R., Weiss, R. A., and Liotta, L. A. ( 1996) Laser capture microdissection. Science 274, 998–1001 [DOI] [PubMed] [Google Scholar]
- 20.Hudelist, G., Singer, C. F., Pischinger, K. I., Kaserer, K., Manavi, M., Kubista, E., and Czerwenka, K. F. ( 2006) Proteomic analysis in human breast cancer: identification of a characteristic protein expression profile of malignant breast epithelium. Proteomics 6, 1989–2002 [DOI] [PubMed] [Google Scholar]
- 21.Neubauer, H., Clare, S. E., Kurek, R., Fehm, T., Wallwiener, D., Sotlar, K., Nordheim, A., Wozny, W., Schwall, G. P., Poznanovij, S., Sastri, C., Hunzinger, C., Stegmann, W., Schrattenholz, A., and Cahill, M. A. ( 2006) Breast cancer proteomics by laser capture microdissection, sample pooling, 54-cm IPG IEF, and differential iodine radioisotope detection. Electrophoresis 27, 1840–1852 [DOI] [PubMed] [Google Scholar]
- 22.Zang, L., Palmer Toy, D., Hancock, W. S., Sgroi, D. C., and Karger, B. L. ( 2004) Proteomic analysis of ductal carcinoma of the breast using laser capture microdissection, LC-MS, and 16O/18O isotopic labeling. J. Proteome Res. 3, 604–612 [DOI] [PubMed] [Google Scholar]
- 23.Umar, A., Luider, T. M., Foekens, J. A., and Pasa-Tolij, L. ( 2007) NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics 7, 323–329 [DOI] [PubMed] [Google Scholar]
- 24.Pasa-Tolij, L., Masselon, C., Barry, R. C., Shen, Y., and Smith, R. D. ( 2004) Proteomic analyses using an accurate mass and time tag strategy. Biotechniques 37, 621–624, 626–633, 636 passim [DOI] [PubMed] [Google Scholar]
- 25.Hayward, J. L., Carbone, P. P., Heuson, J. C., Kumaoka, S., Segaloff, A., and Rubens, R. D. ( 1977) Assessment of response to therapy in advanced breast cancer: a project of the Programme on Clinical Oncology of the International Union Against Cancer, Geneva, Switzerland. Cancer 39, 1289–1294 [DOI] [PubMed] [Google Scholar]
- 26.McShane, L. M., Altman, D. G., Sauerbrei, W., Taube, S. E., Gion, M., Clark, G. M., and Statistics Subcommittee of NCI-EORTC Working Group on Cancer Diagnostics ( 2005) REporting recommendations for tumor MARKer prognostic studies (REMARK). J. Natl. Cancer Inst. 97, 1180–1184 [DOI] [PubMed] [Google Scholar]
- 27.Umar, A., Dalebout, J. C., Timmermans, A. M., Foekens, J. A., and Luider, T. M. ( 2005) Method optimisation for peptide profiling of microdissected breast carcinoma tissue by matrix-assisted laser desorption/ionisation-time of flight and matrix-assisted laser desorption/ionisation-time of flight/time of flight-mass spectrometry. Proteomics 5, 2680–2688 [DOI] [PubMed] [Google Scholar]
- 28.Shen, Y., Tolij, N., Masselon, C., Pasa-Tolij, L., Camp, D. G., 2nd, Hixson, K. K., Zhao, R., Anderson, G. A., and Smith, R. D. ( 2004) Ultrasensitive proteomics using high-efficiency on-line micro-SPE-nanoLC-nanoESI MS and MS/MS. Anal. Chem. 76, 144–154 [DOI] [PubMed] [Google Scholar]
- 29.Kang, H., Pasa-Tolij, L., and Smith, R. D. ( 2007) Targeted tandem mass spectrometry for high-throughput comparative proteomics employing nanoLC-FTICR MS with external ion dissociation. J. Am. Soc. Mass Spectrom. 18, 1332–1343 [DOI] [PubMed] [Google Scholar]
- 30.Shaffer, S. A., Tolmachev, A., Prior, D. C., Anderson, G. A., Udseth, H. R., and Smith, R. D. ( 1999) Characterization of an improved electrodynamic ion funnel interface for electrospray ionization mass spectrometry. Anal. Chem. 71, 2957–2964 [DOI] [PubMed] [Google Scholar]
- 31.Kelly, R. T., Page, J. S., Luo, Q., Moore, R. J., Orton, D. J., Tang, K., and Smith, R. D. ( 2006) Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal. Chem. 78, 7796–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayampurath, A. M., Jaitly, N., Purvine, S. O., Monroe, M. E., Auberry, K. J., Adkins, J. N., and Smith, R. D. ( 2008) DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 24, 1021–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monroe, M. E., Tolij, N., Jaitly, N., Shaw, J. L., Adkins, J. N., and Smith, R. D. ( 2007) VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 34.Petritis, K., Kangas, L. J., Ferguson, P. L., Anderson, G. A., Pasa-Tolij, L., Lipton, M. S., Auberry, K. J., Strittmatter, E. F., Shen, Y., Zhao, R., and Smith, R. D. ( 2003) Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Anal. Chem. 75, 1039–1048 [DOI] [PubMed] [Google Scholar]
- 35.Jacobs, J. M., Mottaz, H. M., Yu, L. R., Anderson, D. J., Moore, R. J., Chen, W. N., Auberry, K. J., Strittmatter, E. F., Monroe, M. E., Thrall, B. D., Camp, D. G., 2nd, and Smith, R. D. ( 2004) Multidimensional proteome analysis of human mammary epithelial cells. J. Proteome Res. 3, 68–75 [DOI] [PubMed] [Google Scholar]
- 36.Wang, H., Qian, W. J., Mottaz, H. M., Clauss, T. R., Anderson, D. J., Moore, R. J., Camp, D. G., 2nd, Khan, A. H., Sforza, D. M., Pallavicini, M., Smith, D. J., and Smith, R. D. ( 2005) Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 4, 2397–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan, A. J., Strittmatter, E. F., Camp, D. G., 2nd, Smith, R. D., and Pallavicini, M. G. ( 2006) Quantitative proteome analysis of breast cancer cell lines using 18O-labeling and an accurate mass and time tag strategy. Proteomics 6, 2903–2915 [DOI] [PubMed] [Google Scholar]
- 38.Jaitly, N., Monroe, M. E., Petyuk, V. A., Clauss, T. R., Adkins, J. N., and Smith, R. D. ( 2006) Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 78, 7397–7409 [DOI] [PubMed] [Google Scholar]
- 39.Elston, C. W., and Ellis, I. O. ( 1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410 [DOI] [PubMed] [Google Scholar]
- 40.Mustafa, D. A., Burgers, P. C., Dekker, L. J., Charif, H., Titulaer, M. K., Smitt, P. A., Luider, T. M., and Kros, J. M. ( 2007) Identification of glioma neovascularization-related proteins by using MALDI-FTMS and nano-LC fractionation to microdissected tumor vessels. Mol. Cell. Proteomics 6, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 41.Nebert, D. W., and Russell, D. W. ( 2002) Clinical importance of the cytochromes P450. Lancet 360, 1155–1162 [DOI] [PubMed] [Google Scholar]
- 42.Tuquet, C., Dupont, J., Mesneau, A., and Roussaux, J. ( 2000) Effects of tamoxifen on the electron transport chain of isolated rat liver mitochondria. Cell Biol. Toxicol. 16, 207–219 [DOI] [PubMed] [Google Scholar]
- 43.Min, J., Van Veldhoven, P. P., Zhang, L., Hanigan, M. H., Alexander, H., and Alexander, S. ( 2005) Sphingosine-1-phosphate lyase regulates sensitivity of human cells to select chemotherapy drugs in a p38-dependent manner. Mol. Cancer Res. 3, 287–296 [DOI] [PubMed] [Google Scholar]
- 44.Oskouian, B., Sooriyakumaran, P., Borowsky, A. D., Crans, A., Dillard-Telm, L., Tam, Y. Y., Bandhuvula, P., and Saba, J. D. ( 2006) Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc. Natl. Acad. Sci. U. S. A. 103, 17384–17389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, L., Welham, K. J., Watson, M. B., Drew, P. J., Lind, M. J., and Cawkwell, L. ( 2007) The proteomic analysis of cisplatin resistance in breast cancer cells. Oncol. Res. 16, 497–506 [DOI] [PubMed] [Google Scholar]
- 46.Arai, K., Takano, S., Teratani, T., Ito, Y., Yamada, T., and Nozawa, R. ( 2008) S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr. Cancer Drug Targets 8, 243–252 [DOI] [PubMed] [Google Scholar]
- 47.Georgescu, M. M. ( 2008) NHERF1: molecular brake on the PI3K pathway in breast cancer. Breast Cancer Res. 10, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osanai, M., Murata, M., Nishikiori, N., Chiba, H., Kojima, T., and Sawada, N. ( 2007) Occludin-mediated premature senescence is a fail-safe mechanism against tumorigenesis in breast carcinoma cells. Cancer Sci. 98, 1027–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salama, I., Malone, P. S., Mihaimeed, F., and Jones, J. L. ( 2008) A review of the S100 proteins in cancer. Eur. J. Surg. Oncol. 34, 357–364 [DOI] [PubMed] [Google Scholar]
- 50.Sanders, M. E., Dias, E. C., Xu, B. J., Mobley, J. A., Billheimer, D., Roder, H., Grigorieva, J., Dowsett, M., Arteaga, C. L., and Caprioli, R. M. ( 2008) Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MS. J. Proteome Res. 7, 1500–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suh, K. S., Mutoh, M., Gerdes, M., and Yuspa, S. H. ( 2005) CLIC4, an intracellular chloride channel protein, is a novel molecular target for cancer therapy. J. Investig. Dermatol. Symp. Proc. 10, 105–109 [DOI] [PubMed] [Google Scholar]
- 52.Bacci, S., De Cosmo, S., Prudente, S., and Trischitta, V. ( 2007) ENPP1 gene, insulin resistance and related clinical outcomes. Curr. Opin. Clin. Nutr. Metab. Care 10, 403–409 [DOI] [PubMed] [Google Scholar]
- 53.Jenkinson, C. P., Coletta, D. K., Flechtner-Mors, M., Hu, S. L., Fourcaudot, M. J., Rodriguez, L. M., Schneider, J., Arya, R., Stern, M. P., Blangero, J., Duggirala, R., and DeFronzo, R. A. ( 2008) Association of genetic variation in ENPP1 with obesity-related phenotypes. Obesity 16, 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dal Maso, L., Zucchetto, A., Talamini, R., Serraino, D., Stocco, C. F., Vercelli, M., Falcini, F., Franceschi, S., and Prospective Analysis of Case-control studies on Environmental factors and health ( PACE) study group (2008) Effect of obesity and other lifestyle factors on mortality in women with breast cancer. Int. J. Cancer 123, 2188–2194 [DOI] [PubMed] [Google Scholar]
- 55.Marchetti, A., Buttitta, F., Pellegrini, S., Bertacca, G., and Callahan, R. ( 2001) Reduced expression of INT-6/eIF3-p48 in human tumors. Int. J. Oncol. 18, 175–179 [DOI] [PubMed] [Google Scholar]
- 56.Traicoff, J. L., Chung, J. Y., Braunschweig, T., Mazo, I., Shu, Y., Ramesh, A., D'Amico, M. W., Galperin, M. M., Knezevic, V., and Hewitt, S. M. ( 2007) Expression of EIF3-p48/INT6, TID1 and Patched in cancer, a profiling of multiple tumor types and correlation of expression. J. Biomed. Sci. 14, 395–405 [DOI] [PubMed] [Google Scholar]
- 57.Chen, L., Uchida, K., Endler, A., and Shibasaki, F. ( 2007) Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2 alpha for degradation by hypoxia- and pVHL-independent regulation. J. Biol. Chem. 282, 12707–12716 [DOI] [PubMed] [Google Scholar]
- 58.Mack, D. L., Boulanger, C. A., Callahan, R., and Smith, G. H. ( 2007) Expression of truncated Int6/eIF3e in mammary alveolar epithelium leads to persistent hyperplasia and tumorigenesis. Breast Cancer Res. 9, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zucker, S., Hymowitz, M., Rollo, E. E., Mann, R., Conner, C. E., Cao, J., Foda, H. D., Tompkins, D. C., and Toole, B. P. ( 2001) Tumorigenic potential of extracellular matrix metalloproteinase inducer. Am. J. Pathol. 158, 1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quemener, C., Gabison, E. E., Naïmi, B., Lescaille, G., Bougatef, F., Podgorniak, M. P., Labarchède, G., Lebbé, C., Calvo, F., Menashi, S., and Mourah, S. ( 2007) Extracellular matrix metalloproteinase inducer up-regulates the urokinase-type plasminogen activator system promoting tumor cell invasion. Cancer Res. 67, 9–15 [DOI] [PubMed] [Google Scholar]
- 61.Reimers, N., Zafrakas, K., Assmann, V., Egen, C., Riethdorf, L., Riethdorf, S., Berger, J., Ebel, S., Jänicke, F., Sauter, G., and Pantel, K. ( 2004) Expression of extracellular matrix metalloproteases inducer on micrometastatic and primary mammary carcinoma cells. Clin. Cancer Res. 10, 3422–3428 [DOI] [PubMed] [Google Scholar]
- 62.Riethdorf, S., Reimers, N., Assmann, V., Kornfeld, J. W., Terracciano, L., Sauter, G., and Pantel, K. ( 2006) High incidence of EMMPRIN expression in human tumors. Int. J. Cancer 119, 1800–1810 [DOI] [PubMed] [Google Scholar]
- 63.Als, A. B., Dyrskjøt, L., von der Maase, H., Koed, K., Mansilla, F., Toldbod, H. E., Jensen, J. L., Ulhøi, B. P., Sengeløv, L., Jensen, K. M., and Orntoft, T. F. ( 2007) Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin. Cancer Res. 13, 4407–4414 [DOI] [PubMed] [Google Scholar]