Abstract
The spread of cancer cells from a primary tumor to form metastasis at distant sites is a complex multistep process. The cancer cell proteins and plasma membrane proteins in particular involved in this process are poorly defined, and a study of the very early events of the metastatic process using clinical samples or in vitro assays is not feasible. We have used a unique model system consisting of two isogenic human breast cancer cell lines that are equally tumorigenic in mice; but although one gives rise to metastasis, the other disseminates single cells that remain dormant at distant organs. Membrane purification and comparative quantitative LC-MS/MS proteomics identified 13 membrane proteins that were expressed at higher levels and three that were underexpressed in the metastatic compared with the non-metastatic cell line from a total of 1919 identified protein entries. Among the proteins were ecto-5′-nucleotidase (CD73), NDRG1, integrin β1, CD44, CD74, and major histocompatibility complex class II proteins. The altered expression levels of proteins identified by LC-MS/MS were validated using flow cytometry, Western blotting, and immunocyto- and immunohistochemistry. Analysis of clinical breast cancer biopsies demonstrated a significant correlation between high ecto-5′-nucleotidase and integrin β1 expression and poor outcome, measured as tumor spread or distant recurrence within a 10-year follow-up. Further the tissue analysis suggested that NDRG1, HLA-DRα, HLA-DRβ, and CD74 were associated with the ER−/PR− phenotype represented by the two cell lines. The study demonstrates a quantitative and comparative proteomics strategy to identify clinically relevant key molecules in the early events of metastasis, some of which may prove to be potential targets for cancer therapy.
Breast cancer is the most common malignant disease among women in Western countries, occurring in approximately one in 11 women (1). In this disease, malignant cells often disseminate to regional lymph nodes and establish distant metastases, preferentially in the bone, lung, and liver, resulting in poor outcome and high mortality (2, 3).
Metastases are established through a complex set of events that is yet not fully understood but requires detachment of single cells from the primary tumor, penetration of the tissue matrix, and migration of these cells to distant locations where they induce angiogenesis and undergo expansive growth (4). Some disseminated cancer cells seem to be capable of maintaining dormancy in distant organs without establishing metastases but may suddenly become activated many years after resection of the primary tumor (5). The dormancy may be caused by environmental signals, either lack of those inducing differentiation or the presence of signals stimulating growth arrest. Cellular factors and changes in the microenvironment, such as inflammation or a change in hormonal status, might eventually induce proliferation, differentiation, and subsequent metastatic growth, whereas other disseminated cancer cells remain dormant for a lifetime (6).
Traditional models of metastasis suggest that a subpopulation of cells in the primary tumor acquire metastatic capacity late in tumorigenesis, but gene expression profiles and cellular studies have recently provided evidence for a possible alternative model that suggests the metastatic capacity is acquired early in tumorigenesis (7). Stem cell populations have been identified in a range of hematopoietic and solid tumors and might represent the cells of origin for these tumors but might also be responsible for metastasis (8). Although a preserved genetic signature between the primary tumor and the metastasis has been found, other studies provide evidence of a gradual acquisition of genomic changes because distant metastases may not uniformly share mutations and often differ extensively from the primary tumor, reflecting the extent of genetic instability of breast cancer (9, 10). Only few studies provide proteomic characteristics of metastatic versus primary tumor of breast cancer because of the difficulties of obtaining high quality human tumor samples with full clinical histories and the absence of directly relevant in vitro assays (11, 12).
The two isogenic cell lines M-4A4 and NM-2C5, which were derived from the MDA-MB-435 cell line and originated from a highly aggressive human invasive ductal carcinoma, provide an interesting model of the metastatic process (13). M-4A4 and NM-2C5, when inoculated into the mammary fat pad of nude mice, showed equal tumorigeneity, but although M-4A4 established easily detectable metastases restricted to lymph nodes and lungs, NM-2C5 cells disseminated to distal organs, but the cells remained dormant and did not establish metastasis (14). There is an ongoing debate on whether the parent cell line MDA-MB-435 can be defined as a breast cancer cell line because it, along with breast- and epithelia-specific markers, also expresses melanoma-specific genes (15). However, MDA-MB-435 can be induced to express breast differentiation-specific proteins and secrete milk lipids as observed in other well established breast cancer cell lines and has therefore been considered as an excellent model of a highly malignant and dedifferentiated breast cancer (16). Regardless of this debate, our model system remains valuable in the context of cancer metastasis, but the results should, as always when using cell line models, be supported by studies of clinically relevant human tissue specimens.
M-4A4 and NM-2C5 have been extensively compared using gene expression analysis identifying a panel of differentially expressed genes (13, 17–20). However, because the proteome is so much more complex than the genome, similar studies at the protein level with special focus on plasma membrane proteins may add valuable biological insight and identify cell surface molecules that might be targeted with drugs or antibodies to inhibit the metastatic process.
Comparative quantitative proteomics using stable isotope labeling with amino acids in cell culture (SILAC)1 and LC-MS/MS allows a study of proteins with quantitatively different expression levels on metastasizing versus non-metastasizing cells. We used this technique to identify a panel of plasma membrane proteins showing altered expression in cells capable of forming metastasis. Validation studies at the protein and RNA expression level of the cell lines indicate that several of the identified proteins may be important for establishing metastasis in distant organs and thus have potential in target-specific therapy. Therefore, to further evaluate the clinical relevance of a selected number of the candidates identified by our analysis, their expression levels were evaluated in a panel of primary breast cancer biopsies and corresponding axillary lymph node metastasis from patients with known clinical outcomes. The results demonstrated the power of this systematic stepwise strategy for identifying targets of potential clinical value.
EXPERIMENTAL PROCEDURES
Cell Lines
M-4A4 and NM-2C5 cell lines were cloned from the parental cell line, MDA-MB-435, by serial dilution and screening by orthotopic implantation into the mammary fat pad of athymic mice (13). Unless otherwise stated, M-4A4, NM-2C5, and MDA-MB-435 cell lines were cultured in DMEM (Invitrogen 31966-021), 10% fetal bovine serum (Invitrogen 10270-106), 1% penicillin/streptomycin (Invitrogen 15140-122), and 1% non-essential amino acids (Invitrogen 11140-035) at 37 °C and 5% CO2. Cells were kept at low passage numbers throughout the experiments (<10 passages) to avoid phenotypic changes.
Purification of Plasma Membrane Proteins and Analysis by LC-MS/MS
SILAC, a quantitative proteomics strategy that metabolically labels the entire proteome allowing quantitative comparison of proteins/peptides by mass spectrometry analysis, was used on M-4A4 and NM-2C5 (21, 22). The cell lines were propagated as described above but with dialyzed FCS (HyClone) and in custom-made DMEM lacking l-arginine, l-lysine, and l-glutamine (JRH Bioscience). M-4A4 was supplemented with glutamine and isotopic forms of “heavy” arginine and lysine (13C6), and NM-2C5 was supplemented with glutamine and “light” arginine and lysine (12C6), allowing complete incorporation in the cells. One-third of the amount of arginine present in normal DMEM was used to avoid arginine to proline conversion. The isotopic labeling did not affect cell growth rates, morphology, or biological activity. Isotope-labeled M-4A4 and unlabeled NM-2C5 cells were combined 1:1 followed by homogenization, fractionation by centrifugation, and Percoll-sucrose density separation of cell contents. The fractions enriched for cell surface proteins and with minimal mitochondrial contamination, as determined by the activities of γ-glutamyl transpeptidase and succinate dehydrogenase, were either pooled or processed individually for further analysis. The purified plasma membrane fractions were enzymatically digested with lysyl endoproteinase and trypsin and analyzed by LC-MS/MS (Q-TOF Micro tandem mass spectrometer, Waters/Micromass, Manchester, UK) using a 2-h stepped linear gradient. Peptides with incorporated heavy arginine and/or heavy lysine had a higher mass of 6 Da per amino acid relative to peptides with normal isotopic arginine and/or lysine. The intensity ratios of the heavy and “normal” peptide peaks, respectively, in a given mass spectrum indicated the relative abundance of proteins in M-4A4 and NM-2C5, respectively. With variations of one parameter at a time, the purification of plasma membrane proteins was repeated 16 times, and each sample was analyzed by LC-MS/MS. The variations included single versus dual isotopic labeling, Percoll-sucrose density separation of crude membranes versus no density separation, and single versus repeated sample analysis to increase the number of proteins identified by LC-MS/MS. Dual isotopic labeling and repeated analysis of a sample with LC-MS/MS (three to four times) increased the percentage of proteins that could be quantified, whereas the use of Percoll-sucrose density separation resulted in enrichment of membrane proteins.
Cell Surface Membrane Purification—
The cells were lysed by incubation in a hypotonic buffer (10 mm Tris base, 1.5 mm MgCl2, 10 mm NaCl, pH 6.8) followed by sedimentation (300 × g for 5 min), resuspension in gradient buffer (0.25 m sucrose, 10 mm HEPES, 100 mm succinic acid, 1 mm EDTA, 2 mm CaCl2, 2 mm MgCl2, pH 7.4), and homogenization using a motor-driven Potter homogenizer (B. Braun Biotech). The homogenate was centrifuged at 1,000 × g for 10 min, and the supernatant, free of cell debris and nuclei, was further centrifuged at 100,000 × g for 30 min in an M150GX ultracentrifuge (Sorvall). The pellet, containing crude membranes, was resuspended in 2 ml of gradient buffer and manually homogenized and mixed with 1.9 ml of Percoll (Amersham Biosciences) containing 10% PBS and 0.19 ml of 2.5 m sucrose in an Easy-Seal™ tube (5 ml; Sorvall). Gradient buffer was used to fill up the excess volume of the tube before centrifugation at 120,000 × g for 15 min. The gradient was fractionated from the top into 10 fractions by displacing it from the bottom with 2 m sucrose. Percoll was removed by centrifugation at 900,000 × g for 15 min.
Plasma membrane fractions were resuspended and incubated in a carbonate buffer (100 mm Na2CO3, 10 mm DTT, pH 11.5) (23) followed by sedimentation and resuspension in a reduction buffer (0.2 m NaBr, 0.2 m KCl, 50 mm Tris-HCl, 10 mm DTT, pH 8.0). Prior to enzymatic digestion, the membranes were solubilized and incubated in urea buffer (6 m urea, 2 m thiourea, 100 mm Tris-HCl, 10 mm DTT, 100 mm iodoacetamide, pH 8.0).
γ-Glutamyl Transpeptidase Activity—
Aliquots of each fraction (50 μl) were transferred to a microtiter plate and mixed with 150 μl of substrate solution (1 mm l-γ-glutamyl-p-nitroanilide and 20 mm glycylglycine in 0.1 m Tris-HCl, pH 7.6), and the relative concentration of 4-nitroaniline was measured 10 times at 405 nm in 15 min at 37 °C using a Victor3 Multilabel Plate Reader (PerkinElmer Life Sciences). The relative γ-glutamyl transpeptidase activity was calculated on a linear segment of the measurements.
Succinate Dehydrogenase Activity—
Aliquots of each fraction (20 μl) were transferred to a microtiter plate and mixed with 80 μl of H2O and 100 μl of substrate solution (0.1 m phosphate buffer, 0.1 m sodium succinate, 0.05 m sucrose, pH 7.4) containing 2 mg/ml p-iodonitrotetrazolium (Sigma-Aldrich). The relative concentration of iodonitrotetrazolium formazan was measured 10 times at 490 nm over 5 min at 37 °C (Victor3 Multilabel Plate Reader), and the relative succinate dehydrogenase activity was calculated on a linear segment of the measurements.
Protein Concentration Measurement—
The protein concentration was determined by a colorimetric, detergent-compatible, Lowry-based assay (DC protein assay, Bio-Rad) in accordance with the manufacturer's protocol using a BSA preparation as standard (Pierce).
Data Analysis, Quantification, and Database Searching
The raw LC-MS/MS data obtained were processed by the MassLynx 4.0 software (Waters, Milford, MA) and analyzed by the freeware VEMS V3.207 (24, 25). The VEMS program ExRaw.exe was used to extract the retention times from the raw LC-MS/MS data files. Data from sequential LC-MS/MS analyses of the technical replicates were analyzed as one batch. The data were compared with all human proteins (n = 66,620) in the International Protein Index (IPI) version 3.23 (26).
Default VEMS settings were used for identification. Computational methods, scoring function, and statistical evaluation are described in detail in Matthiesen et al. (24). In brief, the cutoff score value for accepting individual MS/MS spectra was set to 10 to initially include as many protein identifications as possible. The MS/MS spectra of all single peptide-based protein identifications were manually inspected, and protein identifications based on poor MS/MS spectra (e.g. spectra with few matching b- and y-ions or assignment of both heavy and normal arginine and/or lysine in the same peptide) were excluded. The false discovery rate was calculated as the number of proteins identified using a reversed database (false positives) divided by the sum of proteins identified (true positives identified using the IPI and the false positives identified using IPI reversed) (27). The false discovery rate was estimated to be 3.9% using IPI version 3.23 reversed, made using the Decoy Database Builder software (27).
Mass accuracy of precursor and fragment ions were set to ±0.5 Da, and the mass accuracy of the peptides identified were generally below 50 ppm with an average of ∼30 ppm. Additional lowering of the mass accuracy did not influence the false discovery rate significantly (data not shown). The enzyme specificity setting was cleavage C-terminal to arginine and lysine, and one missed cleavage was allowed. Method-specific settings were as follows: 1) cysteine modification: carbamidomethylation; and 2) variable modifications, whole database: Met oxidation, [13C6]Arg, and [13C6]Lys.
Default VEMS settings were used for quantification. The cutoff score of the peptides used was set to 20, mass accuracy was set to ±0.3 Da, and method-specific settings were “Labeled amino acids”: [13C6]Arg and [13C6]Lys. For a peptide pair to be quantified, at least one peptide had to meet the criteria of the cutoff score, whereas the other peptide should have a count above 5 (or meet the criteria of the cutoff score). Data points with fewer counts than 5 were removed, whereas no outlier data points were removed.
The VEMS peptide quantification values (QVs) were based on calculation of QV for each set of the first three peaks in the light and heavy isotopic clusters.
 |
(Eq. 1) |
The maximum standard deviation of the peptide QV accepted for quantification was 5. The VEMS QV for any given set of peaks was calculated as the peak intensity (I) of the heavy isotope divided by the total peak intensity of the heavy and the light isotope from multiple scans (the number of scans depending on the peak intensity).
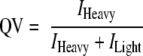 |
(Eq. 2) |
This means that a protein expressed in equal amount by both cell lines has a QV of 0.5 or 50%. For each sample, the QVs of β-actin (UniProt accession number P60709) and Na+/K+-ATPase (UniProt accession number P05023) were examined, and the average QV was calculated. When average QV was more than ±6 from 50%, the quantifications were normalized to assure comparable results between experiments. A QV-dependent normalization function, which is deduced in supplemental Data 1, was calculated for each sample to be normalized.
A protein expressed in only NM-2C5 has a theoretical QV of 0%, and a protein expressed in only M-4A4 has a theoretical QV of 100%. A threshold of 2-fold change in expression levels of proteins in M-4A4 relative to NM-2C5 was chosen to ensure the biological relevance of the markers identified corresponding to QV of 33 and 67%. For marker identification, cutoff QVs of 40 and 60% were used to allow for a general margin of error as the true signal is often underestimated due to signals from background “noise” or saturation of the detector.
For a protein to be considered differentially expressed it should meet the following criteria. 1) The proteins should be identified in two or more samples and be identified by two or more peptides in at least one of these. 2) The protein quantifications should be based on two or more different peptides, and at least one protein QV should be above 60% or below 40%. Further proteins that did not consistently exhibit differential expression despite fulfilling the above criteria were excluded.
The protein list generated by VEMS was transferred to ProteinCenter (Professional Edition, version 1.1.2–1.3.4, Proxeon, Odense, Denmark). To reduce redundancy the identified proteins were clustered into groups according to similarity levels 95, 80, and 65% (SL95, SL80, and SL65, respectively), but all proteins matching the data equally well are reported in supplemental Data 2 and 3. For marker identification, the proteins were sorted according to the number of samples they were identified in and according to the quantification.
Antibodies
The following antibodies were purchased and used for Western blotting (WB), flow cytometry (FACS), and immunohistochemistry (IHC)/immunocytochemistry (ICC) unless otherwise indicated. Anti-Na+/K+ ATPase (ab7671; WB only), anti-CD44 (ab6124), anti-CD74 (ab22603), anti-HLA-DRα (ab20181), anti-HLA-DRβ (ab20183), anti-integrin β1 (ab7168; FACS and WB only) were purchased from Abcam (Cambridge, UK). Anti-annexin A2 (A14020; FACS and WB only) and anti-ecto-5′-NT (550256; FACS only) were purchased from BD Biosciences. Anti-NDRG1 (H00010397M03) was purchased from Abnova (Taipei City, Taiwan). Anti-integrin β1 (AM298-5 M MU298-UC; IHC/ICC only) was purchased from Biogenex (San Ramon, CA). CD44v5 (BMS115; ICC only) and CD44v6 (BMS116; ICC only) were purchased from Bender Medsystems (Vienna, Austria). Two polyclonal anti-ecto-5′-NT antibodies were purchased from Abgent (San Diego, CA) (AP2014b; WB only) and Santa Cruz Biotechnology (Santa Cruz, CA) (sc-14682; IHC/ICC only). The secondary antibodies, HRP-conjugated goat anti-mouse (P0447), FITC-conjugated goat anti-mouse (F0479), and FITC-conjugated swine anti-rabbit (F0054), were all obtained from Dako (Glostrup, Denmark).
Western Blotting
Based on protein concentration measurements, equal amounts of plasma membrane preparations generated from M-4A4 and NM-2C5 cells were resolved by 4–20% SDS-PAGE (Pierce) and electroblotted onto a PVDF membrane. The membranes were blocked in PBS, 0.1% Tween 20, 5% nonfat dry milk powder for 1 h at room temperature and incubated with primary antibody for 16 h at 4 °C followed by washing and incubation with HRP-conjugated secondary antibody for 1 h at room temperature. All antibody incubations and washing steps were carried out in PBS, 0.1% Tween 20. The immunoreactive bands were visualized using an ECL Western blot kit (Amersham Biosciences). Anti-Na+/K+-ATPase was always included in separate lanes with M-4A4 and NM-2C5 membrane lysates to ensure equal loading of the two cell lines.
Flow Cytometry Analysis
Subconfluent cells were washed with PBS and harvested by ESGRO Complete Accutase (Millipore, Danvers, MA) and suspended in complete medium containing enzyme inhibitors (Complete protease inhibitor mixture tablets, Roche Diagnostics GmbH) to a final concentration of 106 cells/ml of medium. The cells were incubated with primary antibody for 60 min, washed twice with complete medium, and pelleted at 300 × g for 5 min at 4 °C. Cell suspensions were then incubated with a FITC-labeled secondary antibody for 60 min, washed, resuspended in PBS, and analyzed on a FACScan (BD Biosciences). Buffer, medium, and cells were kept at 4 °C until analyzed. As a negative control, cells were incubated without primary antibody. The results were interpreted using the ratio of geometric mean values within each cell line, with and without a primary antibody, and between the cell lines as the autofluorescence of the cells without primary antibody or in the presence of an isotype control differed in the two cell lines.
Quantitative Real Time PCR
Relative quantification of gene expression was performed in triplicates using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in accordance with the recommendation of the supplier. The median relative expression levels were normalized using the reference genes TATA box-binding protein, β-actin, and Na+/K+-ATPase. The primers for specific amplification were all obtained from QuantiTect Primer Assay collection (Qiagen): TBP/TATA box-binding protein, QT00000721; ACTB/β-actin, QT00095431; ATP1A1/Na+-K+-ATPase, QT00059962; NDRG1/NDRG1, QT00059990; ITGB1/integrin β1, QT00068124; ITGA6/integrin α6, QT00025725; HLA-DRA/HLA-DRα, QT00089383; HLA-DRB1/HLA-DRβ1, QT00090993; HLA-DRB3/HLA-DRβ3, QT00084056; CD74/CD74 antigen, QT00059402; NT5E/ecto-5′-NT, QT00027279; ANXA2/annexin A2, QT00071365; SLC26A2/sulfate transporter, QT00999761; ARF4/ADP-ribosylation factor 4, QT00024731; ICAM-1/intercellular adhesion molecule 1; APMAP/BSCv protein, QI00066605; ITGAV/integrin αV, QT00051891; SLC3A2/CD98, QT00085897; MCAM/MUC18, QT00085897; and CD44/CD44 antigen, forward TTTGCAGTCAACAGTC and reverse GTTACACCCCAATCTTCATGTCCAC (28).
Tissue Specimens
Selection of breast cancer biopsies was based on the medical history of the patient related to whether (n = 15) or not (n = 15) the patient experienced distant metastases within a 10-year follow-up period and on the subtype: estrogen receptor-positive (ER+), progesterone receptor-positive (PR+), and HER2/neu-negative (HER2−) (n = 10), ER+/PR+/HER2+ (n = 2), ER−/PR−/HER2− (n = 12), or HER2-overexpressing subtype ER−/PR−/HER2+ (n = 6). When axillary lymph node metastasis was present at primary surgery (n = 15), this tissue was included in the tissue array. ER, PR, and HER2 status were re-evaluated according to current technical standards. Normal breast tissues (from breast reductive surgery) were also included in the tissue array (n = 2). Selection and evaluation of cancer tissue were performed by a highly experienced pathologist.
Formalin-fixed paraffin-embedded primary breast tumor biopsies and corresponding axillary lymph node metastasis retrieved at the primary diagnostic surgery were obtained from the tissue bank of Department of Pathology, Odense University Hospital. A representative area of each tumor was selected from a hematoxylin- and eosin-stained tumor slide and punched out from the corresponding paraffin-embedded tumor tissue to form tissue arrays, each containing 15–20 tissue sections with a diameter of 0.4 mm. Based on standard histopathological criteria the immunohistochemical staining results were rated according to percentage of positive tumor cells (0–100%) and staining intensity from +1 to +3 with +3 being the most intense. When a heterogeneous staining pattern was present, the tumor was rated according to the most intense stained area when this was representative of at least 10% of the positive tumor cells.
All samples were coded, and patient confidentiality was maintained. The study was approved by the ethics committee of Funen and Vejle County.
Immunohistochemistry and Immunocytochemistry
Plasma/thrombin cell blocks were generated from the cell lines by adding 50 μl of plasma to 5 × 105 cells followed by 35 μl of bovine thrombin (Biofac A/S, Ejby, Denmark), which leads to the formation of a clot surrounding the cells. The plasma/thrombin cell clots and patient tissue biopsies were fixed in 4% neutral buffered formaldehyde, pH 7.4, for 24 h. Sections (4 μm) were cut from paraffin-embedded tissue blocks, mounted on ChemMate™ Capillary Gap Slides (Dako), dried at 60 °C, deparaffinized, and hydrated. Prior to antigen retrieval, blocking of endogenous peroxidase was performed in 1.5% hydrogen peroxide in TBS buffer, pH 7.4, for 10 min. A panel of antigen retrieval protocols was initially evaluated, including heat-induced epitope retrieval by microwave boiling for 15 min in (i) T-EG buffer (10 mm Tris, 0.5 mm EGTA, pH 9.0), (ii) 10 mm citrate buffer, pH 6.0, or (iii) Dako Target Retrieval Solution (Dako). Slides were heated for 11 min at full power (900 watts) in the microwave oven and then for 15 min at 400 watts. After heating, slides remained in buffer for 15 min. Heat-induced epitope retrieval by microwave boiling in T-EG or Target Retrieval Solution for 15 min proved to be the optimal antigen retrieval methods for a panel of antibodies tested. Sections were subsequently incubated with antibody diluted in antibody diluent (S2022, Dako) for 1 h at room temperature, washed with TNT buffer (0.1 m Tris, 0.15 m NaCl, 0.05% Tween 20, pH 7.5), and immunostained using the EnVision+™ HRP detection system (K4001, Dako) or PowerVision+™ HRP detection system (ImmunoVision Technologies) on the TechMate™ 500 instrument (Dako). 3,3′-Diaminobenzidine was used as the substrate-chromogen system (K3468, Dako). Immunostaining was followed by nuclear counterstaining in Mayer's hematoxylin. Finally coverslips were mounted with AquaTex (Merck). For each experiment, samples with either an isotype-matched antibody or no primary antibody were included as controls. Evaluation of the IHC staining was performed by a pathologist in a blinded setup. To identify proteins that were differentially expressed between any two groups of tissue specimens, a two-tailed Fisher's exact test was applied to identify p values smaller than 0.05. The significance level in the clinical study was calculated based on individual testing of each of the six marker antibodies against the panel of clinical samples. Due to the limited strength of the study (the low sample size in each group) significance was not maintained when using Bonferroni correction for multiple comparisons.
RESULTS
Proteomics—
To identify proteins, particularly plasma membrane proteins that may influence the ability of breast cancer cells to expand and establish metastasis at distant sites, the isogenic cell line pair M-4A4 and NM-2C5 was used as a model system. Comparison of protein expression levels in membrane-enriched preparations of the two cell lines using quantitative LC-MS/MS proteomics identified a total of 1919 protein entries. LC-MS/MS data were processed in VEMS. All data, including protein identifications, quantified peptides, sequence coverage, accuracy, precursor m/z observed, precursor charge observed, scores, and E-values, are supplied in supplemental Data 2 and 3. Assigned and manually verified spectra for all single peptide-based protein identifications are provided in supplemental Data 4. Refining the search to proteins identified by at least two peptides reduced the identified protein entries to 1145 of which 622 were membrane proteins. Thirteen proteins were found to be quantitatively expressed at higher amounts in the metastatic M-4A4 cells compared with the non-metastatic NM-2C5 cells, whereas three proteins were expressed in lower amounts in M-4A4 compared with NM-2C5 cells, and thus all of these proteins may be potential markers of cancer cells with the ability to establish metastasis (Table I). Quantitative LC-MS/MS analysis indicated that CD74 was mainly expressed in M-4A4, whereas HLA-DRα exhibited expression levels in a range from 2- to 26-fold higher in M-4A4 than in NM-2C5. The remaining 14 proteins were expressed 2- to 5-fold higher or lower, respectively, in M-4A4 cells compared with NM-2C5. An arbitrary minimum threshold of a 2-fold change was used and deemed to be biologically relevant.
Table I.
List of plasma membrane proteins that exhibited altered expression level in M-4A4 compared with NM-2C5 as identified by comparative quantitative proteomics
| Protein name | Gene name | UniProt accession no. | Identified/quantified in number of samples | Number of peptides/sample | Sequence coverage % | VEMS QV range | Expression level in M-4A4 vs. NM-2C5a |
|---|---|---|---|---|---|---|---|
| HLA-DRα | HLA-DRA | P01903 | 8/5 | 1–5 | 28.0 | 65–96.4 | ≫2 |
| HLA-DRβ | HLA-DRB1–5 | P04229 | 3/3 | 1–2 | 7.9 | 81.8 | >2 |
| CD74 | CD74 | P04233 | 2/2 | 1–3 | 13.9 | 94.5–99.3 | ≫2 |
| Ecto-5'nucleotidase | NT5E | P21589 | 8/4 | 1–6 | 15.9 | 58.2–79.3 | >2 |
| NDRG1 | NDRG1 | Q92597 | 2/2 | 2–4 | 18.8 | 69.1–70.2 | >2 |
| Integrin β1 | ITGB1 | P05556 | 6/5 | 1–8 | 12.8 | 56.4–77 | >2 |
| Integrin α6 | ITGA6 | P23229 | 5/4 | 1–5 | 5.9 | 61.4–75 | >2 |
| Annexin A2 | ANXA2 | P07355 | 12/10 | 1–16 | 52.6 | 52–65.8 | 2 |
| Sulphate transporter | SLC26A2 | P50443 | 2/2 | 1–2 | 3.4 | 81.5 | >2 |
| ADP-ribosylation factor 4 | ARF4 | P18085 | 2/1 | 1–2 | 14.0 | 66.7 | 2 |
| ICAM-1 | ICAM-1 | P05362 | 9/7 | 1–4 | 9.2 | 50.1–62.4 | 2 |
| BSCv protein | APMAP | Q9HDC9 | 3/3 | 2–4 | 14.2 | 57.8–61.8 | 2 |
| Integrin αV | ITGAV | P06756 | 5/2 | 1–3 | 4.2 | 55.6–60.5 | 2 |
| Protein name | Gene name | UniProt accession no. | Identified/quantified in number of samples | Number of peptides/sample | Sequence coverage % | VEMS QV range | Expression level in NM-2C5 vs. M-M4A4 |
|---|---|---|---|---|---|---|---|
| CD98 | SLC3A2 | P08195 | 13/11 | 2–9 | 24.0 | 30.9–53.3 | 2 |
| CD44 | CD44 | P16070 | 14/14 | 2–6 | 9.4 | 30.9–42.3 | 2 |
| MUC18 | MCAM | P43121 | 5/5 | 1–2 | 3.7 | 28.8–34.1 | >2 |
The relative expression level of proteins on M-4A4 compared with NM-2C5 cells is indicated as 2-fold change (2), 2–5-fold change (>2), or more than 5-fold (≫2). All listed proteins were identified in at least two samples, each run twice or in triplicates.
Biochemical Validation of Altered Protein Expression—
To validate the quantitative proteomic differences observed by mass spectrometry, the expression levels of eight proteins were examined in the two cell lines using immunocytochemistry, flow cytometry, and/or Western blotting.
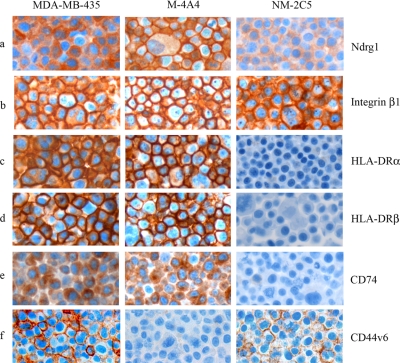
Immunocytochemical analysis of the NM-2C5 and M-4A4 cell lines as well as the “parental” cell line MDA-MB-435 was performed when commercial antibodies were available (Fig. 1). Interestingly NDRG1 showed a distinct translocation from the cytoplasm in NM-2C5 to the plasma membrane in M-4A4 cells (Fig. 1a). In MDA-MB-435 cells, NDRG1 was localized in the cytoplasm in some cells and the plasma membrane in others, corresponding to a mix of non-metastatic and metastatic cells. Analysis of integrin β1 showed more intense and distinct plasma membrane staining in M-4A4 compared with NM-2C5 (Fig. 1b). MDA-MB-435 showed an intermediate of the two expression patterns. Intense cell surface expression of the MHC class II proteins HLA-DRα and HLA-DRβ was observed in M-4A4 cells, whereas no staining of NM-2C5 cells was observed (Fig. 1, c and d). CD74 expression was also restricted to M-4A4 cells, and interestingly, in addition to the traditional cytoplasmic staining pattern of CD74/invariant chain, some staining of the cell surface, where CD74 acts as a migration inhibitor factor (MIF) receptor, was also observed (Fig. 1e) (29). MDA-MB-435 showed a staining pattern similar to that of M-4A4 for HLA-DRα, HLA-DRβ, and CD74 proteins. The expression level of CD44 on M-4A4 compared with NM-2C5 was analyzed using a panel of anti-CD44 antibodies. A CD44v6 antibody, recognizing an epitope encoded by exon v6 on the variant portion of human CD44, exhibited cell surface staining of NM-2C5 and MDA-MB-435, whereas M-4A4 was not stained (Fig. 1f). In contrast, staining using an anti-CD44 antibody recognizing an epitope conserved on all CD44 isoforms showed equal cell surface staining of the three cell lines (data not shown). Further a CD44v5-specific antibody did not stain any of the three cell lines (data not shown).
Fig. 1.
Comparison of the expression patterns of selected candidate proteins in MDA-MB-435, M-4A4, and NM-2C5 as analyzed by immunocytochemistry. Formalin-fixed and paraffin-embedded cells were stained with monoclonal antibodies directed against NDRG1 (a), integrin β1 (b), HLA-DRα (c), HLA-DRβ (d), CD74 (e), and CD44v6 (f).
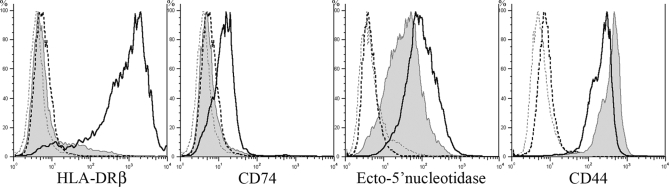
Expression levels of the different proteins were next examined by flow cytometry (Fig. 2). HLA-DRβ exhibited the most distinct difference in expression levels of all proteins examined by flow cytometry with a 65-fold increase in M-4A4 versus NM-2C5. HLA-DRα exhibited a 2-fold higher expression level in M-4A4 versus NM-2C5 by flow cytometry (data not shown), a somewhat lower expression difference than that observed by proteomics analysis (2- to 26-fold) and immunocytochemistry, suggesting that HLA-DRα and -DRβ molecules were presented on the plasma membrane as separate proteins rather than as an active MHC class II complex. CD74 and ecto-5′-NT also showed twice the expression level on M-4A4 compared with NM-2C5, whereas less difference in expression levels was observed for integrin β1, NDRG1, and annexin A2 (data not shown). CD44 showed 2-fold higher expression on NM-2C5 versus M-4A4 in agreement with the proteomics study.
Fig. 2.
Comparison of the cell surface expression of HLA-DRβ, CD74, ecto-5′-nucleotidase, and CD44 on M-4A4 and NM-2C5 cells by flow cytometry. Black and gray lines indicate the level of protein expression in M-4A4 and NM-2C5 cells, respectively. Dashed lines indicate background staining measured without primary antibody. Monoclonal primary antibodies followed by FITC-conjugated secondary antibody were used.
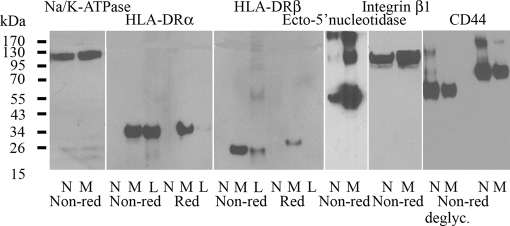
Semiquantitative measurement of the denatured proteins by Western blotting in reduced and non-reduced states was performed using Na+/K+-ATPase, a plasma membrane protein present in quantitatively equal amounts in both cell lines, for normalization (Fig. 3). HLA-DRα (35 kDa) and -DRβ (29 kDa) were detected in M-4A4 and lymphocytes (positive control) but not in NM-2C5. A much more intense band for ecto-5′-NT (58 kDa) was observed with M-4A4 compared with NM-2C5 under non-reducing conditions. Protein bands corresponding to a dimer and a trimer of ecto-5′-NT were also detected. The band for integrin β1 (110 kDa) was also at least twice the intensity in M-4A4 compared with NM-2C5 confirming the proteomics data.
Fig. 3.
Comparison of the expression levels of selected candidate proteins in Western blots of the plasma membrane fraction from NM-2C5 (N) and M-4A4 (M) cells. Lymphocyte lysate (L) was used as a positive control when relevant. Na+/K+-ATPase was used for normalization. HLA-DRα and -DRβ, ecto-5′-nucleotidase, integrin β1, and CD44 proteins were measured in the non-reduced (Non-red) and reduced (Red) states using a panel of monoclonal antibodies. CD44 was further analyzed in its deglycosylated (deglyc.) forms.
Using the anti-CD44 antibody, which recognized all CD44 isoforms, protein bands with molecular masses of 95 and 170 kDa were stained in both cell lines. Comparison of the staining intensity of the bands showed that the amount of the higher molecular mass isoform of CD44 was greater in NM-2C5 than in M-4A4. To further characterize the isoforms recognized in M-4A4 and NM-2C5, cell lysates were treated with the deglycosylation reagent peptide-N-glycosidase F. A shift from 95 to 65 kDa was observed for the lower CD44 band and from 170 to 130 kDa for the higher CD44 band upon deglycosylation. Under these deglycosylation conditions, the intensity of the 130-kDa band was significantly stronger in NM-2C5 than in M-4A4, whereas the intensity of the 65-kDa band was only slightly stronger in NM-2C5 than in M-4A4. Under reducing conditions, CD44, integrin β1, or ecto-5′-NT could not be detected in Western blotting of M-4A4 and NM-2C5 probably due to epitopes including disulfide bonds. Western blotting of NDRG1 and annexin A2 resulted in equally intense bands in M-4A4 and NM-2C5 (data not shown). The discrepancy between the proteomics and Western blot results for NDRG1 and annexin A2 was likely due to the semiquantitative nature of the latter method. Further analysis of the remaining candidate cell surface markers identified by proteomics was not possible because of the lack of suitable reagents for immunohisto-, immunocytochemistry, and/or flow cytometry analysis.
Evaluation of Expression at the Transcriptional Level—
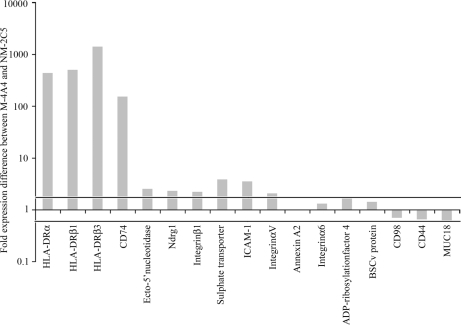
The observed altered protein expression levels corresponded with alterations in mRNA expression levels as measured by quantitative real time PCR, although the magnitude of changes in transcription did not exactly coincide (Fig. 4). The mRNA expression levels of HLA-DRα, -DRβ, and CD74 were approximately 100-fold higher in M-4A4 than in NM-2C5, whereas mRNA expression levels of ecto-5′-NT, NDRG1, integrin β1, integrin αV, sulfate transporter, and ICAM-1 were 5-fold higher or less in M-4A4 compared with NM-2C5. Finally mRNA expression levels of annexin A2, integrin α6, ADP-ribosylation factor 4, and BSCv protein were less than 2-fold higher in M-4A4 compared with NM-2C5. The mRNA expression levels of CD98, CD44, and MUC18 were 2-fold lower or less in M-4A4 compared with NM-2C5.
Fig. 4.
Comparison of the -fold change in expression levels of mRNA in M-4A4 versus NM-2C5 cells as measured by quantitative real time PCR. The number 1 (y axis) corresponds to equal mRNA expression in M-4A4 and NM-2C5. The horizontal lines indicate 2-fold higher and lower expression levels in M-4A4 versus NM-2C5, respectively.
Evaluation of Protein Expression in Clinical Cancer Biopsies—
When grouping the patients based on whether or not the primary tumors had metastasized to axillary lymph nodes or distant organs, a significant correlation between outcome and integrin β1 (p < 0.05) or ecto-5′-NT (p < 0.05) expression was observed (Table II). Similarly a significant correlation between outcome and ecto-5′-NT (p < 0.05) expression was observed when the patients were grouped based on whether or not they developed distant recurrence within a 10-year follow-up period (Table III).
Table II.
Immunohistochemical analysis of protein expression of five candidate proteins in 30 primary breast cancer biopsies stratified according to metastasis and receptor status
Distant metastasis was registered within a 10-year period.
| HLA-DRα/β | CD74 | NDRG1 | Integrin β1 | Ecto-5′-NT | |
|---|---|---|---|---|---|
| ER+/PR+ in cancer cells of primary tumor (n = 12) | |||||
| Node-positive and/or distant metastasis | 14% (1/7a) | 29% (2/7) | 43% (3/7) | 57% (4/7) | 86% (6/7) |
| Node-negative and no distant metastasis | 0% (0/5) | 40% (2/5) | 20% (1/5) | 0% (0/5) | 20% (1/5) |
| ER+/PR+ combined | 8% (1/12)b | 33% (4/12) | 33% (4/12)c | 33% (4/12) | 58% (7/12) |
| ER−/PR− in cancer cells of primary tumor (n = 18) | |||||
| Node-positive and/or distant metastasis | 36% (5/14) | 57% (8/14) | 86% (12/14) | 71% (10/14) | 57% (8/14) |
| Node-negative and no distant metastasis | 100% (4/4) | 75% (3/4) | 75% (3/4) | 50% (2/4) | 25% (1/4) |
| ER−/PR− combined | 50% (9/18)b | 61% (11/18) | 83% (15/18)c | 66% (12/18) | 50% (9/18) |
| ER±/PR± combined (n = 30) | |||||
| Node-positive and/or distant metastasis | 29% (6/21) | 48% (10/21) | 71% (15/21) | 67% (14/21)d | 67% (14/21)e |
| Node-negative and no distant metastasis | 44% (4/9) | 56% (5/9) | 44% (4/9) | 22% (2/9)d | 22% (2/9)e |
1/7 indicates one positive biopsy of a total of seven.
p value <0.05 between groups with identical letters (Fisher's exact test).
Table III.
Immunohistochemical analysis of protein expression of five candidate proteins in 30 primary breast cancer biopsies stratified according to recurrence and receptor status within a 10-year period
| HLA-DRα/β | CD74 | NDRG1 | Integrin β1 | Ecto-5′-NT | |
|---|---|---|---|---|---|
| ER+/PR+ in cancer cells of primary tumor (n = 12) | |||||
| Relapse within a 10-year period | 14% (1/7a) | 29% (2/7) | 43% (3/7) | 57% (4/7) | 71% (5/7) |
| No relapse within a 10-year period | 0% (0/5) | 40% (2/5) | 20% (1/5) | 0% (0/5) | 20% (1/5) |
| ER+/PR+ combined | 8% (1/12)b | 33% (4/12) | 33% (4/12)c | 33% (4/12) | 50% (6/12) |
| ER−/PR− in cancer cells of primary tumor (n = 18) | |||||
| Relapse within a 10-year period | 25% (2/8) | 63% (5/8) | 75% (6/8) | 63% (5/8) | 63% (5/8) |
| No relapse within a 10-year period | 60% (6/10) | 40% (4/10) | 80% (8/10) | 50% (5/10) | 20% (2/10) |
| ER−/PR− combined | 44% (8/18)b | 50% (9/18) | 78% (14/18)c | 56% (10/18) | 39% (7/18) |
| ER±/PR± combined (n = 30) | |||||
| Relapse within a 10-year period | 20% (3/15) | 47% (7/15) | 60% (9/15) | 60% (9/15) | 67% (10/15)d |
| No relapse within a 10-year period | 40% (6/15) | 40% (6/15) | 60% (9/15) | 33% (5/15) | 20% (3/15)d |
1/7 indicates one positive biopsy of a total of seven.
p value <0.05 between groups with identical letters (Fisher's exact test).
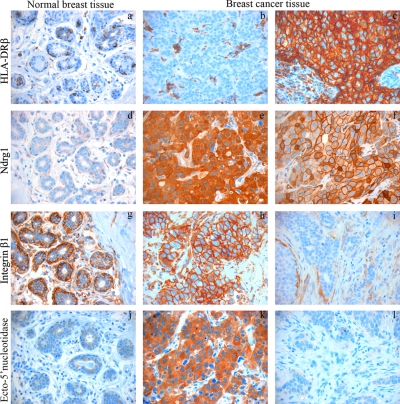
IHC analysis revealed that myoepithelial, but not luminal, cells of normal resting breast epithelial cells showed high expression of integrin β1 (Fig. 5, g–i). High integrin β1 expression was observed in the malignant epithelial cells of biopsies of 67% (n = 14 of 21) of the primary tumors that had developed distant and/or lymph node metastases, whereas only 22% (n = 2 of 9; p < 0.05) of primary tumors with no disseminated disease exhibited integrin β1 expression. Two patients with integrin β1-negative primary tumor exhibited integrin β1 expression in the axillary lymph node metastasis, suggesting that an integrin β1-expressing subclone of the primary tumor was responsible for the axillary lymph node metastasis. Integrin β1 expression seemed to be independent of hormonal receptor and HER2 status (Table IV).
Fig. 5.
Immunohistochemical analysis of staining patterns of anti-HLA-DRβ (a–c), -NDRG1 (d–f), -integrin β1 (g–i), and -ecto-5′-nucleotidase (j–l) antibodies in breast cancer and normal breast (a, d, g, and j) tissue biopsies. Hormonal and recurrence status within a 10-year period is as follows for breast cancer tissue: b, ER+/PR+/+ relapse; c, ER−/PR−/− relapse; e, ER−/PR−/+relapse (cytoplasm staining); f, ER−/PR−/+relapse (membrane staining); h, ER−/PR−/+relapse; i, ER−/PR−/− relapse; k, ER−/PR−/+relapse; l, ER+/PR+/−relapse.
Table IV.
Immunohistochemical analysis of protein expression of five candidate proteins in 30 primary breast cancer biopsies stratified according to HER2 and metastasis status
Distant metastasis was registered within a 10-year period.
| HLA-DRα/β | CD74 | NDRG1 | Integrin β1 | Ecto-5′-NT | |
|---|---|---|---|---|---|
| HER2+(ER+/PR+) in cancer cells of primary tumor (n = 2) | |||||
| Node-positive and/or distant metastasis | 50% (1/2a) | 50% (1/2) | 100% (2/2) | 50% (1/2) | 100% (2/2) |
| Node-negative and no distant metastasis | |||||
| HER2+(ER+/PR+) combined | 50% (1/2) | 50% (1/2) | 100% (2/2) | 50% (1/2) | 100% (2/2) |
| HER2-(ER+/PR+) in cancer cells of primary tumor | |||||
| Node-positive and/or distant metastasis | 0% (0/5) | 20% (1/5) | 20% (1/5) | 60% (3/5) | 80% (4/5) |
| Node-negative and no distant metastasis | 0% (0/5) | 40% (2/5) | 20% (1/5) | 0% (0/5) | 20% (1/5) |
| HER2-(ER+/PR+) combined | 0% (0/10) | 30% (3/10) | 20% (2/10) | 30% (3/10) | 50% (5/10) |
| HER2+(ER−/PR−) in cancer cells of primary tumor | |||||
| Node-positive and/or distant metastasis | 50% (2/4) | 50% (2/4) | 75% (3/4) | 50% (2/4) | 75% (3/4) |
| Node-negative and no distant metastasis | 100% (2/2) | 100% (2/2) | 50% (1/2) | 50% (1/2) | 50% (1/2) |
| HER2+(ER−/PR−) combined | 67% (4/6) | 67% (4/6) | 67% (4/6) | 50% (3/6) | 67% (4/6) |
| HER2−(ER−/PR−) in cancer cells of primary tumor | |||||
| Node-positive and/or distant metastasis | 30% (3/10) | 60% (6/10) | 90% (9/10) | 80% (8/10) | 50% (5/10) |
| Node-negative and no distant metastasis | 100% (2/2) | 50% (1/2) | 100% (2/2) | 50% (1/2) | 0% (0/2) |
| HER2−(ER−/PR−) combined | 42% (5/12) | 58% (7/12) | 92% (11/12) | 75% (9/12) | 42% (5/12) |
1/2 indicates one positive biopsy of a total of two.
IHC analysis showed that normal breast epithelia expressed no or only low amounts of ecto-5′-NT (Fig. 5j). Intense ecto-5′-NT expression was observed in 67% (n = 14 of 21) of the primary tumors that developed distant and/or lymph node metastases but in only 22% (n = 2 of 9; p < 0.05) of primary tumors with no disseminated disease. Similarly high ecto-5′-NT expression was observed in 67% (n = 10 of 15) of primary tumors from patients with relapse within a 10-year period but only in 20% (n = 3 of 15; p < 0.05) of patients without relapse. Further two patients with ecto-5′-NT-negative primary tumors exhibited ecto-5′-NT expression in the axillary lymph node metastasis, suggesting that an ecto-5′-NT-expressing subclone of the primary tumor was responsible for the axillary lymph node metastasis. The ecto-5′-NT expression seemed to be independent of hormonal receptor and HER2 status (Table IV). Complete overlap in the tumors that expressed ecto-5′-NT and integrin β1 expression was not observed, suggesting that they were independent, but potentially additive, factors for development of metastasis.
In contrast to ecto-5′-NT and integrin β1, HLA-DRα, -DRβ, CD74, and NDRG1 expression did not seem to correlate with the presence of metastasis (axillary lymph node or distant) or recurrence but rather to the ER−/PR− phenotype of the M-4A4 and NM-2C5 cell lines (Tables II and III). IHC staining for NDRG1 (Fig. 5, d–f) showed low, predominantly cytoplasmic, expression in normal breast epithelial cells. The tumor cells of ER+/PR+ biopsies were mainly negative (33%; n = 4 of 12) or exhibited only weak staining similar to normal breast epithelial cells regardless of their 10-year metastatic status. In contrast, a significant correlation of NDRG1-positive tumor cells and ER−/PR− biopsies were found (83%; n = 15 of 18; p < 0.05), again regardless of recurrence status. In biopsies from the two patients with relapses of ER+/PR+/HER2+ tumors, NDRG1 was highly expressed in the tumor cells (Table IV). As observed by ICC staining of the cancer cell lines, the subcellular location of NDRG1 varied from tumor to tumor and within tumors and in some cases were mostly cytoplasmic, whereas in others NDRG1 was predominantly present on the plasma membrane or both. No clear correlation between subcellular expression patterns and hormonal or HER2 status or other clinical parameters could be made.
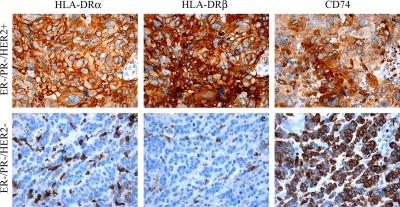
IHC analysis showed that HLA-DRα and -DRβ were expressed in normal resting breast epithelial cells (n = 2) and constitutively expressed by professional antigen-presenting cells (APC), such as dendritic cells, B cells, and macrophages, in cancer and normal tissue (Fig. 5, a–d). The malignant cells in primary breast cancer samples of those that had metastasized within a period of 10 years did not show clear staining differences from those that did not metastasize. However, significant correlations between cancer cell expression of the two proteins and the ER−/PR− phenotype of the biopsies were found regardless of their clinical outcome (p < 0.05). Positive staining of the two proteins were observed in cancer cells of 50% (n = 9 of 18) of ER−/PR− tumors, whereas only 8% (n = 1 of 12) of the ER+/PR+ tumors were stained. The expression of HER2 did not influence the frequency of HLA-DRα and HLA-DRβ expression (Table IV). Staining for CD74 generally followed that of HLA-DRα and HLA-DRβ and was mostly cytoplasmic (Fig. 6, a–c). Interestingly in two cases of triple negative biopsies from patients with node-positive and distant metastasis, CD74 was present in the membrane, whereas HLA-DRα and HLA-DRβ expression was absent (Fig. 6, d–f).
Fig. 6.
Immunohistochemical demonstration that co-expressions of HLA-DRα, HLA-DRβ, and CD74 do not always occur in breast cancer. One breast cancer specimen (ER−/PR−/HER2+, node-negative, and no distant metastasis) co-expressed HLA-DRα, HLA-DRβ, and CD74 (a–c), whereas another (ER−/PR−/HER2−, node-positive, and distant metastasis) only expressed CD74 and not HLA-DRα or HLA-DRβ (d–f).
Analysis of tissues for CD44 identified expression on the surface of all normal breast epithelial cells. In contrast, only a few breast cancers expressed CD44 on the surface (n = 4), whereas the remaining were CD44-negative with the exception of subpopulations of CD44-positive cells that were clustered together. Although an overall lower expression of CD44 was observed in tumors compared with normal tissue, no correlation to relapse or breast cancer subtype could be identified (data not shown).
DISCUSSION
Our quantitative comparative proteomic study revealed 16 membrane proteins expressed at higher or lower levels (2-fold or more) in the metastatic compared with the non-metastatic cell line and our analysis by other biochemical techniques such as flow cytometry, Western blotting and ICC unequivocally confirmed these findings validating the strategy used. These two cell lines are equally tumorigenic in nude mice, but only M-4A4 cells have the capability to establish metastasis, whereas NM-2C5 spread, but do not expand at the distant site. Therefore, the findings were worthy of deeper analysis in human tumor samples, which led to the confirmation that integrin β1 and ecto-5′-NT expression correlate significantly with metastatic performance in human malignant disease.
Ecto-5′-NT, which is a glycosylphosphatidylinositol-anchored component of lipid rafts and an enzyme converting 5′-AMP to extracellular adenosine (30), was one of the proteins identified in our proteomics study to be expressed at a 2- to 5-fold higher level in M-4A4 than in NM-2C5. The role of ecto-5′-NT in cancer has been shown to relate to adenosine generation; adenosine, acting through specific receptors, is an important metabolite released by cancer cells that establishes physiological conditions conducive to tumor progression (31). The relationship of ecto-5′-NT expression, its activity, and adenosine generation to tumor cell biology has not been fully elucidated. Further ecto-5′-NT has been identified as an adhesion molecule that binds to other cells and extracellular matrix proteins such as laminin and fibronectin (32). Our IHC analysis of clinical breast cancer specimens showed significant correlation between ecto-5′-NT expression and tumor spread and recurrence; e.g. high ecto-5′-NT expression was observed in 67% of the primary breast cancer biopsies from patients with recurrence but in only 20% of the biopsies from patients with no recurrence. Recently a study of clinical breast cancer specimens suggested an inverse correlation between ER expression and ecto-5′-NT expression (33); however, our study shows that ecto-5′-NT expression is independent of ER, PR, and HER2 status. Similar results were also reported from an earlier study (34), and careful analysis of the study by Spychala et al. (33) showed that the inverse correlation between ER expression and ecto-5′-NT expression was only found when an arbitrary cutoff at 20% ER+ cancer cells was set. We believe that the observation of ecto-5′-NT in ER− breast cancer cells relates to the aggressiveness of this breast cancer subtype resulting in earlier development of metastasis (33, 35). We propose that increased expression of ecto-5′-NT may facilitate establishment of micrometastases at distant sites either indirectly through adenosine promotion of tumor cell growth and invasiveness or directly by the action of ecto-5′-NT as an adhesion molecule. This theory is supported by studies showing that small interfering RNA against ecto-5′-NT effectively inhibits invasion and migration of the highly aggressive MDA-MB-231 cells and prevents adhesion to the extracellular matrix (36).
Integrins, ICAM-1 (CD54), CD44, and MUC18 (CD146) are all adhesion receptors that link the extracellular matrix to the cytoskeleton and play critical roles in the metastatic progression of breast cancer (37). Tumor cells might use integrins for migration and anchorage at sites of metastasis (38–40). In our proteomics study, we found 2- to 3-fold higher expression levels of integrins β1, α6, and αV and ICAM-1, whereas CD44 and MUC18 exhibited 2- to 3-fold lower expression in M-4A4 versus NM-2C5. Quantitative real time PCR confirmed this at the mRNA level. The higher level of integrin β1 and lower level of CD44 in M-4A4 versus NM-2C5 were also confirmed by Western blotting, and ICC showed a more distinct plasma membrane expression of integrin β1 in M-4A4 versus NM-2C5. Our IHC data showed that 67% of the node-positive patients and/or patients developing recurrent disease within a 10-year period had higher luminal integrin β1 expression compared with 22% in the group of metastasis-free patients, significantly correlating activation of integrins to tumor progression. ER, PR, and HER2 status had no significant influence on the expression of integrin β1. Our data are supported by other studies showing that integrin α6β1, a receptor for laminin, facilitates tumorigenesis and promotes tumor cell survival in distant organs (41). Targeted elimination of integrin α6β1 in a breast carcinoma cell line reduced the size of the primary tumor and the number of metastatic foci in the lungs of experimental mice (42). A high expression level of integrin α6 has also been correlated with reduced survival of women with breast cancer (43), whereas integrin αVβ3 seems to promote spontaneous metastasis of breast cancer to bone (44). It was previously suggested that a network of proteins, including HLA-DR molecules and integrin β1, plays a role in signal transduction, cell adhesion, and motility in melanoma and lymphoid cells (45). Analysis of cell membrane proteins in cell lines derived from metastasis and malignant melanoma has also demonstrated that integrin β1 and integrin αV are coexpressed with ecto-5′-NT (46); this is in agreement with our observation of a significant, but not complete, overlap between integrin β1 and ecto-5′-NT expression in the clinical breast cancer specimens.
CD44 proteins are a family of ubiquitously expressed transmembrane glycoproteins and a major receptor for hyaluronan. All isoforms of the protein are encoded by a single gene but are derived by alternative splicing (v1–v10) and variation in O- and N-glycosylations (47, 48). CD44s, the smallest and most widely expressed isoform, is expressed in ductal epithelial cells and myoepithelial cells in all cases of normal and benign breast tissue; however, the expression of CD44s progressively decreases with increasing deviation from normal histology corresponding to our data (49). The peptides identified by our study were identical to protein sequences in both the N-terminal part (exon 2) and the C-terminal part of CD44, sequences present in all CD44 isoforms. However, our biochemical studies suggest that only some CD44 isoforms exhibit different expression levels in the two cell lines as ICC, using an anti-CD44 antibody recognizing all CD44 isoforms, showed no significant difference in staining patterns or intensity of the two cell lines as also shown by Urquidi et al. (13). In contrast, an antibody against CD44v6 stained NM-2C5 but not M-4A4, whereas none of the cell lines were stained using an antibody against CD44v5. Western blotting of NM-2C5 and M-4A4 cells identified two CD44 protein bands at 95 and 170 kDa that were both glycosylated. The higher molecular mass form seemed to be more highly expressed in NM-2C5 than in M-4A4 cells. IHC staining of CD44 showed a strongly heterogeneous expression pattern (data not shown). This suggests that further studies might identify different isoforms that are associated with metastatic behavior but were not elucidated with the antibodies that we used.
We found a 2- to 3-fold higher expression level of NDRG1 in the plasma membrane fraction of M-4A4 than of NM-2C5. ICC confirmed this difference but revealed an additional difference in the subcellular localization of NDRG1 in the two cell lines: NDRG1 was predominantly cytoplasmic in NM-2C5, whereas the plasma membrane was predominantly stained in M-4A4. Due to the multiple localizations of NDRG1 and the absence of a transmembrane domain in the protein, we suggest that NDRG1 is translocated to the plasma membrane in M-4A4 in the process of metastasis, likely enabling the cells to adhere and proliferate in distant organs. However, this protein, also referred to as CAP43, DRG1, RTP, and RIT42, is expressed in normal tissue, and a strong association of NDRG1 with the plasma membrane in lactating breast epithelial cells has been observed by others, whereas in other cell types the NDRG1 staining was localized in the cytoplasm or nucleus (50). The function of this protein remains unclear, and some controversy regarding its role in cancer exists. Some studies suggest NDRG1 involvement in metastatic suppression, whereas others show higher expression of this protein, likely related to metastasis (51–55). The difference in subcellular localization of NDRG1 may be important for its function, and we therefore remain open to the concept that it plays some role in the development of metastases. Our IHC study on clinical samples showed that NDRG1 expression was significantly correlated with the ER−/PR− status of breast cancer cells because 83% of ER−/PR− biopsies independent of metastatic status showed increased NDRG1 expression compared with only 33% in ER+/PR+ biopsies. In another study, estradiol was shown to induce down-regulation of NDRG1 in ER+ breast cancer cells but not in ER− cells, indicating a hormonal influence on NDRG1 only when the hormone receptors are present, which also could explain some of the discrepancies among the NDRG1 studies (55). NDRG1 expression in ER−/PR−/HER2− but not ER+/PR+/HER2− breast cancer cells may correlate with the ability of the primary tumor cells to metastasize.
MHC class II HLA-DRαβ heterodimers are constitutively expressed by professional APC but, unlike MHC class I antigens, are not present on resting breast epithelial cells. However, MHC class II antigens have been detected in the normal lactating breast as well as in several other carcinomas (56–58). In professional APC, CD74/invariant chain is present in intracellular association with the immature MHC class II molecule blocking the peptide-binding groove; however, a pool of CD74 resides transiently on the cell surface in diverse cells (59). The exact function of cell surface CD74 has not been determined, but its recent identification as a MIF receptor opens a new and likely fruitful area of research. A study by Starlets et al. (29) suggests that CD74 acts as a survival receptor, whereas other studies have identified CD44 as a potential accessory protein required for MIF-CD74 signal transduction (60). MIF, a proinflammatory cytokine, is overexpressed in prostate cancer, but the mechanism by which MIF exerts effects on cancer cells remains undetermined (61).
We found that the expression level of CD74 increased more than 100-fold, HLA-DRα increased up to 26-fold, and HLA-DRβ increased only 5-fold in M-4A4 compared with NM-2C5 in the proteomics study. In flow cytometry, a 65-fold difference in expression of HLA-DRβ was observed between the two cell lines, whereas the differences in HLA-DRα and CD74 expression were 2-fold, suggesting that the proteins are regulated and exist individually rather than in a complex. ICC analysis of HLA-DRα and -DRβ showed distinct cell surface staining of M-4A4 cells, whereas CD74 was present in both the cytoplasm and at the cell surface of M-4A4. In contrast, no staining for the three proteins was observed in NM-2C5. The differences in protein expression level between assays may relate to the fact that some intracellular protein in addition to membrane protein was measured by the proteomics analysis and to the quality of the antibodies used for flow cytometry and ICC. Our IHC data showed that the tumor cells of ER−/PR− breast cancer biopsies, regardless of metastatic status, expressed significantly higher levels of HLA-DRα and -DRβ compared with the ER+/PR+ biopsies. CD74 was present in tumor cells of 61% of ER−/PR− breast cancer biopsies but in only 33% of the ER+/PR+ biopsies. The tumor cells staining for CD74 were mainly cytoplasmic, but importantly, in a few cases CD74 membrane staining was observed. Interestingly CD74 has been suggested as a promising target of cancer therapy in multiple myeloma and exhibits restricted expression in normal tissue (62).
This study indicates the feasibility of using mass spectrometry and other proteomics techniques to find new connections between the ability of cancer cells to establish metastasis in distant organs and altered expression levels of specific proteins. The study was designed to focus on plasma membrane proteins relative to the entire proteome, thereby increasing the likelihood of identifying proteins that could be targeted by antibodies and small molecule drugs and thus inhibit the metastatic process. Several of the plasma membrane proteins, which we identified and characterized, seem to be involved in the metastatic behavior of human breast cancer. This indicates that further investigations using this strategy hold promise for identifying novel mechanistically important targets for preventing metastasis and significantly advancing treatment of breast cancer. Because of novel technical improvements, future studies will also include identification and characterization of proteins with altered expression in other subcellular compartments, thereby bringing further insight to the biology of metastasis.
Acknowledgments
We thank Kate Rafn for expert technical assistance with LC-MS/MS, Lisbeth Mortensen and Ole Nielsen for excellent technical assistance with the immunohistochemical analysis, and M. K. Occhipinti-Bender for editorial assistance.
Footnotes
Published, MCP Papers in Press, March 24, 2009, DOI 10.1074/mcp.M800061-MCP200
The abbreviations used are: SILAC, stable isotope labeling with amino acids in cell culture; APC, antigen-presenting cells; ecto-5′-NT, ecto-5′-nucleotidase; ER, estrogen receptor; HRP, horseradish peroxidase; ICC, immunocytochemistry; IHC, immunohistochemistry; MIF, migration inhibitor factor; PR, progesterone receptor; QV, quantification value; WB, Western blotting; MHC, major histocompatibility complex; DMEM, Dulbecco's modified Eagle's medium; VEMS, Virtual Expert Mass Spectrometrist; IPI, International Protein Index; FACS, fluorescence-activated cell sorting; ICAM-1, intercellular adhesion molecule 1; NDRG1, N-myc downstream regulated gene 1; ICAM, intercellular adhesion molecule; APMAP, adipocyte plasma membrane associated protein.
This work was supported by the Danish Cancer Society, the Danish Cancer Research Foundation, The Danish Ministry of the Interior and Health's special initiative on molecular analysis for individualized cancer treatment, the Dagmar Marshall Foundation, and the Gangsted Foundation.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Parkin, D. M., Bray, F., Ferlay, J., and Pisani, P. ( 2005) Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, D. H., Tai, L. K., Wong, L. L., Sethi, S. K., and Koay, E. S. ( 2005) Proteomics of breast cancer: enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics 5, 1797–1805 [DOI] [PubMed] [Google Scholar]
- 3.Lacroix, M., Toillon, R. A., and Leclercq, G. ( 2004) Stable ‘portrait’ of breast tumors during progression: data from biology, pathology and genetics. Endocr.-Relat. Cancer 11, 497–522 [DOI] [PubMed] [Google Scholar]
- 4.Weigelt, B., Peterse, J. L., and van't Veer, L. J. ( 2005) Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5, 591–602 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki, M., Mose, E. S., Montel, V., and Tarin, D. ( 2006) Dormant cancer cells retrieved from metastasis-free organs regain tumorigenic and metastatic potency. Am. J. Pathol. 169, 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brabletz, T., Jung, A., Spaderna, S., Hlubek, F., and Kirchner, T. ( 2005) Opinion: migrating cancer stem cells-an integrated concept of malignant tumour progression. Nat. Rev. Cancer 5, 744–749 [DOI] [PubMed] [Google Scholar]
- 7.Weigelt, B., Hu, Z., He, X., Livasy, C., Carey, L. A., Ewend, M. G., Glas, A. M., Perou, C. M., and Van't Veer, L. J. ( 2005) Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 65, 9155–9158 [DOI] [PubMed] [Google Scholar]
- 8.Tu, S. M., Lin, S. H., and Logothetis, C. J. ( 2002) Stem-cell origin of metastasis and heterogeneity in solid tumours. Lancet Oncol. 3, 508–513 [DOI] [PubMed] [Google Scholar]
- 9.Suzuki, M., and Tarin, D. ( 2007) Gene expression profiling of human lymph node metastases and matched primary breast carcinomas: clinical implications. Mol. Oncol. 1, 172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt-Kittler, O., Ragg, T., Daskalakis, A., Granzow, M., Ahr, A., Blankenstein, T. J., Kaufmann, M., Diebold, J., Arnholdt, H., Muller, P., Bischoff, J., Harich, D., Schlimok, G., Riethmuller, G., Eils, R., and Klein, C. A. ( 2003) From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc. Natl. Acad. Sci. U. S. A. 100, 7737–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun, B., Zhang, S., Zhang, D., Li, Y., Zhao, X., Luo, Y., and Guo, Y. ( 2008) Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin. Cancer Res. 14, 7050–7059 [DOI] [PubMed] [Google Scholar]
- 12.Goodison, S., and Urquidi, V. ( 2008) Breast tumor metastasis: analysis via proteomic profiling. Expert Rev. Proteomics 5, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urquidi, V., Sloan, D., Kawai, K., Agarwal, D., Woodman, A. C., Tarin, D., and Goodison, S. ( 2002) Contrasting expression of thrombospondin-1 and osteopontin correlates with absence or presence of metastatic phenotype in an isogenic model of spontaneous human breast cancer metastasis. Clin. Cancer Res. 8, 61–74 [PubMed] [Google Scholar]
- 14.Goodison, S., Kawai, K., Hihara, J., Jiang, P., Yang, M., Urquidi, V., Hoffman, R. M., and Tarin, D. ( 2003) Prolonged dormancy and site-specific growth potential of cancer cells spontaneously disseminated from nonmetastatic breast tumors as revealed by labeling with green fluorescent protein. Clin. Cancer Res. 9, 3808–3814 [PubMed] [Google Scholar]
- 15.Rae, J. M., Creighton, C. J., Meck, J. M., Haddad, B. R., and Johnson, M. D. ( 2007) MDA-MB-435 cells are derived from M14 melanoma cells—a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 104, 13–19 [DOI] [PubMed] [Google Scholar]
- 16.Sellappan, S., Grijalva, R., Zhou, X., Yang, W., Eli, M. B., Mills, G. B., and Yu, D. ( 2004) Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 64, 3479–3485 [DOI] [PubMed] [Google Scholar]
- 17.Montel, V., Huang, T. Y., Mose, E., Pestonjamasp, K., and Tarin, D. ( 2005) Expression profiling of primary tumors and matched lymphatic and lung metastases in a xenogeneic breast cancer model. Am. J. Pathol. 166, 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montel, V., Mose, E. S., and Tarin, D. ( 2006) Tumor-stromal interactions reciprocally modulate gene expression patterns during carcinogenesis and metastasis. Int. J. Cancer 119, 251–263 [DOI] [PubMed] [Google Scholar]
- 19.Schwirzke, M., Evtimova, V., Burtscher, H., Jarsch, M., Tarin, D., and Weidle, U. H. ( 2001) Identification of metastasis-associated genes by transcriptional profiling of a pair of metastatic versus non-metastatic human mammary carcinoma cell lines. Anticancer Res. 21, 1771–1776 [PubMed] [Google Scholar]
- 20.Euer, N., Schwirzke, M., Evtimova, V., Burtscher, H., Jarsch, M., Tarin, D., and Weidle, U. H. ( 2002) Identification of genes associated with metastasis of mammary carcinoma in metastatic versus non-metastatic cell lines. Anticancer Res. 22, 733–740 [PubMed] [Google Scholar]
- 21.Ong, S. E., Foster, L. J., and Mann, M. ( 2003) Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 [DOI] [PubMed] [Google Scholar]
- 22.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 23.Hubbard, A. L., and Ma, A. ( 1983) Isolation of rat hepatocyte plasma membranes. II. Identification of membrane-associated cytoskeletal proteins. J. Cell Biol. 96, 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthiesen, R., Bunkenborg, J., Stensballe, A., Jensen, O. N., Welinder, K. G., and Bauw, G. ( 2004) Database-independent, database-dependent, and extended interpretation of peptide mass spectra in VEMS V2.0. Proteomics 4, 2583–2593 [DOI] [PubMed] [Google Scholar]
- 25.Matthiesen, R., Trelle, M. B., Højrup, P., Bunkenborg, J., and Jensen, O. N. ( 2005) VEMS 3.0: algorithms and computational tools for tandem mass spectrometry based identification of post-translational modifications in proteins. J. Proteome Res. 4, 2338–2347 [DOI] [PubMed] [Google Scholar]
- 26.Kersey, P. J., Duarte, J., Williams, A., Karavidopoulou, Y., Birney, E., and Apweiler, R. ( 2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 27.Reidegeld, K. A., Eisenacher, M., Kohl, M., Chamrad, D., Körting, G., Blüggel, M., Meyer, H. E., and Stephan, C. ( 2008) An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications. Proteomics 8, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 28.Hill, A., McFarlane, S., Mulligan, K., Gillespie, H., Draffin, J. E., Trimble, A., Ouhtit, A., Johnston, P. G., Harkin, D. P., McCormick, D., and Waugh, D. J. ( 2006) Cortactin underpins CD44-promoted invasion and adhesion of breast cancer cells to bone marrow endothelial cells. Oncogene 25, 6079–6091 [DOI] [PubMed] [Google Scholar]
- 29.Starlets, D., Gore, Y., Binsky, I., Haran, M., Harpaz, N., Shvidel, L., Becker-Herman, S., Berrebi, A., and Shachar, I. ( 2006) Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood 107, 4807–4816 [DOI] [PubMed] [Google Scholar]
- 30.Hunsucker, S. A., Mitchell, B. S., and Spychala, J. ( 2005) The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol. Ther. 107, 1–30 [DOI] [PubMed] [Google Scholar]
- 31.Spychala, J. ( 2000) Tumor-promoting functions of adenosine. Pharmacol. Ther. 87, 161–173 [DOI] [PubMed] [Google Scholar]
- 32.Airas, L., Niemelä, J., Salmi, M., Puurunen, T., Smith, D. J., and Jalkanen, S. ( 1997) Differential regulation and function of CD73, a glycosyl-phosphatidylinositol-linked 70-kD adhesion molecule, on lymphocytes and endothelial cells. J. Cell Biol. 136, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spychala, J., Lazarowski, E., Ostapkowicz, A., Ayscue, L. H., Jin, A., and Mitchell, B. S. ( 2004) Role of estrogen receptor in the regulation of ecto-5′-nucleotidase and adenosine in breast cancer. Clin. Cancer Res. 10, 708–717 [DOI] [PubMed] [Google Scholar]
- 34.Krüger, K. H., Thompson, L. F., Kaufmann, M., and Möller, P. ( 1991) Expression of ecto-5′-nucleotidase (CD73) in normal mammary gland and in breast carcinoma. Br. J. Cancer 63, 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostapkowicz, A., Inai, K., Smith, L., Kreda, S., and Spychala, J. ( 2006) Lipid rafts remodeling in estrogen receptor-negative breast cancer is reversed by histone deacetylase inhibitor. Mol. Cancer Ther. 5, 238–245 [DOI] [PubMed] [Google Scholar]
- 36.Zhi, X., Chen, S., Zhou, P., Shao, Z., Wang, L., Ou, Z., and Yin, L. ( 2007) RNA interference of ecto-5′-nucleotidase (CD73) inhibits human breast cancer cell growth and invasion. Clin. Exp. Metastasis 24, 439–448 [DOI] [PubMed] [Google Scholar]
- 37.Akiyama, S. K., Olden, K., and Yamada, K. M. ( 1995) Fibronectin and integrins in invasion and metastasis. Cancer Metastasis Rev. 14, 173–189 [DOI] [PubMed] [Google Scholar]
- 38.Taddei, I., Faraldo, M. M., Teulière, J., Deugnier, M. A., Thiery, J. P., and Glukhova, M. A. ( 2003) Integrins in mammary gland development and differentiation of mammary epithelium. J. Mammary Gland Biol. Neoplasia 8, 383–394 [DOI] [PubMed] [Google Scholar]
- 39.Giancotti, F. G., and Ruoslahti, E. ( 1999) Integrin signaling. Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 40.Newham, P., and Humphries, M. J. ( 1996) Integrin adhesion receptors: structure, function and implications for biomedicine. Mol. Med. Today 2, 304–313 [DOI] [PubMed] [Google Scholar]
- 41.Kikkawa, Y., Sanzen, N., Fujiwara, H., Sonnenberg, A., and Sekiguchi, K. ( 2000) Integrin binding specificity of laminin-10/11: laminin-10/11 are recognized by α3β1, α6β1 and α6β4 integrins. J. Cell Sci. 113, 869–876 [DOI] [PubMed] [Google Scholar]
- 42.Wewer, U. M., Shaw, L. M., Albrechtsen, R., and Mercurio, A. M. ( 1997) The integrin α6β1 promotes the survival of metastatic human breast carcinoma cells in mice. Am. J. Pathol. 151, 1191–1198 [PMC free article] [PubMed] [Google Scholar]
- 43.Friedrichs, K., Ruiz, P., Franke, F., Gille, I., Terpe, H. J., and Imhof, B. A. ( 1995) High expression level of α6 integrin in human breast carcinoma is correlated with reduced survival. Cancer Res. 55, 901–906 [PubMed] [Google Scholar]
- 44.Sloan, E. K., Pouliot, N., Stanley, K. L., Chia, J., Moseley, J. M., Hards, D. K., and Anderson, R. L. ( 2006) Tumor-specific expression of αvβ3 integrin promotes spontaneous metastasis of breast cancer to bone. Breast Cancer Res. 8, R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altomonte, M., Fonsatti, E., Visintin, A., and Maio, M. ( 2003) Targeted therapy of solid malignancies via HLA class II antigens: a new biotherapeutic approach? Oncogene 22, 6564–6569 [DOI] [PubMed] [Google Scholar]
- 46.Sadej, R., Spychala, J., and Skladanowski, A. C. ( 2006) Expression of ecto-5′-nucleotidase (eN, CD73) in cell lines from various stages of human melanoma. Melanoma Res. 16, 213–222 [DOI] [PubMed] [Google Scholar]
- 47.Goodison, S., Urquidi, V., and Tarin, D. ( 1999) CD44 cell adhesion molecules. Mol. Pathol. 52, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tölg, C., Hofmann, M., Herrlich, P., and Ponta, H. ( 1993) Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 21, 1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afify, A., McNiel, M. A., Braggin, J., Bailey, H., and Paulino, A. F. ( 2008) Expression of CD44s, CD44v6, and hyaluronan across the spectrum of normal-hyperplasia-carcinoma in breast. Appl. Immunohistochem. Mol. Morphol. 16, 121–127 [DOI] [PubMed] [Google Scholar]
- 50.Lachat, P., Shaw, P., Gebhard, S., van Belzen, N., Chaubert, P., and Bosman, F. T. ( 2002) Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem. Cell Biol. 118, 399–408 [DOI] [PubMed] [Google Scholar]
- 51.Bandyopadhyay, S., Pai, S. K., Hirota, S., Hosobe, S., Takano, Y., Saito, K., Piquemal, D., Commes, T., Watabe, M., Gross, S. C., Wang, Y., Ran, S., and Watabe, K. ( 2004) Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene 23, 5675–5681 [DOI] [PubMed] [Google Scholar]
- 52.Kovacevic, Z., and Richardson, D. R. ( 2006) The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis 27, 2355–2366 [DOI] [PubMed] [Google Scholar]
- 53.Chua, M. S., Sun, H., Cheung, S. T., Mason, V., Higgins, J., Ross, D. T., Fan, S. T., and So, S. ( 2007) Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Mod. Pathol. 20, 76–83 [DOI] [PubMed] [Google Scholar]
- 54.Cangul, H., Salnikow, K., Yee, H., Zagzag, D., Commes, T., and Costa, M. ( 2002) Enhanced expression of a novel protein in human cancer cells: a potential aid to cancer diagnosis. Cell Biol. Toxicol. 18, 87–96 [DOI] [PubMed] [Google Scholar]
- 55.Fotovati, A., Fujii, T., Yamaguchi, M., Kage, M., Shirouzu, K., Oie, S., Basaki, Y., Ono, M., Yamana, H., and Kuwano, M. ( 2006) 17β-Estradiol induces down-regulation of Cap43/NDRG1/Drg-1, a putative differentiation-related and metastasis suppressor gene, in human breast cancer cells. Clin. Cancer Res. 12, 3010–3018 [DOI] [PubMed] [Google Scholar]
- 56.Bártek, J., Petrek, M., Vojtb̆sek, B., Bártková, J., Kovarík, J., and Rejthar, A. ( 1987) HLA-DR antigens on differentiating human mammary gland epithelium and breast tumours. Br. J. Cancer 56, 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Claesson, L., Larhammar, D., Rask, L., and Peterson, P. A. ( 1983) cDNA clone for the human invariant gamma chain of class II histocompatibility antigens and its implications for the protein structure. Proc. Natl. Acad. Sci. U. S. A. 80, 7395–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strubin, M., Mach, B., and Long, E. O. ( 1984) The complete sequence of the mRNA for the HLA-DR-associated invariant chain reveals a polypeptide with an unusual transmembrane polarity. EMBO J. 3, 869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wraight, C. J., van Endert, P., Moller, P., Lipp, J., Ling, N. R., MacLennan, I. C., Koch, N., and Moldenhauer, G. ( 1990) Human major histocompatibility complex class II invariant chain is expressed on the cell surface. J. Biol. Chem. 265, 5787–5792 [PubMed] [Google Scholar]
- 60.Shi, X., Leng, L., Wang, T., Wang, W., Du, X., Li, J., McDonald, C., Chen, Z., Murphy, J. W., Lolis, E., Noble, P., Knudson, W., and Bucala, R. ( 2006) CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 25, 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer-Siegler, K. L., Iczkowski, K. A., Leng, L., Bucala, R., and Vera, P. L. ( 2006) Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 177, 8730–8739 [DOI] [PubMed] [Google Scholar]
- 62.Burton, J. D., Ely, S., Reddy, P. K., Stein, R., Gold, D. V., Cardillo, T. M., and Goldenberg, D. M. ( 2004) CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin. Cancer Res. 10, 6606–6611 [DOI] [PubMed] [Google Scholar]