Abstract
Schistosomes are the causative agents of schistosomiasis, one of the most prevalent and serious of the parasitic diseases that currently infects ∼200 million people worldwide. Schistosome excretory/secretory (ES) proteins have been shown to play important roles in modulating mammalian host immune systems. In our current study, we performed a global proteomics identification of the ES proteins from adult worms of Schistosoma japonicum, one of the three major schistosome species. Our results unambiguously identified 101 proteins, including 53 putatively secreted proteins. By quantitative analysis, we revealed fatty acid-binding protein as a major constituent of the in vitro ES proteome. Strikingly the heat shock proteins HSP70s, HSP90, and HSP97 constituted the largest protein family in the ES proteome, implying a central role for these proteins in immunomodulation in the host-parasite relationship. Other important S. japonicum ES proteins included actins, 14-3-3, aminopeptidase, enolase, and glyceraldehyde-3-phosphate dehydrogenase, some of which have been considered as viable vaccine candidates and therapeutic targets. A comparison with previous studies suggests that 48.5% of S. japonicum ES proteins are common to other parasite ES products, indicating that the molecular mechanisms involved in evading the host immune response may be conserved across different parasites. Interestingly seven host proteins, including antimicrobial protein CAP18, immunoglobulins, and a complement component, were identified among in vitro S. japonicum ES products likely originating from the schistosome tegument or gut, indicating that host innate and acquired immune systems could defend against schistosome invasion. Our present study represents the first attempt at profiling S. japonicum ES proteins, provides an insight into host-parasite interactions, and establishes a resource for the development of diagnostic agents and vaccines for the control of schistosomiasis.
Schistosomes, or blood flukes, are water-borne parasites that are the causative agents of schistosomiasis. An estimated 200 million people worldwide are infected with schistosomes with an additional 650 million people at risk of infection (1). One of the major species of schistosomes, Schistosoma japonicum, is a mammalian parasite endemic in East Asia, especially in China and the Philippines. Schistosomes have complex life cycles. Larval schistosome worms (cercariae) are released by freshwater snails and subsequently invade their definite hosts, human or other mammals, via skin penetration. Once in a host animal, cercariae develop into schistosomula and adult worms, which reside in the portal mesenteric system of the host. When the females lay eggs, some eggs leave the host body and hatch in bodies of water as miracidia. The miracidia seek out and penetrate intermediate host snails, completing the schistosome life cycle.
Schistosome strategies for evasion of the host immune system, which permit extended survival in mammalian hosts, are not well understood. These dominant evasion strategies have been described as a system of mimicry capable of producing antigens that are similar to endogenous host components (2–4), antigen disguise through acquisition of host molecules to cover the outer worm surface (5, 6), and immunological modulation through interference with host immune systems (7–10). Among these strategies, schistosome excretory/secretory (ES)1 products have been shown to elicit host immunological modulation functions (7, 11). Schistosome ES proteins are released or secreted from epithelial surfaces of the gut and/or tegument as well as other specialized ES organs throughout almost all life stages. Schistosoma mansoni primary sporocysts have been reported to synthesize and secrete a wide variety of glycoproteins when cultured in vitro (12, 13). These glycoproteins were shown to have antioxidant activities against potential oxidative killing by mollusk defense systems (14). Similarly ES molecules from schistosome cercariae were also reported to down-regulate host immune responses (7). The anti-inflammatory activity of S. mansoni schistosomula ES products (ESPs) was found to be dominantly associated with Sm16.8 protein (15). In addition, ES proteins from S. mansoni adult worms (16), eggs (17), and miracidia (18) have also been investigated.
Identification of all ES complex components is important for understanding how schistosomes regulate host immune systems to establish chronic infections and also other aspects of parasite-host interaction. Importantly this information can be expected to facilitate the discovery of vaccines and new therapeutic drug targets as well as new diagnostic reagents for schistosomiasis control. Proteomics approaches encompass the most efficient and powerful tools for identification of protein complexes and have been widely used to decipher the ES components of the filarial parasite Brugia malayi (19), Leishmania (Trypanosomatidae) (20), nematodes (21–26), and Trematoda (27–37). For the genus Schistosoma, the ES compositions of S. mansoni have been identified in many developmental life stages, including sporocyst (34), cercaria (35, 36), and egg (37) but have not been characterized in the adult worm.
Characterization of the S. japonicum ES proteome has not been reported. S. japonicum is significantly different from S. mansoni and Schistosoma hematobium in skin invasion, skin migration, and its developmental patterns of swift migration and maturation (38–40). As such, S. japonicum represents a distinct and valuable model for the study of blood fluke immune evasion strategies. Our research group recently generated and reported a large number of S. japonicum protein-coding genes and expressed sequence tags (ESTs) (41, 42). This preliminary work provides important translated protein sequence data resources for mass spectrum data searching. The present study characterized the in vitro ES proteome of adult worms of S. japonicum (43) using a high throughput LC-MS/MS screening. This life stage spans the longest time frame of parasitic interaction with the host that is distinct from previous reports on proteomics identification of ES compositions in other species. Finally the present study confidently identified 101 S. japonicum ES proteins. This information represents substantial progress toward deciphering the worm ES proteome. These new data provide the basis for further investigations into the molecular basis of schistosome modulation of host immunity, increase the possibility of identifying vaccine candidates and new drug targets, and may aid the development of protein probes for selective and sensitive diagnosis of schistosomiasis.
EXPERIMENTAL PROCEDURES
Schistosome Materials—
Individual laboratory rabbits (Oryctolagus cuniculus) were percutaneously infected with 1000 S. japonicum cercariae isolated from field-collected, naturally infected Oncomelania hupensis snails. After cercarial challenge, adult S. japonicum worms were collected by flushing the hepatic portal system and mesenteric veins of infected rabbits at 42–45 days postinfection. The worms were washed at least twice in PBS (pH 7.4) to remove host tissues.
Preparation of S. japonicum ESP Extracts—
To collect S. japonicum ESPs, around 800 mixed sex adult stage worms (∼400 μl) were soaked in 1 ml of PBS (pH 7.4) for 10 min at room temperature under atmospheric conditions. After incubation, the worm mixture was centrifuged at low speed, and the brown supernatant was collected. The above procedure was repeated 10 times until the supernatant became transparent. A flow chart for this procedure is shown in Fig. 1A. The supernatants were combined and stored at −80 °C until use.
Fig. 1.
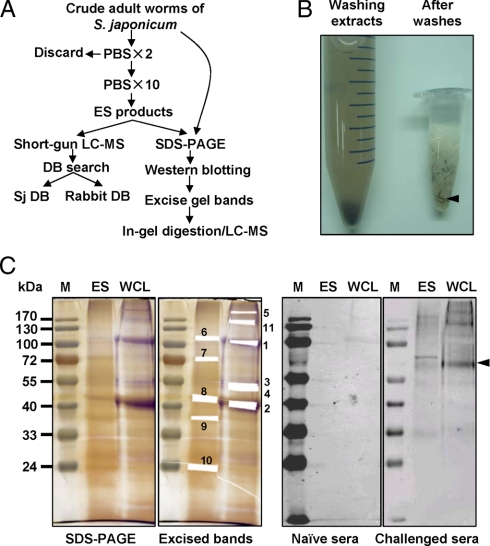
Extraction of ES proteins of adult worms of S. japonicum and immunoblot assay. A, flow chart of the proteomics study of ES products from S. japonicum. The adult worms were prewashed two times with PBS and then further soaked in PBS with 10 buffer changes. The PBS supernatants containing ESPs were collected and used for proteomics analyses by LC-MS followed by schistosome and rabbit DB searches. The extracted protein mixtures from whole adult worms or ESPs were separated by SDS-PAGE followed by Western blotting in parallel with in-gel digestion of select bands and LC-MS analysis. B, isolated S. japonicum adult worms were soaked repeatedly in PBS, and the soaking extracts were collected in a 15-ml tube as shown on the left. Worm appearance after washing is shown on the right. An arrow indicates worm carcasses. C, Western blot analysis of WCL extract and the ES proteome of S. japonicum adult worms. Pictures shown from left to right are a 12% acrylamide silver-stained gel, the gel band excising pattern, immunoblotting with naïve rabbit sera, and immunoblotting with anti-S. japonicum sera. Numbers indicate the antigenic or abundant protein bands that were isolated and subjected to MS analysis. We were not able to analyze one antigenic band that was recognized by challenged rabbit sera because of the difficulty in determining and excising the corresponding gel band. M, protein marker.
Protein Extraction and Digestion—
Proteins in the pooled supernatants were precipitated by addition of 6 volumes of cold acetone mixed with 10% TCA and 20 mm DTT followed by overnight incubation at −20 °C. Subsequently precipitated proteins were pelleted by centrifugation at 12,000 rpm for 20 min. The pellet was washed three times with 90% TCA and then subjected to sonication in a buffer containing 50 mm Tris-HCl (pH 8.3), 5 mm EDTA, 1 mm PMSF, 0.5% SDS, and 0.5 mm DTT. The lysate was further solubilized by the addition of SDS to a final concentration of 0.5%.
After the lysate was centrifuged, the protein mixture from the supernatant was treated with 10 mm DTT and then carboxamidomethylated in 55 mm iodoacetamide. Next the protein mixture was diluted with deionized water and then digested with modified, sequencing grade bovine trypsin (Roche Applied Science) at a substrate:enzyme ratio of 50:1. The concentration of SDS was lower than 0.1% in the mixture to facilitate enzymatic digestion.
Protein Identification by Mass Spectrometry—
The proteome of S. japonicum ESPs was characterized by nanoscale capillary LC-MS/MS (nano-LC-MS/MS) as described previously (42) with minor modifications. The resulting peptide mixture was fractionated into 10 subgroups by strong cation exchange (SCX) chromatography using a 4.0 × 150-mm POROS 50 HS column (MDS Sciex/Applied Biosystems). The volume of each SCX fraction was adjusted to ∼30 μl using a vacuum lyophilizer. SCX fractions with high salt concentrations were desalted using an 8 × 4-mm Dikma EasyGuard short reverse-phase (RP) column (Dikma Technologies) before downstream analysis. Subsequently the peptide mixture from each SCX fraction was loaded onto an RP trap column (C18, 5 μm, 300 Å, 300-μm inner diameter × 5 mm; Dionex/LC Packings) for on-line desalting at a flow rate of 10 μl/min. The trap column was sequentially connected in line with an analytical fused silica nanocolumn of 75 μm × 150-mm inner diameter packed with C18 Acclaim PepMap (3-μm) stationary phase (Dionex/LC Packings). The peptide mixture was eluted into a QSTAR Pulsar I mass spectrometer (MDS Sciex/Applied Biosystems) coupled to a Protana NanoES electrospray ionization source at a flow rate of 200 nl/min. The Agilent 1100 capillary LC system (Agilent Technologies) was used to deliver mobile phases A (0.5% acetic acid in water) and B (0.5% acetic acid in 100% ACN) using a linear gradient of 5–50% B (60 min), 50–90% B (30 min), and then 90% B (15 min). A poly(ether ether ketone) MicroTee (Upchurch Scientific) was used to join the analytical column and a 10-μm-inner diameter PicoTip nanospray emitter (New Objective). The side port of the MicroTee was attached by a platinum wire through which a spray voltage of 2500 V was applied to the emitter for a steady spray. The MS/MS spectra were recorded with information-dependent acquisition and duty cycle enhancement. The nine most intense ions with charges of 2–4 in each survey scan were fragmented with rolling collision energy.
Schistosome Protein Database (DB) Search—
The schistosome protein DB used for mass spectra assignments contains 11,717 sequences retrieved via the Web Entrez search engine at the National Center for Biotechnology Information (NCBI) Web site on October 16, 2008. All mass spectra were calibrated iteratively using known autocleaved peptides of bovine trypsin to achieve a mass measurement accuracy of 20 ± 10 ppm. The peak lists were generated using Mascot.dll (Version 1.6.0.19, MDS Sciex/Applied Biosystems) with default parameter values. The DB search was performed with software MASCOT 1.9.0 (Matrix Science). To obtain an estimate of the false-positive rate (FPR) of peptide identification, we also performed parallel reverse DB searching (43).
Throughout all forward and reverse DB searches, a maximum of two missed cleavage sites was allowed per peptide. The ion type was set as monoisotopic, and peptide charges +1/+2/+3 were taken into account. Modifications were allowed for variable carbamoylmethylation of cysteine and oxidation of methionine. All DB searches were constrained to a mass tolerance of 0.5 Da for both the parent ion and fragment ion.
The primary peptide assignments were filtered according to specific criteria as follows. (a) For a single tandem mass spectrum, the top ranked peptide assignment was considered, whereas the lower ranked peptide assignments were discarded without further consideration. (b) Candidate peptides without internal KR, KK, RK, or RR were accepted (44). (c) Peptides with more than 5 amino acids were accepted. The Mascot score cutoff for peptide identification was dynamically determined for a given FPR, which was calculated based on forward and reverse DB searches (43). In the present study, we used the highest Mascot peptide score returned from a reverse DB search as the cutoff for peptide filtering to achieve an FPR of zero.
Protein identification was based on the peptide assignments with proteins sharing the same peptide grouped together using MassSieve 1.06a (45). Proteins identified by a single spectrum were discarded. Protein abundance was represented by spectral count (SC) defined as the number of fragmentation spectra of the peptides of an identified protein (46). Any trypsin identifications were removed from the final result due to the usage of trypsin as a digestion reagent. Peptides identified from the schistosome DB were further compared with rabbit sequences (protein and DNA) using BLAST programs to confirm schistosome-specific peptides. The available rabbit genome trace sequences were downloaded from the Broad Institute Web site and included 2.5 million assembled contigs, scaffolds, and unplaced sequences. Another rabbit sequencing trace data set was also retrieved from NCBI TraceDB that contained ∼29 million sequencing reads representing 2× coverage of the rabbit genome. In addition, we retrieved 1305 genome survey sequences, 34,922 ESTs, and 3975 mRNA sequences with an organism restriction of O. cuniculus from NCBI via the Entrez system. These DNA sequences were combined for TBLASTN searches with schistosome peptides.
Identification of Host Proteins in S. japonicum ESPs—
To identify the rabbit proteins in the ESP of S. japonicum, we also performed a rabbit protein DB search using the same set of mass data collected in this study. The integrated rabbit protein DB used for DB search contains 9583 sequences, which were collected as follows. First, we downloaded three public DBs, including the NCBI non-redundant DB (6,298,708 entries, dated June 18, 2008), UniProtKB/Swiss-Prot (released version 55.5, 389,046 entries, dated June 10, 2008), and UniProtKB/TrEMBL DBs (released version 38.5, June 10, 2008) from UniProt. From these databases, we extracted all sequences annotated either as “rabbit” or “O. cuniculus.” We collected 4078, 869, and 1217 rabbit sequences, respectively, from the three DBs. Second, we retrieved 7596 rabbit protein sequences via the GenBank™ Entrez search interface with organism restriction to O. cuniculus. Finally the resultant 137,60 rabbit protein sequences were further reduced to 9583 non-redundant sequences. This final set was used for DB searches. The search engine, search parameters, peptide filtering, and protein assignments for mass data interpretation were the same as used for the schistosome DB search. Parallel reverse DB searches were performed that achieved an FPR of zero by setting the score cutoff with no hits in the reverse DB.
We also performed a DB search against Swiss-Prot (released version 55.5, 389,046 entries) to identify host proteins using the procedures and criteria outlined above. The results from rabbit and Swiss-Prot DB searches were combined, and the peptides identified were compared with schistosome sequences (protein and DNA) using BLAST programs. Any peptides matching schistosome sequences were discarded. Additional unpublished schistosome genomic sequences were downloaded for comparison from the Wellcome Trust Sanger Institute (assembly v3.1)2 and Shanghai Center for Life Science and Biotechnology Information (updated on August 29, 2008).3
Prediction of Secreted Proteins—
N-terminal signal sequences were predicted using the SignalP 3.0 Web server (47), and non-classical secretory proteins were determined using SecretomeP 2.0 (48). For SignalP predictions, positive identifications were made when both neural network and hidden Markov model algorithms gave coincident estimations. Non-classical secreted proteins were predicted to obtain a neural network score exceeding the normal threshold of 0.5 but not to contain a signal peptide. Herein to avoid false predictions, we used all protein sequences instead of choosing a representative sequence in the protein groups for secretion prediction. This condition was set because more than one-third of the identifications were of protein groups rather than single proteins. Proteins within the same protein group were expected to have similar characteristics as these proteins are orthologs, paralogs, or members of the same protein family. Thus, if one or more protein sequences from a given protein group were predicted as classical or non-classical secreted protein(s) then this protein group was annotated as secreted.
Evolution Analysis—
Alignment of schistosome fatty acid-binding protein (FABP) sequences was performed using MegAlign 5.01 from the Lasergene suite (DNAStar Inc., Madison, WI) and viewed using GeneDoc 2.7 (49). A phylogenetic tree of the FABP coding sequences was established using Mega 4.0 (50) and the neighbor-joining method with default parameters and was tested using 1000 bootstraps. The schistosome FABP sequences were downloaded from GenBank with the exception of five unpublished S. japonicum cDNA sequences that were downloaded from the Chinese National Human Genome Center at Shanghai.4
SDS-PAGE and Silver Staining—
Equal amounts of protein (25 μg) from whole worm extracts and ES proteins were separated by one-dimensional SDS-PAGE. A repeat sample loading of an equal amount of protein was run in the same gel. After SDS-PAGE, the gel was cut into two pieces with each piece containing identical loaded samples. One piece of gel was used for electrotransfer to nitrocellulose and Western blotting as detailed below, and the other was subjected to silver staining as described previously (51).
Western Blotting and In-gel Tryptic Digestion—
Normal sera were collected from non-exposed rabbits. The proteins in the SDS-PAGE gel were transferred to a Hybond-C nitrocellulose membrane (Amersham Biosciences), which was then probed overnight at 4 °C with schistosome-challenged rabbit sera or naïve rabbit sera at a dilution of 1:500 in PBS containing 0.05% Tween 20 (PBST). After washing in PBST, the membrane was incubated with 1:1000 IRDye800-conjugated affinity-purified donkey anti-rabbit IgG (heavy and light) (Rockland) for 1 h at room temperature. Signals were imaged using an Odyssey Infrared Imaging System (LI-COR Biosciences). Protein gel bands that reacted with schistosome-challenged rabbit sera and other dominant bands were excised from the parallel gel piece and subjected to in-gel tryptic digestion according to an optimized procedure (51).
Mass Spectrometry Analyses of the Protein Gel Bands—
Digested peptide mixtures of protein gel bands were analyzed using an Agilent 1200 Series nano-LC system coupled through an orthogonal nanospray ion source Chip Cube to an Agilent 1200 Series LC/MSD Trap XCT Ultra ion trap mass spectrometer. Sample enrichment, desalting, and peptide separation by RP chromatography (75 μm × 150 mm) were completed within the Chip Cube. The peptides were eluted at a flow rate of 300 nl/min with a gradient beginning with 5% solvent B (0.1% formic acid in acetonitrile) and 95% solvent A (0.1% formic acid in water) followed by a gradient from 5 to 15% B over 2 min, 15–30% B over 13 min, 30–90% B over 10 min, 90–90% for 2 min, and down to 5% B over 3 min.
The LC/MSD Trap XCT Ultra was operated in the standard-enhanced scan mode. The ionization mode was positive. Drying gas flowed at 4 liters/min at a temperature of 325 °C. The capillary was set at 1800 V with the skimmer at 40 V, and the capillary exit was set at 158.5 V. The trap drive was set at 82.9 V. Ion charge control was on with a maximum accumulation time of 200 ms, the smart target was 500,000, and the MS scan range was 400–1800 with averages of 2. Automatic MS/MS was performed in ultrascan mode with the number of precursor ions set at 3, fragmentation amplitude at 1.00 V, active exclusion on (after two spectra for 0.5 min), exclusion of singly charged ions on, an MS/MS scan range of 100–2200, and ultrascan on.
The DB searches against the forward and reverse schistosome DBs were performed using MASCOT 1.90 as described above. Peptides were filtered as described above. The FPR for peptide identification was 1.3% as determined by a reverse DB search.
RESULTS
Preparation of the ESPs of S. japonicum Adult Worms—
To date, most studies have collected ESPs following in vitro culture. However, the excretion and secretion behavior of the parasites will be altered due to the removal from natural host tissues and in vitro maintenance in a chemical mixture (30). Therefore, direct analysis of in vivo ES proteins will likely yield the most informative results. However, isolation of the special host fluid containing ES proteins of S. japonicum is challenging because the parasites live in the portal and mesenteric vein systems of the mammalian hosts. In the present study, we used an alternative strategy of extracting ES proteins by repeatedly soaking the newly isolated adult worm bodies of S. japonicum in buffers, avoiding the extended in vitro culture that can alter S. japonicum secretions. The worms were repeatedly soaked in PBS, and the extracts were harvested until the color of the buffer remained clear (Fig. 1, A and B). The body color of most worms faded following release of the majority of their gut contents, although some of the worm bodies remained dark in color even after the soaking buffer turned clear (Fig. 1B). Microscopic examination was subsequently performed to confirm that the worm bodies were complete and that the tegument remained intact, indicating that the extracts were mainly from the gut or extracellular fluid rather than from detached tissues.
The total amount of extracted proteins from ∼800 S. japonicum adult worms was ∼500 μg. To exclude the possibility that the extracted ESP was from whole worm lysates, we compared the gel patterns of ES proteins with soluble worm extracts. As shown in Fig. 1C, the extracted ESPs and whole worm extracts exhibited clearly distinct protein separation patterns especially with the most abundant protein bands. The S. japonicum ESPs appeared as a complicated mixture with major protein bands ranging from 20 to 72 kDa.
ES Proteome Determination by LC-MS/MS Analysis—
S. japonicum ESPs were analyzed by two-dimensional nano-LC-MS/MS followed by forward and reverse DB searching and further investigations. The raw data files of Mascot were imported into MassSieve from which 174 “parsimony” proteins (groups) were obtained. Following reverse schistosome DB searching, the highest Mowse score of an identified peptide was 29.7, which was subsequently used as the cutoff for peptide filtering to achieve an FPR of zero. Based on this threshold, 1100 mass spectra representing 351 distinct schistosome peptides were identified. Single spectrum identifications were discarded. Finally 291 distinct peptides representing 101 S. japonicum proteins were unequivocally identified based on two or more matching mass spectra (Table I and supplemental Table 1). Sixty-four proteins had clearly distinct protein assignments, whereas the remaining proteins were classified into protein groups based on sharing the same sets of peptides (supplemental Table 1).
Table I.
ES proteins of S. japonicum adult worms
aa, amino acids; Seq cov., sequence coverage.
| Gi (NCBI) no. | Annotationa | aa | SMSb | SCb | Seq cov. | Spec | Nematoded
|
Trematodae
|
Ocf | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ac A | B A | H A | Tc L/A | E1 A | E2 A | Ef A | F1 A | F2 L | F3 A | P A | Sb A | S1 S | S2 C | S3 C | S4 E | ||||||||
| % | |||||||||||||||||||||||
| Fatty acid binding | |||||||||||||||||||||||
| 30144605 | FABP | 132 | 6676.5 | 142 | 70.5 | − | − | − | − | − | − | − | − | + | − | + | − | + | − | + | + | − | 6 |
| Chaperone/heat shock protein | |||||||||||||||||||||||
| 56759038 | HSP90α2 | 719 | 2806 | 68 | 26.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 552242 | HSP70 | 637 | 2776.2 | 59 | 13.7 | − | − | + | − | − | + | − | − | − | − | − | + | − | − | + | − | − | 3 |
| 76155249 | HSP70 | 137 | 1927.6 | 36 | 45.3 | NC | − | + | − | − | + | − | − | − | − | − | + | − | − | + | − | − | 3 |
| 189502946 | HSP70-5 | 648 | 1522.4 | 33 | 11.9 | SP | − | + | − | − | + | − | − | − | − | − | + | − | − | + | − | − | 3 |
| 76155211 | HSP70 | 266 | 860.54 | 19 | 41.7 | − | − | + | − | − | + | − | − | − | − | − | + | − | − | + | − | − | 3 |
| 76156161 | HSP4 | 514 | 631.73 | 16 | 8.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76155264 | HSP97 | 252 | 223.68 | 5 | 12.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56756997 | DnaJ homolog A1 | 400 | 225.93 | 4 | 8.0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76154306 | Chaperonin | 305 | 155.67 | 4 | 11.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 60688010 | HSP70 | 82 | 82.07 | 2 | 20.7 | NC | − | + | − | − | + | − | − | − | − | − | + | − | − | + | − | − | 3 |
| 76157510 | CCT7 | 246 | 77.19 | 2 | 4.1 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56753850 | Endoplasmin | 797 | 98.7 | 2 | 3.0 | SP | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56756959 | DJ-1 family protein | 184 | 94.92 | 2 | 9.2 | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 2 |
| Structure protein | |||||||||||||||||||||||
| 1703114 | Actin-2 | 376 | 1929.4 | 47 | 33.2 | NC | − | + | − | + | + | + | + | + | − | − | − | − | − | + | + | + | 9 |
| 56754341 | Actin-1 | 360 | 1852.2 | 45 | 34.7 | − | − | + | − | + | + | + | + | + | − | − | − | − | − | + | + | + | 9 |
| 56754704 | Actin, cytoplasm | 376 | 1347.2 | 32 | 23.7 | − | − | + | − | + | + | + | + | + | − | − | − | − | − | + | + | + | 9 |
| 30995481 | β-Tubulin | 443 | 753.79 | 17 | 20.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| 161072 | α-Tubulin | 451 | 966.32 | 17 | 13.5 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| 29841388 | α-Tubulin | 327 | 838.89 | 14 | 18.7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| 56758452 | CAP1 | 242 | 96.7 | 2 | 5.8 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Metabolic enzyme | |||||||||||||||||||||||
| 349802 | Enolase | 434 | 1750.4 | 32 | 23.7 | NC | − | + | + | + | + | − | + | + | − | − | − | + | − | + | − | + | 9 |
| 56757978 | Pyruvate kinase | 561 | 1274.8 | 31 | 18.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 160996 | GAPDH | 338 | 1170.5 | 28 | 20.1 | − | − | − | − | − | − | − | + | + | − | − | − | − | + | + | − | + | 5 |
| 95113705 | FBA | 363 | 656.42 | 15 | 30.3 | − | + | − | − | + | − | + | − | − | − | − | − | − | − | + | + | + | 6 |
| 29841448 | Phosphoglycerate kinase | 417 | 615.05 | 14 | 13.4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| 31044498 | LDH | 331 | 320.83 | 6 | 4.5 | NC | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 120709 | GAPDH | 338 | 196.24 | 5 | 7.7 | − | − | − | − | − | − | − | + | + | − | − | − | − | + | + | − | + | 5 |
| 29841160 | Transketolase | 225 | 207.8 | 5 | 13.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | 1 |
| 3122305 | 6-Phosphofructokinase | 781 | 98.09 | 3 | 3.2 | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | 2 |
| 56755469 | Aldehyde dehydrogenase | 245 | 141.57 | 3 | 5.7 | NC | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | 3 |
| 29841244 | Glutamine synthetase | 364 | 149.7 | 3 | 3.3 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56758570 | mMDH | 341 | 125.69 | 3 | 7.0 | NC | + | − | + | − | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| 1255223 | Triose-phosphate isomerase | 252 | 178.35 | 3 | 9.5 | NC | − | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | 2 |
| 56756751 | Glucosidase, α | 443 | 87.42 | 2 | 5.0 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56758850 | UDP-glucose 4-epimerase | 351 | 85.24 | 2 | 4.0 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76156298 | Phosphoglucose isomerase | 274 | 69.82 | 2 | 2.9 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 166159344 | Phosphoglycerate mutase | 250 | 83.68 | 2 | 4.0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | 2 |
| Detoxification protein | |||||||||||||||||||||||
| 60279643 | Thioredoxin peroxidase-2 | 194 | 954.46 | 22 | 34.0 | NC | − | + | − | + | − | − | − | − | − | + | − | − | − | + | − | − | 4 |
| 1389744 | GST | 211 | 834.79 | 20 | 24.6 | − | + | − | + | − | − | − | + | + | − | + | + | + | + | + | + | + | 11 |
| 117380647 | SOD-like protein | 153 | 797.35 | 17 | 32.0 | NC | − | + | + | − | − | + | − | + | + | + | − | + | + | + | − | − | 9 |
| 38259184 | Thioredoxin peroxidase-1 | 184 | 536.66 | 12 | 18.5 | NC | − | + | − | + | − | − | − | − | − | + | − | − | − | + | − | − | 4 |
| 29841216 | GST | 218 | 306.78 | 7 | 17.0 | NC | + | − | + | − | − | − | + | + | − | + | + | + | + | + | + | + | 11 |
| % | |||||||||||||||||||||||
| 198385352 | TGR | 596 | 147.8 | 4 | 2.3 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 3892185 | PDI | 480 | 106.27 | 3 | 6.9 | NC | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + | 3 |
| 76155624 | PDI homolog | 356 | 189.1 | 3 | 3.4 | NC | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + | 3 |
| Muscle contraction | |||||||||||||||||||||||
| 56752573 | Titin | 761 | 1736.9 | 32 | 10.8 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76156843 | Titin-like protein | 369 | 71.5 | 2 | 4.1 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Signal transduction | |||||||||||||||||||||||
| 189502922 | 14-3-3 protein homolog 1 | 254 | 2177.7 | 47 | 61.4 | − | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| 56758294 | 14-3-3 protein | 254 | 822.02 | 21 | 27.2 | − | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| 76155391 | Creatine kinase | 242 | 203.15 | 5 | 15.7 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56757151 | 14-3-3 ε | 251 | 165.22 | 4 | 9.2 | NC | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| 189502932 | 14-3-3 protein homolog 1 | 231 | 88.45 | 2 | 13.4 | NC | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| Proteolysis | |||||||||||||||||||||||
| 56756044 | LAP | 398 | 1362.9 | 32 | 28.1 | NC | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2829279 | Calpain | 760 | 509.03 | 13 | 9.2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 56756901 | Calpain B | 773 | 73.84 | 2 | 5.0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 313121 | ER60 | 484 | 79.36 | 2 | 2.5 | SP | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Protein catabolic process | |||||||||||||||||||||||
| 56753927 | Ubiquitin C | 457 | 756.87 | 18 | 8.8 | NC | − | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + | 3 |
| 32452858 | Ubiquitin-activating enzyme E | 565 | 228.14 | 5 | 1.9 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56756008 | Ornithine aminotransferase | 438 | 82.05 | 2 | 5.9 | SP | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Transcription or translation | |||||||||||||||||||||||
| 76154398 | MF3 protein | 197 | 455.33 | 10 | 28.9 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56759476 | Transcriptional coactivator | 213 | 148.99 | 3 | 8.0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 29841143 | Ras-related protein ORAB-1 | 198 | 115.18 | 3 | 11.6 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 29841162 | RAB5B | 214 | 162.98 | 3 | 7.0 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 33348818 | Elongation factor 1-α | 465 | 84.62 | 2 | 7.1 | − | + | − | − | + | − | − | − | − | − | − | − | − | − | + | − | + | 4 |
| 60692924 | RPS21.2 (putative) | 89 | 61.93 | 2 | 16.9 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Cell cycle | |||||||||||||||||||||||
| 56755261 | Cyclophilin 1 | 164 | 487.79 | 11 | 21.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | 2 |
| Egg protein | |||||||||||||||||||||||
| 23664252 | 21.7-kDa protein | 185 | 395.8 | 8 | 8.6 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 56755421 | Major egg antigen | 354 | 240.41 | 5 | 14.4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | 2 |
| 56752663 | Egg antigen | 467 | 82.98 | 2 | 6.4 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Immune response | |||||||||||||||||||||||
| 56754313 | Annexin | 359 | 326.89 | 7 | 5.0 | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | 1 |
| 56757229 | Annexin B13 | 354 | 312.22 | 7 | 9.0 | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | 1 |
| 56757962 | Immunophilin | 431 | 312.03 | 7 | 20.2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | 1 |
| Muscle protein | |||||||||||||||||||||||
| 4761226 | Paramyosin | 866 | 302.01 | 7 | 8.1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 146741272 | Paramyosin | 866 | 133.72 | 3 | 5.4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 5305387 | Dynein light chain 1 | 89 | 112.74 | 3 | 15.7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 29841466 | Myophilin antigen | 190 | 81.56 | 2 | 7.4 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Protein binding/folding | |||||||||||||||||||||||
| 117380649 | Calreticulin | 396 | 294.29 | 6 | 4.3 | SP | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | + | 3 |
| 29841364 | LBD3 | 120 | 141.83 | 4 | 12.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 29841046 | Endophilin-B1 | 255 | 93.76 | 2 | 5.1 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Protease | |||||||||||||||||||||||
| 11167 | Cathepsin B | 342 | 223.11 | 4 | 3.8 | SP | + | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | 2 |
| % | |||||||||||||||||||||||
| Tegument protein | |||||||||||||||||||||||
| 161127 | Sj22.6 | 191 | 87.46 | 2 | 4.7 | NC | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | 1 |
| 56755910 | Fasciola/Schistosoma cross-reactive protein | 156 | 102.97 | 2 | 11.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Actin binding | |||||||||||||||||||||||
| 56755882 | Actin-binding and severin family group-like protein | 361 | 144.01 | 4 | 5.8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 495668 | Fimbrin | 651 | 95.11 | 3 | 5.5 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 76153279 | Fimbrin | 391 | 131.07 | 3 | 6.1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | 1 |
| 38683290 | Actin-binding/filamin-like protein | 984 | 84.55 | 2 | 2.0 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1469904 | JF-2 | 519 | 99.59 | 2 | 2.9 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76161984 | Thymosin isoform 2 | 91 | 79.3 | 2 | 14.3 | SP | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Nucleotide metabolism | |||||||||||||||||||||||
| 82697983 | Adenosine deaminase | 352 | 81.43 | 2 | 5.7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76156228 | AsnRS | 324 | 120.65 | 3 | 12.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 28916483 | NDK | 157 | 94.67 | 2 | 16.6 | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | 2 |
| 56758560 | MTAP | 299 | 99.46 | 3 | 5.4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Nucleosome assembly | |||||||||||||||||||||||
| 56752775 | Protein SET | 254 | 169.27 | 4 | 17.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Response to stress | |||||||||||||||||||||||
| 56759388 | STIP1 protein | 319 | 232.15 | 5 | 5.3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Others | |||||||||||||||||||||||
| 76155389 | Creatine kinase | 240 | 105.26 | 3 | 7.5 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76154162 | AFFP | 257 | 122.03 | 2 | 6.2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Unknown protein | |||||||||||||||||||||||
| 189503024 | Hypothetical protein | 236 | 166.02 | 4 | 18.6 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 56758512 | Hypothetical protein | 233 | 130.15 | 3 | 4.7 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 29841468 | Hypothetical protein | 420 | 101.57 | 2 | 4.0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 76157439 | Hypothetical protein | 137 | 104.13 | 2 | 10.2 | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
AFFP, actin filament-fragmenting protein; AsnRS, asparaginyl-tRNA synthetase; CCT7, chaperonin containing TCP1, subunit 7 (η); ER60, ER-luminal cysteine protease ER 60; FABP, FABP variant H or F; FBA, fructose-1,6-bisphosphate aldolase; LAP, leucine aminopeptidase; LBD3, LIM domain-binding protein 3; LDH, lactate dehydrogenase; mMDH, mitochondrial malate dehydrogenase; MTAP, methylthioadenosine phosphorylase; NDK, nucleoside-diphosphate kinase; PDI, protein-disulfide isomerase; PRP19, pre-mRNA processing factor 19; RPS21.2, 40 S ribosomal protein S21 isoform 2; Sj22.6, 22.6-kDa tegument-associated antigen; STIP1, stress-induced phosphoprotein 1; TGR, thioredoxin glutathione reductase.
The relative abundance of ES proteins is represented by summed Mowse peptide score (SMS) or SC. The summed Mowse peptide score is the sum of Mascot scores of all peptides including redundant peptides, not just a simple summed Mascot protein score of distinct peptides.
The ES proteins were predicted to be a classical secretory protein containing an SP or NC secretory protein or not secreted (−) by secretion prediction (Spe) using SecretomeP and SignalP. If the prediction of the representative protein failed to obtain a positive result, then the prediction result of the proteins within the same protein group is shown in italic.
The ES proteome of S. japonicum was compared with that of nematodes including A. caninum (Ac, Ref. 25), B. malayi (B, Ref. 19), Haemonchus contortus (H, Ref. 23), and T. circumcincta (Tc, Ref. 26). A, adult; L, larva.
The ES proteome of S. japonicum was compared with that of Trematoda including E. caproni (E1, Ref. 27; E2, Ref. 34), Echinostoma friedi (Ef, Ref. 28), F. hepatica (F1, Ref. 30; F2, Ref. 31; F3, Ref. 29), P. westermani (P, Ref. 32), S. bovis (Sb, Ref. 33) and S. mansoni (S1, Ref. 34; S2, Ref. 35; S3, Ref. 36; S4, Ref. 35). S, sporocyst; C, cercaria; E, egg.
Oc, occurrence (number of times) across the ES proteomes of different parasites.
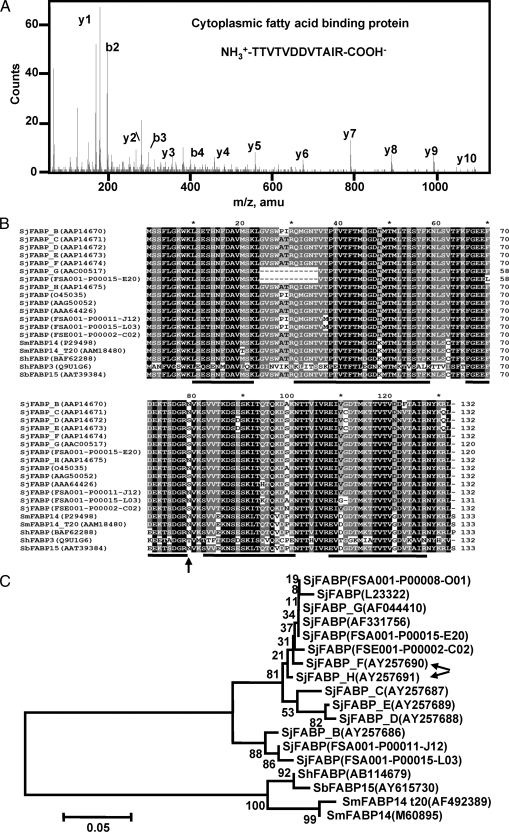
The relative abundance of individual ES protein was represented by SC (46) as well as the summed total Mowse peptide scores. Although this measurement may not precisely define the absolute quantity of proteins, it provides a semiquantitative view of the relative abundance of different proteins in a mixture. Among the 10 most abundant S. japonicum ES proteins, the first was FABP, which had the highest SC of 142 and a summed Mowse peptide score of 6676. One of its peptides, TTVTVDDVTAIR (representative MS/MS in Fig. 2A), was sequenced 91 times, which was more than 3-fold more often than the second most abundant protein (27 times) (supplemental Table 1). Interestingly because multiple SjFABP protein variants were deposited in the data set, the F or H sequence variant of SjFABP proteins cannot be discriminated by proteomics methods. These two protein variants differ by only one amino acid residue (Ser-79/Asn-79), which falls beyond the sensitivity of MS detection (Fig. 2B), although the sequence coverage (70.5%) is the highest. These schistosome FABPs are highly conserved across the full sequences (Fig. 2B) and S. mansoni, Schistosoma bovis, and S. hematobium FABPs are grouped together as orthologs, suggesting that they share a common conserved FABP of ancient origin (Fig. 2C). However, all SjFABPs analyzed were divergent from other schistosome FABPs. By phylogenetic analysis, these SjFABPs were found to be divided into two discrete phylogenetic groups (Fig. 2C) as was also indicated by sequence similarity and restriction site analysis (52).
Fig. 2.
Identification of FABP as the most abundant ES protein of S. japonicum adult worms. As revealed by MS analysis, TTVTVDDVTAIR is the most abundant peptide of FABP, the top abundant ES protein of S. japonicum adult worms. The b and y series of ions are labeled above the peaks. B, sequence alignment of FABPs from S. japonicum and other schistosomes. These FABP sequences were collected from GenBank or from an unpublished S. japonicum cDNA source from the Chinese National Human Genome Center at Shanghai.4 Four-level shading by GeneDoc was used to show the conservation between sequences with black color indicating identity. The accession numbers or full-length cDNA names for each protein sequence are shown in parentheses. The discrete lines below the alignments illustrate the peptides identified by LC-MS. An arrow under the alignment indicates the single differential amino acid between SjFABP variants F (Ser-79) and H (Asn-79). C, the phylogenetic relationship between SjFABPs and other schistosome FABPs was calculated by the neighbor-joining method using the coding sequences of these proteins. The numbers indicate the bootstrap values for each node. The DNA accession numbers or full-length cDNA names of each sequence are listed in parentheses. The SjFABPs are divergent from other schistosome FABPs and form two discrete subgroups. Arrows indicate the variants identified by LC-MS. Sb, S. bovis; Sh, S. hematobium.
The other nine most abundant proteins were heat shock protein 90 kDa α (HSP90α), three isoforms of heat shock protein 70 kDa (HSP70), actin-2, 14-3-3 protein, actin-1, enolase, and titin (Table I). Interestingly according to the relative abundance calculated by the ratio of accumulative summed SC to the total summed SC, these 10 most abundant proteins (only 9% of the total ES proteins) account for half of the quantity of the ESP pool, whereas the low abundance ES proteins (SC 1–3) represented only 10% of the total identified mass spectra (Table I).
To determine whether these peptides originated from the schistosome or the host, the 291 distinct peptides identified from the integrated schistosome DB were used to search against comprehensive rabbit genomic DBs, including genome trace sequences, assembled genomic sequences, cDNAs, and protein sequences using TBLASTN or BLASTP programs. We found that only 36 (12%) peptides were common in rabbit DBs, whereas the majority of the remaining proteins were schistosome-specific (supplemental Table 1). These data suggested that most ES proteins were indeed schistosome proteins rather than the host proteins and that schistosome proteins comprise the majority of ESP components.
Secretory Proteins in the S. japonicum ES Proteome—
The soluble and cyclic ES proteins of schistosome would be expected to be secreted from cells either through an N-terminal secretory signal sequence or via a non-classical secretory pathway (48). To validate the proteomics identifications of the S. japonicum ES proteins isolated in this study, we further analyzed the secretion characteristics of these proteins using SignalP 3.0 (47) and SecretomeP 2.0 (48), which can predict classical and non-classical secreted proteins with an N-terminal signal cleavable peptide, respectively.
Fifty-three of the 101 (52%) ES proteins were predicted to be either a classical or a non-classical secreted protein (Table I). This proportion is in accordance with, although a little lower than, that of egg ES proteome (63%) from S. mansoni (37) and adult worm ES proteome (65%) from B. malayi (19) where both of these samples were prepared following in vitro culture, suggesting that our method for preparation of ESPs was appropriate. The prediction details from SecretomeP and SignalP for all of the proteins in identified protein groups are listed in supplemental Table 2. There were 46 identified protein groups that were predicted to be non-classical (NC) secreted proteins, whereas only seven were considered to be classical secreted proteins with a typical N-terminal signal peptide (SP). These results suggested that the non-classical secretory pathway is the dominant route by which schistosome ES proteins are generated.
On the other hand, 48 proteins (48%) were predicted to be non-secreted products, including DJ-1, elongation factor EF2, and EF1α. However, the latter was repeatedly detected in the various ESPs from different parasites, including Ancylostoma caninum (25), Teladorsagia circumcincta (26), and S. mansoni (35), suggesting that such proteins are purposely released by parasites through an unknown excretory mechanism. Some ES proteins were predicted as non-secreted possibly due to N-terminal sequence truncations, which are common in EST clusters assembled with 3′-mRNA-enriched ESTs (41, 42). This is the case with HSP70 (gi|76155211) for which the N-terminal sequence was truncated. Interestingly one HSP70 isoform (gi|189502946) was identified as having an N-terminal SP.
Both methods failed to predict several enzymes as secreted proteins, and some of these proteins were reported to be cell membrane-associated, extracellular, or secretory. These proteins included adenosine deaminase (53, 54), glucose-6-phosphate isomerase (55), and GST (56). This finding may reflect limitations of the prediction algorisms used by SecretomeP and SignalP.
Comparison with Reported Parasite ES Proteomes—
We compared the ES proteome of S. japonicum with 15 reported ES proteomics identifications from nematodes and trematodes (Table I). Herein the comparison was mainly based on protein name annotation or sequence identity in the case of confusion in protein name. We found that 54 (48.5%) ES proteins of S. japonicum were common in at least one of these ES proteomes. Interestingly among these proteins, GST, enolase, SOD1, actin-2, FABP, actin-1, fructose-1,6-bisphosphate aldolase, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are more than 5-fold over-represented across different ES proteomics identifications (Table I). HSP70s, 14-3-3 proteins, thioredoxin peroxidase-1 (TPX1), EF1α, malate dehydrogenase, and cathepsin L were also over-represented at a somewhat lesser level. This finding strengthens the notion that parasites use a limited and conserved set of ES proteins for evasion from host immune attacks or for modulation of host responses (34). Also this observation illustrates the importance of these abundant proteins as vaccine candidates and drug targets for control of schistosomiasis and other helminthiasis. Among the most abundant S. japonicum ES proteins, HSP90α, leucine aminopeptidase, and titin as well as 44 other ES proteins were only found in the ES proteome of S. japonicum, implying that this parasite may use a species-specific manner of immune evasion.
Host Proteins Identified from the S. japonicum ESPs—
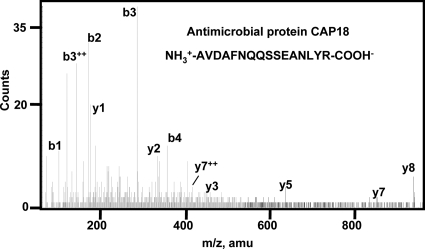
The gut of the schistosome is full of ingested host blood, whereas its body surface is coated with host blood proteins for immune disguise (57, 58) or other host molecules to avoid an immune response. To identify the host proteins in the pool of S. japonicum ESPs, all of the collected mass spectra were used to search an integrated rabbit protein DB and the Swiss-Prot DB. In the reverse rabbit DB search and reverse Swiss-Prot DB search, the highest Mowse peptide scores were 25.11 and 45.91, respectively, which were then used as cutoffs for peptide filtering. Thus a zero FPR was achieved for both DB searches. Finally we unambiguously identified 14 distinct peptides representing seven host proteins based on 26 host-specific mass spectra (Table II and supplemental Table 3). The two most abundant host proteins were antimicrobial protein CAP18 (a component of the host innate immune system) and fibronectin. Representative MS/MS of rabbit antimicrobial protein CAP18 is presented in Fig. 3. Interestingly more than half of the identifications were of host immune molecules such as immunoglobulins and component C3, indicating a close interaction of the host immune system and the worms. It should be pointed out that few host blood proteins were found possibly because the prewash prior to collection of ESPs may have eliminated most of the host proteins. Moreover most of the remaining host proteins may have been below the detection limit of the mass spectrometer.
Table II.
Rabbit (host) proteins identified in ESPs of S. japonicum
SMS, summed Mowse score.
| Accession no. | Annotation | SMS | SC |
|---|---|---|---|
| P25230.1 | Antimicrobial protein CAP18 precursor | 283.16 | 6 |
| P02751 | Fibronectin precursor | 299.25 | 6 |
| P68135 | Actin, α1 | 128.9 | 4 |
| Q2UVX4 | Complement C3 precursor | 159.9 | 3 |
| AAF98337.1 | IgG heavy chain VDJ region | 160.42 | 3 |
| ABD64612.1 | Immunoglobulin γ1 constant region | 90.16 | 2 |
| AAP43445.1 | Immunoglobulin heavy chain variable region | 85.45 | 2 |
Fig. 3.
Representative tandem mass spectra of rabbit antimicrobial protein CAP18 identified in ESPs of adult S. japonicum worms. Peptide AVDAFNQQSSEANLYR of rabbit CAP18 was sequenced five times. The b and y series of ions are labeled above the peaks.
Serum Immunoblot and LC-MS Analyses of Gel Bands—
Western blots were performed with sera from rabbits infected by schistosome, with naïve rabbit sera used as control, to compare the antigenicity patterns of the whole cell lysis (WCL) extract and the S. japonicum ES proteomes (Fig. 1C). As shown by immunoblotting, there were fewer immune antigenic proteins in ES product than in whole worm protein extracts, which is contrary to the notion that ESPs are exposed to the host immune system and enriched in antigenic proteins. This was also true for the ES and tegument proteome of S. bovis adult worms (33). One explanation for this phenomenon is that the challenged sera contained antibodies against ES proteins from eggs or schistosomula but minimal antibodies against the adult worms.
The abundant and antigenic protein bands were excised and subjected to in-gel digestion followed by protein identification by LC-MS. We identified 33 S. japonicum proteins from 11 excised gel bands (Table I and supplemental Table 4). As expected, compared with naïve sera, the schistosome-infected rabbit sera recognized many more antigenic protein bands. In ESPs, one protein reacted strongly with naïve sera (band 7) and was identified as myosin heavy chain (Fig. 1C and Table III). Interestingly band 6 was also revealed to be the same protein but was not antigenic (Fig. 1C). Bands 8 and 9, highly abundant in ESP, were identified as HSP70 and actin-2 in accordance with the results from shotgun LC-MS (Table I). However, band 10 was identified as ATP synthase subunit β, which was not identified by shotgun LC-MS. Although FABP was expected to be an abundant band in the gel, it was not found in the range of about 14 kDa. This absence could be explained by possible protein degradation as the entire ES product lane exhibited a smear pattern.
Table III.
LC-MS analysis of SDS-PAGE gel bands from whole worm extracts and ESPs of S. japonicum
aa, amino acids; Seq, sequence; WCL-abd, abundant in WCL extract of adult worms; ES-abd, abundant in ES products; WCL-ag, antigenic in WCL extract of adult worms; ES-ag, antigenic in ES products; SMS, summed Mowse score.
| Gel band no. | Category | Gi (NCBI) no. | Annotation | Protein length (aa) | Seq coverage | SMS | SC |
|---|---|---|---|---|---|---|---|
| aa | % | ||||||
| 1 | WCL-abd | 1703114 | Actin-2 | 376 | 33.5 | 1381.2 | 27 |
| 1 | WCL-abd | 30995481 | β-Tubulin | 443 | 11.7 | 239.12 | 5 |
| 1 | WCL-abd | 349802 | Enolase | 434 | 2.3 | 225.76 | 4 |
| 1 | WCL-abd | 123599 | HSP70 | 198 | 9.6 | 160.23 | 3 |
| 1 | WCL-abd | 160996 | GAPDH | 338 | 5.9 | 181.04 | 3 |
| 1 | WCL-abd | 161072 | α-Tubulin | 451 | 3.3 | 179.08 | 3 |
| 1 | WCL-abd | 38683290 | Actin-binding/filamin-like protein | 984 | 1.5 | 177.33 | 3 |
| 1 | WCL-abd | 84402 | Glutathione transferase | 219 | 10.5 | 168.27 | 3 |
| 1 | WCL-abd | 95113705 | Fructose-1,6-bisphosphate aldolase | 363 | 6.6 | 193.26 | 3 |
| 2 | WCL-abd | 2829289 | HSP70 | 648 | 15.9 | 708.32 | 11 |
| 2 | WCL-abd | 399939 | HSP70 homolog | 637 | 7.1 | 197.05 | 3 |
| 3 | WCL-abd | 3941320 | Myosin | 802 | 20.0 | 2065.5 | 33 |
| 3 | WCL-abd | 3041711 | Paramyosin (antigen Sj97) | 866 | 20.7 | 328.36 | 6 |
| 4 | WCL-ag | 3041711 | Paramyosin (antigen Sj97) | 866 | 20.7 | 2861.1 | 47 |
| 5 | WCL-ag | 146741272 | Paramyosin | 866 | 10.6 | 474.02 | 8 |
| 5 | WCL-ag | 3041711 | Paramyosin (antigen Sj97) | 866 | 20.7 | 332.14 | 5 |
| 5 | WCL-ag | 56753909 | Oncosphere protein Tso22b | 328 | 12.2 | 322.81 | 5 |
| 5 | WCL-ag | 56759038 | HSP90α2 | 719 | 4.2 | 213.19 | 4 |
| 5 | WCL-ag | 56756044 | His-tagged cytosolic leucine aminopeptidase | 398 | 3.8 | 133.36 | 2 |
| 6 | WCL-abd | 76154815 | Myosin heavy chain | 976 | 22.6 | 710.14 | 13 |
| 6 | WCL-abd | 56758294 | 14-3-3 protein | 254 | 4.7 | 117.71 | 2 |
| 7 | ES-ag | 76154815 | Myosin heavy chain | 976 | 22.6 | 2194.7 | 37 |
| 8 | ES-abd | 76155249 | Heat shock protein 70 | 137 | 21.2 | 201.03 | 4 |
| 8 | ES-abd | 76162587 | Similar to major vault protein | 111 | 12.6 | 308.21 | 4 |
| 8 | ES-abd | 56756008 | Ornithine aminotransferase | 438 | 8.4 | 160.93 | 3 |
| 8 | ES-abd | 56758570 | Mitochondrial malate dehydrogenase | 341 | 7.9 | 107.26 | 2 |
| 8 | ES-abd | 60688010 | Heat shock protein 70 | 82 | 17.1 | 137.26 | 2 |
| 9 | ES-abd | 56754704 | Actin-2 | 376 | 9.3 | 773.07 | 16 |
| 9 | ES-abd | 76154123 | Lamin b2 | 337 | 8.3 | 213.52 | 4 |
| 9 | ES-abd | 76152538 | Nuclear lamin C protein | 266 | 10.2 | 142.92 | 3 |
| 9 | ES-abd | 56757081 | Similar to major vault protein | 157 | 9.6 | 95.88 | 2 |
| 10 | ES-abd | 56758584 | ATP synthase subunit β, mitochondrial precursor | 514 | 11.9 | 296.46 | 6 |
| 10 | ES-abd | 56759082 | Mitochondrial H+-ATPase a subunit | 209 | 18.2 | 255.38 | 5 |
| 10 | ES-abd | 1174754 | Tropomyosin-1 | 284 | 12.0 | 115.56 | 2 |
| 11 | WCL-ag | 1174754 | Tropomyosin-1 | 284 | 12.0 | 2882.6 | 45 |
| 11 | WCL-ag | 1174756 | Tropomyosin-2 (TMII) | 284 | 12.7 | 229.88 | 4 |
| 11 | WCL-ag | 189502922 | 14-3-3 protein homolog 1 | 254 | 9.1 | 151.54 | 2 |
The most abundant proteins in WCL extract showed extremely low antigenicity. These proteins were identified as actin-2 (band 1) and HSP70 (band 2) (Fig. 1C and Table III). The major antigenic proteins in WCL extract were identified to be paramyosin (also termed antigen Sj97) (bands 3, 4, and 5) and tropomyosin (band 11).
DISCUSSION
The ES proteins of schistosomes have been shown to play important roles in immune modulation and immune evasion (7, 11). In the present study, we used for the first time a global proteomics approach to identify the major S. japonicum ES proteins and identified 101 S. japonicum proteins and seven host proteins in the ESPs of adult S. japonicum worms. We also present the results of a comparative study of our data with the ES protein profiles of S. mansoni and other helminth parasites.
Conventional methods for preparation of parasite ESPs involve in vitro culture techniques (19–37). However, in vitro culture in medium is dissimilar in many respects to the in vivo environment, resulting in altered parasite excretion and secretion behaviors (30). Host fluid is full of nutrients, blood cells, immune cells, and molecules including cell factors, hormone molecules, and complement components. These components interact with schistosome worms and possibly serve as inducers for excretion and secretion of macromolecules by the blood flukes. Most importantly, host-borne schistosomes are under high survival pressure due to host immune attack, which might in turn trigger the excretion and secretion of specific schistosome factors for immune evasion. This in vivo environment cannot readily be mimicked by in vitro culture. Therefore, the optimal way to study ES product is to analyze in vivo samples. However, it is difficult to collect and harvest ES proteins from host body fluids notwithstanding the fact that ES products in this environment represent the best materials for investigation of the role of ES proteins in the host-parasite relationship.
In this study, the ES product was gathered by soaking adult worms in buffer for short periods. The transient soaking avoids possible alteration of secretion activities of the parasites, whereas the repetition increases the yield of the ESPs. The proportion of secreted proteins in the present ES samples was similar to proportions reported for ES products prepared by conventional methods (19–37). Furthermore a comprehensive comparison of ES proteomes across different parasites indicated that 45% of the ES proteins of S. japonicum were present in other ES proteomes prepared by conventional methods. Moreover serious cell damage or decay did not occur in the ES products of S. japonicum because abundant intracellular components such as ribosomal and nuclear (histone) proteins were only rarely detected. These findings indicated that the ES product collection method used here was an acceptable alternative to in vitro culture-based procedures. Although contamination of ES proteins from dead worms cannot be avoided in our preparation, the ESPs we collected may mimic the exact components of in vivo ES proteome of schistosome, which is in fact a mixture of ES proteins from alive or dead adult worms or eggs.
Strikingly in the comprehensive comparison study, we found that a small subset of proteins was highly conserved in ES proteomes from various parasites regardless of their genus or species. In particular, GST, enolase, SOD1, actin-2, FABP, actin-1, fructose-1,6-bisphosphate aldolase, and GAPDH were conserved across reported parasite proteomes. This finding suggests that there is a minimum but universal molecular mechanism that has been evolutionally shaped by host-parasite interactions.
In the present study, FABP was demonstrated to be the most abundant S. japonicum ES protein (Table I), highlighting the vital importance of this protein in parasite-host interactions and in particular in the evasion process through immunosuppression of host immune system. However, we did not find an abundant 14-kDa FABP band in the SDS-PAGE gel possibly because of protein degradation. Because schistosomes cannot synthesize fatty acids de novo but instead depend on the host for these nutrients, blocking fatty acid uptake represents a potentially important therapeutic strategy. Therefore, schistosome FABPs, which are capable of binding fatty acids and/or retinoids (59), have been selected by the World Health Organization as one of six antischistosome vaccine candidates (60). This protein has been reported to localize on the subtegumental region and the gut epithelium (61). However, our study indicated it may be excreted or secreted into host serum (Table I); this result strongly argues against the possibility that SjFABP is unlikely to be accessible to the immune system (52). Furthermore the presence of FK506-binding protein in the adult worm ES proteome was also observed in Fasciola hepatica (29, 30), S. bovis (33), and S. mansoni (34, 35), suggesting that its secretion is conserved among trematodes. FABP from F. hepatica induces an immune response in mice that is protective to challenge infection with S. bovis cercariae (62), whereas FABP from S. mansoni induces immune protection against challenge with F. hepatica metacercariae and S. mansoni cercariae (62). Collectively these data suggest that FABP may constitute a potent cross-protective and multipurpose antitrematode vaccine. FABP does not perform well as an antigen for diagnosis of fasciolosis (63), but monoclonal antibodies specific for this antigen detect FABP in Fasciola ES materials (64). This observation raises the possibility of a role for an anti-FABP antibody in serodiagnosis of schistosomiasis japonica that has yet to be evaluated.
Another striking finding from our investigation is that, using shotgun LC-MS and SDS-PAGE, heat shock proteins, including HSP70s, HSP90α, HSP4, and HSP97, form the prominent S. japonicum ES protein family. HSP70 is highly abundant in ES products and WCL extracts of S. japonicum adult worms, but it is not antigenic. HSPs are traditionally considered as cytosolic molecular chaperones. However, recent studies indicate that HSP70s also exists as surface-bound or extracellular proteins (65, 66) and were found to be present in secretory products of the cercariae of S. mansoni (SmHSP70, -86, and -60) (35) and adult worms of B. malayi (19), Echinostoma caproni (27), and Paragonimus westermani (32). Furthermore one S. japonicum HSP70 isoform (gi|189502946) was predicted to have a typical secretory signal peptide (Table I). Interestingly HSPs were found to exert dual immunoregulatory effects, including immunostimulatory and immunosuppressive roles (65, 67). Among host-parasite interactions, HSP70 from Toxoplasma gondii induced immunosuppression by down-regulation of host nitric oxide production and stimulation of continuous production of Th2 cytokines (68).
Our findings also suggest that schistosome HSP70 might play roles in immunomodulation and/or immunoevasion. SmHSP70 elicits an early humoral immune response and may be a valid target for immunodiagnosis (69). However, when used as a diagnostic antigen, SmHSP70 cross-reacted with antibodies in sera from humans infected with schistosomes, filariae, and malaria but did not cross-react with sera from patients infected with S. japonicum (70). Therefore, antibodies against species-specific HSP70 peptides should be evaluated for the diagnosis of schistosomiasis.
Structural proteins, including cytoskeleton and muscle proteins, were identified in S. japonicum ES products. These proteins included actins, tubulins, titins, paramyosin, and myophilin with actins being the most abundant. Although the presence of these proteins may result from possible cell leakage from dead worms, their potential roles in host-parasite interaction should not be ignored. Furthermore muscle or cytoskeleton proteins like actin and paramyosin were located on the schistosome worm surface (71), which may be the reason for their presence in ES products. A recent study indicated that E. caproni actin and HSP70 in ESPs induced an early IgM response; this may point to a role in their early survival within the host (27).
The next most abundant ES protein was 14-3-3 protein, which was reported to localize to the excretory system and tegument of female S. mansoni worms (72) and has been used as a vaccine candidate against schistosomiasis (73). In S. japonicum ESP, several cytosolic glycolytic enzymes were unexpectedly identified, including enolase, pyruvate kinase, and GAPDH. Recent studies indicate that enolase is a multifunctional protein localized on the cell surface (74) and the tegument (75, 76) of helminths. Enolase has been shown to bind plasminogen in prokaryotic and eukaryotic cells and pathogens (74), including schistosomes (75). The S. japonicum secretory enolase may stimulate fibrinolytic activity for parasitic invasion and migration within the host. Similarly pyruvate kinase is also a glycolytic enzyme. However, this enzyme was shown to be one of the immunodominant proteins of Neospora caninum and was recognized by challenged bovine sera (78), suggesting that it might be exposed on the interface during host-parasite interactions. GAPDH is an established cytosolic housekeeping protein. In addition to its classical glycolytic role, GAPDH has recently been revealed to be a multifunctional protein (79) bound to the cell membrane (80). The GAPDH in S. japonicum ESPs may be derived from the tegument (76) or may have been secreted by specific excretory cells of the schistosome as occurs in the ameba Entamoeba histolytica (81), an anaerobic parasitic protozoan.
We detected leucine aminopeptidase in S. japonicum ES products, which is predicted to be a non-secretory protein. However, proteomics approaches revealed that adult worms of the filarial parasite B. malayi also secrete leucine aminopeptidase into the ESPs (19). Furthermore leucine aminopeptidase from the fluke F. hepatica was characterized as the immunodominant antigen in ESPs from human infection (77), suggesting that this protein is secretory or excretory in these parasites and that it plays an important role in host-parasite interactions.
The S. japonicum ES proteome also contains several detoxification molecules, including TPX1, TPX2, SOD, GSTs, and thioredoxin glutathione reductase. These observations highlight the importance of detoxification in protection of schistosome against the mammalian host immune response. Thus these proteins have been tested comprehensively as potential drug targets. For example, thioredoxin glutathione reductase from S. mansoni was recently reported to be an interesting drug target for schistosome treatment (82). GST is also a vaccine candidate proposed by the World Health Organization (60). We found that GST has the highest occurrence across various ESPs from different parasites, suggesting that this protein is among the most conserved ES proteins and that it likely plays a vital role in host-parasite interactions.
Among the antigenic proteins of S. japonicum adult worms were myosin heavy chain in the ES proteome and paramyosin and tropomyosin in WCL extract. These proteins are known antigenic proteins and have been used for vaccine studies (83–85).
The host proteins identified in S. japonicum ESPs comprise only a small minority of the total number of identified ES proteins; this is likely a reflection of the low abundance in the ESPs. Interestingly essential blood components such as host albumin and hemoglobin, the most abundant proteins in host serum, were not identified. If nonspecific adsorption of host proteins by schistosome were to prevail then albumin and hemoglobin would be expected to be the dominant host proteins in our results. This was not the case, raising the possibility that most, if not all, of these host proteins identified in ESPs of S. japonicum were selectively adsorbed by schistosome worms with specific tegument receptors (61, 86). Therefore, these findings illustrate a potential molecular mechanism for host-parasite interplay: the schistosome specifically adsorbs host Igs and fibronectins to prevent host immune detections and attacks, while the host tries to stimulate its innate immune reactive molecules (antimicrobial protein CAP18) and acquired immune (Igs and complement C3) systems against schistosome invasion. It is possible that these host proteins were from the schistosome gut. Host albumin and hemoglobin are also expected to be the dominant markers for gut-releasing components. Their absence in the present results could simply indicate the complete digestion of ingested host blood components (87, 88).
In conclusion, our study has highlighted the principal proteins released by the S. japonicum parasite for interaction with the host immune system. We anticipate that this information will stimulate development of candidate antigens for vaccination against S. japonicum and novel immunomodulatory compounds with therapeutic potential.
Acknowledgments
We thank Professor Paul J. Brindley at George Washington University Medical Center for assistance with revision of the manuscript.
Footnotes
Published, MCP Papers in Press, March 18, 2009, DOI 10.1074/mcp.M800538-MCP200
The abbreviations used are: ES, excretory/secretory; CAP, cytoskeleton-associated protein; DB, database; ESP, ES product; EST, expressed sequence tag; FABP, fatty acid-binding protein; FPR, false-positive rate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSP, heat shock protein; Ig, immunoglobulin; nano-LC-MS/MS, nanoscale capillary LC-MS/MS; NC, non-classical; RP, reverse-phase; SC, spectral count; SCX, strong cation exchange; SOD, superoxide dismutase; SP, signal peptide; TPX, thioredoxin peroxidase; WCL, whole cell lysis; BLAST, basic local alignment search tool; Mowse, molecular weight search; Sj, S. japonicum; Sm, S. mansoni; MSD, mass selective detector; XCT, X-ray computed tomography.
M. Berriman, unpublished data.
Z. Chen, unpublished data.
S.-Y. Wang, unpublished data.
This work was supported by Chinese National Key Program on Basic Research (973) Grants 2007CB513100 and 2006CB708510, Chinese High-Tech Research and Development Program (863) Grants 2006AA02Z318 and 2007AA02Z153, the National Foundation for Excellence Doctoral Project, Shanghai Commission for Science and Technology Grants 06JC14059 and 05DZ22201, the China Postdoctoral Science Foundation, and Shanghai Rising-Star Program Grant 07QA14043.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.World Health Organization ( 2006) Preventive Chemotherapy in Human Helminthiasis. Coordinated Use of Anthelminthic Drugs in Control Interventions—Guidelines for Health Professionals and Programme Managers, World Health Organization, Geneva
- 2.Yoshino, T. P., and Bayne, C. J. ( 1983) Mimicry of snail host antigens by miracidia and primary sporocysts of Schistosoma mansoni. Parasite Immunol. 5, 317–328 [DOI] [PubMed] [Google Scholar]
- 3.Damian, R. T. ( 1997) Parasite immune evasion and exploitation: reflections and projections. Parasitology 115, (suppl.) S169–S175 [DOI] [PubMed] [Google Scholar]
- 4.Salzet, M., Capron, A., and Stefano, G. B. ( 2000) Molecular crosstalk in host-parasite relationships: schistosome- and leech-host interactions. Parasitol Today 16, 536–540 [DOI] [PubMed] [Google Scholar]
- 5.McLaren, D. J. ( 1984) Disguise as an evasive stratagem of parasitic organisms. Parasitology 88, 597–611 [DOI] [PubMed] [Google Scholar]
- 6.McLaren, D. J., and Terry, R. J. ( 1982) The protective role of acquired host antigens during schistosome maturation. Parasite Immunol. 4, 129–148 [DOI] [PubMed] [Google Scholar]
- 7.Jenkins, S. J., Hewitson, J. P., Jenkins, G. R., and Mountford, A. P. ( 2005) Modulation of the host's immune response by schistosome larvae. Parasite Immunol. 27, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallon, P. G., and Mangan, N. E. ( 2007) Suppression of TH2-type allergic reactions by helminth infection. Nat. Rev. Immunol. 7, 220–230 [DOI] [PubMed] [Google Scholar]
- 9.van Riet, E., Hartgers, F. C., and Yazdanbakhsh, M. ( 2007) Chronic helminth infections induce immunomodulation: consequences and mechanisms. Immunobiology 212, 475–490 [DOI] [PubMed] [Google Scholar]
- 10.Arruda, L. K., and Santos, A. B. ( 2005) Immunologic responses to common antigens in helminthic infections and allergic disease. Curr. Opin. Allergy Clin. Immunol. 5, 399–402 [DOI] [PubMed] [Google Scholar]
- 11.Lightowlers, M. W., and Rickard, M. D. ( 1988) Excretory-secretory products of helminth parasites: effects on host immune responses. Parasitology 96, (suppl.) S123–S166 [DOI] [PubMed] [Google Scholar]
- 12.Lodes, M. J., and Yoshino, T. P. ( 1989) Characterization of excretory-secretory proteins synthesized in vitro by Schistosoma mansoni primary sporocysts. J. Parasitol. 75, 853–862 [PubMed] [Google Scholar]
- 13.Crews-Oyen, A. E., and Yoshino, T. P. ( 1995) Schistosoma mansoni: characterization of excretory-secretory polypeptides synthesized in vitro by daughter sporocysts. Exp. Parasitol. 80, 27–35 [DOI] [PubMed] [Google Scholar]
- 14.Connors, V. A., Lodes, M. J., and Yoshino, T. P. ( 1991) Identification of a Schistosoma mansoni sporocyst excretory-secretory antioxidant molecule and its effect on superoxide production by Biomphalaria glabrata hemocytes. J. Invertebr. Pathol. 58, 387–395 [DOI] [PubMed] [Google Scholar]
- 15.Ramaswamy, K., Salafsky, B., Potluri, S., He, Y. X., Li, J. W., and Shibuya, T. ( 1995. –1996) Secretion of an anti-inflammatory, immunomodulatory factor by Schistosomulae of Schistosoma mansoni. J. Inflamm. 46, 13–22 [PubMed] [Google Scholar]
- 16.Call, J. L., Pilcher, J. B., Freeman, G. L., Jr., and Tsang, V. C. ( 1995) Serum-free culturing of adult Schistosoma mansoni in dialysis bags for the production of excretory/secretory antigens. J. Parasitol. 81, 742–746 [PubMed] [Google Scholar]
- 17.Kawanaka, M., and Carter, C. E. ( 1992) Schistosoma japonicum: excretory-secretory products of the eggs during miracidial development. Exp. Parasitol. 74, 143–150 [DOI] [PubMed] [Google Scholar]
- 18.Yoshino, T. P., Lodes, M. J., Rege, A. A., and Chappell, C. L. ( 1993) Proteinase activity in miracidia, transformation excretory-secretory products, and primary sporocysts of Schistosoma mansoni. J. Parasitol. 79, 23–31 [PubMed] [Google Scholar]
- 19.Hewitson, J. P., Harcus, Y. M., Curwen, R. S., Dowle, A. A., Atmadja, A. K., Ashton, P. D., Wilson, A., and Maizels, R. M. ( 2008) The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 160, 8–21 [DOI] [PubMed] [Google Scholar]
- 20.Silverman, J. M., Chan, S. K., Robinson, D. P., Dwyer, D. M., Nandan, D., Foster, L. J., and Reiner, N. E. ( 2008) Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 9, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson, M. W., Greig, R., Beattie, K. A., Lamont, D. J., and Connolly, B. ( 2007) Comparative analysis of the excretory-secretory proteome of the muscle larva of Trichinella pseudospiralis and Trichinella spiralis. Int. J. Parasitol. 37, 139–148 [DOI] [PubMed] [Google Scholar]
- 22.Robinson, M. W., and Connolly, B. ( 2005) Proteomic analysis of the excretory-secretory proteins of the Trichinella spiralis L1 larva, a nematode parasite of skeletal muscle. Proteomics 5, 4525–4532 [DOI] [PubMed] [Google Scholar]
- 23.Yatsuda, A. P., Krijgsveld, J., Cornelissen, A. W., Heck, A. J., and de Vries, E. ( 2003) Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J. Biol. Chem. 278, 16941–16951 [DOI] [PubMed] [Google Scholar]
- 24.Matthews, J. B., Davidson, A. J., and Beynon, R. J. ( 2004) The application of mass spectrometry to identify immunogenic components of excretory/secretory products from adult Dictyocaulus viviparus. Parasitology 128, Suppl. 1, S43–S47 [DOI] [PubMed] [Google Scholar]
- 25.Mulvenna, J., Hamilton, B., Nagaraj, S. H., Smyth, D., Loukas, A., and Gorman, J. J. ( 2009) Proteomic analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell. Proteomics 8, 109–121 [DOI] [PubMed] [Google Scholar]
- 26.Craig, H., Wastling, J. M., and Knox, D. P. ( 2006) A preliminary proteomic survey of the in vitro excretory/secretory products of fourth-stage larval and adult Teladorsagia circumcincta. Parasitology 132, 535–543 [DOI] [PubMed] [Google Scholar]
- 27.Sotillo, J., Valero, L., Sánchez, Del, Pino, M. M., Fried, B., Esteban, J. G., Marcilla, A., and Toledo, R. ( 2008) Identification of antigenic proteins from Echinostoma caproni (Trematoda) recognized by mouse immunoglobulins M, A and G using an immunoproteomic approach. Parasite Immunol. 30, 271–279 [DOI] [PubMed] [Google Scholar]
- 28.Bernal, D., Carpena, I., Espert, A. M., De la Rubia, J. E., Esteban, J. G., Toledo, R., and Marcilla, A. ( 2006) Identification of proteins in excretory/secretory extracts of Echinostoma friedi (Trematoda) from chronic and acute infections. Proteomics 6, 2835–2843 [DOI] [PubMed] [Google Scholar]
- 29.Jefferies, J. R., Campbell, A. M., van Rossum, A. J., Barrett, J., and Brophy, P. M. ( 2001) Proteomic analysis of Fasciola hepatica excretory-secretory products. Proteomics 1, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 30.Morphew, R. M., Wright, H. A., LaCourse, E. J., Woods, D. J., and Brophy, P. M. ( 2007) Comparative proteomics of excretory-secretory proteins released by the liver fluke Fasciola hepatica in sheep host bile and during in vitro culture ex host. Mol. Cell. Proteomics 6, 963–972 [DOI] [PubMed] [Google Scholar]
- 31.Gourbal, B. E., Guillou, F., Mitta, G., Sibille, P., Thèron, A., Pointier, J. P., and Coustau, C. ( 2008) Excretory-secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Mol. Biochem. Parasitol. 161, 63–66 [DOI] [PubMed] [Google Scholar]
- 32.Lee, E. G., Na, B. K., Bae, Y. A., Kim, S. H., Je, E. Y., Ju, J. W., Cho, S. H., Kim, T. S., Kang, S. Y., Cho, S. Y., and Kong, Y. ( 2006) Identification of immunodominant excretory-secretory cysteine proteases of adult Paragonimus westermani by proteome analysis. Proteomics 6, 1290–1300 [DOI] [PubMed] [Google Scholar]
- 33.Pérez-Sánchez, R., Ramajo-Hernández, A., Ramajo-Martín, V., and Oleaga, A. ( 2006) Proteomic analysis of the tegument and excretory-secretory products of adult Schistosoma bovis worms. Proteomics 6, Suppl. 1, S226–S236 [DOI] [PubMed] [Google Scholar]
- 34.Guillou, F., Roger, E., Moné, Y., Rognon, A., Grunau, C., Théron, A., Mitta, G., Coustau, C., and Gourbal, B. E. ( 2007) Excretory-secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol. Biochem. Parasitol. 155, 45–56 [DOI] [PubMed] [Google Scholar]
- 35.Knudsen, G. M., Medzihradszky, K. F., Lim, K. C., Hansell, E., and McKerrow, J. H. ( 2005) Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol. Cell. Proteomics 4, 1862–1875 [DOI] [PubMed] [Google Scholar]
- 36.Curwen, R. S., Ashton, P. D., Sundaralingam, S., and Wilson, R. A. ( 2006) Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol. Cell. Proteomics 5, 835–844 [DOI] [PubMed] [Google Scholar]
- 37.Cass, C. L., Johnson, J. R., Califf, L. L., Xu, T., Hernandez, H. J., Stadecker, M. J., Yates, J. R., III, and Williams, D. L. ( 2007) Proteomic analysis of Schistosoma mansoni egg secretions. Mol. Biochem. Parasitol. 155, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He, Y. X., Salafsky, B., and Ramaswamy, K. ( 2005) Comparison of skin invasion among three major species of Schistosoma. Trends Parasitol. 21, 201–203 [DOI] [PubMed] [Google Scholar]
- 39.Rheinberg, C. E., Moné, H., Caffrey, C. R., Imbert-Establet, D., Jourdane, J., and Ruppel, A. ( 1998) Schistosoma haematobium, S. intercalatum, S. japonicum, S. mansoni, and S. rodhaini in mice: relationship between patterns of lung migration by schistosomula and perfusion recovery of adult worms. Parasitol. Res. 84, 338–342 [DOI] [PubMed] [Google Scholar]
- 40.He, Y. X., Chen, L., and Ramaswamy, K. ( 2002) Schistosoma mansoni, S. haematobium, and S. japonicum: early events associated with penetration and migration of schistosomula through human skin. Exp. Parasitol. 102, 99–108 [DOI] [PubMed] [Google Scholar]
- 41.Hu, W., Yan, Q., Shen, D. K., Liu, F., Zhu, Z. D., Song, H. D., Xu, X. R., Wang, Z. J., Rong, Y. P., Zeng, L. C., Wu, J., Zhang, X., Wang, J. J., Xu, X. N., Wang, S. Y., Fu, G., Zhang, X. L., Wang, Z. Q., Brindley, P. J., McManus, D. P., Xue, C. L., Feng, Z., Chen, Z., and Han, Z. G. ( 2003) Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat. Genet. 35, 139–147 [DOI] [PubMed] [Google Scholar]
- 42.Liu, F., Lu, J., Hu, W., Wang, S. Y., Cui, S. J., Chi, M., Yan, Q., Wang, X. R., Song, H. D., Xu, X. N., Wang, J. J., Zhang, X. L., Zhang, X., Wang, Z. Q., Xue, C. L., Brindley, P. J., McManus, D. P., Yang, P. Y., Feng, Z., Chen, Z., and Han, Z. G. ( 2006) New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 44.Resing, K. A., Meyer-Arendt, K., Mendoza, A. M., Aveline-Wolf, L. D., Jonscher, K. R., Pierce, K. G., Old, W. M., Cheung, H. T., Russell, S., Wattawa, J. L., Goehle, G. R., Knight, R. D., and Ahn, N. G. ( 2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem. 76, 3556–3568 [DOI] [PubMed] [Google Scholar]
- 45.Slotta, D. J., McFarland, M., Makusky, A., and Markey, S. ( 2007) P18-T MassSieve: a new tool for mass spectrometry-based proteomics. J. Biomol. Tech. 18, 7 [Google Scholar]
- 46.Dong, M. Q., Venable, J. D., Au, N., Xu, T., Park, S. K., Cociorva, D., Johnson, J. R., Dillin, A., and Yates, J. R., III ( 2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- 47.Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. ( 2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 48.Bendtsen, J. D., Jensen, L. J., Blom, N., Von Heijne, G., and Brunak, S. ( 2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Select. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 49.Nicholas, K. B., Nicholas, H. B., Jr., and Deerfield, D. W., II ( 1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News 4, 1–4 [Google Scholar]
- 50.Tamura, K., Dudley, J., Nei, M., and Kumar, S. ( 2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 52.Scott, J. C., Kennedy, M. W., and McManus, D. P. ( 2000) Molecular and immunological characterisation of a polymorphic cytosolic fatty acid binding protein from the human blood fluke of humans, Schistosoma japonicum. Biochim. Biophys. Acta 1517, 53–62 [DOI] [PubMed] [Google Scholar]
- 53.Martín, M., Aran, J. M., Colomer, D., Huguet, J., Centelles, J. J., Vives-Corrons, J. L., and Franco, R. ( 1995) Surface adenosine deaminase. A novel B-cell marker in chronic lymphocytic leukemia. Hum. Immunol. 42, 265–273 [DOI] [PubMed] [Google Scholar]
- 54.Eltzschig, H. K., Faigle, M., Knapp, S., Karhausen, J., Ibla, J., Rosenberger, P., Odegard, K. C., Laussen, P. C., Thompson, L. F., and Colgan, S. P. ( 2006) Endothelial catabolism of extracellular adenosine during hypoxia: the role of surface adenosine deaminase and CD26. Blood 108, 1602–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumoto, I., Maccioni, M., Lee, D. M., Maurice, M., Simmons, B., Brenner, M., Mathis, D., and Benoist, C. ( 2002) How antibodies to a ubiquitous cytoplasmic enzyme may provoke joint-specific autoimmune disease. Nat Immunol. 3, 360–365 [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee, S. B., Aravinda, S., Gopalakrishnan, B., Nagpal, S., Salunke, D. M., and Shaha, C. ( 1999) Secretion of glutathione S-transferase isoforms in the seminiferous tubular fluid, tissue distribution and sex steroid binding by rat GSTM1. Biochem. J. 340, 309–320 [PMC free article] [PubMed] [Google Scholar]
- 57.Loukas, A., Jones, M. K., King, L. T., Brindley, P. J., and McManus, D. P. ( 2001) Receptor for Fc on the Surfaces of Schistosomes. Infect. Immun. 69, 3646–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gearner, G. W., and Kemp, W. M. ( 1994) Electrophoretic and serological analysis of host antigens associated with the adult Schistosoma mansoni tegument. J. Parasitol. 80, 275–283 [PubMed] [Google Scholar]
- 59.Kennedy, M. W., Scott, J. C., Lo, S., Beauchamp, J., and McManus, D. P. ( 2000) Sj-FABPc fatty-acid-binding protein of the human blood fluke Schistosoma japonicum: structural and functional characterization and unusual solvent exposure of a portal-proximal tryptophan residue. Biochem. J. 349, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergquist, N. R., and Colley, D. G. ( 1998) Schistosomiasis vaccines: research to development. Parasitol. Today 14, 99–104 [DOI] [PubMed] [Google Scholar]
- 61.Brito, C. F., Oliveira, G. C., Oliveira, S. C., Street, M., Riengrojpitak, S., Wilson, R. A., Simpson, A. J., and Correa-Oliveira, R. ( 2002) Sm14 gene expression in different stages of the Schistosoma mansoni life cycle and immunolocalization of the Sm14 protein within the adult worm. Braz. J. Med. Biol. Res. 35, 377–381 [DOI] [PubMed] [Google Scholar]
- 62.Abáné, J. L., Oleaga, A., Ramajo, V., Casanueva, P., Arellano, J. L., Hillyer, G. V., and Muro, A. ( 2000) Vaccination of mice against Schistosoma bovis with a recombinant fatty acid binding protein from Fasciola hepatica. Vet. Parasitol. 91, 33–42 [DOI] [PubMed] [Google Scholar]
- 63.Raina, O. K., Sriveny, D., and Yadav, S. C. ( 2004) Humoral immune response against Fasciola gigantica fatty acid binding protein. Vet. Parasitol. 124, 65–72 [DOI] [PubMed] [Google Scholar]
- 64.Sirisriro, A., Grams, R., Vichasri-Grams, S., Ardseungneon, P., Pankao, V., Meepool, A., Chaithirayanon, K., Viyanant, V., Tan-Ariya, P., Upatham, E. S., and Sobhon, P. ( 2002) Production and characterization of a monoclonal antibody against recombinant fatty acid binding protein of Fasciola gigantica. Vet. Parasitol. 105, 119–129 [DOI] [PubMed] [Google Scholar]
- 65.Asea, A. ( 2008) Heat shock proteins and toll-like receptors. Handb. Exp. Pharmacol. 183, 111–127 [DOI] [PubMed] [Google Scholar]
- 66.Mambula, S. S., Stevenson, M. A., Ogawa, K., and Calderwood, S. K. ( 2007) Mechanisms for Hsp70 secretion: crossing membranes without a leader. Methods 43, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pockley, A. G., Muthana, M., and Calderwood, S. K. ( 2008) The dual immunoregulatory roles of stress proteins. Trends Biochem. Sci. 33, 71–79 [DOI] [PubMed] [Google Scholar]
- 68.Ahmed, A. K., Mun, H. S., Aosai, F., Piao, L. X., Fang, H., Norose, K., and Yano, A. ( 2004) Roles of Toxoplasma gondii-derived heat shock protein 70 in host defense against T. gondii infection. Microbiol. Immunol. 48, 911–915 [DOI] [PubMed] [Google Scholar]
- 69.Kanamura, H. Y., Hancock, K., Rodrigues, V., and Damian, R. T. ( 2002) Schistosoma mansoni heat shock protein 70 elicits an early humoral immune response in S. mansoni infected baboons. Mem. Inst. Oswaldo Cruz 97, 711–716 [DOI] [PubMed] [Google Scholar]
- 70.Moser, D., Doumbo, O., and Klinkert, M. Q. ( 1990) The humoral response to heat shock protein 70 in human and murine Schistosomiasis mansoni. Parasite Immunol. 12, 341–352 [DOI] [PubMed] [Google Scholar]
- 71.Matsumoto, Y., Perry, G., Levine, R. J., Blanton, R., Mahmoud, A. A., and Aikawa, M. ( 1988) Paramyosin and actin in schistosomal teguments. Nature 333, 76–78 [DOI] [PubMed] [Google Scholar]
- 72.Schechtman, D., Winnen, R., Tarrab-Hazdai, R., Ram, D., Shinder, V., Grevelding, C. G., Kunz, W., and Arnon, R. ( 2001) Expression and immunolocalization of the 14-3-3 protein of Schistosoma mansoni. Parasitology 123, 573–582 [DOI] [PubMed] [Google Scholar]
- 73.Schechtman, D., Tarrab-Hazdai, R., and Arnon, R. ( 2001) The 14-3-3 protein as a vaccine candidate against schistosomiasis. Parasite Immunol. 23, 213–217 [DOI] [PubMed] [Google Scholar]
- 74.Seweryn, E., Pietkiewicz, J., Szamborska, A., and Gamian, A. ( 2007) Enolase on the surface of prokaryotic and eukaryotic cells is a receptor for human plasminogen. Postepy Hig. Med. Dosw. (Online) 61, 672–682 [PubMed] [Google Scholar]
- 75.Ramajo-Hernández, A., Pérez-Sánchez, R., Ramajo-Martín, V., and Oleaga, A. ( 2007) Schistosoma bovis: plasminogen binding in adults and the identification of plasminogen-binding proteins from the worm tegument. Exp. Parasitol. 115, 83–91 [DOI] [PubMed] [Google Scholar]
- 76.Pérez-Sánchez, R., Valero, M. L., Ramajo-Hernández, A., Siles-Lucas, M., Ramajo-Martín, V., and Oleaga, A. ( 2008) A proteomic approach to the identification of tegumental proteins of male and female Schistosoma bovis worms. Mol. Biochem. Parasitol. 161, 112–123 [DOI] [PubMed] [Google Scholar]
- 77.Marcilla, A., De la Rubia, J. E., Sotillo, J., Bernal, D., Carmona, C., Villavicencio, Z., Acosta, D., Tort, J., Bornay, F. J., Esteban, J. G., and Toledo, R. ( 2008) Leucine aminopeptidase is an immunodominant antigen of Fasciola hepatica excretory and secretory products in human infections. Clin. Vaccine Immunol. 15, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin, Y. S., Lee, E. G., Shin, G. W., Kim, Y. R., Lee, E. Y., Kim, J. H., Jang, H., Gershwin, L. J., Kim, D. Y., Kim, Y. H., Kim, G. S., Suh, M. D., and Jung, T. S. ( 2004) Identification of antigenic proteins from Neospora caninum recognized by bovine immunoglobulins M, E, A and G using immunoproteomics. Proteomics 4, 3600–3609 [DOI] [PubMed] [Google Scholar]
- 79.Sirover, M. A. ( 1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432, 159–184 [DOI] [PubMed] [Google Scholar]
- 80.Heard, K. S., Diguette, M., Heard, A. C., and Carruthers, A. ( 1998) Membrane-bound glyceraldehyde-3-phosphate dehydrogenase and multiphasic erythrocyte sugar transport. Exp. Physiol. 83, 195–202 [DOI] [PubMed] [Google Scholar]
- 81.Alvarez, A. H., Martinez-Cadena, G., Silva, M. E., Saavedra, E., and Avila, E. E. ( 2007) Entamoeba histolytica: ADP-ribosylation of secreted glyceraldehyde-3-phosphate dehydrogenase. Exp. Parasitol. 117, 349–356 [DOI] [PubMed] [Google Scholar]
- 82.Kuntz, A. N., Davioud-Charvet, E., Sayed, A. A., Califf, L. L., Dessolin, J., Arnér, E. S., and Williams, D. L. ( 2007) Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target. PLoS Med. 4, e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pearce, E. J., James, S. L., Hieny, S., Lanar, D. E., and Sher, A. ( 1988) Induction of protective immunity against Schistosoma mansoni by vaccination with schistosome paramyosin (Sm97), a nonsurface parasite antigen. Proc. Natl. Acad. Sci. U. S. A. 85, 5678–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu, H., Rekosh, D. M., Andrews, W., Higashi, G. I., Nicholson, L., and Loverde, P. T. ( 1991) Schistosoma mansoni tropomyosin: production and purification of the recombinant protein and studies on its immunodiagnostic potential. Am. J. Trop. Med. Hyg. 45, 121–131 [DOI] [PubMed] [Google Scholar]
- 85.Eberl, M., Mountford, A. P., Jankovic, D., and Beck, E. ( 1999) Isolation of T-cell antigens by using a recombinant protein library and its application to the identification of novel vaccine candidates against schistosomiasis. Infect. Immun. 67, 3383–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tarleton, R. L., and Kemp, W. M. ( 1981) Demonstration of IgG-Fc and C3 receptors on adult Schistosoma mansoni. J. Immunol. 126, 379–384 [PubMed] [Google Scholar]
- 87.Brindley, P. J., Kalinna, B. H., Dalton, J. P., Day, S. R., Wong, J. Y., Smythe, M. L., and McManus, D. P. ( 1997) Proteolytic degradation of host hemoglobin by schistosomes. Mol. Biochem. Parasitol. 89, 1–9 [DOI] [PubMed] [Google Scholar]
- 88.Halton, D. W. ( 1997) Nutritional adaptations to parasitism within the Platyhelminthes. Int. J. Parasitol. 27, 693–704 [DOI] [PubMed] [Google Scholar]