Abstract
Decomposing variation in population growth into contributions from both ecological and evolutionary processes is of fundamental concern, particularly in a world characterized by rapid responses to anthropogenic threats. Although the impact of ecological change on evolutionary response has long been acknowledged, the converse has predominantly been neglected, especially empirically. By applying a recently published conceptual framework, we assess and contrast the relative importance of phenotypic and environmental variability on annual population growth in five ungulate populations. In four of the five populations, the contribution of phenotypic variability was greater than the contribution of environmental variability, although not significantly so. The similarity in the contributions of environment and phenotype suggests that neither is worthy of neglect. Population growth is a consequence of multiple processes, which strengthens arguments advocating integrated approaches to assess how populations respond to their environments.
Keywords: eco-evolutionary dynamics, ecology, evolution, phenotype, population
1. Introduction
Although the link between natural selection and demography has long been recognized, only recently have biologists begun to appreciate that ecological and evolutionary changes can occur on the same time scale (Thompson 1998; Sinervo et al. 2000; Yoshida et al. 2003; Hairston et al. 2005; Hanski & Saccheri 2006; Kinnison & Hairston 2007; Pelletier et al. 2007a). The traditional view argued that natural selection determines which phenotypes persist or go extinct, while density-dependent and stochastic factors determine population growth (Saccheri & Hanski 2006). Population biologists have typically ignored the potential feedback of change in phenotypic distribution on population processes (Slobodkin 1961). Many techniques in evolutionary ecology assume environmental consistency (Falconer & Mackay 1996). The assumption of environmental constancy has recently been challenged by several studies, demonstrating that, under certain circumstances, evolution can occur on contemporary time scales (Hendry & Kinnison 1999; Kinnison & Hendry 2001) and that evolutionary processes can have quantifiable effects on ecological dynamics (Sinervo et al. 2000; Yoshida et al. 2003; Hairston et al. 2005; Pelletier et al. 2007a). It is therefore necessary to consider ecological variation in evolutionary studies, as well as evolutionary responses in ecological studies, to understand comprehensively the interplay between these processes (Fussmann et al. 2007).
A first step in understanding the interactions between ecology and evolution is to explore the links between different levels of biological organization (Pelletier et al. 2009). At the population level, the fundamental processes of birth and death link natural selection and population dynamics. It is therefore necessary to consider the possibility of an eco-evolutionary feedback between phenotypic traits and demography (Ricklefs & Wikelski 2002; Coulson et al. 2006). This feedback arises because a change in any environmental variable can alter selective pressures. One consequence is a new phenotype distribution, which in turn affects density that then affects (partially) all subsequent phenotype distributions (Coulson et al. 2006; Kokko & Lopez-Sepulcre 2007). Applying an evolutionary demography approach to reanalyse the exceptional long-term sequence of beak shape in a Darwin's finch species (Geospiza fortis) on Galapagos, Hairston et al. (2005) showed that an adaptive response in beak size contributed twice as much to variation in population size as ecological processes. A few notable exceptions apart (Hairston et al. 2005; Pelletier et al. 2007a), the effect of changes in phenotypic trait distributions on ecological processes has rarely been investigated under non-laboratory conditions (Saccheri & Hanski 2006). As an illustration, it is not known how environmental conditions, phenotypic change and population dynamics are influenced by concurrent selection or how emergent changes determine their dynamics. Initial attempts to address this topic might therefore ask: does annual population growth vary the most as a direct result of environmental change, or is it more strongly related to phenotypic variability?
One way to address this question empirically is to explore how a trait distribution in 1 year influences subsequent vital rates (here, survival and recruitment), and hence population growth. The association between a trait distribution and population growth is also probably affected by environmental conditions (Pelletier et al. 2007a) and/or predation pressure (Yoshida et al. 2003; Jones et al. 2009). There is a need to quantify the interplay between phenotypic distributions and population growth across a range of environmental conditions and model species. Using longitudinal monitoring (between 16 and 33 years) of five ungulate populations exposed to disparate environmental conditions, the aim of this paper is to disentangle the effects of environmental and phenotypic changes on population dynamics. An essential step in the assessment of population fluctuations is to understand how the dynamics of heritable phenotypic traits influence population processes (Coulson & Tuljapurkar 2008). Evaluating the relative importance of different processes on population growth is fundamental to population and evolutionary biology, especially given the increasing number of studies showing rapid phenotypic changes to anthropogenic threats (reviewed by Parmesan & Yohe 2003; Bradshaw & Holzapfel 2006; Gienapp et al. 2008; Hendry et al. 2008).
2. Conceptual framework
We begin by outlining Hairston et al.'s (2005) framework for comparing ecological and evolutionary dynamics, before progressing to discuss the details of its application here. Hairston et al. (2005) aimed to compare ‘ecological and evolutionary dynamics’. Their rationale was that temporal changes in some attribute of population dynamics—say population growth from 1 year to the next, hereafter simply population growth—are the result of temporal changes in ecological and evolutionary processes. Expressed mathematically, this statement is
where X is the attribute of population dynamics; k is an ecological variable; and z is an evolutionary variable. Hairston et al (2005) considered summary statistics (e.g. mean beak shape) as evolutionary variables and demonstrated applications in discrete and continuous time. The discrete-time analogue of the continuous expression above is
where h is some interval of time between consecutive measures. Our explanation of the framework is restricted to the discrete case, since it represents the data collection protocol of the study populations as well as their biology (they are the so-called ‘birth-pulse’ populations). Application requires three steps: (i) derive statistical relationships between X and z independent of k and between X and k independent of z, (ii) calculate changes in z and k across the time interval, and (iii) compare the contributions of z and k to X. More than one z and k can be incorporated in a multivariate version, but we do not expand that argument due to our aim of application across multiple systems.
Ecological and evolutionary variables are rather broad terms; changes in either result from multiple underlying causes. Changes in ‘evolutionary variables’ (as defined by Hairston et al. 2005) are not necessarily examples of evolutionary dynamics. Suppose that z is a phenotype. Changes to z across an interval might result from, for example, phenotypic plasticity (Pigliucci 2001; Nussey et al. 2005a,b) or changes in the age structure of the population (Coulson & Tuljapurkar 2008). Henceforth, we refer to z as phenotypic and k as environmental variables rather than evolutionary and ecological, respectively. No attempt is made to decompose causes of observed variation in z and k in this first empirical application of the framework. To reiterate, the aim here is to apply the method and hence disentangle the effects of changes in the environment and phenotype distributions on population dynamics.
3. Material and methods
(a) Study populations
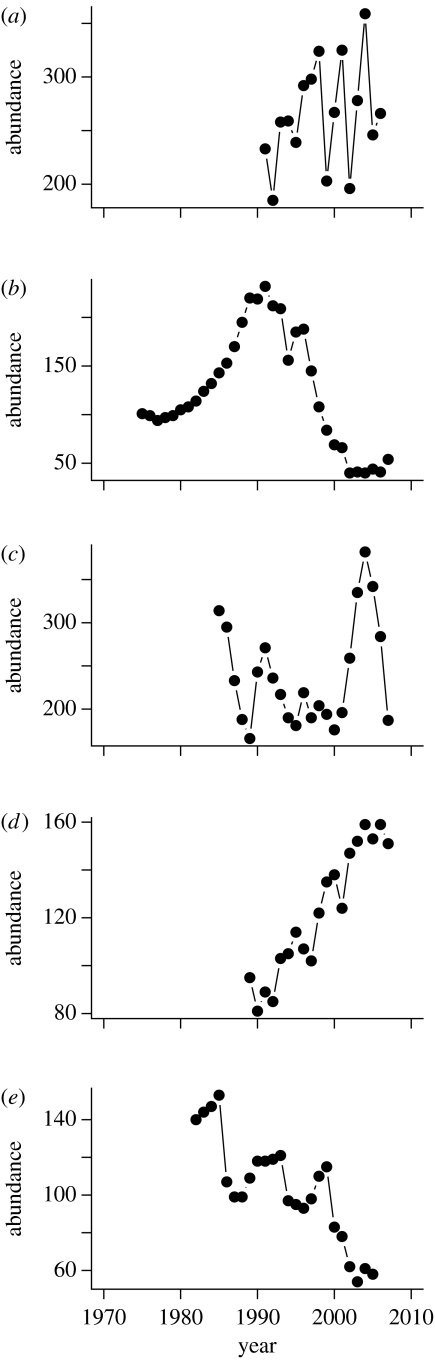
The data used here are taken from five longitudinal studies of marked, long-lived ungulates in temperate areas of the Northern Hemisphere. Although these species have broadly similar life history, their dynamics fluctuate widely (figure l) and they occupy very different habitats. The Soay sheep population (Ovis aries) lives on a Hebridean island devoid of trees, whereas forests with isolated meadows dominate the habitat of roe deer (Capreolus capreolus) in Trois-Fontaines, France. The three Canadian populations (two bighorn sheep (Ovis canadensis) and one mountain goat (Oreamnos americanus)) occupy alpine regions of the Rockies. A summary of available data and key references is given in table 1; further information on data collection protocols is given in the electronic supplementary material 1. We define population growth from year t to t+1 as the number of individuals in the study area in year t+1 divided by the corresponding number in year t.
Figure 1.
Although all five ungulate populations are of similar life history, their population dynamics vary markedly. Time series are shown for the periods of each population analysed. Study populations: (a) Soay sheep; (b) bighorn sheep at Ram Mountain; (c) roe deer; (d) mountain goats; and (e) bighorn sheep at Sheep River.
Table 1.
Summary of study populations, with dates between extreme measurements of population growth (as observed annual population growth, X), environmental variables (k), phenotypic variables (z) and key references for further information. (Commas indicate two separate series of years when data requirements were met. If no date is given when mass was measured, the data were collected from individuals within 48 hours of birth. Note that all study populations used regression methods to estimate birth mass (except Sheep River, see §3). INDVI, integrated normalized difference vegetation index; PDO, Pacific Decadal Oscillation. The biological impacts of these environmental variables are discussed at length in the appropriate references. Further details on these populations can be found in the electronic supplementary material 1.)
| population | location | number of years | X | k | z |
|---|---|---|---|---|---|
| Soay sheep | St Kilda, Scotland | 16 | λt (Aug) | Sward height in March (Crawley et al. 2004) | birth mass (Clutton-Brock et al. 1992) |
| mountain goats | Caw Ridge, Canada | 9, 8 | λt (Jun) | maximum rate of increase in INDVI+standing INDVI (Pettorelli et al. 2007) | kid mass on 15 July (Côté & Festa-Bianchet 2001) |
| roe deer | Trois-Fontaines, France | 21 | λt (Mar) | June rainfall (Gaillard et al. 1997) | birth mass (Gaillard et al. 1993) |
| bighorn sheep | Ram Mountain, Canada | 18, 12 | λt (May) | PDO (Zhang et al. 1997); density (Festa-Bianchet et al. 1998) | mass on 5 June (Festa-Bianchet et al. 1996) |
| bighorn sheep | Sheep river, Canada | 33 | λt (Mar) | PDO2 (Zhang et al. 1997); presence of predators (Festa-Bianchet et al. 2006) | chest circumference at six months of age (Pelletier et al. 2005) |
Mass early in life is considered generally to be a key phenotypic trait (Albon et al. 1987; Clutton-Brock et al. 1987; Metcalfe & Monaghan 2001) with a heritable component (Coltman et al. 2005; Wilson et al. 2005). Birth weight is an important determinant of neonatal survival in several species of mammals (Albon et al. 1987; Clutton-Brock 1991; Côté & Festa-Bianchet 2001), and early conditions can have lasting effects on multiple fitness components (Lummaa & Clutton-Brock 1998; Lindström 1999). Data on mass early in life, which we refer to as ‘juvenile mass’, are available in all populations except bighorn sheep at Sheep River. At Sheep River, chest circumference at six months of age is available and correlates very highly with autumn weight (r=0.90, p<0.001, Pelletier et al. 2005). Juvenile mass values were adjusted for date of capture in a given year using regression techniques in all populations.
As different research groups have monitored different study sites, they have different sampling protocols (table 1). All populations are furthermore influenced to various extents by different environmental variables. For the purposes of our analysis, we used the environmental variable identified as the best correlate of annual growth in each population. Environmental variables, where high values indicate harsh conditions, were reflected around zero for ease of (statistical and visual) comparison. For example, high values of the Pacific Decadal Oscillation (PDO, Zhang et al. 1997) were correlated with decreases in population growth in bighorn sheep, whereas high values of sward height (a vegetation index; Crawley et al. 2004) correlated with increases in population growth in Soay sheep. A PDO of, say, −4 was therefore reflected to be +4 in analyses. This process ensured that increasing environmental variables correlated with increasing population growth in all populations.
(b) Statistical analysis
The first step in application of the framework is to derive coefficients for expected population growth following observed phenotypic and environmental changes. These coefficients were obtained from generalized linear models (Fox 2002). Different populations required different models: coefficients for the Soay sheep and both the bighorn sheep populations were estimated using standard linear models. Standard linear models were not appropriate for the roe deer and mountain goat populations since the variance around population growth increased nonlinearly with the mean. Gamma-type models (where variance increases as a function of the mean2), but with an identity link function, were used for these populations. Additionally, density at Ram Mountain (Festa-Bianchet et al. 1998) and predator presence in both the bighorn sheep populations (Festa-Bianchet et al. 2006) were controlled for. Significant outliers (identified using ‘cook's distance’; Fox 2002, p. 206) were removed to achieve diagnostic plots that did not reveal systematic bias (see the electronic supplementary material 1).
Once the coefficients have been determined, they can be used to hypothesize the effect of environmental or phenotypic change on population growth. Since different populations have different dynamics (figure 1), and therefore different types of within-population variations, mixed-effect models were used to assess these effects statistically. Mixed-effect models are appropriate tools for analysis when some level of structure—e.g. experimental units or repeated measurements on experimental subjects—is apparent in the data (so-called grouped data; see Pinheiro & Bates 2000). Mixed-effect models consist of fixed and random effects. Fixed effects are parameters associated with global trends, modelling patterns of variation common to all experimental units (Pinheiro & Bates 2000, p. 3). Random effects model the correlation in residuals caused by the structure of the data (Diggle et al. 2002, p. 82). In the simplest case, different experimental subjects are distinguished using different intercepts. In more complex cases, the residuals in the different experimental subjects might vary systematically and be modelled by different slopes and different intercepts. If random slopes are necessary, then the responses to changes in the explanatory variables vary in direction between the experimental subjects. Here, the five populations are the experimental subjects. Modelling the correlation in residuals (using random effects) is critical here because the number of experimental subjects is small (5) and less than the number of observations per subject (mean 23.4) (Diggle et al. 2002). It is also consistent with our motivation of assessing differences in the relative importance of environmental and phenotypic fluctuations across the five populations.
There are five ways of combining phenotypic and environmental variability as fixed effects (both with interactions, both without interactions, phenotype only, environment only and neither), and five ways of combining phenotype and environmental variability as random effects (population-level variability in gradients for both, for environment only, for phenotype only, in intercepts only and not at all). These combinations of explanatory variables were regressed against population growth (differenced to remove temporal autocorrelation) across the same time interval. The model with most support was chosen from the complete set of potential models (Whittingham et al. 2006) and differentiated using the Akaike Information Criterion (AIC). Information criteria (Burnham & Anderson 2002) provide a compromise between number of parameters used and model likelihood. The model with most support has the lowest AIC value. Models within two AIC values of the minimum AIC value have ‘substantial’ support, and only when this difference is greater than four does the model's support become ‘considerably less’ (Burnham & Anderson 2002, p. 71). We also calculated Akaike weights (Burnham & Anderson 2002), which can be interpreted as the likelihood of a particular model being the best, given the set of models used. Another interpretation of model weight is that it quantifies the weight of evidence in favour of a particular model (Burnham & Anderson 2004). We do not calculate AIC values for models without random effects, since comparisons are only reasonable between models with and without random effects if they have the same fixed effects. This comparison was made using likelihood ratio tests.
(c) Individual-based method comparison
The Soay sheep population was previously the subject of an individual-based analysis, which found that the distribution of weights in the population contributed significantly to the impact that each individual makes to annual population growth (Pelletier et al. 2007a). Pelletier et al.'s (2007a) approach was a retrospective decomposition of population growth into contributions from phenotype. Hairston et al. (2005) calculated hypothetical changes in population growth from observed phenotypic variability. Although both methods attempt to do very different things, they are linked. When there is viability or fertility selection on a trait, then Pelletier et al.'s (2007a) method will lead to observed population growth being strongly influenced by the distribution of the trait value. This selection (assuming principally that traits are heritable and phenotypic plasticity is low) will generate a change in the mean value of the trait over a time step. This is how Hairston et al.'s (2005) framework defines phenotypic change. To illustrate the ability of Hairston et al.'s (2005) framework to approximate results from more involved methods, we regressed results from the framework applied here against the individual-based one (see fig. 2c in Pelletier et al. 2007a). A quasibinomial model (with logit link function) was used since contribution to population growth is bounded between 0 and 1 (Fox 2002).
Mixed-effect models were fitted using the lmer function in the lme4 package (v. 0.999375-24, Bates 2005) in the R environment (v. 2.7.1, R Development Core Team 2008). The Laplacian approximation to maximum likelihood was used.
4. Results
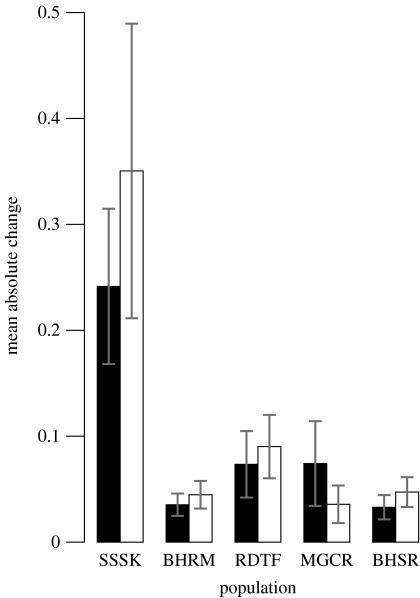
Phenotypic and environmental changes were of similar orders of magnitude: phenotypic change had a slightly greater effect in four of the five populations (the exception being mountain goats), but was not statistically significant (using one- or two-tailed t- or Wilcoxon signed-rank tests; figure 2). The Soay sheep population fluctuated the most with environmental and phenotypic changes: absolute rates of year-to-year change were three times greater in this population than in the others (figure 2). The ratio of phenotypic to environmental change was the lowest for mountain goats (0.48) and varied between 1.45 (bighorn sheep at Sheep River) and 1.22 for the remainder.
Figure 2.
Mean absolute change of environment (black bars) and phenotype (white bars) with 95% parametric confidence intervals for each population, calculated following eqn (8) by Hairston et al. (2005). The change quantifies the effects on population growth of environmental and phenotypic changes, respectively. Absolute rates are presented since changes in environmental and phenotypic variables can be negative as well as positive. Study population codes: SSSK, Soay sheep; BHRM, bighorn sheep at Ram Mountain; RDTF, roe deer; MGCR, mountain goats; BHSR, bighorn sheep at Sheep River.
The model with the most support featured global effects of environmental and phenotypic changes as well as their interaction. This model had the lowest AIC value, but a model weight of only 0.567 (table 2). A model that featured environmental change only also had some support (ΔAIC was 3, model weight 0.123) and is especially worthy of consideration given that it is more parsimonious, containing two fewer parameters. The relative similarity in directional trends of changes in population size with changes in phenotype and environment (see the electronic supplementary material 2) means that there were no statistical reasons to differentiate regression slopes (table 2) or intercepts (likelihood ratio test: p-value on >0.05) between the five populations.
Table 2.
Akaike Information Criterion (AIC, with model weights in brackets) for all possible models. AIC values are given to one decimal place, AIC weights to three decimal places. (Each row and column combination represents one model, e.g. the cell [2,2] (the AIC is −31.3, the model weight is 0.013) is for a model containing global effects of k and z and population-level variability in gradients and intercepts for z. The minimum AIC value denotes the model with most support (denoted in bold), which contains effects of environmental (k) and phenotypic (z) changes as well as their interaction. This model does not differ significantly (according to likelihood ratio tests) from one without random intercepts.)
| population-level variability in gradients and intercepts for both k and z | population-level variability in gradients and intercepts for z only | population-level variability in gradients and intercepts for k only | population-level variability in intercepts only | |
|---|---|---|---|---|
| global effect of k, z and their interaction | −28.9 (0.004) | −34.9 (0.077) | −34.9 (0.077) | −38.9 (0.567) |
| global effect of k and z | −25.3 (0.001) | −31.3 (0.013) | −31.3 (0.013) | −35.3 (0.093) |
| global effect of z only | −16.4 (0.000) | 3.0 (0.000) | −22.4 (0.000) | −1.0 (0.000) |
| global effect of k only | −25.9 (0.001) | −31.9 (0.017) | −31.9 (0.017) | −35.9 (0.123) |
| no global effect | −17.5 (0.000) | 4.5 (0.000) | −23.5 (0.000) | 1.5 (0.000) |
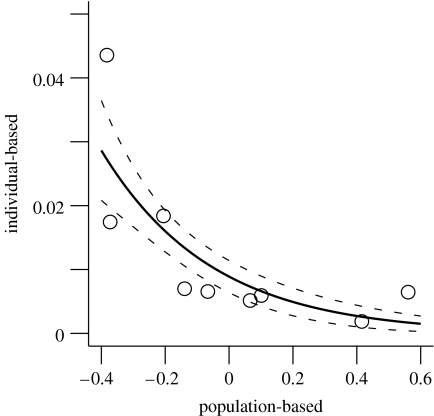
The conceptual framework applied here was compared with a more involved method that linked phenotypic traits to population growth at the individual level, applied previously to the Soay sheep population. The summed individual contributions of juvenile mass to annual population growth correlated significantly with the phenotypic rates of change estimated here (on the scale of the logit link function: β=−2.975, s.e.=0.971, p<0.05, r2=0.687; figure 3). We stress again that the two approaches attempt to do very different things (see §§3 and 5 for more).
Figure 3.
The correlation between the Hairston et al. (2005) population-based method and Pelletier et al.'s (2007a) individual-based method was fairly strong: years when mean birth weight decreases the most are also years when the summed individual contributions to population growth are the largest in the Soay sheep population. The methods differ markedly, being linked by a chain of responses (see main text for details). The individual-based method is quantified as the summed contribution of juvenile mass to population growth; the Hairston et al. (2005) method is the absolute rate of change in mean juvenile mass.
5. Discussion
In this paper, we applied a framework that aims to disentangle phenotypic, environmental and population fluctuations. The framework quantifies a link between phenotype, environment and some measure of population performance. Our results suggest that (i) phenotypic and environmental variability make statistically indistinguishable contributions to population growth (figure 2), (ii) the model for population growth with the most support featured environmental and phenotypic changes as well as their interaction as explanatory variables (table 2; see the electronic supplementary material 2), and (iii) the method correlated fairly well with a more data-intensive, individual-based approach (figure 3).
Is the overall similarity in trends between environmental and phenotypic variables with population growth to be expected, given the broadly similar life history of the case studies analysed? Possibly, although the five populations experience different climatic regimes, they have variable age at first breeding (between 1 and 4), variable generation times (between 4 and 9) and highly contrasting population dynamics (figure 1). The Soay sheep population has the lowest age at first breeding and generation time, and fluctuates substantially around the population's average trajectory (figure 1a), all of which are potential reasons for the differences in mean absolute rates of phenotypic and environmental changes (figure 2). We do not explore these patterns further due, in part, to the relatively low number of populations analysed. The most likely model (given those assessed) for annual population growth contains phenotypic and environmental changes as well as their interaction (figure 2). The support for this model strengthens the argument that phenotypic and environmental distributions fluctuate on a similar time scale, which was the pivot of Hairston et al.'s (2005) framework. These fluctuations were statistically identical across the five populations (table 2; see the electronic supplementary material 2). The feedback from the population on the environmental variables is negligible, except for the Soay sheep on St Kilda where sward height (Crawley et al. 2004) could conceivably show an evolutionary response to grazing pressure. Eco-evolutionary feedbacks are clearly important in many systems (Post & Palkovacs 2009), as is variable plasticity in phenotypic responses (Pigliucci 2001), but we followed Hairston et al. (2005) in assuming here that evolutionary responses in environmental variables used in our study are negligible.
Drastic phenotypic change might not result in changes in population growth. For example, the bighorn sheep populations have experienced evolutionary change in horn size and body weight in response to trophy hunting (Coltman et al. 2003, 2005). Juvenile mass is known to affect juvenile survival and therefore population growth in ungulates and mammals (Albon et al. 1987; Clutton-Brock et al. 1987; Metcalfe & Monaghan 2001). Our analyses only considered the weight distribution in one demographic class, which might hamper our ability to detect an eco-evolutionary feedback (Post & Palkovacs 2009). The extent to which juvenile mass affects population growth differs between populations: the r2 values of models used to calculate the hypothetical changes in z and k ranged between 0.574 for Soay sheep and 0.163 for bighorn sheep at Sheep River. Although the same trait is used throughout, its impact is not uniform across the five populations; neither is the impact of the environmental variables. Inclusion of additional phenotypes or environmental variables could decrease the unexplained variation since an individual is neither defined by one trait alone nor affected by a singular environmental variable. Different populations are probably affected by different combinations of phenotypic traits, which would hamper our aim of quantifying the effects of phenotypic and environmental changes across multiple contrasting populations.
In the univariate case, does the inability to detect population-level variability in how phenotypic variability affects population growth (table 2), despite differences in effect magnitude (figure 2), suggest that similarity in evolutionary direction (Schluter et al. 2004) plays a critical role in the impact of juvenile mass on population growth? Although mean absolute rates of change were similar (figure 2), changes in population size are predicted more accurately by environmental fluctuations: the massive change in model likelihood when environment is removed suggests a lack of predictive power based on phenotype alone (table 2). A model only featuring environmental fluctuations had non-negligible model weight (table 2). The phenotypic change might mirror (or exaggerate) environmental fluctuations (Pelletier et al. 2007b), yet be a relatively poor predictor of population dynamics. Changes in phenotype are not independent of changes in environment and might arise from, for example, phenotypic plasticity (Pigliucci 2001). Future developments could perhaps extend this framework to a hierarchical approach that links phenotype, demography and population growth (Coulson et al. 2003).
It is uncontested that alternative approaches can yield contrasting results. Comparison of data-intensive approaches with those that do not rely on individual-based data remains valuable. Pelletier et al. (2007a) linked phenotypes at the individual level to variation in annual population growth and found that body weight contributed up to 18 per cent of total variation in annual population growth of the Soay sheep population. Thus, in similar fashion to Hairston et al. (2005), Pelletier et al. (2007a, p. 1571) concluded that ‘there is substantial opportunity for evolutionary dynamics to leave an ecological signature and vice versa’. The correlation between their individual-based method and the framework applied here was fairly strong: years when mean birth weight decreases the most are also years when the summed individual contributions to population growth are the largest (figure 3). When food is scarce, density is often high (Crawley et al. 2004), which causes decreases in individual condition and consequently increased mortality (Pelletier et al. 2007a). Assuming that birth weight is a trait under heritable selection and low phenotypic plasticity, it is anticipated that selection will be stronger when environmental conditions are harsh (Wilson et al. 2006). The trait then contributes more to population growth (Pelletier et al. 2007a). This will generate a change in the mean value of the trait over a time step, which, coupled with the change in population growth, will give a large absolute value of ∂X/∂z in Hairston et al.'s (2005) conceptual framework. As mentioned previously, no attempt is made here to decompose causes of observed variation in population growth into its environmental or phenotypic drivers, or to consider the effect of environmental change on phenotypic expression. The method of Hairston et al. (2005) does not directly characterize evolutionary dynamics and uses hypothetical rather than observed changes in population size. It neither considers the differences between individuals that are a prerequisite for evolutionary change nor those between different parts (e.g. age and sex classes) of the population that are masked by taking averages. It summarizes many processes, but can nevertheless elucidate population-level trends where exhaustive data are unavailable.
The conceptual framework proposed by Hairston et al. (2005) has prompted an explosion (Hanski & Saccheri 2006; Saccheri & Hanski 2006; Kinnison & Hairston 2007; Pelletier et al. 2007a) of interest in how ecological and evolutionary processes interact. This first published application of their method emphasizes the importance of considering how populations respond to changes in their environmental and phenotypic distributions. There was insufficient evidence to determine whether phenotypic or environmental fluctuations contributed more, in the statistical sense, to annual population growth (figure 2; table 2). Does the similarity hold across the entire life-history spectrum? Differences across a range of species, life histories and climates in population-level consequences of phenotypic and environmental variability could prove insightful for demographic inference generally, including conservation applications. Hairston et al.'s (2005) framework is a simplified, but nevertheless useful, tool to quantify environmental and phenotypic impact on population dynamics because it enables analysis in less data-rich systems than those used here. Our results stress the importance of developing and applying approaches that reflect the interdependence of environmental, phenotypic and population dynamics.
Acknowledgements
The authors would like to thank Luca Börger, Tim Clutton-Brock, Tim Coulson, Marco Festa-Bianchet, Jean-Michel Gaillard, Andrew Hendry, Michael Kinnison, Josephine Pemberton and an anonymous reviewer for making data available and providing comments that markedly improved earlier drafts. T.H.G.E. is funded by a Natural Environment Research Council grant NE/E015956/1 to Andy Purvis and Paul Pearson. F.P. is funded by the NERC Centre for Population Biology and a grant from the Natural Sciences and Engineering Research Council of Canada. Finally, we want to thank the funding agencies, logistical support networks and, especially, the many volunteers on the five populations.
Footnotes
One contribution of 14 to a Theme Issue ‘Eco-evolutionary dynamics’.
Supplementary Material
Additional details on each study population used in analysis
Three-dimensional regression planes per population
References
- Albon S.D., Clutton-Brock T.H., Guinness F.E. Early development and population dynamics in red deer. II. Density-independent effects of cohort variation. J. Anim. Ecol. 1987;56:69–81. doi:10.2307/4800 [Google Scholar]
- Bates D.M. Fitting linear mixed models in R. R News. 2005;5:27–31. [Google Scholar]
- Bradshaw W.E., Holzapfel C.M. Climate change—evolutionary response to rapid climate change. Science. 2006;312:1477–1478. doi: 10.1126/science.1127000. doi:10.1126/science.1127000 [DOI] [PubMed] [Google Scholar]
- Burnham K.P., Anderson D.R. Springer; New York, NY: 2002. Model selection and multimodel inference: a practical information–theoretical approach. [Google Scholar]
- Burnham K.P., Anderson D.R. Multimodel inference: understanding AIC and BIC in model selection. Sociol. Methods Res. 2004;33:261–304. doi:10.1177/0049124104268644 [Google Scholar]
- Clutton-Brock T.H. Princeton University Press; Princeton, NJ: 1991. The evolution of parental care. [Google Scholar]
- Clutton-Brock T.H., Major M., Albon S.D., Guinness F.E. Early development and population dynamics in red deer. I. Density-dependent effects on juvenile survival. J. Anim. Ecol. 1987;56:53–64. doi:10.2307/4799 [Google Scholar]
- Clutton-Brock T.H., Price O., Albon S., Jewell P. Early development and population fluctuations in Soay sheep. J. Anim. Ecol. 1992;61:381–396. doi:10.2307/5330 [Google Scholar]
- Coltman D.W., O'Donoghue P., Jorgenson J.T., Hogg J.T., Strobeck C., Festa-Bianchet M. Undesirable evolutionary consequences of trophy hunting. Nature. 2003;426:655–658. doi: 10.1038/nature02177. doi:10.1038/nature02177 [DOI] [PubMed] [Google Scholar]
- Coltman D.W., O'Donoghue P., Hogg J.T., Festa-Bianchet M. Selection and genetic (CO)variance in bighorn sheep. Evolution. 2005;59:1372–1382. doi:10.1111/j.0014-3820.2005.tb01786.x [PubMed] [Google Scholar]
- Côté S.D., Festa-Bianchet M. Birthdate, mass and survival in mountain goat kids: effects of maternal characteristics and forage quality. Oecologia. 2001;127:230–238. doi: 10.1007/s004420000584. doi:10.1007/s004420000584 [DOI] [PubMed] [Google Scholar]
- Coulson T., Tuljapurkar S. The dynamics of a quantitative trait in an age-structured population living in a variable environment. Am. Nat. 2008;172:599–612. doi: 10.1086/591693. doi:10.1086/591693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson T., Kruuk L.E.B., Tavecchia G., Pemberton J.M., Clutton-Brock T.H. Estimating selection on neonatal traits in red deer using elasticity path analysis. Evolution. 2003;57:2879–2892. doi: 10.1111/j.0014-3820.2003.tb01528.x. doi:10.1111/j.0014-3820.2003.tb01528.x [DOI] [PubMed] [Google Scholar]
- Coulson T., Benton T.G., Lundberg P., Dall S.R.X., Kendall B.E. Putting evolutionary biology back in the ecological theatre: a demographic framework mapping genes to communities. Evol. Ecol. Res. 2006;8:1155–1171. [Google Scholar]
- Crawley M.J., Albon S.D., Bazely D.R., Milner J.M., Pilkington J.G., Tuke A.L. Vegetation and sheep population dynamics. In: Clutton-Brock T., Pemberton J., editors. Soay sheep. Cambridge University Press; Cambridge, UK: 2004. pp. 89–112. [Google Scholar]
- Diggle P.J., Liang K.-Y., Zeger S.L. Oxford University Press; New York, NY: 2002. Analysis of longitudinal data. [Google Scholar]
- Falconer D.S., Mackay T.F.C. Pearson Prentice Hall; Harlow, UK: 1996. Introduction to quantitative genetics. [Google Scholar]
- Festa-Bianchet M., Jorgenson J.T., King W.J., Smith K.G., Wishart W.D. The development of sexual dimorphism: seasonal and lifetime mass changes in bighorn sheep. Can. J. Zool. 1996;74:330–342. doi:10.1139/z96-041 [Google Scholar]
- Festa-Bianchet M., Gaillard J.M., Jorgenson J.T. Mass- and density-dependent reproductive success and reproductive costs in a capital breeder. Am. Nat. 1998;152:367–379. doi: 10.1086/286175. doi:10.1086/286175 [DOI] [PubMed] [Google Scholar]
- Festa-Bianchet M., Coulson T., Gaillard J.M., Hogg J.T., Pelletier F. Stochastic predation events and population persistence in bighorn sheep. Proc. R. Soc. B. 2006;273:1537–1543. doi: 10.1098/rspb.2006.3467. doi:10.1098/rspb.2006.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.A. Sage Publications; Thousand Oaks, CA: 2002. An R and S-PLUS companion to applied regression. [Google Scholar]
- Fussmann G.F., Loreau M., Abrams P.A. Eco-evolutionary dynamics of communities and ecosystems. Funct. Ecol. 2007;21:465–477. doi:10.1111/j.1365-2435.2007.01275.x [Google Scholar]
- Gaillard J.M., Delorme D., Jullien J.M. Effects of cohort, sex, and birth date on body development of roe deer (Capreolus capreolus) fawns. Oecologia. 1993;94:57–61. doi: 10.1007/BF00317301. doi:10.1007/BF00317301 [DOI] [PubMed] [Google Scholar]
- Gaillard J.-M., Boutin J.-M., Delorme D., Van Laere G., Duncan P., Lebreton J.-D. Early survival in roe deer: causes and consequences of cohort variation in two contrasted populations. Oecologia. 1997;112:502–513. doi: 10.1007/s004420050338. doi:10.1007/s004420050338 [DOI] [PubMed] [Google Scholar]
- Gienapp P., Teplitsky C., Alho J.S., Mills J.A., Merilä J. Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. doi:10.1111/j.1365-294X.2007.03413.x [DOI] [PubMed] [Google Scholar]
- Hairston N.G., Jr, Ellner S.P., Geber M.A., Yoshida T., Fox J.A. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 2005;8:1114–1127. doi:10.1111/j.1461-0248.2005.00812.x [Google Scholar]
- Hanski I., Saccheri I. Molecular-level variation affects population growth in a butterfly metapopulation. PLoS Biol. 2006;4:719–726. doi: 10.1371/journal.pbio.0040129. doi:10.1371/journal.pbio.0040129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry A.P., Kinnison M.T. The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. doi:10.2307/2640428 [DOI] [PubMed] [Google Scholar]
- Hendry A.P., Farrugia T., Kinnison M.T. Human influences on rates of phenotypic change in wild animal populations. Mol. Ecol. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. doi:10.1111/j.1365-294X.2007.03428.x [DOI] [PubMed] [Google Scholar]
- Jones L.E., Becks L., Ellner S.P., Hairston N.G., Jr, Yoshida T., Fussman G.F. Rapid contemporary evolution and clonal food web dynamics. Phil. Trans. R. Soc. B. 2009;364:1579–1591. doi: 10.1098/rstb.2009.0004. doi:10.1098/rstb.2009.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnison M.T., Hairston N.G., Jr Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Funct. Ecol. 2007;21:444–454. doi:10.1111/j.1365-2435.2007.01278.x [Google Scholar]
- Kinnison M.T., Hendry A.P. The pace of modern life II: from rates of contemporary microevolution to pattern and process. Genetica. 2001;112–113:145–164. doi:10.1023/A:1013375419520 [PubMed] [Google Scholar]
- Kokko H., Lopez-Sepulcre A. The ecogenetic link between demography and evolution: can we bridge the gap between theory and data? Ecol. Lett. 2007;10:773–782. doi: 10.1111/j.1461-0248.2007.01086.x. doi:10.1111/j.1461-0248.2007.01086.x [DOI] [PubMed] [Google Scholar]
- Lindström J. Early development and fitness in birds and mammals. Trends Ecol. Evol. 1999;14:343–348. doi: 10.1016/s0169-5347(99)01639-0. doi:10.1016/S0169-5347(99)01639-0 [DOI] [PubMed] [Google Scholar]
- Lummaa V., Clutton-Brock T.H. Early development, survival and reproduction in humans. Trends Ecol. Evol. 1998;13:141–147. doi:10.1016/S0169-5347(01)02414-4 [Google Scholar]
- Metcalfe N.B., Monaghan P. Compensation for a bad start: grow now, pay later? Trends. Ecol. Evol. 2001;16:254–260. doi: 10.1016/s0169-5347(01)02124-3. doi:10.1016/S0169-5347(01)02124-3 [DOI] [PubMed] [Google Scholar]
- Nussey D.H., Clutton-Brock T.H., Elston D.A., Albon S.D., Kruuk L.E.B. Phenotypic plasticity in a maternal trait in red deer. J. Anim. Ecol. 2005a;74:387–396. doi:10.1111/j.1365-2656.2005.00941.x [Google Scholar]
- Nussey D.H., Postma E., Gienapp P., Visser M.E. Selection on heritable phenotypic plasticity in a wild bird population. Science. 2005b;310:304–306. doi: 10.1126/science.1117004. doi:10.1126/science.1117004 [DOI] [PubMed] [Google Scholar]
- Parmesan C., Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. doi:10.1038/nature01286 [DOI] [PubMed] [Google Scholar]
- Pelletier F., Page K.A., Ostiguy T., Festa-Bianchet M. Fecal counts of lungworm larvae and reproductive effort in bighorn sheep (Ovis canadensis) Oikos. 2005;110:473–480. doi:10.1111/j.0030-1299.2005.14120.x [Google Scholar]
- Pelletier F., Clutton-Brock T., Pemberton J., Tuljapurkar S., Coulson T. The evolutionary demography of ecological change: linking trait variation and population growth. Science. 2007a;315:1571–1574. doi: 10.1126/science.1139024. doi:10.1126/science.1139024 [DOI] [PubMed] [Google Scholar]
- Pelletier F., Reale D., Garant D., Coltman D.W., Festa-Bianchet M. Selection on heritable seasonal phenotypic plasticity of body mass. Evolution. 2007b;61:1969–1979. doi: 10.1111/j.1558-5646.2007.00160.x. doi:10.1111/j.1558-5646.2007.00160.x [DOI] [PubMed] [Google Scholar]
- Pelletier F., Garant D., Hendry A.P. Eco-evolutionary dynamics. Phil. Trans. R. Soc. B. 2009;364:1483–1489. doi: 10.1098/rstb.2009.0027. doi:10.1098/rstb.2009.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettorelli N., Pelletier F., von Hardenberg A., Festa-Bianchet M., Cote S.D. Early onset of vegetation growth vs. rapid green-up: impacts on juvenile mountain ungulates. Ecology. 2007;88:381–390. doi: 10.1890/06-0875. doi:10.1890/06-0875 [DOI] [PubMed] [Google Scholar]
- Pigliucci M. John Hopkins University Press; Baltimore, MD: 2001. Phenotypic plasticity. [Google Scholar]
- Pinheiro J.C., Bates D.M. Mixed-effects models in S and S-plus. Springer-Verlag; New York, NY: 2000. [Google Scholar]
- Post D.M., Palkovacs E.P. Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Phil. Trans. R. Soc. B. 2009;364:1629–1640. doi: 10.1098/rstb.2009.0012. doi:10.1098/rstb.2009.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2008. R: a language and environment for statistical computing. [Google Scholar]
- Ricklefs R.E., Wikelski M. The physiology/life-history nexus. Trends Ecol. Evol. 2002;17:462–468. doi:10.1016/S0169-5347(02)02578-8 [Google Scholar]
- Saccheri I., Hanski I. Natural selection and population dynamics. Trends Ecol. Evol. 2006;21:341–347. doi: 10.1016/j.tree.2006.03.018. doi:10.1016/j.tree.2006.03.018 [DOI] [PubMed] [Google Scholar]
- Schluter D., Clifford E.A., Nemethy M., McKinnon J.S. Parallel evolution and inheritance of quantitative traits. Am. Nat. 2004;163:809–822. doi: 10.1086/383621. doi:10.1086/383621 [DOI] [PubMed] [Google Scholar]
- Sinervo B., Svensson E., Comendant T. Density cycles and an offspring quantity and quality game driven by natural selection. Nature. 2000;406:985–988. doi: 10.1038/35023149. doi:10.1038/35023149 [DOI] [PubMed] [Google Scholar]
- Slobodkin L.B. Rinehart and Winston; New York, NY: 1961. Growth and regulation of animal populations. [Google Scholar]
- Thompson J.N. Rapid evolution as an ecological process. Trends Ecol. Evol. 1998;13:329–332. doi: 10.1016/s0169-5347(98)01378-0. doi:10.1016/S0169-5347(98)01378-0 [DOI] [PubMed] [Google Scholar]
- Whittingham M.J., Stephens P.A., Bradbury R.B., Freckleton R.P. Why do we still use stepwise modelling in ecology and behaviour? J. Anim. Ecol. 2006;75:1182–1189. doi: 10.1111/j.1365-2656.2006.01141.x. doi:10.1111/j.1365-2656.2006.01141.x [DOI] [PubMed] [Google Scholar]
- Wilson A.J., Pilkington J., Pemberton J., Coltman D.W., Overall A.D.J., Byrne K.A., Kruuk L.E.B. Selection on mothers and offspring: whose phenotype is it and does it matter? Evolution. 2005;59:451–463. doi:10.1111/j.0014-3820.2005.tb01003.x [PubMed] [Google Scholar]
- Wilson A.J., Pemberton J.M., Pilkington J.G., Coltman D.W., Mifsud D.V., Clutton-Brock T.H., Kruuk L.E.B. Environmental coupling of selection and heritability limits phenotypic evolution. PLoS Biol. 2006;4:1270–1275. doi: 10.1371/journal.pbio.0040216. doi:10.1371/journal.pbio.0040216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Jones L.E., Ellner S.P., Fussmann G.F., Hairston N.G., Jr Rapid evolution drives ecological dynamics in a predator–prey system. Nature. 2003;424:303–306. doi: 10.1038/nature01767. doi:10.1038/nature01767 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wallace J.M., Battisti D.S. ENSO-like interdecadal variability: 1900–93. J. Climate. 1997;10:1004–1020. doi:10.1175/1520-0442(1997)010<1004:ELIV>2.0.CO;2 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional details on each study population used in analysis
Three-dimensional regression planes per population